Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV)/human herpesvirus 8 is a tumorigenic rhadinovirus that is associated with all forms of Kaposi's sarcoma. Current serological detection of KSHV is based on enzyme-linked immunosorbent or immunofluorescence assays that suffer from a variety of problems, including the lack of defined standards for test comparison. While KSHV is the only known human rhadinovirus, two lineages of KSHV-like rhadinoviruses are found in Old World primates: the RV1 lineage includes KSHV and retroperitoneal fibromatosis herpesvirus (RFHV) in macaques, and the RV2 lineage includes RRV and MneRV2 from different macaque species. To develop animal models of KSHV-associated diseases, we developed quantitative multiplex bead-based serological assays to detect antibodies against rhadinovirus antigens. Proteins from KSHV (RV1) and MneRV2 (RV2) virions were coupled to spectrally distinct fluorescent beads and used in Luminex flow cytometry-based assays to detect immune responses in macaques. Both assays showed large dynamic ranges with high levels of seroreactivity to both KSHV and MneRV2 proteins. A large set of macaque serum samples from the Washington National Primate Research Center was screened, and most of the samples (82%) were positive in both assays, consistent with the high level of RV1-RV2 coinfection detected by PCR. The macaque sera showed broad, variable, and unique serological responses to the different viral antigens, allowing an initial seroprevalence to be determined for the macaque viruses. The Luminex assays offer a novel multiplexed approach to assess rhadinovirus infection patterns in both humans and nonhuman primates. This will help advance our understanding of rhadinovirus biology and associated host immunological responses.
INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV)/human herpesvirus 8, a member of the rhadinovirus genus of gammaherpesviruses, was first identified in 1994 in Kaposi's sarcoma (KS) lesions in human immunodeficiency virus (HIV)-infected patients with AIDS (1). Since then, KSHV has been causally linked to all types of KS, including HIV-negative classic KS; endemic, epidemic (AIDS-related), and iatrogenic KS; as well as several lymphoproliferative diseases, including primary effusion lymphoma (PEL) and multicentric Castleman's disease (2). KSHV has a genome of approximately 165 kb, which contains more than 90 different genes (3). As with other herpesviruses, the KSHV genes are expressed at different stages of the virus life cycle and are generally categorized as either latent or lytic. Relatively few genes are expressed during viral latency, allowing the virus to minimize its exposure to the host immune system. After activation of the virus from latency, a large number of lytic genes are expressed, including all of the genes necessary for virus replication and production of infectious virions. Serological assays for KSHV have been developed to detect immune responses against both lytic- and latency-associated antigens. Most assay development has targeted the latency-associated nuclear antigen (LANA), the virion-associated open reading frame 65 (ORF65) capsid protein, and the K8.1 virion glycoprotein (4–7). Diagnosis of KSHV infection has proved problematic due to discordance between serological tests for different viral antigens and difficulties in establishing positive and negative reference populations (8–10). Low viral loads in blood or saliva limit the ability of even sensitive PCR-based approaches to be used for diagnosis (11, 12). Several multiantigen tests have been recently developed in order to have a wide-based screen for serological evidence of virus infection (13–15).
In 1997, we identified the macaque homolog of KSHV, the retroperitoneal fibromatosis herpesvirus (RFHV), in retroperitoneal fibromatosis (RF) lesions, a KS-like tumor present in rhesus and pig-tailed macaques with simian AIDS, at the Washington National Primate Research Center (WaNPRC) (16). Using real-time quantitative PCR (qPCR) assays specific for RFHV, we detected high levels of RFHV in RF lesions, suggesting an important causal association (17). Approximately two RFHV genomes per cell were detected in these lesions, and the RFHV LANA homolog was detected in the nuclei of nearly every RF tumor cell (18, 19). These studies suggested that macaque RFHV represents a close animal model of KSHV transmission and pathogenesis. Subsequently, another herpesvirus, the rhesus rhadinovirus (RRV), was detected in rhesus macaques at the New England National Primate Research Center (20). Sequence analysis revealed a strong genetic similarity between RRV and KSHV, with conservation of most of the lytic and latent genes of KSHV (21, 22). Further studies, using the consensus-degenerate hybrid oligonucleotide primer (CODEHOP) PCR approach, revealed the presence of rhadinoviruses related to both KSHV and RRV in many Old World nonhuman primate species, including drills, mandrills, baboons, gorillas, and chimpanzees (see reference 23). Phylogenetic analysis of available DNA sequences revealed that Old World primates are host to two divergent rhadinovirus lineages (24, 25). KSHV, RFHV, and other homologs of KSHV group together within the RV1 lineage of Old World primate rhadinoviruses, while RRV and other closely related viruses group together within a second RV2 lineage. Although only one human rhadinovirus, i.e., KSHV, has been identified at present, these studies suggest the existence of a second, as-yet-undiscovered, human rhadinovirus belonging to the RV2 rhadinovirus lineage.
PCR-based studies have shown that the macaque RV1 and RV2 rhadinoviruses are highly prevalent in the National Primate Research Centers (20, 26, 27). However, there has been little development of serological assays to detect rhadinovirus infection or distinguish between RV1 and RV2 rhadinovirus infections. In previous studies, RRV virions purified from infected cultures of primary rhesus fetal fibroblasts or other virus-infected cells were disrupted and used in enzyme-linked immunosorbent assay (ELISA)-based assays to screen for serological reactivity in macaques at the New England and Oregon National Primate Research Centers (20, 28). Although these studies detected antibody reactivity in more than 90% of the tested animals, no attempt was made to distinguish RV1 and RV2 seroreactivity. In order to further study the infection, transmission, activation, and pathogenesis of RV1 and RV2 rhadinoviruses in macaques, we have developed high-throughput Luminex-based assays to detect and discriminate serological reactivity to multiple RV1 and RV2 rhadinovirus antigens. Unlike most Luminex assays developed for other antigens, the assays described here were developed de novo without highly characterized ELISA-based assays for comparison. We have used the Luminex-based assays to screen for rhadinovirus infection in macaques at the WaNPRC and have followed the immunoreactivity to multiple viral antigens longitudinally during natural infections.
MATERIALS AND METHODS
Serum and plasma samples.
Plasma samples were obtained during the biannual health screening of macaques at the WaNPRC. Additional samples were obtained from stored aliquots of serum and plasma obtained from animals in previous studies. A juvenile macaque, M04203, had been experimentally infected with saliva from an animal with high levels of RFHV, and blood was collected prior to and after infection. Additional serum samples were available from three macaques, M02298, M03126, and M03240, that had been part of a simian immunodeficiency virus (SIV) vaccine study. Serum samples from two KSHV-positive and three KSHV-negative individuals were from a cohort described previously (29).
Cells.
The pleural effusion lymphoma cell line, BCBL-1, which is persistently infected with KSHV, was obtained from D. Ganem. Cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum, penicillin, streptomycin, glutamine, and 2-mercaptoethanol. Rhesus primary fetal fibroblasts (RPFF), kindly provided by M. Axthelm, were cultured in complete Dulbecco's modified Eagle's medium (DMEM) with 10% cosmic calf serum, penicillin, streptomycin, and HEPES.
Antibody reagents.
Alexa-tagged goat anti-human IgG (Invitrogen) was used to detect IgG in macaque serum in the BCBL-1 immunofluorescence assays. The mouse monoclonal antibody to the KSHV ORF59 lytic marker (ABI) and Alexa-tagged goat-anti-mouse IgG (Invitrogen) were used to detect lytically activated BCBL-1 cells. Phycoerythrin-labeled anti-human IgG (Jackson ImmunoResearch Laboratories) was used to detect IgG in macaque serum in the Luminex assays.
Immunofluorescence assay.
KSHV-infected BCBL-1 cells were incubated with or without the phorbol ester, tetradecanoyl phorbol acetate (TPA), for 48 h and then immobilized onto poly-l-lysine slides. Cells were fixed, permeabilized, and incubated with serum (1:25) from macaque M04203, obtained before and after RFHV infection. Bound serum antibodies were detected with goat anti-human Alexa antibody, diluted 1:500, in blocker with 10% goat serum. Cells were counterstained with mouse anti-ORF59 antibody to KSHV ORF59 and goat anti-mouse IgG-Alexa (1:500). Cells were analyzed and imaged on an LSM-5 Pascal confocal fluorescence microscope (Zeiss).
Virion purification.
BCBL-1 cells latently infected with KSHV were expanded in roller bottles and treated with TPA for 4 days to induce virion production. RPFF cells were expanded in roller bottles and lytically infected with MneRV2 (isolate MneJ97167, described previously [30]) at 60% confluence to achieve optimal virus production. Supernatants from both cultures were harvested, and large cellular material was pelleted at 4,000 × g. The supernatant was then filtered using a 0.45-μm-pore-size acetate filter unit (Corning) to remove cellular debris. Virions were pelleted at 18,000 × g for 90 min on a 20% glycerol cushion with a type 19 rotor (Beckman), resuspended in RPMI, and frozen in aliquots. Virus protein concentration was determined by micro-bicinchoninic acid (BCA) assay (Pierce) and characterized by SDS-PAGE gel to verify the presence of viral proteins and to confirm the absence of contaminating serum proteins.
Coupling of disrupted virion proteins to carboxylated beads.
Purified KSHV and MneRV2 virions were disrupted in 0.033% Triton X-100 (Sigma) for 1 h at 37°C as previously described (31), and protein levels were quantified by micro-BCA assay (Pierce). The disrupted virion proteins were coupled to different bead sets by using a two-step carbodiimide reaction (31). Briefly, 1.25 × 106 microsphere beads (Luminex) were activated with 1-ethyl-1-3-(3-dimethylaminopropyl)-carbodimide hydrochloride (Pierce) and N-hydroxysuflosuccinimide (Pierce) in 100 μl of activation buffer (Bio-Rad) for 20 min at room temperature. Activated beads were washed 3 times with phosphate-buffered saline (PBS), pH 7.4, and incubated with different concentrations (5 to 12 μg/106 beads) of KSHV (bead set 60) or MneRV2 (bead set 61) virion proteins. Coupled beads were washed, incubated with microsphere blocking buffer (Bio-Rad), and then washed again and resuspended in storage buffer and stored at 4°C.
Validation of bead coupling.
Beads coupled to the RV1 or RV2 virion proteins were tested using several macaque and human plasma samples that had been determined to be negative or positive for antibodies to the corresponding viral antigens by immunofluorescence and SDS-PAGE Western blot analysis. Briefly, the RV1 and RV2 virion-coupled bead sets were incubated with either positive or negative plasma at a 1:100 dilution in Blotto for 1 h at room temperature and then washed (3 times with PBS). Bound antibody was detected with the phycoerythrin-tagged mouse anti-human IgG monoclonal antibody using the Luminex 200HT system (Luminex). Median fluorescent intensity (MFI) was determined. Protein (12 μg) was determined to be optimal in the coupling reactions, allowing near saturation of the beads. This provides a large dynamic range of binding, with maximal levels of 16,848 and 18,448 MFI observed with the strongest positive serum tested in the Luminex RV1 and RV2 virion assays, respectively. The inter- and intraassay variability was tested using the control serums for the RV1 and RV2 virion protein assays.
Multiplex Luminex assays.
For high-throughput multiplex assays, the bead sets coupled to the RV1 and RV2 virion proteins were individually vortexed and sonicated for 30 s to create a single bead suspension. Bead concentration was determined by counting beads on the Luminex instrument. A bead master mix was created containing approximately 200 beads per set in a final volume of 50 μl aliquoted into each well of a prewetted 96-well filter plate (Millipore or Pall). An equal volume of serum or plasma diluted 1:50 in PBS was incubated with the bead mix for 1 h at room temperature. Plates were then washed 3 times by adding PBS and aspirating with a vacuum manifold. Vacuum manifold pressure was adjusted so that evacuation of wells was accomplished by slow drip to avoid bead loss in the filter. Bound antibody was detected using the phycoerythrin-labeled mouse anti-human IgG monoclonal antibody in the Luminex 200HT system, as described above.
Assay variability.
The inter- and intraassay variability was determined from multiplex assays performed in triplicate with three positive- and two negative-control samples. This was repeated on three separate days. Intraassay variability was determined from the means and standard deviations from the three replicates in each assay for all three assays. Interassay variability was calculated by comparing the means and standard deviations from the three different assays to measure plate-to-plate consistency. The percent relative standard deviation (%RSD) was determined to be 8.1% for both inter- and intraassays for the RV1 virion assay and 8.1 and 8.4% for the inter- and intraassays for the RV2 virion assay, respectively.
Data analysis.
The GraphPad Prism software (San Diego, CA) was used for statistical analysis. The paired t test (two-tailed) was used to compare the means of the RV1 and RV2 seroreactivities obtained from the same animal in the multiplexed assays, the unpaired t test (two-tailed) was used to compare the RV1 or RV2 seroreactivities between specific groups of macaques, while the Kruskal-Wallis analysis using Dunn's multiple comparison test was used to compare the seroreactivities across the three macaque species. A cutoff limit of 58 MFI (RV1 assay) and 95 MFI (RV2 assay) was derived from the mean of the nonreactive juvenile sera (RV1 = 9 serum samples; RV2 = 10 serum samples) plus 5 standard deviations. These serum samples were obtained from juvenile animals less than 13 months of age from the WaNPRC infant colony and had been separated from adult animals at birth.
RESULTS
Seroreactivity of pig-tailed macaques against RRV-infected cell cultures by ELISA.
In initial studies, we screened sera from 51 pig-tailed macaques from an outdoor breeding colony of the WaNPRC for seroreactivity to RV2 lineage rhadinoviruses, using an ELISA-based assay targeting antigens present in lysates of RRV-infected rhesus fibroblast cell cultures, similar to previous studies (28). Although we have now isolated a pig-tailed macaque rhadinovirus, MneRV2, these first studies were done looking for antibodies in pig-tailed macaques that crossed to the closely related rhadinovirus, RRV, from rhesus macaques. Both RRV and MneRV2 infections of RPFF are permissive with expression of the early and late lytic antigens and production of infectious virions (20, 28, 30). Thus, this ELISA can detect antibody responses to both cell-associated viral antigens as well as virion antigens. Sera obtained from the macaque breeding colony were tested at a dilution of 1:100 in the ELISA. The majority of macaques tested were older than 4 years and showed moderate to strong seroreactivity to the RRV antigens (Fig. 1). Macaques younger than 6 months of age showed little reactivity, while those between 6 to 12 months showed mixed reactivity. This study confirms previous results obtained at the New England Primate Research Center, which showed serological evidence for widespread rhadinovirus infections in captive populations of adult macaques (20). Since subsequent studies identified significant levels of coinfections with both RV1 and RV2 lineage rhadinoviruses in captive macaque populations (27), the specificities of the ELISAs used in these initial studies are unknown.
Fig 1.
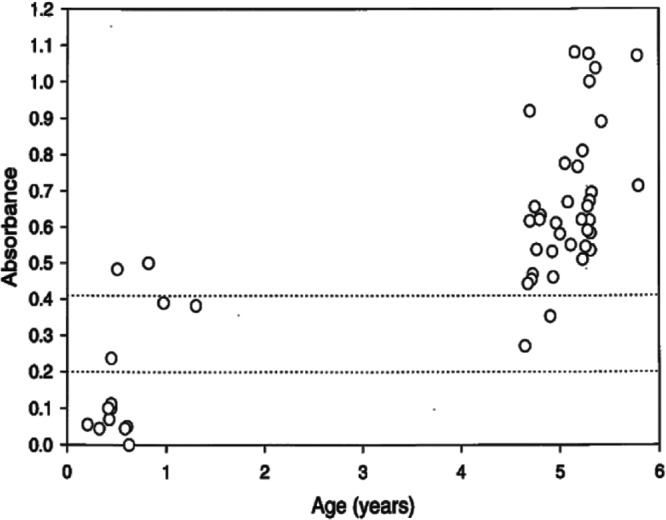
RRV ELISA of macaque sera. Serum samples from an outdoor colony of pig-tailed macaques were assayed by ELISA for reactivity to disrupted RRV-infected rhesus fibroblast cultures, as described in Materials and Methods. The dotted lines bracket indeterminate serum levels.
Seroreactivity of pig-tailed macaques against KSHV-infected cell cultures by IFA.
Although RFHV, the macaque RV1 rhadinovirus homolog of KSHV, has been detected by PCR and significant regions of its genome have been sequenced, no infectious clone has yet been isolated. Therefore, to determine the seroreactivity of pig-tailed macaques for RV1 lineage rhadinoviruses, we initially tested for cross-reactivity of macaque serum to KSHV antigens using the gold standard immunofluorescence assay (IFA) based on KSHV-infected BCBL cells. BCBL cells are latently infected with KSHV and can be activated to express early and late lytic antigens and produce infectious virions by treatment with the phorbol ester TPA or sodium butyrate. TPA-treated BCBL cells have been widely used to determine the seroprevalence of antibodies to lytic cycle KSHV antigens in human populations, as discussed above. Serum from a juvenile macaque, M04320, showed little reactivity against latent and lytically activated KSHV-infected BCBL cells (Fig. 2a and b, respectively). Serum was obtained at a later time point from this macaque after an experimental infection with saliva from an RFHV-infected macaque. While this serum showed very little reactivity to the untreated latent BCBL cells (Fig. 2c), it reacted strongly with the TPA-treated BCBL cells (Fig. 2d). To confirm that this reactivity was specific for KSHV-infected lytically activated cells, the TPA-treated BCBL cells were double stained with the macaque serum and a mouse monoclonal antibody against KSHV ORF59, a nuclear marker of lytic cycle activation. As shown in Fig. 2e, the cells that reacted strongly with the macaque serum (red) costained for nuclear KSHV ORF59 expression (green). Similar reactivity was detected with other adult macaque sera, demonstrating that macaque rhadinoviruses elicit IgG antibodies that cross-react with KSHV antigens.
Fig 2.
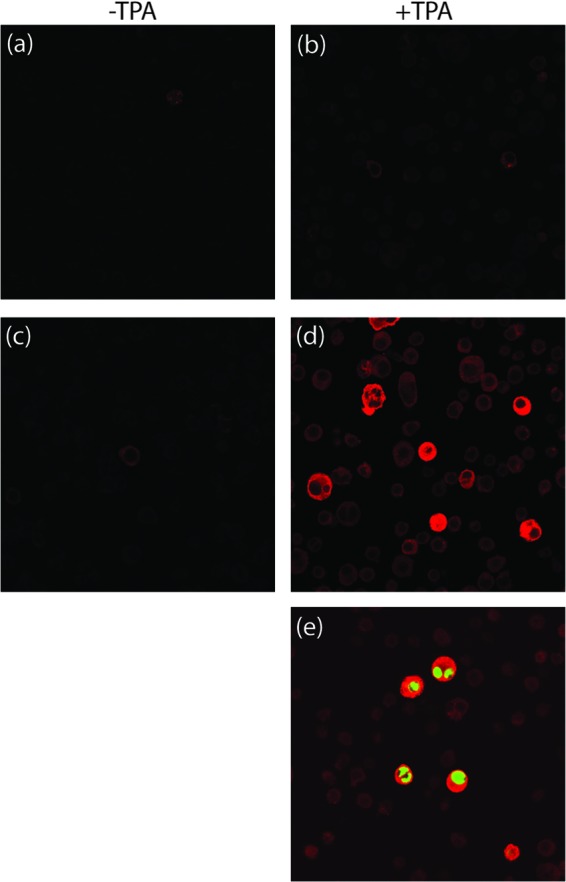
KSHV IFA of macaque serum. Latent (a and c) or TPA-treated (b, d, and e) KSHV-infected BCBL-1 cells were incubated with serum from macaque M04230 obtained before (a, b) and after (c, d, and e) infection with RFHV, as described in Materials and Methods. (e) TPA-treated cells were double stained with mouse anti-KSHV ORF59 monoclonal antibody to show colocalization of macaque serum antibodies with lytically activated cells. Bound antibody was detected with goat-anti-human IgG-Alexa, which cross-reacts with macaque IgG, or goat anti-mouse IgG-Alexa using a Zeiss confocal microscope.
Development of Luminex-based serology assays to detect antibodies to RV1 and RV2 rhadinovirus virion antigens.
The RV2 ELISA and RV1 IFA studies demonstrated that macaques in the WaNPRC were infected with KSHV-like rhadinoviruses that had elicited serological antibody responses. However, the specificity of these antibodies was not clear. Therefore, serological assays based on the Luminex bead technology were developed to examine the antibody responses to the different macaque rhadinoviruses. The Luminex technology uses polystyrene beads filled with different ratios of two fluorescent dyes which results in 100 different bead sets, each with its own spectral signature. The surface chemistry of the beads allows for covalent coupling of different antigens. Antibody reactivity to these antigens is detected using a secondary antibody coupled to phycoerythrin, a common reporter fluorochrome. The Luminex instrument uses two lasers to detect both the bead spectral signature as well as the phycoerythrin-coupled secondary antibody levels. Both recombinant protein and crude viral preparations have been successfully used as capture antigens, both singularly as well as in multiplexed assays using bead sets with different spectral signatures (31, 32). Such assays have shown great potential for fast, inexpensive multiplexed serological analysis with greater sensitivity and better performance than ELISA. In order to develop a Luminex-based serological assay to detect antibodies to rhadinovirus antigens, we coupled antigens from either KSHV virions purified from TPA-induced BCBL cell supernatants (RV1 assay) or MneRV2 virions purified from RPFF cell cultures lytically infected with an MneRV2 strain that we isolated from a pig-tailed macaque (RV2 assay) (30). The KSHV and MneRV2 virion antigens were solubilized with Triton X-100 and coupled to polystyrene beads with different spectral signatures, Luminex bead sets 60 and 61, respectively, as described in Materials and Methods.
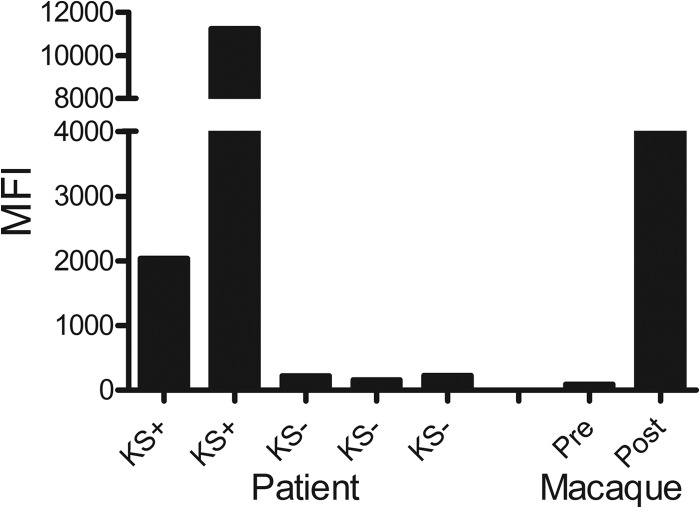
Initially, the RV1 bead assay, using KSHV virion antigens, was tested using sera from two individuals known to have strong seroreactivity to KSHV antigens by IFA. Sera from three individuals with low seroreactivity to KSHV antigens were used as negative controls. The RV1-coupled beads were incubated with serum samples diluted 1:100, as described in Materials and Methods. The beads were washed and incubated with a secondary anti-human IgG antibody coupled to phycoerythrin, which also reacts with macaque IgG antibodies. Finally, the beads were washed again, and bound phycoerythrin was quantitated for each bead analyzed. One hundred beads were read in the Luminex 200HT system, and the median fluorescence intensity (MFI) was determined for each sample. As shown in Fig. 3, sera from KSHV-positive individuals showed moderate to strong reactivity in the RV1 assay, with MFI levels ranging from 2,000 to nearly 12,000. In contrast, sera from KSHV-negative individuals showed low MFI values (164 to 229). To determine whether macaque antibodies would cross-react with the KSHV virion antigens, we tested the serum from the infected macaque, M04320, that gave a positive signal in the KSHV IFA assay (Fig. 2). As shown in Fig. 3, serum obtained from this macaque at 6 months of age prior to infection gave an MFI of 36, less than that seen with the KSHV-negative human controls. This serum was also negative in the KSHV IFA (Fig. 2a and b). In contrast, serum obtained from this macaque at 2 years of age after experimental infection with RFHV, which was strongly positive for lytic KSHV antigens in the IFA (Fig. 2d and e), was also strongly reactive with the KSHV virion antigens coupled to Luminex beads (Fig. 3). The reactivity of the macaque serum (∼4,000 MFI) was intermediate between the reactivities detected with the two KSHV-positive sera (2,000 and 12,000 MFI). These results suggest that the RV1 assay using KSHV virion antigens can detect macaque antibodies to macaque rhadinovirus virion antigens at levels comparable to those of antibodies elicited by KSHV in humans.
Fig 3.
Initial test of the KSHV-based RV1 virion Luminex assay. Sera from negative controls (KS−) or individuals seroreactive to KSHV (KS+) were assayed using the Luminex platform for reactivity to fluorescent bead set 60 coupled to proteins from disrupted KSHV virions to validate reactivity of the coupled KSHV antigens. Sera from the macaque M04230 obtained before and after RFHV infection were similarly assayed to detect macaque antibodies cross-reactive with the KSHV-coupled virion beads.
Colony-wide screen of RV1 and RV2 seroreactivity.
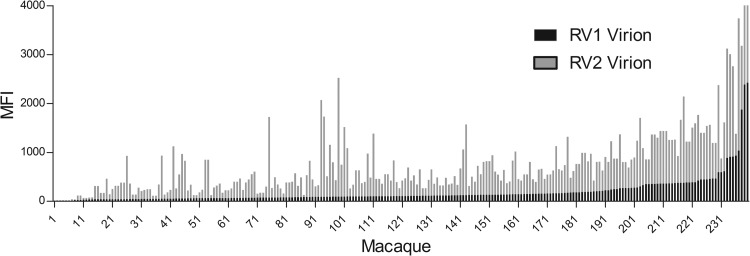
In order to identify positive macaque sera that could be used in further development and evaluation of the RV1 and RV2 Luminex assays, we screened more than 200 macaque sera obtained during a biannual colony health screen at the WaNPRC. These macaques were a mixture of species, including pig-tailed, rhesus, and cynomolgus macaques. The RV1 and RV2 assays were performed as a biplexed assay by incubating the RV1 antigen-coupled beads (KSHV virion antigens; bead set 60) and the RV2 antigen-coupled beads (MneRV2 virion antigens; bead set 61) simultaneously with the diluted serum sample. The MFI values for the RV1 and RV2 assay for each macaque serum sample were sorted by RV1 reactivity (Fig. 4). The RV1 values ranged from 9 to 2,425 MFI, with a median of 108 MFI, while the RV2 values ranged from 4 to 2,705, with a median of 401 MFI. A total of 164 of the 208 macaques had RV1 MFI values below 200. Only 11 animals had RV1 MFI values above 500. In contrast, only 43 animals had RV2 values below 200 MFI, with 83 animals having MFI values above 500. Positive and negative sera were identified and used to determine the dynamic range and variability of the assay, as described in Materials and Methods. The dynamic range of both assays was high, >16,000 MFI, and the interassay and intraassay variability was approximately 8%.
Fig 4.
RV1 and RV2 virion multiplexed Luminex assay screen of macaque sera. Macaque sera from an annual health screen of the macaque colony at the Washington National Primate Research Center were assayed for seroreactivity in the RV1 and RV2 virion multiplex Luminex assay. The reactivity of each macaque serum for both RV1 and RV2 virion antigens is shown sorted by RV1 reactivity. MFI, median fluorescent intensity.
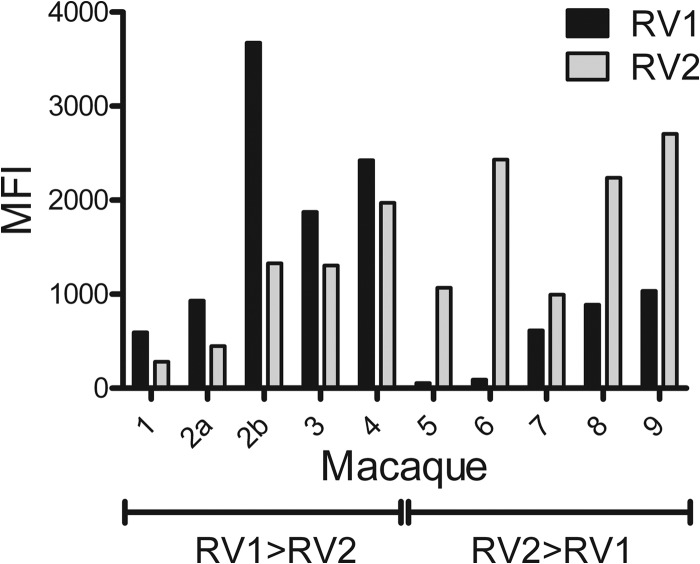
A cross-section of representative RV1 and RV2 serological data from individual macaques is shown in Fig. 5. A small number of animals (7/208) had seroreactivities similar to those seen with macaques 1 to 4, in which the antibodies to the RV1 virion proteins were higher than the antibodies to RV2 virion proteins. An additional serum sample was available from macaque 2 obtained during an SIV vaccine study, which showed a strong induction of anti-RV1 antibodies with a nearly 3-fold-higher level than that of anti-RV2 antibody. The majority of the macaques (179/208) had seroreactivities similar to those seen with macaques 5 to 9, in which the RV2 antibodies were higher than the RV1 antibodies. A small number of animals (6/208) gave RV1 and RV2 seroreactivities that were approximately equivalent. These results indicate that the RV1 and RV2 assays have a high degree of specificity and that the majority of animals are coinfected with both viruses. Sixteen animals, of which 10 were identified as juveniles less than 13 months of age from the infant colony, had very low seroreactivities in both assays.
Fig 5.
RV1 and RV2 virion seroreactivity of selected macaques. Selected data from the RV1 and RV2 virion multiplex Luminex assay shown in Fig. 4 are shown for specific macaques to illustrate the specificity of the two assays. An additional sample (2b) was available for macaque 2 (M03240) that had been part of an SIV vaccine study. Sera from macaques 1 to 4 show RV1 reactivity that is stronger than RV2 reactivity (RV1>RV2). Sera from macaques 5 to 9 show RV2 reactivity that is stronger than RV1 reactivity (RV2>RV1). MFI, median fluorescent intensity.
Seroreactivity of specific groups of captive macaques.
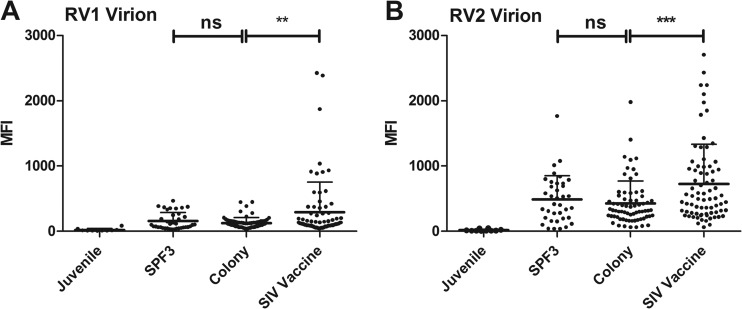
To further delineate the antirhadinovirus seroreactivity, the RV1 and RV2 assay results were divided into specific groups corresponding to animals in the WaNPRC that were (i) juveniles (6 to 13 months old); (ii) in the specific pathogen-free (SPF3) colony that have been screened for absence of D-type simian retrovirus, simian T-cell leukemia virus, and cercopithecine herpesvirus 1 (CeHV-1; the macaque homolog of herpes simplex virus) (31); (iii) in the general breeding colony; or (iv) part of an SIV vaccine study (S. L. Hu, personal communication). The sera from 10 juvenile macaques less than 13 months of age had very low seroreactivity to both the RV1 and RV2 virion antigens, with no significant difference (Fig. 6A and B). All 10 samples showed a baseline seroreactivity in the RV2 assay with a mean of 16 MFI and a range of 4 to 45 ( Table 1). A five-standard-deviation positivity cutoff at 95 MFI was determined for this assay. In the RV1 assay, nine of 10 serum samples had similar baseline seroreactivities, with a mean of 16 MFI and a range of 9 to 36. A five-standard-deviation positivity cutoff was determined at 58 MFI. The tenth serum sample, from an 8-month-old macaque, had an RV1 seroreactivity of 84 MFI, which was above the positivity cutoff of 58 MFI (Table 1), and an RV2 seroreactivity of 45 MFI, below the cutoff.
Fig 6.
RV1 and RV2 virion seroreactivity of specific groups of macaques. Data from the RV1 and RV2 virion multiplex Luminex assay of Fig. 4 are shown for specific groups of macaques, including (i) juveniles (less than 13 months of age); (ii) macaques from a specific pathogen-free (SPF3) colony that have been screened for absence of D-type simian retrovirus, simian T-cell leukemia virus, and cercopithecine herpesvirus 1 (the macaque homolog of herpes simplex virus); (iii) macaques from the general colony; and (iv) macaques participating in SIV vaccine studies. The seroreactivity of the SIV vaccine animals was significantly higher than that of the general colony animals in both assays. (A) RV1 virion assay; **, P = 0.0033; (B) RV2 virion assay; ***, P = 0.0005, according to the unpaired t test, two-tailed. The seroreactivities of the SPF3 and the general colony animals were not significantly different in either assay. The RV2 seroreactivity was significantly greater than the RV1 seroreactivity within the SPF3, general colony, and SIV vaccine groups (P < 0.0001), using the paired t test, two-tailed. No difference was detected between the RV1 and RV2 seroreactivity in the juveniles. The means and standard deviation are shown.
Table 1.
Comparison of the RV1 and RV2 virion antibody reactivities in specific groups of captive macaques at the Washington National Primate Research Center
| Lineagea | Animalb | Mean | Median | Range | No. (%) positive |
|---|---|---|---|---|---|
| RV1 (cutoff = 58 MFI) | Juvenile | 22 | 13 | 9–84 | 1/10 (10) |
| SPF3 | 157 | 99 | 30–465 | 28/37 (76) | |
| Colony | 125 | 108 | 49–448 | 58/67 (87) | |
| SIV | 3,656 | 123 | 45–2,425 | 40/45 (89) | |
| RV2 (cutoff = 95 MFI) | Juvenile | 16 | 7 | 5–45 | 0/10 (0) |
| SPF3 | 477 | 475 | 42–1,079 | 33/37 (89) | |
| Colony | 513 | 350 | 163–1,980 | 60/67 (94) | |
| SIV | 17,626 | 1,973 | 447–2,705 | 44/45 (98) |
Cutoff, mean of nonreactive juvenile sera plus 5 standard deviations; MFI, median fluorescent intensity.
Juvenile, hand-reared juvenile animals from 6 to 13 months of age; SPF3, animals from a specific pathogen-free colony that have been screened for absence of simian retrovirus 2, simian T-cell leukemia virus, and cercopithecine herpesvirus 1 (the macaque homolog of herpes simplex virus); colony, general colony born and raised animals; SIV, animals in SIV vaccine studies. The seroreactivities in the SIV vaccine study animals were significantly higher than in the general colony animals for both RV1 (P = 0.0033) and RV2 (P = 0.0005), as shown in Fig. 6.
Overall, in the adult macaques, 84% of the serum samples were positive in the RV1 assay, 91% of the serum samples were positive in the RV2 assay, and 82% of the serum samples were positive in both assays. The RV2 seroreactivity was significantly higher than the RV1 seroreactivity (P < 0.0001), with mean differences ranging from 388 MFI in the general colony animals to 1,397 MFI in the SIV vaccine group (Table 1 and Fig. 6). Although the levels were different, the percentages of macaques seropositive for either virus in the different groups were quite similar (SPF3, 76 to 89%; colony, 87 to 94%; SIV, 89 to 98%) (Table 1). No significant differences in seroreactivity to either virus were detected between the SPF3 and general colony animals (mean RV1, 157 versus 125 MFI; mean RV2, 475 versus 513 MFI, respectively; Fig. 6 and Table 1). However, the RV1 and RV2 seroreactivities in the SIV vaccine group were significantly greater than the seroreactivities in the other groups (Table 1; Fig. 6).
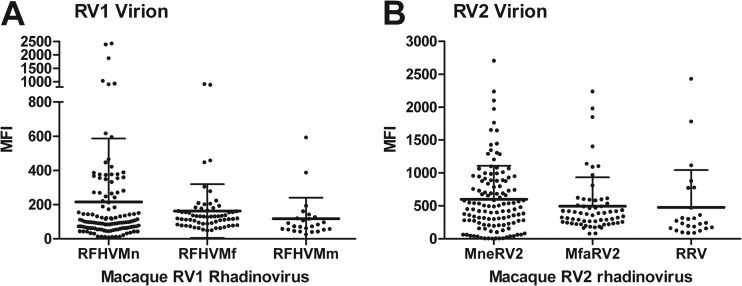
We examined the ability of the KSHV (host species, human) virion antigens coupled to bead set 60 in the RV1 Luminex assay and the MneRV2 (host species, pig-tailed macaque) virion antigens coupled to bead set 61 in the RV2 Luminex assay to react with antiviral antibodies elicited in the different macaque species against the host-specific rhadinoviruses (RFHVMn and MneRV2 in pig-tailed macaques, RFHVMm and RRV in rhesus macaques, RFHVMf and MfaRV2 in cynomolgus macaques). Data from animals for which host species information was available were compared. No significant difference was detected in the ability of the RV1 assay to detect seroreactivity in the different macaque species to the different macaque RV1 rhadinoviruses, RFHVMn, RFHVMf, or RFHVMm (Fig. 7A). Similarly, no significant difference was detected in the ability of the RV2 assay to detect seroreactivity in the different macaque species to the different macaque RV2 rhadinoviruses, MneRV2, MfaRV2, or RRV (Fig. 7B). In each macaque species, the RV2 seroreactivity was significantly higher than the RV1 seroreactivity (Fig. 7). More data points were available from the pig-tailed macaques due to the higher prevalence of this macaque species in the WaNPRC colony.
Fig 7.
RV1 and RV2 virion seroreactivity elicited by the rhadinoviruses present in the different macaque species. The RV1 and RV2 virion multiplex Luminex assay data of Fig. 4 were analyzed according to existing information regarding macaque species of the serum sample tested. The seroreactivity to the virus eliciting these antibodies in the different macaque species was compared: pig-tailed macaques, RFHVMn (RV1) and MneRV2 (RV2); cynomolgus macaques, RFHVMf (RV1) and MfaRV2 (RV2); rhesus macaques, RFHVMm (RV1) and RRV (RV2). No significant difference was detected in the ability of the RV1 assay (A) to detect antibodies elicited against RFHVMn compared to RFHVMm or RFHVMf or the RV2 assay (B) to detect antibodies elicited against MneRV2 compared to RRV or MfaRV2, according to the Kruskal-Wallis test. In each macaque species, the RV2 seroreactivity was significantly higher than the RV1 seroreactivity (RFHVMn versus MneRV2, P < 0.0001; RFHVMf versus MfaRV2, P < 0.0001; RFHVMm versus RRV, P = 0.0017), using the paired t test (two-tailed). The means and standard deviations are shown.
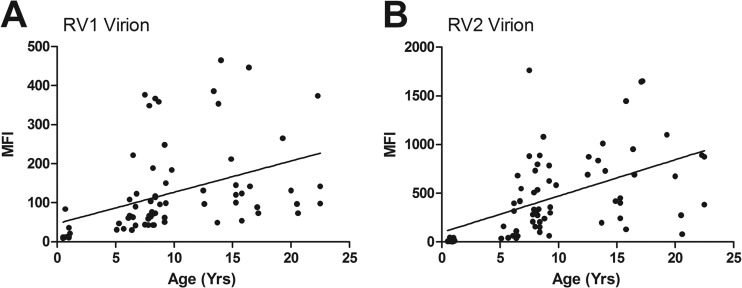
The RV1 and RV2 seroreactivities were compared to the age of the macaque in samples with an available birthdate. The animals in the SIV vaccine studies were excluded from the analysis. In the animals with age data, the RV1 seroreactivity ranged from 9 MFI in a negative 0.5-year-old juvenile to 465 MFI in a 14-year-old animal in the SPF3 colony (Fig. 8A). The RV2 seroreactivity ranged from 4 MFI in a negative 0.9-year-old juvenile to 1,763 MFI in a 7.5-year-old animal in the SPF3 colony (Fig. 8B). A strong correlation was detected between seroreactivity and age (RV1, P = 0.0004; RV2, P < 0.0001), with older animals showing a higher seroreactivity. No significant difference was detected in the RV1 and RV2 seroreactivities of males and females (data not shown).
Fig 8.
RV1 and RV2 virion seroreactivity related to age of the macaques. Data from the RV1 and RV2 virion multiplex Luminex assay of Fig. 4 were analyzed according to age of the macaque at the time of testing, excluding animals from the SIV vaccine studies. The Pearson's correlation analysis was employed to determine the correlation between seroreactivity and age; n = 71 serum samples. (A) RV1 virion assay, y axis range = 0 to 500 MFI, R = 0.4069, P = 0.0004 (two-tailed), significant at the 0.05 level; (B) RV2 virion assay, y axis range = 0 to 2,000 MFI, R = 0.5117, P < 0.0001 (two-tailed), significant at the 0.05 level.
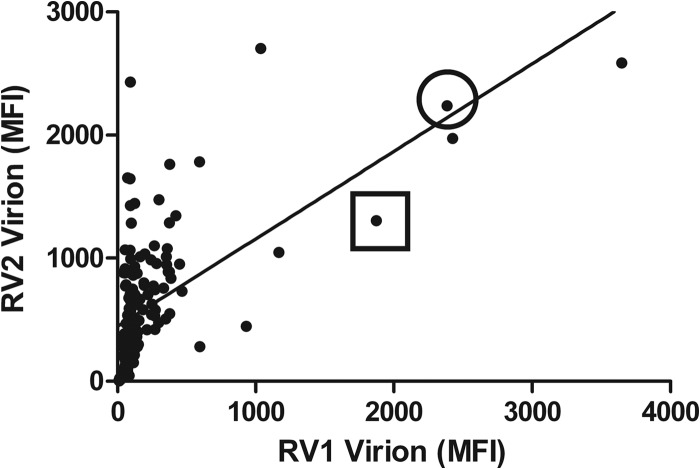
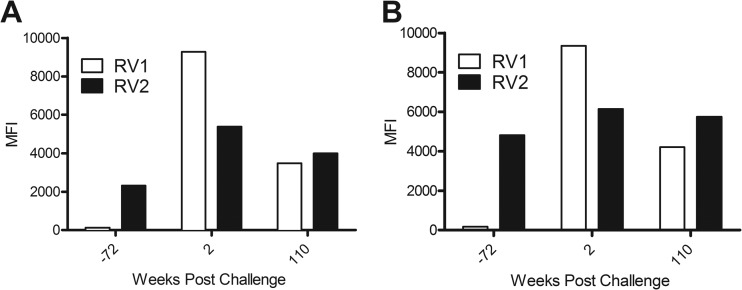
Overall analysis of the seroreactivities for all of the macaque sera revealed a strong correlation between the RV1 and RV2 seroreactivities (P < 0.0001; R = 0.5939) (Fig. 9). In general, those sera with the highest seroreactivity to the RV1 antigens also had the highest seroreactivity to the RV2 antigens. The animals with high seroreactivities to both viruses were all young adults (∼5 to 10 years of age) and had been part of an SIV vaccine study, in which the animals had been initially immunized with DNA of plasmids expressing SIV gp140 and Gag-Pol and then subsequently boosted with SIV gp140 and Gag-Pol proteins. The animals had been challenged with simian-human immunodeficiency virus (SHIV)-F162P4, an SIV-HIV hybrid virus (S. L. Hu, personal communication). Two animals, M02298 and M03126, which had shown high RV1 and RV2 seroreactivities in the initial Luminex screen (see Fig. 9), were chosen for further study. An initial longitudinal analysis of these macaques was performed using serum samples from colony health screens taken before, during, and after the vaccine study. Eighteen months prior to entry into the study, both animals had very low seroreactivities to the RV1 virion proteins (117 and 170 MFI), just above the cutoff level, and moderately high seroreactivities to the RV2 virion proteins (2,310 and 4,800 MFI) (Fig. 10), indicating that both animals were naturally coinfected with RV1 and RV2 rhadinovirus prior to entry into the study. As with the other animals in the larger colony screen, the RV2 seroreactivities at this point were substantially higher than the RV1 seroreactivities in both animals, with titer differences of 2,196 and 4,631 MFI, respectively. Two weeks after SHIV challenge, the RV1 seroreactivities had dramatically increased (79-fold in M02298 and 55-fold in M03126), with RV1 titer differences greater than 9,000 MFI for both animals. Although the RV2 seroreactivity also increased after SHIV challenge in both animals, this change was relatively modest (1.3- to 2.3-fold, respectively). More than 2 years postchallenge, the RV1 and RV2 seroreactivities had moderated but remained strongly positive in both macaques, with the RV2 seroreactivity again higher than the RV1 seroreactivity. These data suggest that the natural latent RV1 and RV2 rhadinovirus infections in these animals were differentially reactivated after the SHIV challenge and that this activation could be detected with the multiplex RV1 and RV2 virion Luminex assays.
Fig 9.
Correlation of RV1 and RV2 virion seroreactivity. The RV1 and RV2 virion results from the multiplex Luminex assay of Fig. 4 were compared. The Pearson's correlation analysis was employed to determine the correlation between the RV1 and RV2 seroreactivity. n = 212 serum samples, R = 0.5939, P < 0.0001 (two-tailed), significant at the 0.05 level. Two animals from the SIV vaccine study, M02298 (square) and M03126 (circle), were chosen for longitudinal analysis in Fig. 10.
Fig 10.
Longitudinal analysis of two SIV vaccine study macaques. Two macaques, M02298 (A) and M03126 (B), with high RV1 and RV2 seroreactivity were chosen for further longitudinal study from the SIV vaccine study (see Fig. 9). Three serum samples were identified in the colony health records for both animals, 72 weeks before SHIV challenge, 2 weeks postchallenge, and 110 weeks postchallenge, and were tested in the RV1 and RV2 virion multiplex Luminex assay.
DISCUSSION
In order to develop animal models of KSHV-associated diseases and aid in further studies of KSHV infection, transmission, and pathology, we have developed Luminex-based quantitative multiplex serological assays to detect antibodies elicited against macaque rhadinovirus virion antigens. Since macaques are frequently coinfected with viruses from both RV1 and RV2 rhadinovirus lineages, we developed separate assays for both viral lineages. For the RV2 assay, we used purified virions of MneRV2, the pig-tailed macaque RV2 rhadinovirus, since the majority of macaques at the Washington National Primate Research Center (WaNPRC) are pig-tailed macaques. We have recently sequenced the entire genome of MneRV2 and found that the virion proteins are highly conserved with other macaque RV2 rhadinoviruses, such as RRV, with sequence similarities up to 95% (unpublished results). As we are unable to culture RFHV, the macaque RV1 rhadinovirus, we developed a Luminex assay against virion antigens of KSHV, the human RV1 rhadinovirus. Since KSHV virions contain more than 20 different virion proteins with different levels of sequence similarity to the corresponding RFHV virion proteins, it was expected that the KSHV-based RV1 assay would not be as sensitive for macaque antibodies as would an RFHV-based assay. However, we found that macaques infected with RFHV developed high titers of antibodies reactive with KSHV virion proteins, showing the utility of the KSHV-based RV1 assay for serological screening of macaque RV1 rhadinovirus infections. An additional advantage of the KSHV-based RV1 assay is that the assay and protocols developed for screening macaque sera can be used directly for screening of human sera for antibodies to KSHV. Initially, we tested the RV1 assay with sera from individuals with and without high-titer antibodies to KSHV to verify that the beads coupled to KSHV virion proteins would be reactive with sera known to be positive for antibodies to KSHV. In this small test, there was a strong correlation between the RV1 Luminex assay and previous IFA studies. However, additional studies are needed to further characterize the utility of the KSHV virion-based Luminex assay to follow infections in KS patients.
To investigate the utility of the RV1 and RV2 Luminex assays, we screened a large set of 208 macaque sera obtained during a colony health screen at the WaNPRC. Most of the macaque sera (82%) showed reactivity toward both RV1 and RV2 antigens consistent with the high level of RV1 and RV2 rhadinovirus coinfections detected by PCR in this and other captive macaque populations (26, 27). Since the RV1 and RV2 rhadinoviruses are related and show significant protein sequence similarity (19, 24, 30, 33), we expected that antibodies elicited against a member of one rhadinovirus lineage might cross-react with antigens from the other rhadinovirus lineage. However, numerous sera in our study showed distinct reactivities in one or the other assay, suggesting that the assays were specifically detecting antibodies elicited by either an RV1 or an RV2 rhadinovirus. Additionally, the longitudinal studies showed a lack of correlation between the RV1 and RV2 seroreactivities over time, further demonstrating assay specificity. The specificity may be due to the choice of virion-associated proteins as antigenic targets in our assays, since the virion proteins are usually less well conserved than the basic herpesvirus proteins involved in replication. Additionally, the use of KSHV virions as the antigen source for the RV1 assay may have limited the amount of epitopes cross-reacting with the macaque sera. Whether macaques can develop antibodies to epitopes that are cross-reactive in the two assays remains to be determined.
The RV1 and RV2 seroreactivities were analyzed separately for juveniles, general colony animals, selected animals in a specific-pathogen free (SPF3) colony, and animals from vaccine trials that had been challenged with SHIV. The SPF3 colony at the WaNPRC was developed to eliminate three specific pathogens, cercopithecine herpesvirus 1 (the macaque homolog of herpes simplex virus 1), the D-type simian retrovirus 2 (SRV-2), and simian T-cell leukemia virus (31). We found no significant difference between the RV1 and RV2 seroreactivities in macaques from the SPF3 colony compared to general colony animals. This demonstrates that the screening used to establish the SPF3 colony did not eliminate animals infected with the macaque rhadinoviruses, which were not specifically targeted during the screening.
Our study showed a strong elevation of both RV1 and RV2 seroreactivity in animals that had participated in SIV vaccine studies. These animals had received different vaccination regimens followed by challenge with highly infectious SHIV isolates. Some of these animals became infected with SHIV and either succumbed to simian AIDS or became long-term nonprogressors. Other animals were protected from the SHIV challenge by the vaccination protocol and showed no evidence of SHIV infection or development of AIDS (S. L. Hu, personal communication). No obvious correlation was seen between rhadinovirus seroreactivity and either active SIV infection or obvious immunodeficiency, making it unclear why these animals had significantly higher RV1 and RV2 antibody levels than other colony animals. We are currently studying these animals to determine the characteristics and cause of this effect.
We detected an 84% seroprevalence of the RV1 lineage macaque viruses in the WaNPRC macaque colony, including RFHVMn in pig-tailed macaques, RFHVMm in rhesus macaques, and RFHVMf in cynomolgus macaques, and a 94% seroprevalence of the RV2 lineage macaque viruses, including RRV in rhesus, MneRV2 in pig-tailed, and MfaRV2 in cynomolgus macaques. This closely matches the 98% seroprevalence of rhadinoviruses in rhesus macaques at the California National Primate Research Center determined with an immunofluorescence assay against rhesus fibroblasts lytically infected with RRV (27). Since the immunofluorescence assay detects antibodies against all lytic RRV proteins in the infected cell, including virion and replication proteins, the specificity of this assay for RV1 or RV2 rhadinoviruses is not known. Interestingly, this study also examined the rhadinovirus seroprevalence in a specific pathogen-free colony similar to that derived in Washington, except that the screening targeted three additional viruses, including RRV, rhesus cytomegalovirus, and simian foamy virus. In their study, none of the animals in the California pathogen-free colony showed evidence of seroreactivity in the RRV-based immunofluorescence assay, indicating that the colony was free of RRV. In contrast, there was no significant difference in RV1 and RV2 seroreactivity between the macaques in the SPF3 colony and those in the general colony at the WaNPRC since the pathogen screening had not targeted the rhadinoviruses.
We detected a significant correlation between the RV1 and RV2 seroreactivity levels in the macaque serum samples. At first take, this could suggest cross-reactivity between the two assays. However, as discussed above, the lack of correlation between the assays for specific serum samples, especially the longitudinal samples, supports the specificity of the assays. This specificity was further confirmed in subsequent studies of the one juvenile macaque that had a low but positive RV1 seroreactivity of 84 MFI and a negative RV2 seroreactivity. Serum from this animal was tested by Western immunoblot analysis for reactivity to recombinant RFHV ORF59 and MneRV2 ORF59, markers of RV1 and RV2 lytic reactivation, respectively. While the serum showed no reactivity with MneRV2 ORF59, a strong reactivity to the RFHV ORF59 was observed, confirming both the RV1 positivity of the juvenile sample and the specificity of the Luminex assays.
We observed that the RV2 seroreactivity was higher than the RV1 seroreactivity in many of the macaques in the colony. While this could be due to our use of cross-reactive KSHV antigens in the RV1 bead set, it is clear that macaques have very strong seroreactivities to the human virus, as some of the highest seroreactivity levels (>9,000 MFI) were detected in the RV1 virion assay. Our serology data mirror previous PCR data, which showed that the levels of RV2 DNA were higher than the RV1 DNA in saliva in infected rhesus macaques (34). The increased RV2 virus levels in these animals could be responsible for the increased anti-RV2 immune response detected in our assay. This supports the current perception that the RV1 rhadinoviruses, including KSHV and macaque RFHV, have a typically latent phenotype, as both viruses are associated with latent infections in in vivo (17, 35) and in vitro (36) tissue culture systems. In contrast, the RV2 rhadinoviruses, including RRV and MneRV2, show a strong replicative phenotype in in vivo (37) and in vitro (20, 30) tissue culture systems. It is interesting to note that the RV1 rhadinovirus genomes contain viral homologs of the MARCH family of immunomodulatory immune evasion genes that are believed to play a role in maintaining latency (33, 38, 39). The RV2 rhadinoviruses lack a homolog of this gene (20, 22) (unpublished observations), suggesting that the RV2 lineage has developed a more lytic phenotype to maintain long-term infections in the host.
Our serological screen of animals in the WaNPRC showed that the RV1 Luminex assay was capable of detecting antibody responses to the macaque RV1 rhadinoviruses from different macaque species, including RFHVMn from pig-tailed macaques, RFHVMm from rhesus macaques, and RFHVMf from cynomolgus macaques. We also showed that the RV2 Luminex assay was capable of detecting antibody responses to the different macaque RV2 rhadinoviruses, including MneRV2 from pig-tailed macaques, RRV from rhesus macaques, and MfaRV2 from cynomolgus macaques. No significant difference in RV1 and RV2 seroreactivity was detected between males and females. When we excluded animals that had been in SIV vaccine studies, we detected a significant correlation between seroreactivity and age. Both assays showed minimal seroreactivities in juvenile animals less than 1 year old. The level of seroreactivity in both assays increased with age, although the RV2 seroreactivity was significantly higher than the RV1 seroreactivity. Interestingly, one animal had a positive RV1 seroreactivity (84 MFI) at 8 months of age, indicating that initial RV1 infections can occur at an early age. At this time point, the serum was negative in the RV2 assay, suggesting that coinfections do not necessarily happen simultaneously. As discussed above, the positive RV1 seroreactivity in this juvenile was confirmed by Western blotting, which showed a strong reaction with the RFHV ORF59 DNA polymerase processivity factor, which is an RV1 lytic cycle marker. No reactivity was detected with MneRV2 ORF59, an RV2 lytic cycle marker, confirming the specificity of the RV1 and RV2 Luminex assays.
Current serological assays to detect KSHV infections rely upon ELISA-based or IFA-based analysis using different antigens or antigen combinations (10, 11, 13). Typical assays target the latency-associated nuclear antigen (LANA), the virion glycoprotein K8.1, and/or the ORF65 capsid protein (7, 40). Discordance between these assays and the lack of “gold standard” reference samples have hindered the understanding of the prevalence, transmission, and natural history of KSHV infection (8). Furthermore, studies have shown that the humoral response to viruses such as KSHV is complex (15), and appropriate serological assays may require approaches that detect multiple antigens simultaneously. Other proteins, such as the complement inhibitory protein (KCP) and viral cyclin, have been shown to be useful targets for serological screening of KSHV infections (14, 41). The Luminex assays that we have developed use a complex mixture of more than 19 viral antigens present in purified virions, including the KSHV ORF65 capsid protein and the K8.1 virion glycoprotein in the RV1 assay and the MneRV2 homologs of these proteins in the RV2 assay. Unlike previous KSHV serological assays which rely to a large extent on detecting antibodies to LANA, the major latency-associated protein, our RV1 and RV2 assays target lytic cycle virion proteins. Our current assays are designed to detect antibodies to the late lytic antigens, allowing detection of immune responses following both de novo permissive infections and reactivation of latent infections. We are currently developing additional Luminex assays in which specific recombinant proteins, such as LANA, can be coupled to a different Luminex bead set. This would provide a multiplex system for detecting antibodies to both lytic and latent antigens simultaneously, allowing us to follow de novo infections and reactivation as well as the establishment of latency and disease processes associated with latency.
Recently, other high-throughput multiplexed assays based on the Luminex platform have been developed to detect antibodies against a variety of viral pathogens (32, 42, 43). Luminex-based assays have shown greater sensitivity and better performance than ELISA with fast turnaround time and high throughput (31). Since standardized ELISA or IFA had not been developed previously for the detection of the antibodies to the macaque rhadinoviruses, we had to bootstrap the development of the Luminex assays from the beginning. The initial assays were developed with serum known or presumed to be reactive (individuals with KS, experimentally infected macaques) or nonreactive (juveniles, KSHV-negative individuals), and substantial assay optimization was performed to minimize background fluorescence in the Luminex system and maximize the dynamic range of the assays. Since the target virion antigens are expressed during the virus replicative stage that occurs after de novo permissive infection or after reactivation of latent infections, our assays can detect and quantitate the immune response that follows these biological events. This is shown in our initial longitudinal assays in the SIV vaccine animals. Our ability to discriminate between the antibody response to coinfecting RV1 and RV2 rhadinoviruses has allowed us to determine an initial seroprevalence for viruses in these two lineages that naturally infect different macaque species. These assays offer a novel way to quantitatively assess RV1 and RV2 infection patterns to help in the understanding of herpesvirus biology and associated host immunological responses.
ACKNOWLEDGMENTS
This project was supported by the National Center for Research Resources (NCRR) and the Office of Research Infrastructure Programs (ORIP) of the National Institutes of Health (NIH) through grant number RR023343.
We acknowledge the important contributions of LaRene Kuller (WaNPRC), who helped with the development of the Luminex assays and provided the serum samples from the colony health screen, Mark Clarke (WaNPRC), who helped with access of information from the WaNPRC database, Margaret Thouless, who initiated the first ELISA-based rhadinovirus screen at the WaNPRC, and Allison Ankrom, who performed the confocal microscopy.
Footnotes
Published ahead of print 23 January 2013
REFERENCES
- 1.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865–1869 [DOI] [PubMed] [Google Scholar]
- 2.Antman K, Chang Y. 2000. Kaposi's sarcoma. N. Engl. J. Med. 342:1027–1038 [DOI] [PubMed] [Google Scholar]
- 3.Russo JJ, Bohenzky RA, Chien MC, Chen J, Yan M, Maddalena D, Parry JP, Peruzzi D, Edelman IS, Chang Y, Moore PS. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. U. S. A. 93:14862–14867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simpson GR, Schulz TF, Whitby D, Cook PM, Boshoff C, Rainbow L, Howard MR, Gao SJ, Bohenzky RA, Simmonds P, Lee C, de Ruiter A, Hatzakis A, Tedder RS, Weller IV, Weiss RA, Moore PS. 1996. Prevalence of Kaposi's sarcoma associated herpesvirus infection measured by antibodies to recombinant capsid protein and latent immunofluorescence antigen. Lancet 348:1133–1138 [DOI] [PubMed] [Google Scholar]
- 5.Pau CP, Lam LL, Spira TJ, Black JB, Stewart JA, Pellett PE, Respess RA. 1998. Mapping and serodiagnostic application of a dominant epitope within the human herpesvirus 8 ORF 65-encoded protein. J. Clin. Microbiol. 36:1574–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katano H, Iwasaki T, Baba N, Terai M, Mori S, Iwamoto A, Kurata T, Sata T. 2000. Identification of antigenic proteins encoded by human herpesvirus 8 and seroprevalence in the general population and among patients with and without Kaposi's sarcoma. J. Virol. 74:3478–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mbisa GL, Miley W, Gamache CJ, Gillette WK, Esposito D, Hopkins R, Busch MP, Schreiber GB, Little RF, Yarchoan R, Ortiz-Conde BA, Labo N, Whitby D. 2010. Detection of antibodies to Kaposi's sarcoma-associated herpesvirus: a new approach using K8.1 ELISA and a newly developed recombinant LANA ELISA. J. Immunol. Methods 356:39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engels EA, Sinclair MD, Biggar RJ, Whitby D, Ebbesen P, Goedert JJ, Gastwirth JL. 2000. Latent class analysis of human herpesvirus 8 assay performance and infection prevalence in sub-Saharan Africa and Malta. Int. J. Cancer 88:1003–1008 [DOI] [PubMed] [Google Scholar]
- 9.Sergerie Y, Abed Y, Roy J, Boivin G. 2004. Comparative evaluation of three serological methods for detection of human herpesvirus 8-specific antibodies in Canadian allogeneic stem cell transplant recipients. J. Clin. Microbiol. 42:2663–2667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nascimento MC, de Souza VA, Sumita LM, Freire W, Munoz F, Kim J, Pannuti CS, Mayaud P. 2007. Comparative study of Kaposi's sarcoma-associated herpesvirus serological assays using clinically and serologically defined reference standards and latent class analysis. J. Clin. Microbiol. 45:715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pellett PE, Wright DJ, Engels EA, Ablashi DV, Dollard SC, Forghani B, Glynn SA, Goedert JJ, Jenkins FJ, Lee TH, Neipel F, Todd DS, Whitby D, Nemo GJ, Busch MP. 2003. Multicenter comparison of serologic assays and estimation of human herpesvirus 8 seroprevalence among US blood donors. Transfusion 43:1260–1268 [DOI] [PubMed] [Google Scholar]
- 12.Albrecht D, Meyer T, Lorenzen T, Stoehr A, Arndt R, Plettenberg A. 2004. Epidemiology of HHV-8 infection in HIV-positive patients with and without Kaposi sarcoma: diagnostic relevance of serology and PCR. J. Clin. Virol. 30:145–149 [DOI] [PubMed] [Google Scholar]
- 13.Laney AS, Peters JS, Manzi SM, Kingsley LA, Chang Y, Moore PS. 2006. Use of a multiantigen detection algorithm for diagnosis of Kaposi's sarcoma-associated herpesvirus infection. J. Clin. Microbiol. 44:3734–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burbelo PD, Leahy HP, Groot S, Bishop LR, Miley W, Iadarola MJ, Whitby D, Kovacs JA. 2009. Four-antigen mixture containing v-cyclin for serological screening of human herpesvirus 8 infection. Clin. Vaccine Immunol. 16:621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng D, Wan J, Cho YG, Wang L, Chiou CJ, Pai S, Woodard C, Zhu J, Liao G, Martinez-Maza O, Qian J, Zhu H, Hayward GS, Ambinder RF, Hayward SD. 2011. Comparison of humoral immune responses to Epstein-Barr virus and Kaposi's sarcoma-associated herpesvirus using a viral proteome microarray. J. Infect. Dis. 204:1683–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rose TM, Strand KB, Schultz ER, Schaefer G, Rankin GW, Jr, Thouless ME, Tsai CC, Bosch ML. 1997. Identification of two homologs of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in retroperitoneal fibromatosis of different macaque species. J. Virol. 71:4138–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruce AG, Bakke AM, Bielefeldt-Ohmann H, Ryan JT, Thouless ME, Tsai CC, Rose TM. 2006. High levels of retroperitoneal fibromatosis (RF)-associated herpesvirus in RF lesions in macaques are associated with ORF73 LANA expression in spindleoid tumour cells. J. Gen. Virol. 87:3529–3538 [DOI] [PubMed] [Google Scholar]
- 18.Bielefeldt-Ohmann H, Barouch DH, Bakke AM, Bruce AG, Durning M, Grant R, Letvin NL, Ryan JT, Schmidt A, Thouless ME, Rose TM. 2005. Intestinal stromal tumors in a simian immunodeficiency virus-infected, simian retrovirus-2 negative rhesus macaque (Macaca mulatta). Vet. Pathol. 42:391–396 [DOI] [PubMed] [Google Scholar]
- 19.Burnside KL, Ryan JT, Bielefeldt-Ohmann H, Gregory Bruce A, Thouless ME, Tsai CC, Rose TM. 2006. RFHVMn ORF73 is structurally related to the KSHV ORF73 latency-associated nuclear antigen (LANA) and is expressed in retroperitoneal fibromatosis (RF) tumor cells. Virology 354:103–115 [DOI] [PubMed] [Google Scholar]
- 20.Desrosiers RC, Sasseville VG, Czajak SC, Zhang X, Mansfield KG, Kaur A, Johnson RP, Lackner AA, Jung JU. 1997. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J. Virol. 71:9764–9769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexander L, Denekamp L, Knapp A, Auerbach MR, Damania B, Desrosiers RC. 2000. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J. Virol. 74:3388–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Searles RP, Bergquam EP, Axthelm MK, Wong SW. 1999. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 73:3040–3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rose TM. 2005. CODEHOP-mediated PCR—a powerful technique for the identification and characterization of viral genomes. Virol. J. 2:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schultz ER, Rankin GW, Jr, Blanc MP, Raden BW, Tsai CC, Rose TM. 2000. Characterization of two divergent lineages of macaque rhadinoviruses related to Kaposi's sarcoma-associated herpesvirus. J. Virol. 74:4919–4928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greensill J, Sheldon JA, Renwick NM, Beer BE, Norley S, Goudsmit J, Schulz TF. 2000. Two distinct gamma-2 herpesviruses in African green monkeys: a second gamma-2 herpesvirus lineage among old world primates? J. Virol. 74:1572–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruce AG, Bakke AM, Thouless ME, Rose TM. 2005. Development of a real-time QPCR assay for the detection of RV2 lineage-specific rhadinoviruses in macaques and baboons. Virol. J. 2:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White JA, Todd PA, Yee JL, Kalman-Bowlus A, Rodgers KS, Yang X, Wong SW, Barry P, Lerche NW. 2009. Prevalence of viremia and oral shedding of rhesus rhadinovirus and retroperitoneal fibromatosis herpesvirus in large age-structured breeding groups of rhesus macaques (Macaca mulatta). Comp. Med. 59:383–390 [PMC free article] [PubMed] [Google Scholar]
- 28.Wong SW, Bergquam EP, Swanson RM, Lee FW, Shiigi SM, Avery NA, Fanton JW, Axthelm MK. 1999. Induction of B cell hyperplasia in simian immunodeficiency virus-infected rhesus macaques with the simian homologue of Kaposi's sarcoma-associated herpesvirus. J. Exp. Med. 190:827–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barcy S, De Rosa SC, Vieira J, Diem K, Ikoma M, Casper C, Corey L. 2008. Gamma delta+ T cells involvement in viral immune control of chronic human herpesvirus 8 infection. J. Immunol. 180:3417–3425 [DOI] [PubMed] [Google Scholar]
- 30.Bruce AG, Bakke AM, Gravett CA, DeMaster LK, Bielefeldt-Ohmann H, Burnside KL, Rose TM. 2009. The ORF59 DNA polymerase processivity factor homologs of Old World primate RV2 rhadinoviruses are highly conserved nuclear antigens expressed in differentiated epithelium in infected macaques. Virol. J. 6:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuller L, Watanabe R, Anderson D, Grant R. 2005. Development of a whole-virus multiplex flow cytometric assay for antibody screening of a specific pathogen-free primate colony. Diagn. Microbiol. Infect. Dis. 53:185–193 [DOI] [PubMed] [Google Scholar]
- 32.Dias D, Van Doren J, Schlottmann S, Kelly S, Puchalski D, Ruiz W, Boerckel P, Kessler J, Antonello JM, Green T, Brown M, Smith J, Chirmule N, Barr E, Jansen KU, Esser MT. 2005. Optimization and validation of a multiplexed Luminex assay to quantify antibodies to neutralizing epitopes on human papillomaviruses 6, 11, 16, and 18. Clin. Diagn. Lab. Immunol. 12:959–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rose TM, Ryan JT, Schultz ER, Raden BW, Tsai CC. 2003. Analysis of 4.3 kilobases of divergent locus B of macaque retroperitoneal fibromatosis-associated herpesvirus reveals a close similarity in gene sequence and genome organization to Kaposi's sarcoma-associated herpesvirus. J. Virol. 77:5084–5097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White JA, Yang X, Todd PA, Lerche NW. 2011. Longitudinal patterns of viremia and oral shedding of rhesus rhadinovirus and retroperitoneal fibromatosis herpesviruses in age-structured captive breeding populations of rhesus macaques (Macaca mulatta). Comp. Med. 61:60–70 [PMC free article] [PubMed] [Google Scholar]
- 35.Boshoff C, Schulz TF, Kennedy MM, Graham AK, Fisher C, Thomas A, McGee JO, Weiss RA, O'Leary JJ. 1995. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat. Med. 1:1274–1278 [DOI] [PubMed] [Google Scholar]
- 36.Bechtel JT, Liang Y, Hvidding J, Ganem D. 2003. Host range of Kaposi's sarcoma-associated herpesvirus in cultured cells. J. Virol. 77:6474–6481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruce AG, Bielefeldt-Ohmann H, Barcy S, Bakke AM, Lewis P, Tsai CC, Murnane RD, Rose TM. 2012. Macaque homologs of EBV and KSHV show uniquely different associations with simian AIDS-related lymphomas. PLoS Pathog. 8:e1002962 doi:10.1371/journal.ppat.1002962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coscoy L, Ganem D. 2000. Kaposi's sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc. Natl. Acad. Sci. U. S. A. 97:8051–8056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris S, Lang SM, Means RE. 2010. Characterization of the rhesus fibromatosis herpesvirus MARCH family member rfK3. Virology 398:214–223 [DOI] [PubMed] [Google Scholar]
- 40.Casper C, Krantz E, Taylor H, Dalessio J, Carrell D, Wald A, Corey L, Ashley R. 2002. Assessment of a combined testing strategy for detection of antibodies to human herpesvirus 8 (HHV-8) in persons with Kaposi's sarcoma, persons with asymptomatic HHV-8 infection, and persons at low risk for HHV-8 infection. J. Clin. Microbiol. 40:3822–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okroj M, Tedeschi R, Mancuso R, Brambilla L, Tourlaki A, Dillner J, Blom AM. 2011. Prevalence of antibodies against Kaposi's sarcoma associated herpes virus (KSHV) complement inhibitory protein (KCP) in KSHV-related diseases and their correlation with clinical parameters. Vaccine 29:1129–1134 [DOI] [PubMed] [Google Scholar]
- 42.Anderson S, Wakeley P, Wibberley G, Webster K, Sawyer J. 2011. Development and evaluation of a Luminex multiplex serology assay to detect antibodies to bovine herpes virus 1, parainfluenza 3 virus, bovine viral diarrhoea virus, and bovine respiratory syncytial virus, with comparison to existing ELISA detection methods. J. Immunol. Methods 366:79–88 [DOI] [PubMed] [Google Scholar]
- 43.Piriou E, Kimmel R, Chelimo K, Middeldorp JM, Odada PS, Ploutz-Snyder R, Moormann AM, Rochford R. 2009. Serological evidence for long-term Epstein-Barr virus reactivation in children living in a holoendemic malaria region of Kenya. J. Med. Virol. 81:1088–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]