Abstract
Recent studies have uncovered dozens of regulatory small RNAs in bacteria. A large number of these small RNAs act by pairing to their target mRNAs. The outcome of pairing can be either stimulation or inhibition of translation. Pairing in vivo frequently depends on the RNA binding proteinn Hfq. Synthesis of these sRNAs is tightly regulated at the level of transcription; many of the well-studied stress response regulons have now been found to include a regulatory RNA. Expression of the small RNA can help the cell cope with environmental stress by redirecting cellular metabolism, exemplified by RyhB, a small RNA expressed upon iron starvation. While small RNAs found in E. coli can usually be identified by sequence comparison to closely related enterobacteria, other approaches are necessary to find the equivalent RNAs in other bacterial species. Nonetheless, it is becoming increasingly clear that many if not all bacteria encode significant numbers of these important regulators. Tracing their evolution through bacterial genomes remains a challenge.
The bacterial genomes of organisms such as E. coli contain the information to allow the bacteria to thrive in a variety of conditions, both inside mammalian hosts and in the external environment. This requires systems to sense, respond to, and recover from a variety of stressful treatments and changes in nutrient availability. Our understanding of these systems has expanded rapidly, enhanced by the sequencing of multiple bacterial genomes. Transcriptional regulators and changes in the basic transcription machinery via use of alternative sigma factors provide appropriate regulated expression of a variety of repair and recovery genes.
In recent years, it has become increasingly clear that, in addition to these transcriptional regulatory programs, stress responses also involve small regulatory RNAs that play important roles in the post-transcriptional regulation of many genes. These RNAs are frequently regulated at the level of synthesis, as part of larger regulons, and may play roles in the immediate response to stress and/or the recovery from stress. As with eukaryotic microRNAs and small interfering RNAs, these bacterial regulatory RNAs, called sRNAs here, frequently act by pairing with specific target mRNAs to change their translation and/or stability. Other sRNAs act to influence the activity of proteins; the function of many others is still unknown [reviewed in (Gottesman 2004; Storz et al. 2005; Storz and Gottesman 2006) and not discussed further here]. Global searches for small RNAs in E. coli, expanding studies on these RNAs and their function, and extension of findings to other bacterial species have provided us with an understanding of how these sRNAs contribute to bacterial physiology and stress responses.
PAIRING RNAS: MODE OF ACTION OF A “TYPICAL” BACTERIAL SMALL RNA
The most studied and, thus far, most numerous family of regulatory sRNAs act by basepairing with specific mRNAs, and, in E. coli, are almost always associated with the RNA chaperone Hfq (see below). The steps in the function of sRNAs of this family are outlined in Figure 1 and discussed in some detail below.
Figure 1.
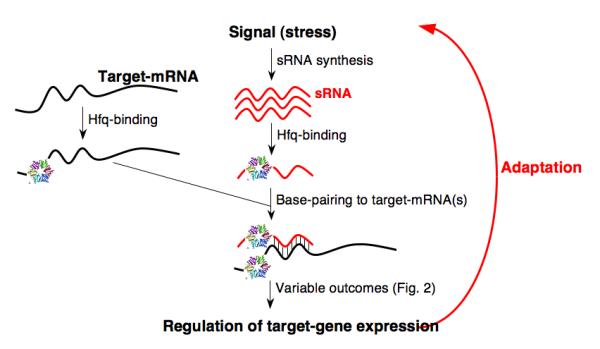
Cycle of activity of Hfq-dependent regulatory RNAs
Control of synthesis as a primary regulatory step
For any regulator, an important issue is how regulation occurs at the right time and place. For regulatory sRNAs, the primary response to environmental cues occurs at the level of synthesis. When the regulatory sRNA is made at high levels, biological effects rapidly follow. Promoters for bacterial regulatory RNAs do not differ qualitatively from promoters for protein coding genes. However, the strength of the sRNA promoter when fully expressed and the extent of regulation may both be particularly stringent. sRNAs belong to well-characterized regulons such as those responding to oxidative stress (Altuvia et al. 1997), osmotic stress (Guillier and Gottesman 2006) and iron starvation (Massé and Gottesman 2002). Thus, the complex network of bacterial transcriptional regulators and their mechanisms for sensing cues are put to use to synthesize these small RNAs.
Some of the regulatory proteins that have been demonstrated to regulate sRNAs are summarized in Table 1. It seems likely that other global regulators will also prove to include non-coding RNAs in their regulons. Only SgrR, newly discovered via its role in regulating the sRNA SgrS (Vanderpool and Gottesman 2004), has not been found to control a large regulon in addition to the small RNA (C. Vanderpool and S. Gottesman, in preparation). The genes encoding SgrR, as well as the LysR family regulators GcvA and OxyR are next to but transcribed divergently from the small RNA that they regulate. The intergenic regions encoding the promoters and regulatory sites for the transcriptional regulator and sRNA thus overlap, allowing for coordinate regulation of the regulatory protein and the small RNA, and suggests the possibility of co-evolution during any horizontal transfer events. In all the other cases listed in Table 1, the regulatory proteins and regulated non-coding RNAs are encoded far from each other.
Table 1.
Regulatory circuits with Hfq-dependent small RNAs
| Regulatory Protein family |
Regulatory Protein |
Inducing Signal |
Small RNAsa |
Targetsb | References |
|---|---|---|---|---|---|
| LysR | OxyR | Oxidative stress |
OxyS |
fhlA;
yobF, wrbA, ybaY |
(Altuvia et al. 1997); (Argaman and Altuvia 2000); (Tjaden et al. 2006) |
| GcvA | GcvB |
dpp,
opp |
(Urbanowski et al. 2000); (McArthur et al. 2006) |
||
| Two component |
OmpR | Osmotic shock |
OmrA, OmrB |
ompT, cirA, other cell surface genes |
(Guillier and Gottesman 2006) |
| RcsB | Cell surface stress |
RprA | rpoS | (Majdalani et al. 2001) | |
| LuxO | Quorum sensing |
Qrr1-4 (Vc) |
hapR,
luxR |
(Lenz et al. 2004) | |
| Sigma Factor |
SigE | Periplasmic stress |
MicA | ompA | (Udekwu et al. 2005); (Johansen et al. 2006); |
| RybB | sigE | (Johansen et al. 2006); Thompson and Gottesman, in preparation |
|||
| Fur Repressor |
Fur | Iron limitation |
RyhB (Ec) (Vc) PrrF (Pa) |
sodB sdh Fe- binding proteins |
(Massé and Gottesman 2002); (Wilderman et al. 2004; Davis et al. 2005; Mey et al. 2005) |
| Mar family | Mar, SoxS, Rob |
Oxidative stress, antibiotic stress |
MicFc | ompF | (Delihas and Forst 2001) |
| Sugar binding, novel |
SgrR | Glucose- phosphate accumulation |
SgrS | ptsG | (Vanderpool and Gottesman 2004) |
| CRP | CRP | Glucose limitation |
Spot 42 | galK | (Møller et al. 2002b) |
Small RNAs were identified in E. coli (Ec) unless otherwise indicated: (Vc: Vibrio cholerae; Pa: Pseudomonas aeruginosa).
Targets are negatively regulated except for rpoS, which is positively regulated.
MicF has also been reported to be regulated by OmpR under some conditions (Ramani et al. 1994).
A given regulatory protein can regulate more than one sRNA. Sigma E controls expression of the apparently unrelated sRNAs MicA and RybB (Table 1). There are a number of examples of closely related RNAs, presumably evolved through duplication events, whose synthesis is controlled by the same regulatory protein, including the four Qrr RNAs in Vibrio cholerae, the duplicate PrrF RNAs in Pseudomonas, and OmrA and B in E. coli. Duplicated RNAs may have evolved somewhat different targets, or their synthesis may respond with different thresholds to inducing signals.
The primary transcript of the sRNA is generally active for regulation, without any requirement for processing, in contrast to the essential processing steps for microRNAs in eukaryotic cells [reviewed in (Cullen 2004; Gottesman 2005)]. While processed transcripts are sometimes seen (Argaman et al. 2001; Repoila and Gottesman 2001; Sledjeski et al. 2001; Vogel et al. 2003; Opkyke et al. 2004), there is little or no information that addresses whether these are active or, if active, whether processing is essential for activity. In immunoprecipitation experiments with the RNA chaperone Hfq, the primary transcript is found to bind this protein (see below) (Wassarman et al. 2001; Zhang et al. 2003).
Finding the regulators of a small RNA
Given that the major regulatory step for most sRNAs is at the level of transcription, identifying the transcriptional regulators that control sRNA expression is a key step to understanding the physiological importance of the sRNA. Analysis of expression of the RNA by Northern blot under various growth conditions is frequently the first step; computational approaches or other methods may provide clues to conditions to test [for instance, see (Massé and Gottesman 2002; Johansen et al. 2006)]. For more precise information, or in the absence of clues, the same sorts of approaches that can be used to define the promoters of any gene can be used to find regulators of these RNAs – deletion analysis of promoters using transcriptional fusions, and genetic screens and selections using the fusions to define trans-acting regulators (Majdalani et al. 2002; Guillier and Gottesman 2006).
Hfq binding and the pairing of the regulatory sRNAs with mRNAs
In vivo roles of Hfq
The RNA binding protein Hfq was first identified biochemically by its role in the in vitro replication of the RNA phage Qβ (Blumenthal and Carmichael 1979). Later studies of hfq mutants demonstrated that cells that are devoid of Hfq grow slowly and have very low levels of RpoS (Tsui et al. 1994; Muffler et al. 1996). The finding that Hfq was also involved in the action of some sRNAs provided a possible explanation for these phenotypes (Zhang et al. 1998), (Sledjeski et al. 2001).
In immunoprecipitation experiments using an anti-Hfq antibody, the sRNAs that use Hfq are significantly enriched in the immunoprecipitate, and can be detected even when not specifically induced (Wassarman et al. 2001; Zhang et al. 2003). This tight binding to Hfq defines the family of Hfq-binding sRNAs. In many cases, the Hfq-binding sRNAs are significantly less stable in the absence of Hfq, and, consequently accumulate to lower levels (Sledjeski et al. 2001; Møller et al. 2002a; Massé et al. 2003; Zhang et al. 2003; Antal et al. 2005). Thus, it is generally assumed that Hfq rapidly binds and protects sRNAs of this class, and that it is the Hfq-bound form which is active in vivo for pairing with target mRNAs (Figure 1), but this has not been directly demonstrated.
The biological effects of these Hfq-binding sRNAs are absent in hfq mutants [see, for example (Zhang et al. 1998; Sledjeski et al. 2001; Massé and Gottesman 2002; Møller et al. 2002b)]. For instance, translation of the stationary sigma factor, RpoS, is dramatically reduced in E. coli hfq mutants; mutations that increase translation disrupt an inhibitory hairpin in the rpoS leader mRNA (Brown and Elliott 1997). At least two small RNAs, and probably others, stimulate translation of rpoS by interacting with and opening the inhibitory hairpin [reviewed in (Repoila et al. 2003)]. In an hfq mutant, these small RNAs can no longer stimulate translation and RpoS is not made. Another example is provided by the phenotype of fur mutants of E. coli. Fur represses RyhB, and RyhB down-regulates the genes encoding succinate dehydrogenase (see below). As a result, fur mutants cannot grow on succinate. Mutations in either ryhB or hfq can restore growth (Massé and Gottesman 2002). hfq mutants of Vibrio and Brucella are avirulent (Robertson and Roop 1999; Ding et al. 2004) and hfq mutants of Vibrio and Pseudomonas have defects in the quorum sensing pathway (Lenz et al. 2004; Sonnleitner et al. 2006), although it has not been demonstrated in all of these cases that the phenotypes are due to loss of function of sRNAs and not some other role of Hfq.
In vitro activities of Hfq
Rapid turnover of the small regulatory RNAs in vivo in the absence of Hfq might be a sufficient explanation for loss of function of these sRNAs, but a variety of in vitro experiments suggest that Hfq has a more direct role as an RNA chaperone. Specifically, interactions of an sRNA and target mRNA are promoted by the presence of Hfq in vitro (Møller et al. 2002a; Zhang et al. 2002; Lease and Woodson 2004; Kawamoto et al. 2006). In these and other experiments, Hfq binds the target mRNA as well as the regulatory RNA (Geissmann and Touati 2004). What proportion of the population of a given mRNA or sRNA is bound at any time has not been examined in vivo.
How does binding of Hfq to both mRNA and regulatory RNA promote pairing? Hfq subunits assemble into a hexameric ring (Figure 1); a model oligonucleotide has been shown to bind to the inner surface of the ring, and evidence for secondary RNA binding sites have also been described (Schumacher et al. 2002; Sauter et al. 2003; Mikulecky et al. 2004; Valentin-Hansen et al. 2004). Models depending upon pairing of Hfq rings or binding of both RNAs to a single ring have been suggested (Storz et al. 2004), and may be important in stabilizing RNA:RNA interactions once they have been initiated. However, most bacterial species, including E. coli, encode only one hfq gene; thus, Hfq must be able to promote interactions between many sRNAs that pair with different mRNAs. This implies that Hfq does not itself provide specificity of pairing. Instead, pairing may proceed more rapidly when the RNA secondary or tertiary structure is remodeled by Hfq binding (Moll et al. 2003b; Geissmann and Touati 2004; Lease and Woodson 2004; Antal et al. 2005), and, once pairing is initiated, it may be extended and/or stabilized by interactions with Hfq. Thus, while complementarity between the sRNA and the target mRNA may be essential for pairing (Kawamoto et al. 2006), it is apparently not sufficient in most cases. High affinity Hfq binding sites on both the sRNA and the target mRNA provide additional necessary elements. Computational approaches that do not take this in to account may predict pairing between sRNAs and target mRNAs do not occur in vivo.
Outcomes of Pairing
Pairing of the small RNA to its target mRNA can result in positive or negative effects on translation. In the best-studied case of stimulation of translation, two different small RNAs, DsrA and RprA, bind to counteract the formation of an inhibitory hairpin in the mRNA of rpoS that blocks translation (Figure 2A) (Majdalani et al. 1998; Majdalani et al. 2002). In many cases, negative effects are associated with the small RNA binding close to the ribosome binding site. Binding may both inhibit translation (under conditions where mRNA degradation is blocked) and lead to rapid mRNA degradation (Morita et al. 2006) (Figure 2B). It is not yet clear whether mRNA degradation is secondary to blocking ribosome access.
Figure 2. Outcomes of pairing for Hfq-dependent regulatory RNAs.
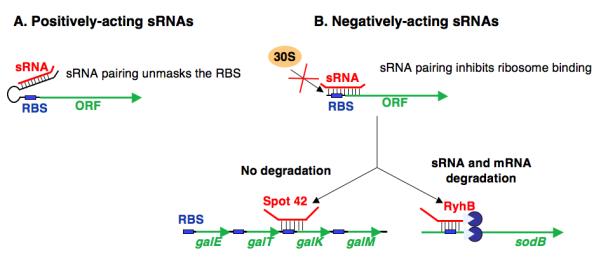
A. Positively-acting RNAs are exemplified by DsrA and RprA action in stimulating rpoS translation [reviewed in (Repoila et al. 2003)]. B. Negatively-acting sRNAs generally pair with target mRNAs near the ribosome binding site, but it is not yet clear what distinguishes cases where there is degradation of the mRNA (RyhB and sodB in the figure) and those where there is no degradation (Spot 42 and galK in the figure).
Translation inhibition and mRNA degradation
For most of the cases of sRNA-dependent mRNA degradation, the target message disappears rapidly (within 2-3 minutes) and completely after induction of the regulatory RNA (Massé and Gottesman 2002; Massé et al. 2005; Rasmussen et al. 2005; Udekwu et al. 2005; Guillier and Gottesman 2006); in some cases, much smaller degradation products can be detected (Morita et al. 2003; Vanderpool and Gottesman 2004). Degradation is generally due to the activity of RNase E (Massé et al. 2003; Afonyushkin et al. 2005; Morita et al. 2005). RNase E is an essential endonuclease that cleaves at single-stranded AU-rich RNA [reviewed in (Kennell 2002)]. Presumably, endonucleolytic cleavage by RNase E serves as a primary step, followed by degradation by a variety of exonucleases. Absence of translation can uncover RNase E cleavage sites internal to a coding region that are otherwise inefficiently cleaved (Joyce and Dreyfus 1998; Baker and Mackie 2003) [reviewed in (Deana and Belasco 2005)]. Thus, an sRNA might promote rapid degradation of a message by blocking ribosome access, thereby allowing access of RNase E to recognition sites internal to the mRNA normally masked by translating ribosomes. If this is the primary cause of degradation of mRNAs targeted by sRNAs, the absence of an internal RNase E site on a given mRNA would allow regulation of translation without degradation of that mRNA. Regulation of translation without significant mRNA degradation has been described for Spot 42 regulation of translation in the gal operon (Figure 2B) (Møller et al. 2002b), although the presence or absence of RNase E sites has not been addressed. Alternatively, the act of pairing may by itself make the mRNA accessible to RNase E, by changing the structure of the RNA. There is evidence that RNase E activity is affected by secondary structure [reviewed in (Kennell 2002)]; possibly the paired RNAs can mimic secondary structure elements in stimulating recognition or cleavage of the mRNA. Finally, recent work suggests that Hfq and RNase E are physically associated in the cell (Morita et al. 2005). If so, the Hfq binding, which is also found to be at AU-rich single-stranded regions [reviewed in (Valentin-Hansen et al. 2004)], may in itself recruit RNase E, bypassing the need for a high affinity site for RNase E binding to promote efficient degradation. However, Hfq binds to both mRNA and regulatory RNA in the absence of pairing, while degradation is only triggered upon pairing, and does not occur in the case of positive regulation, a number of aspects of the mechanism remains to be explained.
Fate of small RNA
A second issue must be considered in thinking about how sRNAs stimulate RNase E-dependent cleavage. How does this pairing stimulate degradation of the regulatory RNA itself? The evidence that there is coupled degradation of the mRNA and the regulatory RNA is not yet complete, but is nevertheless fairly compelling. A variety of sRNAs are relatively stable when turnover is measured in the presence of a general inhibitor of transcription such as rifampicin (Sledjeski et al. 2001; Vogel et al. 2003); however, the same regulatory RNAs are significantly more unstable when synthesis is shut down in a specific fashion (i.e., by turning down the promoter for the sRNA) (Massé et al. 2003). This has been interpreted to mean that the small RNA is degraded upon pairing to its target, which will be rapidly depleted after rifampicin treatment. The recruitment of RNase E by Hfq binding would provide part of the explanation for such coupled degradation, assuming, as noted above, that the act of pairing in itself helps to stimulate the degradation. RNase E endonuclease activity has been shown to be stimulated by a 5′ monophosphate end in vitro (Jiang and Belasco 2004). Such a 5′ monophosphate would be the expected primary product of cleavage by RNase E (Kennell 2002); thus is it possible that a cut within the mRNA may directly stimulate RNase E to cleave the paired sRNA. However, in the absence of Hfq, and therefore presumably in the absence of pairing, the sRNAs are also rapidly degraded and RNase E seems to be critical for this degradation (Massé et al. 2003), suggesting that pairing and initial cleavage of the mRNA is not the sole mechanism for RNase E to obtain access to these sRNAs. It is intriguing that Hfq and RNase E both share a preference for single-stranded AU-rich RNA. Thus, absence of Hfq binding may uncover RNase E cleavage sites. This has been directly demonstrated in vitro for the sodB message, and suggested in other cases (Moll et al. 2003a; Zhang et al. 2003; Afonyushkin et al. 2005).
Further work will be necessary to identify the basis for mRNA degradation and how translational inhibition can lead to mRNA degradation in some cases but not others. However, based on work done thus far, rapid degradation of a target mRNA is a useful and easily detectable outcome of pairing by many small RNAs, allowing the use of approaches, such as microarrays, RT-PCR, as well as Northern blots, to define targets for these small RNAs (discussed further below).
Defining Targets
Hfq-binding sRNAs thus far all act by complementary base-pairing to target mRNAs. Finding these targets continues to pose a challenge. Our ability to predict targets based on the expected complementarity is improving, although it still fails to find many targets (Tjaden et al. 2002). The level of false positive predictions (good pairing predicted, but no regulation found) is also high; possibly these potential targets do not contain Hfq binding sites, a characteristic that is not yet integrated into search programs. It is also unclear how many targets a given sRNA will have. At least one sRNA, RyhB, has been found to regulate dozens of target mRNAs (Massé et al. 2005), while others have been described as regulating only one or a small number of targets (see Table 1 for examples of known targets).
One experimental approach that has been widely used is examination of the effects of small RNA expression by microarray; the success of this method depends upon both detectable expression levels for the mRNA, and the degradation of target mRNAs upon pairing (Davis et al. 2005; Massé et al. 2005; Mey et al. 2005; Guillier and Gottesman 2006; Tjaden et al. 2006). This approach is most likely to uncover direct rather than indirect effects if changes in mRNA levels are examined a short time (5 minutes or less) after expression of the regulatory RNA (Massé et al. 2005). Other approaches require direct interactions to capture target mRNAs, but may be most appropriate for well-expressed messages. Affinity purification of mRNAs binding to an sRNA immobilized on a column has been successful in a number of cases (Antal et al. 2005; Douchin et al. 2006).
Regulatory steps beyond synthesis
As we learn more about how the Hfq-binding sRNAs act, other possible steps for regulation of their activity, in addition to regulation of synthesis, can be identified, although most have not been fully explored. Competition for Hfq has been suggested as a mechanism for negative regulation of RpoS translation by OxyS (Zhang et al. 1998). The intrinsic stability of the sRNA will of course contribute to its accumulation, and this can be affected by Hfq binding as well. Competition between target mRNAs for a given regulatory RNA may be an important point of control if the sRNA is limiting and used stoichiometrically, which is probably the case if degradation of the sRNA is coupled with pairing. Under the strongest induction conditions, sRNA levels may not be a limiting factor. However, sRNA levels may well be limiting when there is only a basal level of induction or during recovery from an inducing stress, when synthesis of the sRNA has slowed or stopped.
FINDING SMALL RNAS AND THEIR TARGETS: EXPANDING OPTIONS
The existence of some species of small RNAs in bacteria have been known since the early days of identifying novel stable RNAs, but in many cases their function took many years to uncover. In recent years, a combination of the availability of sequence information for many bacterial genomes, as well as the development of microarray-based approaches for studying gene expression have allowed both computational and experimentally based discovery of many additional small RNAs, and the growing understanding of their specific expression and function. Approaches for finding small RNAs have been recently reviewed (Gottesman 2004; Huttenhofer and Vogel 2006). The result of these genome-wide approaches has defined more than 80 non-coding RNAs in E. coli, with at least 20 of them binding to and stabilized in vivo by Hfq (Zhang et al. 2003). Of these RNAs, studies on 14 have now been published (see Table 1).
The methods for searching for regulatory RNAs in E. coli have been extrapolated to other organisms. Start and stop sites are useful landmarks for some of these searches. Flanking an sRNA, one can expect to find promoter elements and/or Rho-independent terminators with a characteristic stem-loop followed by a run of Us as a stop signal. Combining conservation and the presence of a promoter and Rho-independent terminator has been developed into a program, sRNAPredict2, recently used to predict sRNAs in a variety of pathogens; a number of these predictions have been confirmed (Livny et al. 2006).
More specific predictions can be made by assuming that a stress and the physiologic response mediated by a sRNA are conserved from one species to another. Under such conditions, one can search for the regulatory sites, in combination with a terminator, for instance, within a given intergenic region. Quorum sensing small RNAs were identified in Vibrio by this approach (Lenz et al. 2004), as were iron-regulated small RNAs, described below.
Regulating iron homeostasis with small RNAs
All organisms, from bacteria to mammals, need to carefully regulate their intracellular iron (Fe) pools. Fe is an essential cofactor for many enzymes; at the same time, iron can be quite toxic, causing damage to proteins and nucleic acids (Imlay 2003). Therefore, organisms generally need to minimize the accumulation of free iron; iron acquisition may be regulated so that intracellular iron is kept to the minimum needed to satisfy requirements, and iron storage systems that sequester excess iron exist in all organisms. It has been known for decades that iron storage is regulated in mammalian cells by an intriguing post-transcriptional mechanism, in which the aconitase enzyme, an Fe-binding enzyme of the TCA cycle, also acts as a regulator, binding an RNA element at the 3′ end of some mRNAs to both positively and negatively regulate translation (Rouault 2002).
Bacteria inside mammalian hosts need to obtain Fe from their host, therefore requiring that they develop methods for iron acquisition that can effectively compete for bound iron with the host. The Fur repressor is used by many bacteria, both gram positive and gram negative, to keep iron acquisition systems under control until they are needed (Hantke 2001). Fur directly binds Fe2+, and acts as a repressor only in the Fe-bound form. RyhB, a small non-coding RNA made in E. coli, acts to regulate internal Fe use, by targeting for degradation the mRNAs of as many as 16 operons encoding Fe-binding proteins (Massé and Gottesman 2002; Massé et al. 2005). RyhB is directly repressed by the Fur repressor, and rapidly induced when Fe is removed from Fur. It was proposed that RyhB helps to prioritize intracellular iron use under iron-limiting conditions by sparing iron for essential functions (Massé et al. 2005), although genes involved in biofilm formation, acid resistance, and others have also been found to be regulated by RyhB as well (Davis et al. 2005; Mey et al. 2005; Oglesby et al. 2005).
The biology underlying the use of Fur and RyhB to regulate intracellular iron utilization is likely to extend to many other bacteria as well. Is a small RNA also used in these bacteria? RyhB homologs can easily be found in other enterobacteria, including Salmonella, Shigella, Klebsiella, Photorhabdus, and Yersinia (Figure 3A); (Massé and Gottesman 2002). Both Yersinia and Salmonella have two copies of an RNA similar to RyhB; one is in the context of the E. coli gene (location A), while the second copy is in a different chromosomal context (location B for Yersinia and location C for Salmonella) (Figure 3B). Evolutionary trees of these RyhB molecules suggest a possible early duplication event, to give the A and B locations; subsequently the B copy may have been lost in E. coli and Salmonella. The RyhB at the C locus in Salmonella is the most divergent of this set, and might have been acquired from elsewhere by horizontal transfer. In all cases, the RyhB homolog is preceded by a predicted Fur binding site (not shown). Vibrio cholerae and other Vibrio species have a single ryhB gene with some distinct characteristics. While still regulated by Fur, the ryhB gene is in yet another genome context and is significantly longer than the E. coli RNA (Davis et al. 2005; Mey et al. 2005).
Figure 3. RyhB homologs in Enterobacteriacae.
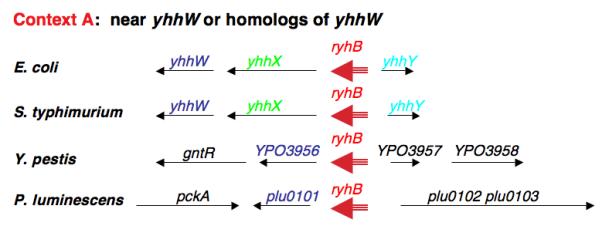
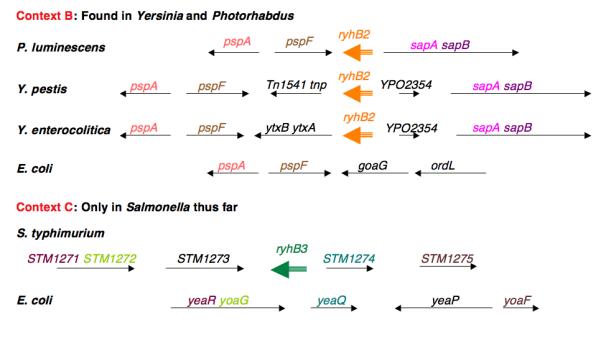
Homologous genes are similarly color-coded. The three contexts shown are far from each other on the bacterial chromosome. We have arbitrarily named those RNAs in the same context as E. coli ryhB and second genes elsewhere ryhB2 or ryhB3 (depending on the context). These are not meant to be permanent names. Information on genes is adapted from information provided on the Comprehensive Microbial Resource of TIGR (The Institute for Genome Research) [http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi],(Peterson et al. 2001).
In organisms more evolutionarily distant from E. coli, RyhB cannot be found by sequence homology. For instance, a search of Pseudomonas aeruginosa sequences failed to reveal a sequence related to RyhB. However, Pseudomonads contain a Fur repressor and have a similar requirement to regulate Fe homeostasis. A computational search for a Fur-regulated small RNA (searching intergenic regions for RNA secondary structure within an appropriate distance from a Fur binding site) revealed a pair of closely related RNAs that were shown to play a similar role in Pseudomonas to E. coli RyhB, and that have been called PrrF1 and PrrF2 (Pseudomonas regulatory RNA involving Fe) (Wilderman et al. 2004). Comparison to other Pseudomonads demonstrated conservation of PrrF-like RNAs. In fact, all sequenced Pseudomonads have two PrrF sRNAs, although only P. aeruginosa has two in tandem; the others have one at the same position as the P. aeruginosa sRNAs while the other is elsewhere (Figure 4). An examination of the evolution of PrrF in the Pseudomonads would suggest that, as for E. coli and its close relatives, PrrF duplicated at some point in the past. The second copy (context B in Figure 4) of PrrF may have been lost from P. aeruginosa; a later duplication of the remaining prrF gene yielded tandem genes that are more similar to each other than they are to any of the other Pseudomonad PrrFs. No similarity in sequence or genome context is obvious between the PrrF sRNAs and RyhB sRNAs in E. coli or its close relatives. At least one target, sodB, is shared by the PrrF and RyhB sRNAs.
Figure 4. PrrF homologs in Pseudomonads.
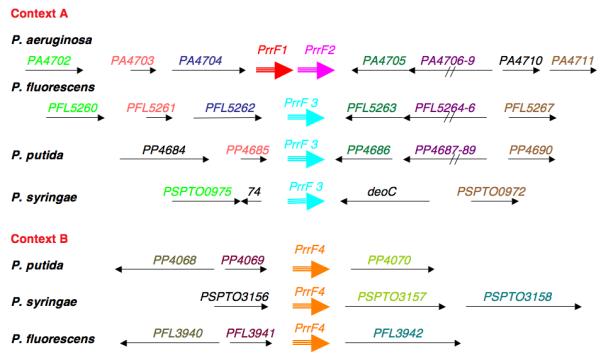
Homologous genes are similarly color-coded. We have arbitrarily named the PrrF like RNAs in context A PrrF3 and those in Context B PrrF4.
Thus, as with RyhB in Yersinia and Salmonella, it would seem that multiple independent duplication events have occurred during evolution, suggesting a requirement for more than one RyhB gene. Exactly why this would be is not yet clear. Mutation of one or the other P. aeruginosa prrF genes gives a partial phenotype in down-regulation of target mRNAs, suggesting that two may be necessary to achieve enough PrrF RNA for full control under severe iron depletion conditions (Wilderman et al. 2004). Alternatively, subtle differences in expression patterns and differences in sequence and therefore pairing might be driving the duplication of these loci.
It seems highly likely that the E. coli and Pseudomonad RyhB-like RNAs are evolved from a common, unidentified ancestor. The lack of sequence homology in these RNAs between Pseudomonas and E. coli, coupled with the conservation of promoter sequences, suggests that RNA genes can change relatively rapidly, possibly as their targets change or new targets are acquired. As more bacterial genomes are sequenced, it should become possible to fill in some of the missing steps and better understand how these RNAs are evolving.
Genome context as a landmark for small RNA discovery
Given the relatively rapid evolution of the RyhB-like non-coding RNAs, tracing these RNAs through evolution can be easier if a protein-coding gene, with its slower rate of evolution, can be used as a genome marker for the RNA. For instance, the 6S RNA, a regulator of RNA polymerase, is processed from a longer message that includes an ORF in many organisms, and by identifying the ORF, the linked but not always homologous 6S RNA was identified (and found to have a conserved structure and function) (Barrick et al. 2005; Trotochaud and Wassarman 2005).
The Hfq-binding small RNAs have generally been found to be free-standing transcripts, not processed from a longer RNA. However, as noted above, for a few small RNAs (SgrS, OxyS, and GcvB), the transcriptional regulator is encoded next to and divergently from the RNA (Table 1). Thus, this protein can serve as a marker for the small RNA (McArthur et al. 2006).
A unique genome neighborhood for Spot 42
A more general investigation of the genome context of the small RNAs has suggested that, at least in a few cases, genes for small RNAs flanked by genes for highly conserved proteins can be found in a similar neighborhood even when the small RNAs themselves have diverged. In particular, this appears to be the case for the genome context of the Hfq-binding Spot 42 RNA.
In E. coli, Spot 42 has been found to be responsible for polarity in the gal operon; it negatively regulates the translation of the third gene in the operon (Figure 2B) (Møller et al. 2002b). Sequence comparisons show that Spot 42 is also present in Salmonella, Erwinia, Yersinia, and Vibrio [see RFAM, (Griffiths-Jones et al. 2005)].
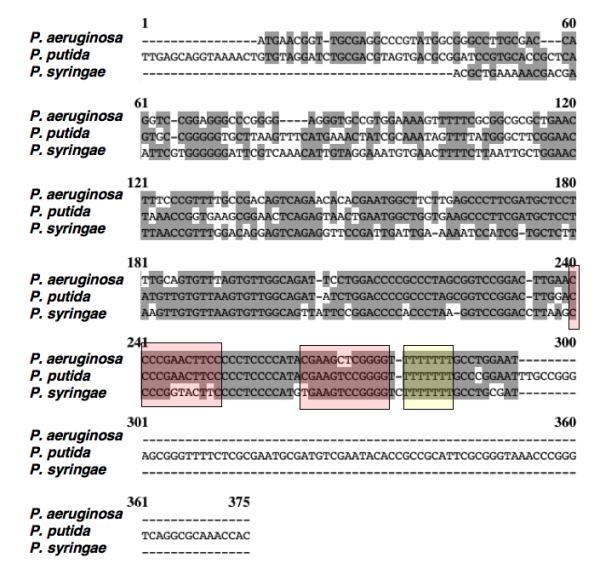
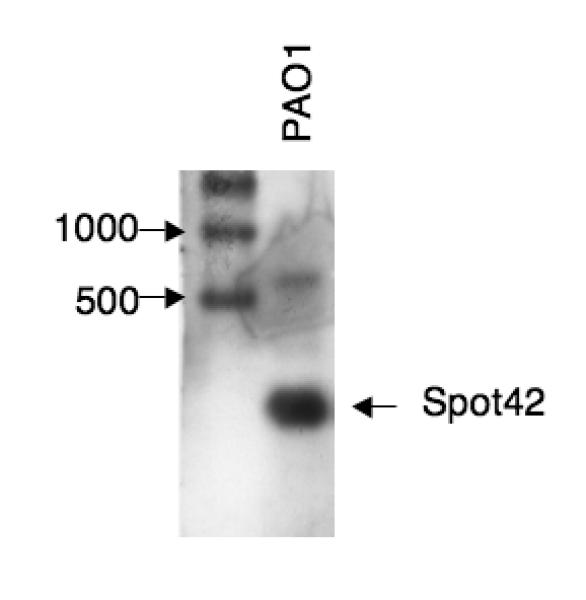
What is notable is the genome location of this RNA (Figure 5A). It is found between polA, encoding DNA polymerase I, a protein found widely in bacteria, and yihA (engB), a conserved GTPase of unknown function. These two genes are also near each other in Pseudomonas aeruginosa, and the sequences in the intergenic region are conserved in other Pseudomonads and are reminiscent of those expected for a non-coding RNA (Figure 5B). In fact, an sRNA is expressed from this locus (Figure 5C).
Figure 5. A possible Pseudomonas Spot 42 RNA.
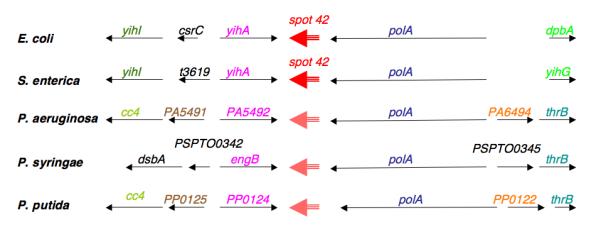
A. Conserved genome context around an sRNA in E. coli, Salmonella, and Pseudomonads. Homologous genes are similarly color-coded. polA: DNA polymerase I; engB: GTP-binding protein.
B. Multiple sequence alignment of Pseudomonas intergenic region between polA and engB. The sequences overlapping the putative sRNAs are shown, with conserved regions in grey and the putative terminator stem-loop boxed in color. Sequences are from Genbank files AE004091 (P. aeruginosa PA01), AE015451 (P. putida KT2440), and AE016853 (P. syringae pv tomato str. DC3000). Other sequenced Pseudomonads are also conserved in this intergenic region.
C. Detection of a Spot 42 sRNA in P. aeruginosa. RNA extracted from P. aeruginosa strain PA01 was probed for an RNA of the sequence predicted in Fig. 5B. The RNA isolation and Northern blot procedure is as previously described (Wilderman et al. 2004).
As with the RyhB-like RNAs, no obvious sequence similarity can be found between Spot 42 from enterobacteria and the Pseudomonas small RNA. In addition, the gal operon, the major demonstrated target for Spot 42 in E. coli (Møller et al. 2002b), is not found in Pseudomonas aeruginosa, suggesting that either this is an RNA with divergent function, in the same site, or that the major (conserved) function of Spot 42 has not yet been described. Further analysis of its function in Pseudomonas as well as further examination of the evolution of this and other small RNAs will clearly be fruitful.
Prospects for the Future
Our understanding of the role of small regulatory RNAs in bacterial physiology has grown significantly over the last decade. Discoveries and approaches developed initially in E. coli are now being applied to a variety of bacteria, including pathogens. It seems highly likely that this once ignored level of regulation will provide new insights into bacterial growth and adaptation to stress and new targets for future antibiotics.
Acknowledgements
The work described in this manuscript was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
REFERENCES
- Afonyushkin T, Vecerek B, Moll I, Blasi U, Kaberdin VR. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res. 2005;33:1678–1689. doi: 10.1093/nar/gki313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- Antal M, Bordeau V, Douchin V, Felden B. A small bacterial RNA regulates a putative ABC transporter. J. Biol. Chem. 2005;280:7901–7908. doi: 10.1074/jbc.M413071200. [DOI] [PubMed] [Google Scholar]
- Argaman L, Altuvia S. fhlA repression by OxyS RNA: Kissing complex formation at two sites results in a stable antisense-target RNA complex. J. Molec. Biol. 2000;300:1101–1112. doi: 10.1006/jmbi.2000.3942. [DOI] [PubMed] [Google Scholar]
- Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EGH, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic region of Escherichia coli. Curr. Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- Baker KE, Mackie GA. Ectopic RNase E sites promote bypass of 5′-end-dependent mRNA decay in Escherichia coli. Molec. Microbiol. 2003;47:75–88. doi: 10.1046/j.1365-2958.2003.03292.x. [DOI] [PubMed] [Google Scholar]
- Barrick JE, Sudarsan N, Weinberg Z, Ruzzo WL, Breaker RR. 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA. 2005;11:774–784. doi: 10.1261/rna.7286705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T, Carmichael GG. RNA replication: function and structure of Qß-replicase. Annu. Rev. Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- Brown L, Elliott T. Mutations that increase expression of the rpoS gene and decrease its dependence on hfq function in Salmonella typhimurium. Journal of Bacteriology. 1997;179:656–662. doi: 10.1128/jb.179.3.656-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. Transcription and processing of human microRNA precursors. Molec. Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Davis BM, Quinones M, Pratt J, Ding Y, Waldor MK. Characterization of the small untranslated RNA RyhB and its regulon in Vibrio cholerae. Journal of Bacteriology. 2005;187:4005–4014. doi: 10.1128/JB.187.12.4005-4014.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deana A, Belasco JG. Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes & Develop. 2005;19:2526–2533. doi: 10.1101/gad.1348805. [DOI] [PubMed] [Google Scholar]
- Delihas N, Forst S. MicF: An antisense RNA gene involved in response of Escherichia coli to global stress factors. J. Molec. Biol. 2001;313:1–12. doi: 10.1006/jmbi.2001.5029. [DOI] [PubMed] [Google Scholar]
- Ding Y, Davis BM, Waldor MK. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol. Microbiol. 2004;53:345–354. doi: 10.1111/j.1365-2958.2004.04142.x. [DOI] [PubMed] [Google Scholar]
- Douchin V, Bohn C, Bouloc P. Down-regulation of porins by a small RNA bypasses the essentiality of the RIP protease RseP in E. coli. J Biol Chem. 2006;281:12253–12259. doi: 10.1074/jbc.M600819200. [DOI] [PubMed] [Google Scholar]
- Geissmann, Touati D. Hfq, a new chaperoning role: binding to messanger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. The Small RNA Regulators of Escherichia coli: Roles and Mechanisms. Annu Rev Microbiol. 2004;58:273–301. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillier M, Gottesman S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Molec. Microbiol. 2006;59:231–247. doi: 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- Hantke K. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 2001;4:172–177. doi: 10.1016/s1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- Huttenhofer A, Vogel J. Experimental approaches to identify non-coding RNAs. Nucleic Acids Res. 2006;2:635–646. doi: 10.1093/nar/gkj469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA. Pathways of oxidative damage. Annu. Rev. Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- Jiang X, Belasco JG. Catalytic activation of multimeric RNase E and RNase G by 5′-monophosphorylated RNA. Proc Natl Acad Sci U S A. 2004;101:9211–9216. doi: 10.1073/pnas.0401382101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen J, Rasmussen AA, Overgaard M, Valentin-Hansen P. Conserved small non-coding RNAs that belong to the σE regulon: Role in down-regulation of outer membrane proteins. J. Mol. Bio. 2006 doi: 10.1016/j.jmb.2006.09.004. in press. [DOI] [PubMed] [Google Scholar]
- Joyce SA, Dreyfus M. In the absence of translation, RNase E can bypass 5′ mRNA stabilizers in Escherichia coli. J. Mol. Bio. 1998;282:241–254. doi: 10.1006/jmbi.1998.2027. [DOI] [PubMed] [Google Scholar]
- Kawamoto H, Koide Y, Morita T, Aiba H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Molec. Microbiol. 2006;61:1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- Kennell D. Processing endoribonucleases and mRNA degradation in bacteria. Journal of Bacteriology. 2002;184:4645–4657. doi: 10.1128/JB.184.17.4645-4657.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease RA, Woodson SA. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J. Molec. Biol. 2004;344:1211–1223. doi: 10.1016/j.jmb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The Small RNA Chaperone Hfq and Multiple Small RNAs Control Quorum Sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Livny J, Brencic A, Lory S, Waldor MK. Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res. 2006;34:3484–3493. doi: 10.1093/nar/gkl453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Chen S, Murrow J, St. John K, Gottesman S. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 2001;39:1382–1394. doi: 10.1111/j.1365-2958.2001.02329.x. [DOI] [PubMed] [Google Scholar]
- Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Hernandez D, Gottesman S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 2002;46:813–826. doi: 10.1046/j.1365-2958.2002.03203.x. [DOI] [PubMed] [Google Scholar]
- Massé E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. Journal of Bacteriology. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur SD, Pulvermacher SC, Stauffer GV. The Yersinia pestis gcvB gene encodes two small regulatory RNA molecules. BMC Microbiology. 2006;6:52. doi: 10.1186/1471-2180-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mey AR, Craig SA, Payne SM. Characterization of Vibrio cholerae RyhB: the RyhB regulon and role of ryhB in biofilm formation. Infect Immun. 2005;73:5706–5719. doi: 10.1128/IAI.73.9.5706-5719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulecky PJ, Meenakshi K, Brescia CC, Takach JC, Sledjeski D, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A)RNAs. Nature Structural and Molecular Biology. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll I, Afonyushkin T, Vytvytska O, Kaberdin VR, Blasi U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA. 2003a;9:1308–1314. doi: 10.1261/rna.5850703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll I, Leitsch D, Steinhauser T, Blasi U. RNA chaperone activity of the Sm-like Hfq protein. EMBO Reports. 2003b;4:284–289. doi: 10.1038/sj.embor.embor772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, El-Kazzaz W, Tanaka Y, Inada T, Aiba H. Accumulation of glucose 6-phosphate or fructose 6-phosphate is responsible for destabilization of glucose transporter mRNA in Escherichia coli. J Biol Chem. 2003;278:15608–15614. doi: 10.1074/jbc.M300177200. [DOI] [PubMed] [Google Scholar]
- Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes:mechanical basis of mRNA stabilization mediated by bacterial noncoding RNAs. Genes & Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Mochizuki Y, Aiba H. Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc Natl Acad Sci U S A. 2006;103:4858–4863. doi: 10.1073/pnas.0509638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffler A, Fischer D, Hengge-Aronis R. The RNA-binding protein HF-I, known as a host factor for phage Qß RNA replication, is essential for rpoS translation in Escherichia coli. Genes & Dev. 1996;10:1143–1151. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- Møller T, Franch T, Hojrup P, Keene DR, Bachinger HP, Brennan R, Valentin-Hansen P. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell. 2002a;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- Møller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes & Dev. 2002b;16:1696–1706. doi: 10.1101/gad.231702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oglesby AG, Murphy ER, Iyer VR, Payne SM. Fur regulates acid resistance in Shigella flexneri via RyhB and ydeP. Molec. Microbiol. 2005;58:1354–1367. doi: 10.1111/j.1365-2958.2005.04920.x. [DOI] [PubMed] [Google Scholar]
- Opkyke JA, Kang J-G, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. Journal of Bacteriology. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson JD, Umayam LA, Dickinson TM, Hickey EK, White O. The Comprehensive Microbial Resource. Nucleic Acids Res. 2001;29:123–125. doi: 10.1093/nar/29.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani N, Hedeshian M, Freundlich M. micF antisense RNA has a major role in osmoregulation of OmpF in Escherichia coli. J Bacteriol. 1994;176:5005–5010. doi: 10.1128/jb.176.16.5005-5010.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen AA, Eriksen M, Gilany K, Udesen C, Franch T, Petersen C, Valentin-Hansen P. Regulation of ompA mRNA stability: the role of a small regulatory RNA in growth phase-dependent control. Mol Microbiol. 2005;58:1421–1429. doi: 10.1111/j.1365-2958.2005.04911.x. [DOI] [PubMed] [Google Scholar]
- Repoila F, Gottesman S. Signal transduction cascade for regulation of RpoS: Temperature regulation of DsrA. Journal of Bacteriology. 2001;183:4012–4023. doi: 10.1128/JB.183.13.4012-4023.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repoila F, Majdalani N, Gottesman S. Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: the RpoS paradigm. Mol. Microbiol. 2003;48:855–861. doi: 10.1046/j.1365-2958.2003.03454.x. [DOI] [PubMed] [Google Scholar]
- Robertson GT, Roop RMJ. The Brucella abortus host factor I (HF-1) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol. Microbiol. 1999;34:690–700. doi: 10.1046/j.1365-2958.1999.01629.x. [DOI] [PubMed] [Google Scholar]
- Rouault TA. Post-transcriptional regulation of human iron metabolism by iron regulatory proteins. Blood Cells Mol. Dis. 2002;29:309–314. doi: 10.1006/bcmd.2002.0571. [DOI] [PubMed] [Google Scholar]
- Sauter C, Basquin J, Suck D. Sm-like proteins in eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res. 2003;31:4091–4098. doi: 10.1093/nar/gkg480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA, Pearson RF, Møller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledjeski DD, Whitman C, Zhang A. Hfq is necessary for regulation by the untranslated RNA DsrA. Journal of Bacteriology. 2001;183:1997–2005. doi: 10.1128/JB.183.6.1997-2005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnleitner E, Schuster M, Sorger-Domenigg T, Greenberg EP, Blasi U. Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Mol. Microbiol. 2006;59:1542–1558. doi: 10.1111/j.1365-2958.2006.05032.x. [DOI] [PubMed] [Google Scholar]
- Storz G, Altuvia S, Wassarman KM. An abundance of RNA regulators. Annu. Rev. Biochem. 2005;74:199–217. doi: 10.1146/annurev.biochem.74.082803.133136. [DOI] [PubMed] [Google Scholar]
- Storz G, Gottesman S. Versatile roles of small RNA regulators in bacteria. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 3rd edition Cold Spring Harbor Laboratory Press; 2006. pp. 567–594. [Google Scholar]
- Storz G, Opdyke JA, Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr Opin Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- Tjaden B, Goodwin SS, Opkyke JA, Guillier M, Fu DX, Gottesman S, Storz G. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res. 2006;34:2791–2802. doi: 10.1093/nar/gkl356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden B, Saxena RM, Stolyar S, Haynor DR, Kolker E, Rosenow C. Transcriptome analysis of Escherichia coli using high density oligonucleotide probe arrays. Nucleic Acids Res. 2002;30:3732–3738. doi: 10.1093/nar/gkf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotochaud AE, Wassarman KM. A highly conserved 6S RNA structure is required for regulation of transcription. Nat. Struct. Mol. Biol. 2005;12:774–784. doi: 10.1038/nsmb917. [DOI] [PubMed] [Google Scholar]
- Tsui H-CT, Leung H-CE, Winkler ME. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- Udekwu KI, Darfeuille F, Vogel J, Reimegard J, Holmqvist E, Wagner EG. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes & Dev. 2005;19:2355–2366. doi: 10.1101/gad.354405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski ML, Stauffer LT, Stauffer GV. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol. Microbiol. 2000;37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- Vanderpool CK, Gottesman S. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Molec. Microbiol. 2004;54:1076–1089. doi: 10.1111/j.1365-2958.2004.04348.x. [DOI] [PubMed] [Google Scholar]
- Vogel J, Bartels V, Tang HH, Churakov G, Slagter-Jager JG, Huttenhofer A, Wagner EGH. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes & Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA, Vasil ML. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc Natl Acad Sci U S A. 2004;101:9792–9797. doi: 10.1073/pnas.0403423101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. The oxyS regulatory RNA represses rpoS translation by binding Hfq (HF-1) protein. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Molec. Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]


