Abstract
Lyme disease is the fastest-growing zoonotic disease in North America. Current methods for detection of Borrelia burgdorferi infection are challenged by analysis subjectivity and standardization of antigen source. In the present study, we developed an immuno-PCR (iPCR)-based approach employing recombinant in vivo-expressed B. burgdorferi antigens for objective detection of a host immune response to B. burgdorferi infection. iPCR is a liquid-phase protein detection method that combines the sensitivity of PCR with the specificity and versatility of immunoassay-based protocols. Use of magnetic beads coated with intact spirochetes provided effective antigen presentation and allowed detection of host-generated antibodies in experimentally infected mice at day 11 postinoculation, whereas host-generated antibodies were detected at day 14 by enzyme-linked immunosorbent assay (ELISA) and day 21 by immunoblotting. Furthermore, magnetic beads coated with recombinant B. burgdorferi in vivo-expressed antigen OspC or BmpA demonstrated positive detection of host-generated antibodies in mice at day 7 postinoculation with markedly increased iPCR signals above the background, with the quantification cycle (Cq) value for each sample minus the mean background Cq plus 3 standard deviations (ΔCq) being 4 to 10, whereas ΔCq was 2.5 for intact spirochete-coated beads. iPCR demonstrated a strong correlation (Spearman rank correlation = 0.895, P < 0.0001) with a commercial ELISA for detection of host antibodies in human Lyme disease patient sera using the B. burgdorferi VlsE C6 peptide. In addition, iPCR showed potential applicability for direct detection of spirochetes in blood. The results presented here indicate that our iPCR assay has the potential to provide an objective format that can be used for sensitive detection of multiple host response antibodies and isotypes to B. burgdorferi infection.
INTRODUCTION
Lyme disease is the leading vector-borne bacterial disease in the world, with approximately 30,000 cases reported in the United States alone each year (http://www.cdc.gov/lyme/). Lyme disease has been characterized as the fastest-growing zoonotic disease in North America. According to the Centers for Disease Control and Prevention (CDC), the number of clinical cases of Lyme disease has more than doubled over the past 10 years, making this emerging infectious disease a major public health concern (http://www.cdc.gov/lyme/). Accurate diagnosis is currently the greatest challenge for the clinical management of Lyme disease. Misdiagnosis is common, as the clinical manifestations of the disease are not unique and detection of a Borrelia burgdorferi infection is difficult and prone to misinterpretation (1, 2). Different approaches for laboratory testing, such as microscopy, genomic DNA amplification, and serology, have been examined, with currently accepted laboratory diagnostics primarily relying on detection of a serological response to B. burgdorferi antigens (1, 3, 4).
Current methods for detection of Lyme disease in a clinical setting approved by the CDC entail a two-tiered approach using a first-tier enzyme immunoassay (EIA) followed by a second-tier immunoblot assay for both IgM and IgG B. burgdorferi-specific antibodies using whole-cell B. burgdorferi lysates, recombinant antigens, or various combinations, depending on the commercial kit used (1). Although adequate, the approach suffers from certain drawbacks, including the subjectivity of immunoblot analysis and the lack of standardization of antigen source and lysate preparations. These challenges have resulted in discordant results between test strategies for detection of host antibodies on the basis of the kit used (5) largely due to lysate/antigen reagent variability (1). The most effective approach appears to be the use of a combination of recombinant antigens to replace whole-organism sonicates, as no single antigen has been found to be sufficient for accurate diagnosis (1).
Other methods for detection of Lyme disease include live culture and approaches employing PCR. Live culture has shown limited success in a clinical setting, is time-consuming, and requires complex media that have a limited commercial supply (1). PCR appears to be the most promising method for direct detection of spirochetes but has not been widely accepted for laboratory diagnosis due to low sensitivity in cerebrospinal fluid and blood and the potential for false-positive results due to accidental laboratory contamination of samples with small quantities of target DNA (6). An improved approach would be to utilize the sensitivity of PCR combined with an antigen-based detection system that is much less susceptible to false-positive results.
Immuno-PCR (iPCR) was first introduced by Sano et al. in 1992 (7) and combines the amplification power of PCR with the versatility of EIA, resulting in improved conventional antigen detection sensitivity. Using iPCR, a typical 100- to 10,000-fold improvement over the detection limit of the EIA has been obtained in almost all applications (8). iPCR has been used to detect viral antigens (9), bacterial antigens (10), prions (11), and bacterial toxins (12). There has also been a limited application of iPCR for antibody detection, such as the measurement of mumps virus-specific immunoglobulin G in human serum (13).
The combination of an iPCR approach and recombinant B. burgdorferi in vivo-expressed antigens has the potential to alleviate a number of the issues posed by Lyme disease diagnostics. Recombinant antigens not only have the potential to standardize the reagents used for Lyme disease diagnostics but also provide the opportunity to combine antigens from multiple strains/species. The sensitivity, ease of use, objective analysis, and multiplex capabilities of iPCR (14) also make it an ideal platform for Lyme disease detection. Furthermore, iPCR has the ability to be translated to an automated point-of-care diagnostic platform using microfluidics (15) that may allow routine, high-throughput, and affordable diagnostic testing of patients for Lyme disease. The goal of this research was to explore the initial application of iPCR using recombinant antigens for either detection of host-generated antibodies or direct detection of spirochetes in B. burgdorferi-infected samples.
MATERIALS AND METHODS
Bacterial strains.
B. burgdorferi clones B31 A3 (16) and B31 A34/pBSV2G-loxP-flaBp-gfp (17) were used in these studies. Spirochetes were grown in liquid Barbour-Stoenner-Kelly (BSK) II medium supplemented with gelatin and 6% rabbit serum (18) and plated in solid BSK medium as previously described (19). All spirochete cultures were grown at 35°C and supplemented with 2.5% CO2. Gentamicin was used at 40 μg/ml. Escherichia coli strains DH5α and BL21 (Novagen, Billerica, MA) were grown in LB broth, on LB agar plates, or in Magic Media (Invitrogen, Carlsbad, CA) containing 100 μg/ml ampicillin.
Mouse infections.
The University of Central Florida (UCF) is accredited by the International Association for Assessment and Accreditation of Laboratory Animal Care. Protocols for all animal experiments were prepared according to the guidelines of the National Institutes of Health and approved by UCF's Institutional Animal Care and Use Committee. For the serological detection experiments, the hair on the upper backs of three mice (6- to 8-week-old C3H/HeN females; Harlan Laboratories, Inc., Dublin, VA) was removed by shaving, and the mice were needle inoculated intradermally on the upper back with B. burgdorferi strain B31 A3 at a dose of 1 × 105 spirochetes divided between two 50-μl inoculations. The number of spirochetes inoculated into mice was determined using a Petroff-Hausser counting chamber and verified by CFU counts in solid BSK medium. The total plasmid content of each inoculum was confirmed to be as expected (20). Whole-blood samples were collected from the three inoculated mice as well as one noninoculated mouse by submandibular bleed preinoculation and at days 1, 3, 4, 7, 9, 11, 14, 16, 18, and 21 postinfection. The coagulated blood was spun at 4,000 × g for 9 min to prepare serum. For the spirochete detection experiments, six mice (6- to 8-week-old C3H/HeN females; Harlan Laboratories, Inc., Dublin, VA) were inoculated intradermally with B. burgdorferi strain B31 A3 at a dose of 1 × 105 spirochetes. Approximately 50 μl of blood was collected by submandibular bleed from all mice prior to inoculation. Subsequently, to prevent complications due to oversampling, approximately 50 μl of blood/mouse was collected every day from groups of two mice so that each group of two mice was bled every 3 days over a time period of 14 days. All blood samples (pre- and postinoculation) were supplemented with an equal volume of 0.5 M sodium EDTA to prevent coagulation.
Similar to plating of in vitro-grown B. burgdorferi, 50 μl of blood from each mouse was combined with BSK plating medium (19) supplemented with a Borrelia antibiotic cocktail consisting of 20 μl/ml phosphomycin (MP Biomedicals, Santa Ana, CA), 50 μl/ml rifampin (Fisher Scientific, Waltham, MA), and 2.5 μl/ml amphotericin B (Fisher Scientific, Waltham, MA), all solubilized in 20% dimethyl sulfoxide. The mixture was poured into sterile petri plates, allowed to solidify, and incubated as indicated above for approximately 7 days, until B. burgdorferi colonies were visible in the solid medium.
Immunoblotting and C6 peptide ELISA.
Total B. burgdorferi lysate for immunoblot analysis was prepared from a 500-ml culture of 1 × 108/ml B. burgdorferi B31 A3. Spirochetes were harvested by centrifugation and washed two times in 30 ml phosphate-buffered saline (PBS; pH 7.4). Washed cells were resuspended in 30 ml PBS and disrupted by sonication on ice using a Misonix model S-4000 sonicator at 40% amplitude for four repetitions of 20 s each. Total protein in the sonicate was normalized to 1 mg/ml with PBS on the basis of the absorbance at 280 nm, and 75 μg of protein was separated by 12.5% polyacrylamide gel electrophoresis. Following protein transfer, nitrocellulose membranes were incubated for 1 h with pre- and postinoculation mouse serum diluted 1:200 in Tris-buffered saline–0.05% Tween (TBST; pH 7.6), washed twice with TBST, incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG/IgM (Chemicon International, Billerica, MA) for 1 h, and washed twice with TBST; and the signal was detected using a SuperSignal West Pico chemiluminescent substrate kit (Thermo Scientific, Rockford, IL). The C6 peptide B. burgdorferi enzyme-linked immunosorbent assay (ELISA) was performed according to the manufacturer's protocol (Immunetics, Boston, MA), with the exception of the use of HRP-conjugated goat anti-mouse IgG/IgM secondary antibody (Chemicon International, Billerica, MA) at a 1:5,000 dilution in place of the anti-human reporter antibody provided with the kit when mouse sera were analyzed.
Cloning and expression of recombinant GST-tagged antigens.
In-frame glutathione S-transferase (GST) fusion proteins for OspC, BmpA, and the VlsE C6 peptide were generated by PCR amplifying the corresponding coding regions without the signal sequences from B. burgdorferi genomic DNA using primer pairs P1 and P2 (OspC), P3 and P4 (BmpA), or P5 and P6 (VlsE C6) engineered with a BamHI or SalI restriction site (Table 1) and Phusion polymerase (New England BioLabs, Ipswich, MA). PCR products were purified (Qiagen, Valencia, CA), digested with restriction enzymes (New England BioLabs, Ipswich, MA), and cloned into BamHI/SalI-digested pGEX-6P-1 (GE Healthcare, Piscataway, NJ) to generate translational fusions with GST at the N terminus. Subsequent clones were selected and sequence confirmed by dideoxy sequencing. Hemagglutinin (HA; OspC) and c-Myc (BmpA) tags were included at the C terminus for determination of protein purity by immunoblotting. pGEX-6P-1 plasmids carrying ospC, bmpA, or vlsE c6 were transformed into a BL21 strain of E. coli (Novagen, Billerica, MA). Protein expression and purification were performed according to the procedures outlined in the Bulk GST purification module (GE Health Sciences, Piscataway, NJ).
Table 1.
iPCR DNA oligonucleotide sequences used in this study
| Oligonucleotide no. | Oligonucleotide identification | Sequence (5′–3′)a |
|---|---|---|
| T1 | Template 1 (IgG coupled) | Biotin-AGCCTCAGACCAAGCCAGACAACTGCCTCGTGACGTTGCTGCCCCTACCAACGTACCCCTACGAGTCC |
| T1F | Template 1 forward | AGCCTCAGACCAAGCCAGAC |
| T1R | Template 1 reverse | GGACTCGTAGGGGTACGTTGG |
| T1P | Template 1 probe | FAM-ACTGCCTCGTGACGTTGCTGCCCCT-BHQ1 |
| T2 | Template 2 (IgM coupled) | Biotin-AGGAGGAGGGTCAAGTCACCAACGCTGCTCCAGGCCATCGTGCTGATCTGGACCCTGGATCGAGTGA |
| T2F | Template 2 forward | AGGAGGAGGGTCAAGTCACC |
| T2R | Template 2 reverse | TCACTCGATCCAGGGTCCAG |
| T2P | Template 2 probe | MAX-ACGCTGCTCCAGGCCATCGTGCTGA-BHQ1 |
| P1 | OspC partial HA forward | cgggatcccatatgTGTAATAATTCAGGGAAAGATGG |
| P2 | OspC HA reverse | acgcgtcgacTTAcgcataatccggcacatcatacggataAGGTTTTTTTGGACTTTCTGC |
| P3 | BmpA partial myc forward | cgggatcccatATGTGTAGTGGTAAAGGTAGTCTTG |
| P4 | BmpA myc reverse | acgcgtcgacTTAcagatcttcttcagaaataagtttttgttcAATAAATTCTTTAAGAAACTTCTCATAAC |
| P5 | C6 Bb forward | cgggatcccatATGAAGAAGGATGATCAGATTG |
| P6 | C6 Bb reverse | acgcgtcgacTTACTTCACAGCAAACTTTCCATC |
Lowercase letters indicate nontemplate sequence used for addition of terminal restriction sites and/or epitope tags. BHQ1, black hole quencher 1.
iPCR reagent preparation.
iPCR assays were assembled in a two-sided (sandwich) manner for both host antibody (Fig. 1A and B) and spirochete capture (Fig. 1C). Whole-cell lysate used for immunoblot analysis (preparation described above) and GST-fusion recombinant antigens were used to coat magnetic beads for host antibody capture using a Dynabeads antibody coupling kit (Invitrogen, Carlsbad, CA). Bead-coupling reactions were performed overnight according to the manufacturer's protocol using 20 to 30 μg antigen(s) per mg Dynabeads M-270 epoxy. The primary antibody used for spirochete capture consisted of protein A-purified anti-B. burgdorferi polyclonal antibody raised in rabbits against whole-cell preparations of B. burgdorferi clone B31 ATCC 35210 (Acris Antibodies, San Diego, CA) and was coupled to magnetic beads as described above. Protein-coated beads were stored at 4°C. The streptavidin-conjugated reporter antibodies were prepared using a Lightning-Link streptavidin conjugation kit (Innova Biosciences, Cambridge, United Kingdom) and polyclonal anti-B. burgdorferi (Acris Antibodies, San Diego, CA), goat anti-mouse IgM/IgG (Sigma-Aldrich, St. Louis, MO), goat anti-human IgG (Invitrogen, Carlsbad, CA), or goat-anti-human IgM (Invitrogen, Carlsbad, CA) antibody according to the manufacturer's protocols using an overnight incubation. Following conjugation, 10 μl of streptavidin-labeled antibody was diluted 1:50 in TBST, 100 nM single-stranded biotin-labeled oligonucleotide template was added, and the mixture was rotated at room temperature for 30 min for antibody-oligonucleotide conjugation. Oligonucleotide sequences T1 (IgG coupled) and T2 (IgM coupled) used for tagging are listed in Table 1. The oligonucleotide-linked streptavidin-conjugated antibody was then diluted to a 1:100 working stock (final dilution, 1:5,000) and stored at 4°C.
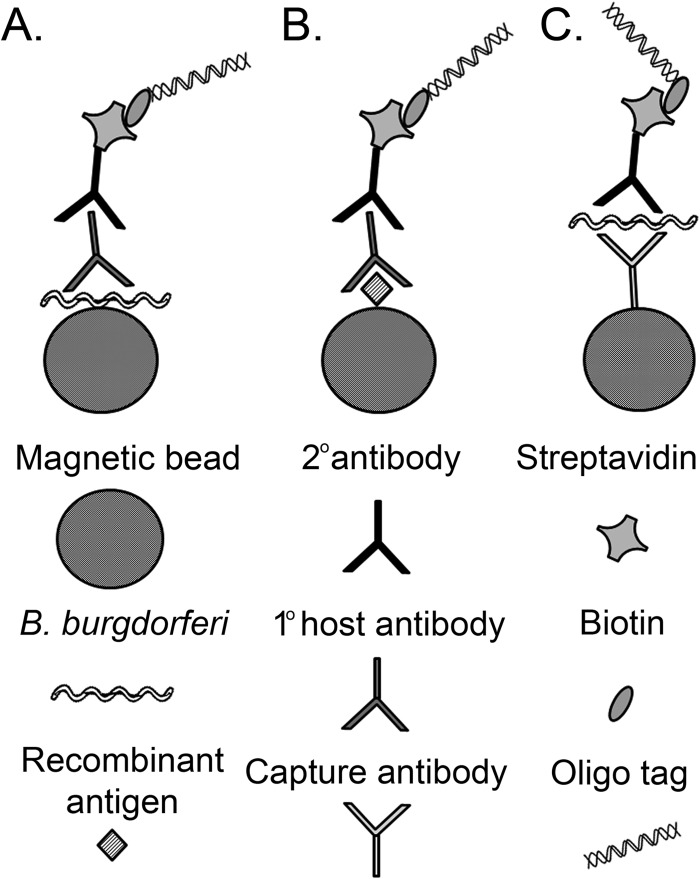
Fig 1.
Schematic representation of iPCR assay for detection of Lyme disease biomarkers. Intact spirochete (A) or recombinant protein antigen coupled to magnetic beads (B) was used to capture B. burgdorferi-specific host-generated antibodies. A biotinylated DNA oligonucleotide reporter molecule coupled to a streptavidin-conjugated reporter antibody was amplified by qPCR for detection and quantification. (C) Anti-B. burgdorferi antibody coupled to magnetic beads was used for spirochete capture, with detection accomplished by qPCR amplification of the DNA oligonucleotide-coupled reporter antibody, similar to detection of host antibody.
iPCR assay.
Following reagent preparation, 10 μl of antigen- or antibody-coated magnetic beads was incubated in 500 μl TBST for 30 min at 25°C on a rotator. Following preliminary washing, beads were resuspended in 500 μl TBST and 5 μl serum (mouse or human), 10 μl spirochetes was suspended in HN buffer (50 mM HEPES, 50 mM NaCl; pH 7.5) or blood (1 × 108 to 1 × 104/ml B. burgdorferi B31 A3) or no serum/spirochete was used (negative control), and the mixture was incubated at 25°C with rotation for 30 min. Beads were subsequently washed and resuspended in 300 μl TBST with the addition of 100 μl each of IgG and IgM diluted (1:5,000) biotinylated oligonucleotide streptavidin-coupled reporter antibody (anti-mouse IgM/IgG, anti-human IgG, anti-human IgM, or anti-B. burgdorferi), and the mixture was incubated at 25°C with rotation for 30 min. Following assembly of the immune complex, beads were washed three times with 900 μl TBST, followed by magnetic bead capture. Washed immune complex-coupled beads were resuspended in 20 μl TBST for subsequent PCR amplification.
Signal amplification by real-time PCR.
To amplify the signal of the immune complex, real-time PCR was performed using an Applied Biosystems 7900 HT system (Life Technologies, Grand Island, NY) and IQ Supermix (Bio-Rad, Hercules, CA) supplemented with synthetic primers and probes T1F/T1R/T1P (IgG detection) or T2F/T2R/T2P (IgM detection) (Table 1). Duplicate reaction mixtures were prepared in 20-μl volumes containing 5 μl of iPCR assay-processed beads as the template, 10 μl of 2× reaction mix, 0.2 μM each primer, and 0.4 μM fluorophore-labeled probe. Cycle parameters included a preliminary denaturation (95°C, 20 s), followed by 40 cycles of denaturation (95°C, 1 s) and annealing/extension (60°C, 20 s). The fluorescent signal was collected at the 6-carboxyfluorescein (FAM) wavelength for IgG reactions and MAX (NHS ester; Integrated DNA Technologies, Coralville, IA) wavelength for IgM reactions. The quantification cycle (Cq) for each reaction was determined using automatic baseline and threshold settings. The average and standard deviation for uninfected/healthy samples were used to determine the background level of amplification, as is commonly observed for iPCR protocols. Positive threshold values were established at three times the standard deviation for background levels.
Human sera.
Retrospective, deidentified human Lyme disease and healthy control serum samples were kindly supplied by Martin Schriefer (Centers for Disease Control and Prevention, Fort Collins, CO). Sera were collected from 18 Lyme disease patients from regions where Lyme disease is endemic upon the initial visit to a physician and 10 days after the initial visit (n = 36). According to CDC's two-tiered serological analysis of the samples, 5 of the patients were two-tiered analysis positive at both the initial and follow-up time points, 3 of the patients were two-tiered analysis negative at both time points, and 10 of the patients were two-tiered analysis negative at the initial visit but two-tiered analysis positive 10 days later. Human control samples consisted of sera collected from healthy blood donors living in areas where Lyme disease is not endemic (n = 5).
Statistical analysis.
Spearman rank correlation analyses were performed using GraphPad Prism software, version 5.0.
RESULTS
iPCR using intact spirochetes provided earlier detection of host response than immunoblotting and C6 ELISA in a murine model.
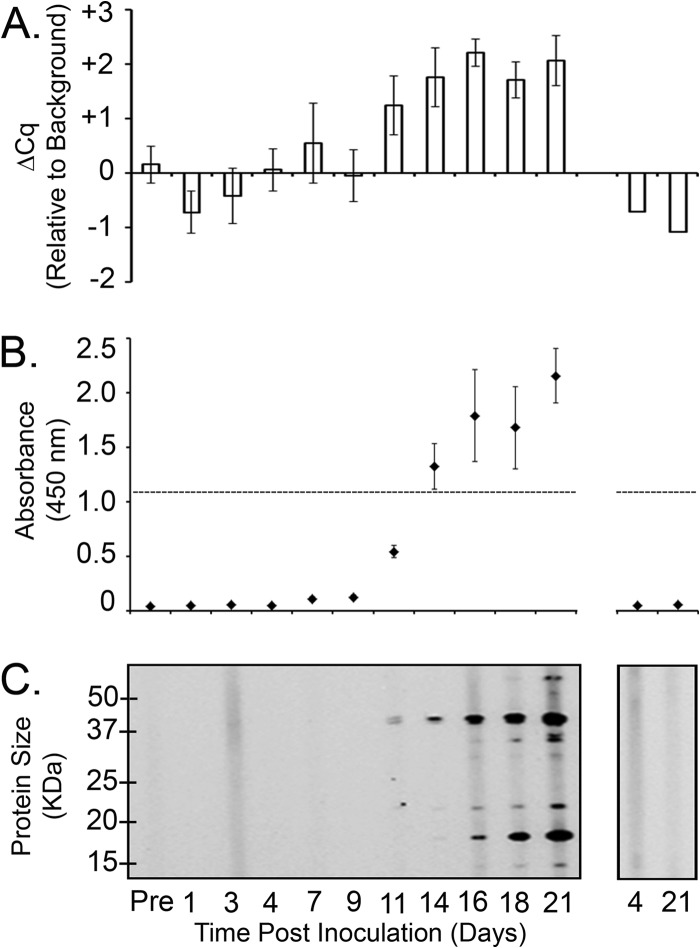
The general approach for detection of a host antibody immune response by immunoassay is to use sonicated or otherwise disrupted organisms to generate protein antigens for antibody capture and subsequent detection. However, we hypothesized that this approach may have limited success for effectively capturing anti-B. burgdorferi antibodies in experimentally infected mouse serum, as the majority of the B. burgdorferi proteins in the total cell lysate are not likely to be immunogenic. Although B. burgdorferi lysate is known to harbor antigenic proteins recognized by mouse and human immune sera, these proteins represent a small percentage of the total proteins in the lysate and therefore may not provide an improved sensitivity of detection of an immune response to B. burgdorferi infection. In an effort to develop a sensitive, objective method for detection of host antibodies against B. burgdorferi antigens, magnetic beads were coated with a polyclonal anti-B. burgdorferi antibody in order to capture formalin-fixed intact spirochetes, resulting in the generation of magnetic beads coated with intact spirochetes (Fig. 2). We predicted that this strategy would result in magnetic beads coated in an enriched pool of spirochete antigenic outer surface proteins capable of interacting specifically with host antibodies produced in response to a B. burgdorferi infection. The sensitivity of iPCR using intact spirochetes to capture host antibodies was compared to the sensitivities of preexisting diagnostic methods, including a commercial C6 ELISA and immunoblotting, using an in vivo murine model. iPCR resulted in the earliest objective detection of a positive infection on day 11 postinoculation (Fig. 3A). In comparison, C6 ELISA and immunoblotting exhibited positive detection of anti-B. burgdorferi antibodies at day 14 and day 21 postinoculation, respectively (Fig. 3B and C). The approximate molecular masses of the immunodominant proteins detected on the immunoblot included 18 kDa, 23 kDa, 33 kDa, 39 kDa, and 66 kDa, which are consistent with the sizes of the bands typically present on a Lyme disease diagnostic immunoblot (21, 22). Uninfected mouse serum was negative by all three methods at all time points tested (Fig. 3).
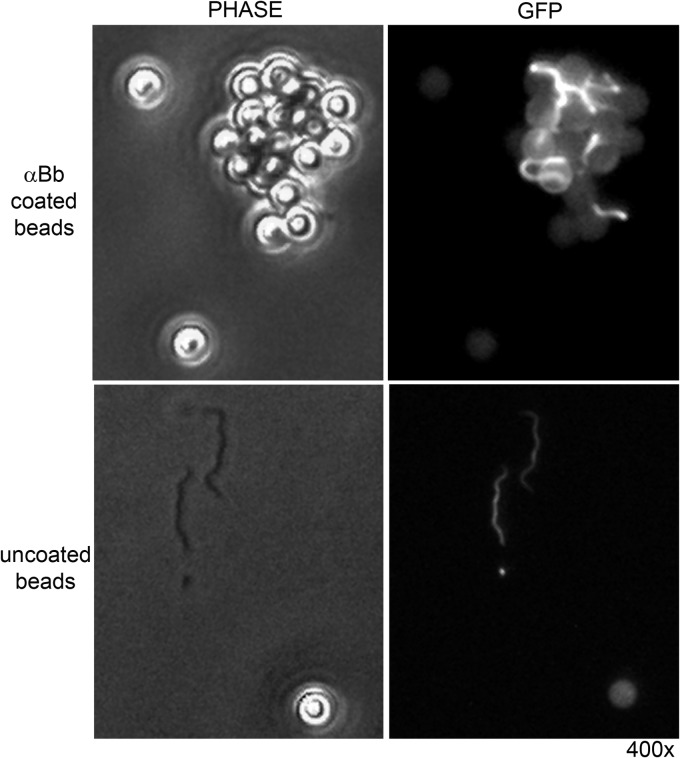
Fig 2.
B. burgdorferi captured on magnetic beads provides a reagent for host antibody detection by iPCR. Phase-contrast microscopy and fluorescence microscopy at 510 nm (GFP [green fluorescent protein]) and ×400 magnification (400×) were used to determine capture of formalin-fixed B. burgdorferi expressing green fluorescent protein on beads coated with anti-B. burgdorferi (αBb) polyclonal antibodies (top) or uncoated beads (bottom).
Fig 3.
iPCR demonstrated earlier detection of host response antibodies in B. burgdorferi-infected mice than C6 ELISA and immunoblotting. Mouse sera were collected prior to inoculation (Pre), on specific days after intradermal inoculation with 1 × 105 B. burgdorferi B31 A3 spirochetes (left), or from uninfected mice (right) over the course of 21 days. (A) Undiluted sera were analyzed for detection of B. burgdorferi IgG antibodies using iPCR. A closed-system, real-time PCR of the DNA reporter molecule was performed using a TaqMan-based fluorescent probe assay. The mean Cq background signal, determined using uninfected sera plus 3 standard deviations, was designated the call threshold for a positive detection event and indicated here as a ΔCq of 0. Data are shown as the Cq value for each sample minus the mean background Cq plus 3 standard deviations (ΔCq). Each data point represents the average of three mice, and the standard deviation between samples is shown. (B) C6 ELISA (Immunetics, Inc., Boston, MA) was performed according to the manufacturer's instructions, with the exception that the secondary antibody was peroxidase-conjugated goat anti-mouse IgM/IgG (1:5,000). The threshold absorbance for the test is indicated (horizontal broken line). Each point represents the average of three mice, and the standard deviation between samples is shown. (C) Total B. burgdorferi sonicate was separated by SDS-PAGE and analyzed by IgM/IgG immunoblotting using immune and preimmune mouse sera diluted 1:200. The positions of the protein standards depict molecular masses (in kilodaltons). Data are representative of three mice analyzed.
iPCR using recombinant GST-OspC and GST-BmpA provided improved sensitivity of detection of murine host antibodies.
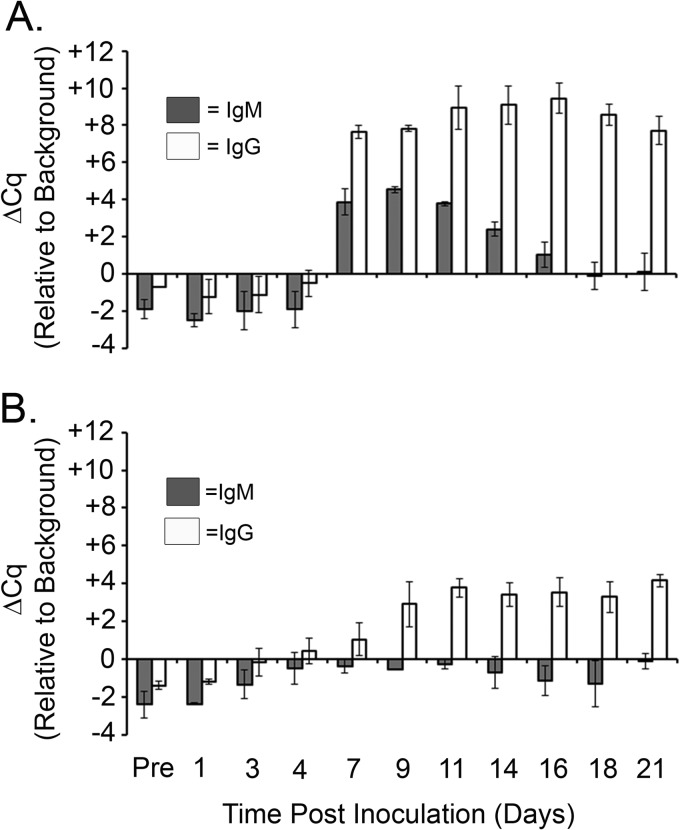
Although beads coated with intact in vitro-grown spirochetes provided early detection of anti-B. burgdorferi antibodies compared to the time to detection by C6 ELISA and immunoblotting (Fig. 3), we hypothesized that the use of specific recombinant antigens known to be actively expressed during murine infection could potentially result in a more sensitive approach. Known B. burgdorferi in vivo-expressed antigens OspC and BmpA (1) were produced and purified as recombinant N-terminal GST-tagged fusion proteins in E. coli. Magnetic beads coated with either recombinant protein were used to capture host antibodies generated against OspC or BmpA, respectively, and IgM and IgG antibodies against each protein were individually quantitated using our iPCR assay. The use of GST-OspC-coated beads resulted in a marked increase in detection of host antibodies starting at day 7 postinoculation for both IgG and IgM (Fig. 4A), with a gradual decrease in the IgM signal back to baseline by day 21 and a minimal decrease in the IgG signal to the same time point. GST-OspC-coated beads provided a dramatic increase in the level of IgG detection (Cq value for each sample minus the mean background Cq plus 3 standard deviations [ΔCq] = 10) compared to the level of iPCR detection of host antibodies using intact spirochete-coated beads (ΔCq = 2.5). GST-BmpA-coated beads provided robust positive detection of IgG antibodies beginning at day 9, followed by a minimal decrease in the detection signal out to day 21 (Fig. 4B). IgM antibodies directed against BmpA demonstrated a slight increase in signal over the 21-day time course of infection but were not detected at levels significantly above background levels, suggesting that BmpA does not elicit a serodiagnostic IgM response. Together these data suggest that the use of magnetic beads coated with specific recombinant B. burgdorferi in vivo-expressed antigens results in robust iPCR detection of a humoral response in mice experimentally infected with B. burgdorferi and that development of an iPCR assay that quantitates the host response to multiple B. burgdorferi antigens may result in an improved diagnostic method.
Fig 4.
iPCR using recombinant antigens OspC and BmpA provided enhanced sensitivity for detection of both IgG and IgM isotypes in a murine model of infection. Magnetic beads coated with either purified recombinant GST-OspC (A) or GST-BmpA (B) protein were used to capture host response antibodies from preimmune (pre) or postimmune mouse sera collected over a time period of 21 days. IgM- and IgG-specific reporter antibody-DNA conjugates detected anti-B. burgdorferi antibodies captured by each set of antigen-coated beads. The IgM and IgG response to each antigen was determined for each mouse by multiplex quantitative PCR using distinct probes specific for the IgM- and IgG-specific DNA reporter molecules. The mean Cq background signal, determined using uninfected sera, plus 3 standard deviations was designated the call threshold for a positive detection event and indicated here as a ΔCq of 0. Data are shown as the Cq value for each sample minus the mean background Cq plus 3 standard deviations (ΔCq). Each data point represents the average of two mice, and the standard deviation between samples is shown.
iPCR demonstrated a strong correlation with a commercial ELISA for detection of host antibodies in human serum using the VlsE C6 peptide.
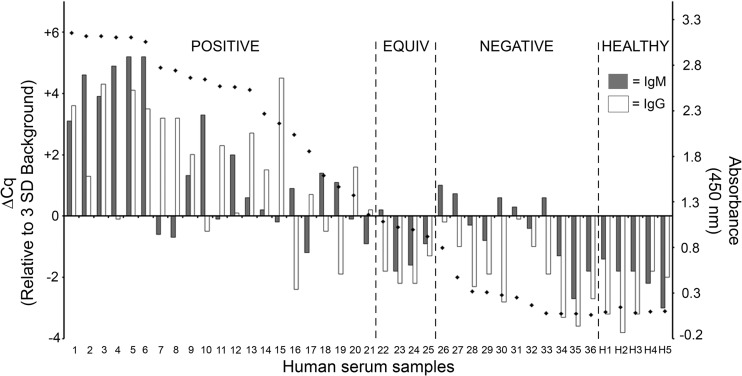
As recommended by the CDC, the first step of two-tier testing for Lyme disease is the use of a sensitive enzyme immunoassay. Although a number of commercial kits exist for testing, the C6 peptide of the VlsE locus has been shown to be a sensitive and effective predictor for follow-up testing by immunoblotting and is available as a commercial testing kit. In order to directly compare the ability of our iPCR assay to detect human antibodies produced against the VlsE C6 peptide with that of an FDA-approved C6 antibody detection method, a panel of serum samples from 18 individuals collected at both an initial visit to the clinic and a 10-day follow-up appointment (n = 36) along with sera collected from 5 healthy patients from areas where Lyme disease is not endemic was analyzed by iPCR and the C6 Lyme disease ELISA (Immunetics, Inc., Boston, MA). iPCR detection of C6-specific host antibodies demonstrated a strong correlation with detection by the commercial C6 ELISA (Spearman rank correlation [rs] = 0. 895, P < 0.0001) (Fig. 5). The iPCR assay differed from the C6 ELISA in that the iPCR assay provided a separate measurement of C6 IgM and C6 IgG antibodies, whereas the C6 ELISA quantitated a combined value for both C6 IgM and C6 IgG antibodies. Therefore, the iPCR result was considered positive if C6 IgM and/or C6 IgG antibodies were detected at or above the established call threshold. All 21 samples that demonstrated a positive result by the C6 ELISA were also positive according to the C6 iPCR (Fig. 5). Of the four serum samples determined to be equivocal by the C6 ELISA, three were found to be negative by C6 iPCR, whereas one sample tested positive for IgM using this method. Furthermore, of the 11 serum samples that tested negative by the C6 ELISA, 5 resulted in positive detection of IgM by C6 iPCR. Of note, all iPCR-positive samples in this group had ΔCq values of 1 or below. All serum samples collected from known healthy individuals tested negative by both the C6 ELISA and C6 iPCR. Together these results suggest that iPCR may have an improved ability to detect host antibodies to the VlsE C6 peptide compared to that of a current commercial method.
Fig 5.
Recombinant antigen iPCR successfully quantified B. burgdorferi VlsE C6 peptide antibodies in human serum samples. Results are for 36 serum samples from 18 Lyme disease patients collected upon the initial visit to a clinic and at a 10-day follow-up visit and 5 healthy controls using a multiplex iPCR protocol to quantitate both IgM and IgG isotypes using recombinant B. burgdorferi VlsE C6 peptide-coated magnetic beads. A call threshold (ΔCq = 0) was assigned at greater than or equal to 3 standard deviations above the mean background signal determined using sera from healthy individuals. Serum samples were also tested using a commercial C6 ELISA (Immunetics, Boston, MA) (diamonds), which was performed according to the manufacturer's protocol, with a call threshold for an absorbance (450 nm) of 1.1 used according to the manufacturer's protocol. The C6 ELISA value represents combined measurement of C6 IgM and IgG antibodies. The data for patients 1 to 36 are grouped into three categories: positive, equivocal (equiv), and negative according to the C6 ELISA values. Samples H1 to H5 correspond to the sera collected from the healthy controls and are grouped accordingly (healthy). The calculated rs values were 0.734 (P < 0.0001) for C6 iPCR IgM versus C6 ELISA, 0.826 (P < 0.0001) for C6 iPCR IgG versus C6 ELISA, and 0.895 (P < 0.0001) for C6 iPCR IgM and/or IgG versus C6 ELISA.
iPCR directly detected B. burgdorferi in blood.
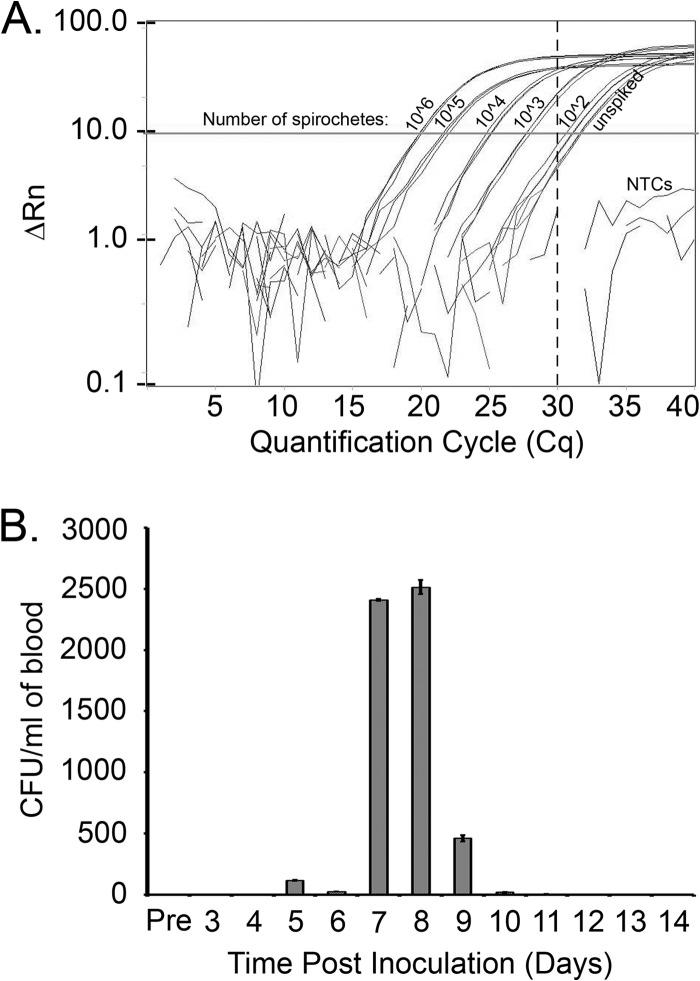
The demonstrated power of iPCR to detect ultralow protein levels (8) suggests that this method may be a promising tool for direct detection of B. burgdorferi in clinical samples. iPCR was shown to be successful for capture of live B. burgdorferi using magnetic beads coated with polyclonal anti-B. burgdorferi antibodies (Fig. 2). This finding suggested the potential for iPCR to directly quantitate spirochetes from within patient samples. To test the sensitivity of iPCR detection of spirochetes, in vitro-grown B. burgdorferi was serially diluted in HN buffer (106 to 102 spirochetes). iPCR detection of spirochetes demonstrated a robust dilution curve and a level of detection of less than 1,000 organisms (Fig. 6A). Detection of in vitro-grown B. burgdorferi spiked into whole uninfected mouse blood resulted in a 10-fold lower limit of detection of 10,000 spirochetes (data not shown), suggesting that components of the blood may have an inhibitory effect on the function of the iPCR assay. To correlate the sensitivity of iPCR detection of spirochetes in blood with quantitation of the number of spirochetes present in the blood of infected mice, cohorts of mice were infected with 1 × 105 B. burgdorferi B31 A3 spirochetes and blood samples were collected every 24 h for a period of 14 days. The number of spirochetes/ml of blood, as determined by CFU counts on solid medium, was found to increase over the first week of infection and reached a peak number of approximately 2,500 spirochetes/ml of blood on day 8 postinoculation (Fig. 6B). The B. burgdorferi colonies that grew out of the infected blood within the solid BSK medium demonstrated a morphology and a growth pattern similar to those that are typically observed for spirochete colonies derived from in vitro-grown cultures (data not shown). Together, these data suggest that although iPCR is a promising method for direct detection of spirochetes in B. burgdorferi-infected samples, the sensitivity of the method is currently below the required level of detection.
Fig 6.
iPCR has the potential to directly detect B. burgdorferi in infected samples. (A) Live spirochetes were serially diluted in HN buffer (106 to 102 spirochetes) and tested in triplicate using iPCR to detect organism capture using anti-B. burgdorferi antibody-coated magnetic beads. A call threshold was assigned at greater than or equal to five times the standard deviation (Cq = 30, vertical broken line) above the mean background signal, as determined using HN buffer alone (unspiked). PCR nontemplate controls (NTCs) included water and TBST used during the iPCR protocol. ΔRn, normalized change in fluorescence. (B) Six mice were prebled (Pre) and inoculated intradermally with 1 × 105 B. burgdorferi strain B31 A3 spirochetes. Approximately 50 μl of blood/mouse was collected every day from groups of two mice so that each group of two mice was bled every 3 days over a time period of 14 days. Blood collected from each mouse was plated in solid medium using 50 μl of blood and supplemented with a Borrelia antibiotic cocktail (see Materials and Methods for details), and the number of CFU per ml of blood was determined. Data shown are the average number of CFU/ml for the two mice sampled at each time point.
DISCUSSION
There is a critical need for development of innovative methods for improved diagnosis of Lyme disease. Because of its immunological specificity, signal amplification power, and potential for high-throughput automation, iPCR is a strong candidate for development as a robust method to overcome the challenges of diagnosis of Lyme disease. We have demonstrated the first application of iPCR for detection of host antibodies against B. burgdorferi in both a murine model and human serum.
iPCR using recombinant B. burgdorferi in vivo-expressed antigens is a sensitive method for detection of host response antibodies in infected mice.
An iPCR assay that incorporated attachment of intact spirochetes to magnetic beads provided sensitivity approximately equivalent to the sensitivities of current diagnostic methods, including C6 ELISA and immunoblotting, when tested in a murine model. However, it is well-known that B. burgdorferi can alter its surface protein expression on the basis of its environment (23–25). These data have led to the conclusion that in vitro-grown spirochetes likely do not present amounts and types of surface proteins equivalent to those that would be encountered by the host immune system in an active B. burgdorferi infection and suggest that the use of multiple in vivo-expressed recombinant antigens may improve assay sensitivity (26).
B. burgdorferi has been shown to express a number of antigens during an active infection, including OspC (27) and BmpA (28), that can be utilized as recombinant antigens to detect host antibodies against B. burgdorferi (21). We hypothesized that saturating the magnetic beads with recombinant in vivo-expressed antigenic proteins would provide more binding targets and hence a higher sensitivity than intact spirochetes. This was evidenced by the fact that active infection was detected on days 7 to 9 postinoculation using recombinant antigen-coated beads but on day 11 postinoculation using intact spirochete-coated beads and the fact that the signal above the background was stronger with recombinant antigen-coated beads, with ΔCq values being 4 to 10 and 2.5, respectively. This approach also provides the opportunity to utilize multiple specific antigens in either a combined or an individual assay that can be objectively quantified by quantitative PCR (qPCR).
Recombinant antigen iPCR successfully quantified B. burgdorferi VlsE C6 peptide antibodies in human serum samples.
The immunodominant C6 peptide domain of the VlsE protein has proven successful as a diagnostic antigen (29) and has become a popular choice for first-tier testing prior to follow-up immunoblot testing (30). An iPCR assay employing a recombinant C6 peptide was developed and compared to an existing commercial kit that uses the same antigen. iPCR detection of C6 antibodies in human serum demonstrated a strong correlation with detection by the commercial C6 Lyme disease ELISA. The C6 ELISA results in a combined score for detection of both IgG and IgM isotypes. To provide an additional level of discrimination, the iPCR protocol separately quantitates IgG and IgM antibodies using distinct qPCR template tags and fluorophores, resulting in an individual IgG and IgM iPCR score for each serum sample. All C6 ELISA-positive serum samples were found to be positive for IgG and/or IgM C6 antibodies by iPCR. The added ability of the iPCR assay to differentially quantitate antibody isotypes for a specific antigen of interest in a single sample may provide important information regarding the disease stage at the time of testing, as IgM is typically produced early in infection, with IgG being produced later and for longer durations (21, 31).
Of the serum samples that were found to be equivocal or negative by C6 ELISA, a subset of samples in each category was found to be positive by the C6 iPCR assay. These results imply that the iPCR assay may have an increased sensitivity of detection over the C6 ELISA; however, further analysis of a larger panel of serum samples is required to fully support this finding. Serum samples from healthy individuals with no known exposure to B. burgdorferi tested negative by both C6 ELISA and iPCR, suggesting equivalent specificities for the two methods. However, considering the small sample size (n = 5), additional samples need to be tested to confirm this result.
iPCR has the potential for direct detection of spirochetes in infected samples.
In an effort to test the applicability of iPCR for direct detection of spirochetes within a sample, it was determined that 1,000 spirochetes were needed in buffer and 10,000 organisms were needed in blood. In the murine model used for development of the protocol, the maximum spirochete load in blood was measured to be approximately 2,500 spirochetes/ml. Therefore, the current protocol is unable to directly detect spirochetes during an active murine infection. It has been estimated that the average number of cultivable B. burgdorferi cells per ml of whole blood in humans is approximately 0.1 spirochete per ml, and therefore, reisolation of spirochetes from blood has been demonstrated to have limited efficacy when using small volumes of blood (32). Hence, an alternative approach has been proposed to sample blood cultures and test by qPCR for increasing amounts of spirochete DNA (33). While an enrichment step is practical, the use of qPCR has the potential to introduce false-positive results from contaminating B. burgdorferi template DNA in the laboratory and typically requires additional protocol steps for nucleic acid purification. iPCR, which herein has demonstrated successful detection of spirochetes directly from whole blood and is much less prone to the same contamination issues, as the PCR template is unrelated to B. burgdorferi and human DNA, could effectively be used to make a more rapid diagnosis from B. burgdorferi-infected blood cultures. Future work will focus on improving the limit of iPCR direct detection of spirochetes in blood to achieve a detection sensitivity of 1 to 10 organisms, as has been demonstrated for other microbial pathogens (10, 34–36). Furthermore, as B. burgdorferi is transiently present in the blood of infected patients, the iPCR method may also be adapted for direct detection of spirochetes in synovial fluid and/or cerebrospinal fluid. Direct detection of spirochetes in patient samples is not anticipated to serve as the sole method for diagnosis of Lyme disease; rather, it is anticipated to serve in conjunction with the sensitive and specific detection of B. burgdorferi antibodies.
Contributions of an iPCR-based approach using recombinant antigens to future automated Lyme disease diagnostics.
The field of Lyme disease diagnostics is challenged by two main issues: a lack of consistent reagents and the need for a more simplified objective form of testing (1). There are currently multiple commercial assays that use a range of antigen types, from single recombinant antigens to multiple antigens to whole sonicated organisms. One principal focus for the field has been on the use of purified, recombinant, or synthetic peptides as the source of antigens in immunoassays (1). Unfortunately, no single antigen has demonstrated sufficient sensitivity and specificity to warrant replacing two-tier testing (1). Protein expression differences among species and the temporal appearance of relevant antibodies to different antigens at various stages of Lyme disease make the choice of a single antigen a difficult task and make the combined use of antigens an attractive alternative (1). The results presented here suggest that iPCR combined with the use of recombinant B. burgdorferi in vivo-expressed antigens has the potential to provide improved sensitivity of detection in an objective format that can be used to detect multiple host response antibodies and isotypes. Moreover, future translation of this method to an automated point-of-care platform will allow objective routine testing of Lyme disease patients.
ACKNOWLEDGMENTS
Many thanks go to Martin Schriefer at the Centers for Disease Control and Prevention, Fort Collins, CO, for the human serum samples. We thank T. Jewett for insightful comments and suggestions.
This work was supported by UCF startup funds and a UCF College of Medicine competitive research award to M.W.J.
Footnotes
Published ahead of print 9 January 2013
REFERENCES
- 1.Aguero-Rosenfeld ME, Wang G, Schwartz I, Wormser GP. 2005. Diagnosis of lyme borreliosis. Clin. Microbiol. Rev. 18:484–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murray TS, Shapiro ED. 2010. Lyme disease. Clin. Lab. Med. 30:311–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brouqui P, Bacellar F, Baranton G, Birtles RJ, Bjoersdorff A, Blanco JR, Caruso G, Cinco M, Fournier PE, Francavilla E, Jensenius M, Kazar J, Laferl H, Lakos A, Lotric Furlan S, Maurin M, Oteo JA, Parola P, Perez-Eid C, Peter O, Postic D, Raoult D, Tellez A, Tselentis Y, Wilske B. 2004. Guidelines for the diagnosis of tick-borne bacterial diseases in Europe. Clin. Microbiol. Infect. 10:1108–1132 [DOI] [PubMed] [Google Scholar]
- 4.Brown SL, Hansen SL, Langone JJ. 1999. Role of serology in the diagnosis of Lyme disease. JAMA 282:62–66 [DOI] [PubMed] [Google Scholar]
- 5.Ang CW, Notermans DW, Hommes M, Simoons-Smit AM, Herremans T. 2011. Large differences between test strategies for the detection of anti-Borrelia antibodies are revealed by comparing eight ELISAs and five immunoblots. Eur. J. Clin. Microbiol. Infect. Dis. 30:1027–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Molloy PJ, Persing DH, Berardi VP. 2001. False-positive results of PCR testing for Lyme disease. Clin. Infect. Dis. 33:412–413 [DOI] [PubMed] [Google Scholar]
- 7.Sano T, Smith CL, Cantor CR. 1992. Immuno-PCR: very sensitive antigen detection by means of specific antibody-DNA conjugates. Science 258:120–122 [DOI] [PubMed] [Google Scholar]
- 8.Niemeyer CM, Adler M, Wacker R. 2005. Immuno-PCR: high sensitivity detection of proteins by nucleic acid amplification. Trends Biotechnol. 23:208–216 [DOI] [PubMed] [Google Scholar]
- 9.Barletta J, Bartolome A, Constantine NT. 2009. Immunomagnetic quantitative immuno-PCR for detection of less than one HIV-1 virion. J. Virol. Methods 157:122–132 [DOI] [PubMed] [Google Scholar]
- 10.Liang H, Cordova SE, Kieft TL, Rogelj S. 2003. A highly sensitive immuno-PCR assay for detecting group A Streptococcus. J. Immunol. Methods 279:101–110 [DOI] [PubMed] [Google Scholar]
- 11.Gofflot S, Deprez M, el Moualij B, Osman A, Thonnart JF, Hougrand O, Heinen E, Zorzi W. 2005. Immunoquantitative PCR for prion protein detection in sporadic Creutzfeldt-Jakob disease. Clin. Chem. 51:1605–1611 [DOI] [PubMed] [Google Scholar]
- 12.Zhang W, Bielaszewska M, Pulz M, Becker K, Friedrich AW, Karch H, Kuczius T. 2008. New immuno-PCR assay for detection of low concentrations of Shiga toxin 2 and its variants. J. Clin. Microbiol. 46:1292–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKie A, Samuel D, Cohen B, Saunders NA. 2002. Development of a quantitative immuno-PCR assay and its use to detect mumps-specific IgG in serum. J. Immunol. Methods 261:167–175 [DOI] [PubMed] [Google Scholar]
- 14.Malou N, Raoult D. 2011. Immuno-PCR: a promising ultrasensitive diagnostic method to detect antigens and antibodies. Trends Microbiol. 19:295–302 [DOI] [PubMed] [Google Scholar]
- 15.Zhao X, Dong T, Pires N, Yang Z, Hjelseth S. 2011. Ultra-sensitive microfluidic system based on IMRAMP assay to quantify hormones in blood, p 1192–1195. In Biomedical engineering and informatics (BMEI). Proceedings of the 4th International Conference on Biomedical Engineering and Informatics, Shanghai, China [Google Scholar]
- 16.Elias AF, Stewart PE, Grimm D, Caimano MJ, Eggers CH, Tilly K, Bono JL, Akins DR, Radolf JD, Schwan TG, Rosa P. 2002. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect. Immun. 70:2139–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bestor A, Stewart PE, Jewett MW, Sarkar A, Tilly K, Rosa PA. 2010. Use of the Cre-lox recombination system to investigate the lp54 gene requirement in the infectious cycle of Borrelia burgdorferi. Infect. Immun. 78:2397–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbour AG. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57:521–525 [PMC free article] [PubMed] [Google Scholar]
- 19.Rosa PA, Hogan D. 1992. Colony formation by Borrelia burgdorferi in solid medium: clonal analysis of osp locus variants. In Munderloh UG, Kurtti TJ. (ed), Proceedings of the First International Conference on Tick Borne Pathogens at the Host-Vector Interface. University of Minnesota, St. Paul, MN [Google Scholar]
- 20.Jewett MW, Lawrence K, Bestor AC, Tilly K, Grimm D, Shaw P, VanRaden M, Gherardini F, Rosa PA. 2007. The critical role of the linear plasmid lp36 in the infectious cycle of Borrelia burgdorferi. Mol. Microbiol. 64:1358–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dressler F, Whalen JA, Reinhardt BN, Steere AC. 1993. Western blotting in the serodiagnosis of Lyme disease. J. Infect. Dis. 167:392–400 [DOI] [PubMed] [Google Scholar]
- 22.Engstrom SM, Shoop E, Johnson RC. 1995. Immunoblot interpretation criteria for serodiagnosis of early Lyme disease. J. Clin. Microbiol. 33:419–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Silva AM, Fikrig E. 1997. Borrelia burgdorferi genes selectively expressed in ticks and mammals. Parasitol. Today 13:267–270 [DOI] [PubMed] [Google Scholar]
- 24.Fikrig E, Feng W, Aversa J, Schoen RT, Flavell RA. 1998. Differential expression of Borrelia burgdorferi genes during erythema migrans and Lyme arthritis. J. Infect. Dis. 178:1198–1201 [DOI] [PubMed] [Google Scholar]
- 25.Schwan TG, Piesman J. 2002. Vector interactions and molecular adaptations of Lyme disease and relapsing fever spirochetes associated with transmission by ticks. Emerg. Infect. Dis. 8:115–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stanek G, Strle F. 2009. Lyme borreliosis: a European perspective on diagnosis and clinical management. Curr. Opin. Infect. Dis. 22:450–454 [DOI] [PubMed] [Google Scholar]
- 27.Gerber MA, Shapiro ED, Bell GL, Sampieri A, Padula SJ. 1995. Recombinant outer surface protein C ELISA for the diagnosis of early Lyme disease. J. Infect. Dis. 171:724–727 [DOI] [PubMed] [Google Scholar]
- 28.Fawcett PT, Rose C, Gibney KM, Chase CA, Kiehl B, Doughty RA. 1993. Detection of antibodies to the recombinant P39 protein of Borrelia burgdorferi using enzyme immunoassay and immunoblotting. J. Rheumatol. 20:734–738 [PubMed] [Google Scholar]
- 29.Lawrenz MB, Hardham JM, Owens RT, Nowakowski J, Steere AC, Wormser GP, Norris SJ. 1999. Human antibody responses to VlsE antigenic variation protein of Borrelia burgdorferi. J. Clin. Microbiol. 37:3997–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bacon RM, Biggerstaff BJ, Schriefer ME, Gilmore RD, Jr, Philipp MT, Steere AC, Wormser GP, Marques AR, Johnson BJ. 2003. Serodiagnosis of Lyme disease by kinetic enzyme-linked immunosorbent assay using recombinant VlsE1 or peptide antigens of Borrelia burgdorferi compared with 2-tiered testing using whole-cell lysates. J. Infect. Dis. 187:1187–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilske B, Fingerle V, Schulte-Spechtel U. 2007. Microbiological and serological diagnosis of Lyme borreliosis. FEMS Immunol. Med. Microbiol. 49:13–21 [DOI] [PubMed] [Google Scholar]
- 32.Wormser GP, Bittker S, Cooper D, Nowakowski J, Nadelman RB, Pavia C. 2001. Yield of large-volume blood cultures in patients with early Lyme disease. J. Infect. Dis. 184:1070–1072 [DOI] [PubMed] [Google Scholar]
- 33.Schwartz I, Bittker S, Bowen SL, Cooper D, Pavia C, Wormser GP. 1993. Polymerase chain reaction amplification of culture supernatants for rapid detection of Borrelia burgdorferi. Eur. J. Clin. Microbiol. Infect. Dis. 12:879–882 [DOI] [PubMed] [Google Scholar]
- 34.Barletta JM, Edelman DC, Constantine NT. 2004. Lowering the detection limits of HIV-1 viral load using real-time immuno-PCR for HIV-1 p24 antigen. Am. J. Clin. Pathol. 122:20–27 [DOI] [PubMed] [Google Scholar]
- 35.Huang SH, Chang TC. 2004. Detection of Staphylococcus aureus by a sensitive immuno-PCR assay. Clin. Chem. 50:1673–1674 [DOI] [PubMed] [Google Scholar]
- 36.Tian P, Mandrell R. 2006. Detection of norovirus capsid proteins in faecal and food samples by a real time immuno-PCR method. J. Appl. Microbiol. 100:564–574 [DOI] [PubMed] [Google Scholar]