Abstract
Protein phosphorylation is a reversible post-translational modification known to regulate protein function, subcellular localization, complex formation, and protein degradation. Detailed phosphoproteomic information is critical to kinomic studies of signal transduction and for elucidation of cancer biomarkers, such as in non-small cell lung adenocarcinoma, where phosphorylation is commonly dysregulated. However, the collection and analysis of phosphorylation data remains a difficult problem. The low concentrations of phosphopeptides in complex biological mixtures as well as challenges inherent in their chemical nature have limited phosphoproteomic characterization and some phosphorylation sites are inaccessible by traditional workflows. We developed a sequential digestion method using complementary proteases, Glu-C and trypsin, to increase phosphoproteomic coverage and supplement traditional approaches. The sequential digestion method is more productive than workflows utilizing only Glu-C and we evaluated the orthogonality of the sequential digestion method relative to replicate trypsin-based analyses. Finally, we demonstrate the ability of the sequential digestion method to access new regions of the phosphoproteome by comparison to existing public phosphoproteomic databases. Our approach increases coverage of the human lung cancer phosphoproteome by accessing both new phosphoproteins and novel phosphorylation site information.
Keywords: Genomics / Proteomics, Mass spectrometry / ICP-MS, Bioanalytical methods
Introduction
Due to the essentiality of phosphorylation in regulating virtually all biochemical systems, broad efforts have been made to characterize the location and frequency of phosphorylation events [1]. In particular, the promise for phosphoproteomics in cancer biomarker discovery is high, given the universal dysregulation of cellular signaling pathways that occurs in human cancers [2,3]. However, despite these rich prospects for translational cancer research, analytical challenges such as stoichiometric limitations and increased fragmentation complexity of modified species have impeded phosphorylation site identification [4]. To address some these limitations, current techniques attempt to access a greater portion of the cellular phosphoproteome through the coupling of peptide fractionation methods to the penultimate analysis by tandem mass spectrometry (LC-MS/MS).
In this now standard phosphoproteomics workflow, proteins are obtained from cell lysates and digested by the protease trypsin. Reduction of sample complexity is effected by separation of trypsinized peptides via their solution charge state at acidic pH into fractions using strong cation exchange chromatography (SCX) [5,6]. Next, these fractions are individually incubated with titanium dioxide (TiO2) microspheres to enrich for phosphopeptides by chelation of the phosphate groups with the surface of the TiO2 resin [7–9]. Finally, each fraction is analyzed by LC-MS/MS and phosphorylated peptides are identified from spectra using searching algorithms, such as SEQUEST [10,11], which often results in the confident identification of more than ten thousand unique phosphorylation sites in a single experiment [12–14] (Fig. 1a). Despite the strengths of this method, trypsin-based approaches do not grant access to all regions of the phosphoproteome, necessitating the development of complimentary workflows.
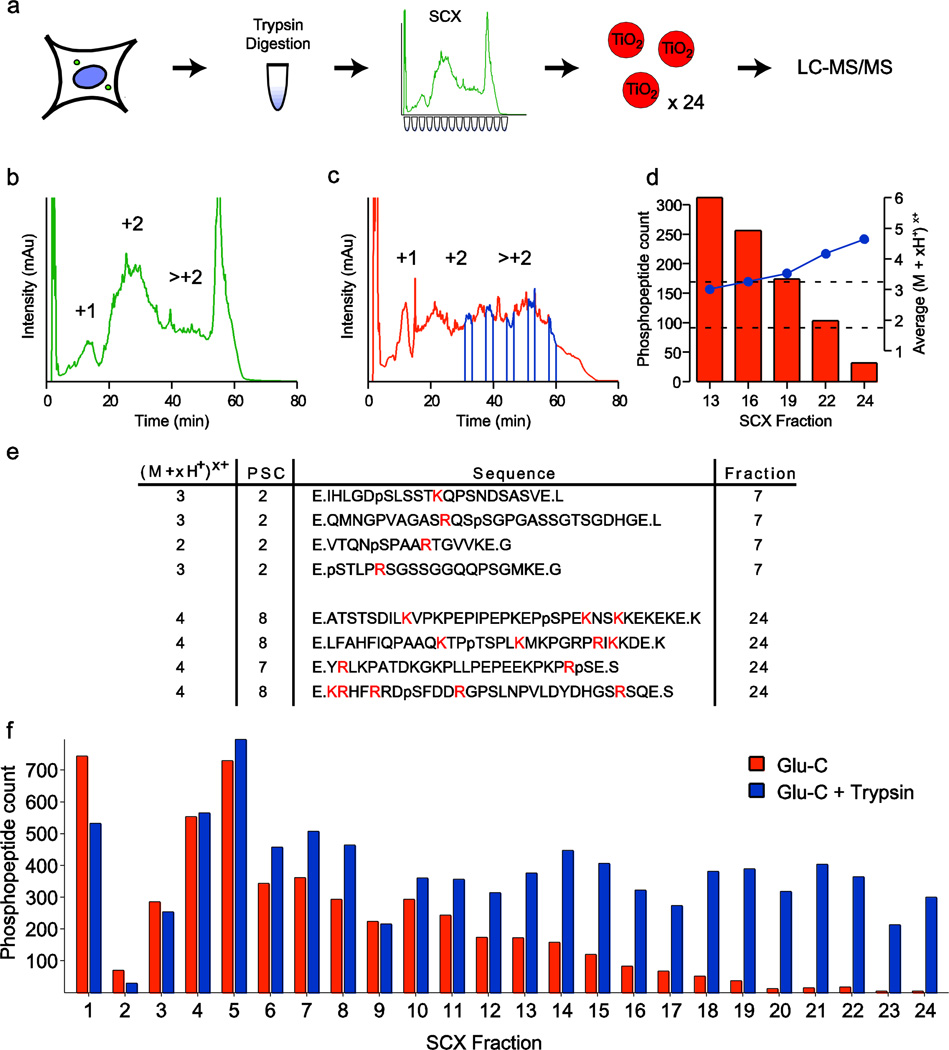
Fig. 1.
Evaluation of a Glu-C only based phosphoproteomic workflow. (a) In the traditional phosphoproteomic workflow, proteins are obtained from cell lysates and proteolytically digested with trypsin. The resultant peptides are separated into fractions via strong cation exchange (SCX) chromatography, and each fraction is treated with titanium dioxide microspheres to enrich for phosphopeptides prior to analysis by LC-MS/MS. SCX chromatography for (b) trypsin and (c) Glu-C based workflows exhibit protease-dependant peptide separation. Fractions 13, 16, 19, 22, and 24 are indicated in blue. (d) The number of phosphopeptide identifications declines steeply in mid to late fractions. Average gas phase peptide charge state (blue) concurrently rises outside the ideal range for peptide identification by CID (+2 and +3 charge states). (e) Phosphorylated peptides in early fractions (upper) have lengths and gas phase charge states amenable to LC-MS/MS analysis, while phosphopeptides from later fractions are longer and contain more trypsin-cleavable basic residues (highlighted). (f) Phosphopeptide count by fraction for Glu-C digested (orange) and subsequently trypsin digested peptides (blue) suggest that these intractable phosphopeptides can be successfully recovered and identified
Trypsin cleaves proteins at the basic residues lysine and arginine, and is widely used in proteomics because the resulting pool of peptides of are of intermediate length and exhibit a gas-phase charge state distribution and localization that is ideal for collision-induced dissociation (CID)-based LC-MS/MS identification. In addition, trypsin digestion creates a large pool of peptides with basic residues at their N- (free H2N-) and C- (Arg & Lys) termini, which results in a predominant peptide solution charge state (PSC) of +2 ([15], Fig. 1b). Missed cleavages, uncleavable sequences and histidines shift this PSC distribution to higher values, while N-terminal acetylation or addition of a phosphate group, for example, lead to lower PSC values [16]. Several other proteases are commercially available such as Lys-N, Lys-C, and Glu-C, the former two of which have been co-opted for use in phosphoproteomics [17,18]. Since the cleavage specificities of these proteases differ from that of trypsin, protein digestions with each reagent will generate distinct peptide pools that may afford access to additional phosphorylation sites that are difficult or impossible to detect using trypsin-based methods alone [19,20]. However, the peptides produced by these alternate digestions are not always amenable to analysis by standard shotgun sequencing platforms due to the relative (in)frequency of these alternate cleavage sites and the uneven distribution of basic amino acids in the resulting peptide pool. To address this limitation, multiple enzyme digests, performed separately and in parallel, can afterwards be pooled to increase protein and post-translational modification coverage [21]. Furthermore, Glu-C has been shown to increase protein coverage beyond that of other proteases such as chymotrypsin and Arg-C [22]. In the same study, a sequential digestion approach was evaluated using Glu-C and Arg-C proteases in succession to diversify peptide pools, and these were found to be less productive than either protease alone, albeit for proteins and not for phosphoproteins.
In the present work, we develop and test alternative workflows for phosphoproteomics based on the combination of Glu-C and trypsin that leverage the strengths of the respective proteases and address the limitations of using Glu-C alone. We show here that when this combination of complimentary proteases is used in place of trypsin alone in the traditional workflow, we observe phosphorylation sites that are inaccessible via the trypsin based approach. Taken together, our results describe a novel workflow that reveals regions of the phosphoproteome that are refractory to standard methods.
Experimental Procedures
Materials
Modified trypsin was from Promega (Madison, WI); Glu-C protease was from Worthington Biochemicals (Lakewood, NJ). Urea, Tris-HCl, CaCl2, ammonium bicarbonate (NH4HCO3), sodium fluoride (NaCl), potassium fluoride (KCl), potassium phosphate (KH2PO4), phosphoric acid, sodium ortho-vanadate, sodium molybdate, sodium tartrate, beta-glycerophosphate, DL-dithiothreitol, iodoacetamide were from Sigma-Aldrich (St. Louis, MO). Acetonitrile (ACN), trifluoroacetic acid (TFA) and water were from Honeywell Burdick and Jackson (Morristown, NH). Methanol was from Fisher (Pittsburgh, PA). High-purity formic acid was from EMD (Gibbstown, NJ). SepPak C18 solid-phase extraction cartridges and Oasis HLB vacuum extraction plates were from Waters Corporation (Milford, MA). Lactic acid was from Lee BioSolutions, Inc. (St. Louis, MO). TiO2 beads were from GL Sciences (Tokyo, Japan). Dulbecco’s modified Eagle’s medium (DMEM), RPMI, PBS, penicillin and streptomycin were from Invitrogen (Carlsbad, CA). Fetal bovine serum (Hyclone) was purchased from ThermoFisher Scientific (Pittsburgh, PA). NCI-H23 cells were obtained from the American Type Culture Collection (ATCC; Manassas, VA).
Sequential digestion workflows
We tested three experimental workflows which differed only in the order of sample preparation steps (Fig 2a). Briefly, a single, large pool of NCI-H23 cells were lysed in 8M urea, and their proteins were digested using Glu-C protease, desalted, and lyophilized in three equal (5mg) aliquots. For method one, hereafter “GSPT” (Glu-C digestion, SCX, Phosphopeptide enrichment (TiO2), and trypsin digestion), one 5mg aliquot of the Glu-C digested peptides was separated by SCX chromatography using a 9.4mm inner diameter (I.D.) column, 24 fractions were collected and individually enriched for phosphopeptides using TiO2, and each fraction was individually digested with trypsin before analysis by LC-MS/MS. The second method, hereafter “GPST” (Glu-C digestion, Phosphopeptide enrichment (TiO2), SCX, and trypsin digestion), took advantage of a single-stage phosphopeptide enrichment step directly on an unfractionated 5mg aliquot of NCI-H23 peptide digest, followed by SCX chromatography using a 2.1 mm I.D. column into 24 fractions, and finally individual trypsin digestion of those fractions. The third method, hereafter “GPTS” (Glu-C digestion, Phosphopeptide enrichment (TiO2), trypsin digestion, and SCX), used the same single-stage phosphopeptide enrichment step on 5mg of Glu-C digested peptides, but then utilized a single trypsin digestion of the resulting phosphopeptides before separation by SCX chromatography on a 2.1 mm column, fraction collection and LC-MS/MS analysis. Alternatively, lysed and denatured NCI-H23 protein lysates were digested with trypsin and analyzed as described previously [23].
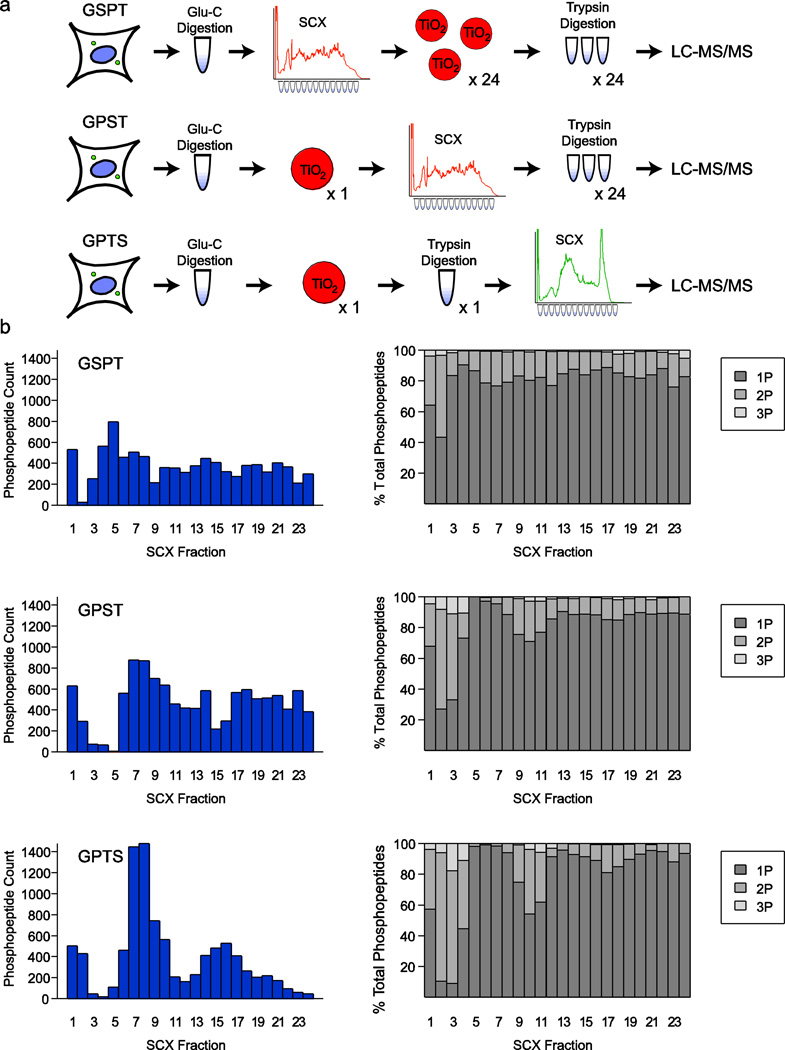
Fig. 2.
Experimental design and analysis of sequential digestion workflows. (a) Schematic diagram of the three sequential digestion approaches. GSPT closely resembles the traditional workflow with substitution of Glu-C for initial digestion and trypsin digestion immediately prior to LC-MS/MS. GPST and GPTS take advantage of a single-stage of phosphopeptide purification, and GPTS utilizes only a single trypsin digestion before SCX separation rather than trypsin digestion of individual fractions. (b) Phosphopeptide count and phosphate distribution across peptides are shown for each approach. Solution charge reduction of tryptic phosphopeptides in SCX causes uneven distribution across fractions for GPTS
Lysis and Digestion of NCI-H23 cells
NCI-H23 cells were grown as adherent cultures in RPMI, respectively, supplemented with 10% FBS and penicillin and streptomycin. For harvesting, cells were collected, washed with PBS and frozen in liquid nitrogen. For lysis, cells were thawed on ice and lysed in lysis buffer (8 M urea, 25 mM Tris-HCl, 150 mM NaCl, phosphatase inhibitors (2.5 mM beta-glycerophosphate, 1 mM sodium fluoride, 1 mM sodium orthovanadate, 1 mM sodium molybdate, 1 mM sodium tartrate) and protease inhibitors (1 mini-Complete EDTA-free tablet per 10 ml lysis buffer; Roche Life Sciences, Mannheim, Germany). The lysate was sonicated three times at 30 – 40% power for 15 sec each with intermittent cooling on ice, followed by centrifugation at 15,000 × g for 30 min at 4 °C to clarify the lysate. The lysate was then reduced with DTT at a final concentration of 5 mM and incubated for 30 min at 55 °C. Afterwards, the lysate was thoroughly cooled to room temperature (~22 °C) and alkylated with 15 mM iodoacetamide at room temperature for 45 min. The alkylation was then quenched by the addition of an additional 5 mM DTT. After 6-fold dilution with 25 mM Tris-HCl pH 8 and 1 mM CaCl2, the sample was digested overnight at 37 °C with 2.5% (w/w) trypsin or Glu-C. The next day, the digest was stopped by the addition of 0.25% TFA (final v/v), centrifuged at 3500 × g for 30 min at room temperature to pellet precipitated lipids, and desalted on a SepPak C18 cartridge (Waters). Desalted peptides were lyophilized and stored at −80 °C until further use.
SCX and Phosphopeptide Enrichment
5 milligrams of lyophilized peptides were resuspended in SCX buffer A (7 mM KH2PO4, pH 2.65 / 30% ACN) and separated per injection on a SCX column (PolySULFOETHYL A 200 × 9.4 mm, 5 µm 200 Å pore, item# 209SE0502; PolyLC Inc, Columbia, MD). For trypsin samples, a gradient of 0 to 10 % SCX buffer B (350 mM KCl / 7 mM KH2PO4, pH 2.65 / 30% ACN) over 10 minutes, 10% to 17% SCX buffer B over 17 minutes, 17% to 32% SCX buffer B over 13 minutes, 32% to 60% SCX buffer B over 10 minutes, 60% to 100% SCX buffer B over 2 minutes, holding at 100% SCX buffer B for 5 minutes, from 100% to 0% SCX buffer B over 2 minutes, and equilibration at 0% SCX buffer B for 65 minutes, all at a flow rate of 2.5 ml/min; for the GSPT sample, a gradient of 0 to 10% SCX buffer B over 5 minutes, 10 to 25% SCX buffer B over 15 minutes, 25 to 55% SCX buffer B over 22 minutes, 55 to 100% SCX buffer B over 13 minutes, holding at 100% SCX buffer B for 10 minutes, 100 to 0% SCX buffer B over 2 minutes, and equilibration at 0% SCX buffer B for 65 minutes, also all at a flow rate of 2.5 ml/min. For the GPTS and GPST samples, identical gradients were run on a 2.1 mm PolySULFOETHYL A column at 0.2 ml/min using the trypsin and GSPT gradients above, respectively. After a full blank injection of the same program was run to equilibrate the column, a 5 mg sample of either digest type or desalted phosphopeptide aliquot in 100 µl of 100% SCX buffer A was injected on to the HPLC, and 24 fractions were collected from the onset of the void volume (2.2 minutes) until the elution of strongly basic peptides at 100% SCX buffer B (52 minutes), at 2.075-minute intervals, for the appropriate HPLC method. After separation, the SCX fractions were lyophilized and desalted using a 60-mg OASIS C18 96-well desalting plate and manifold (Waters, Milford MA). For phosphopeptide purification, peptides were resuspended in 100 µl 2 M lactic acid in 50% ACN (“binding solution”), with the 400 µg titanium dioxide microspheres, and vortexed by affixing to the top of a vortex mixer on the highest speed setting at room temperature (~ 21 °C) for 45 minutes. Afterwards, the beads were washed twice with 50 µl of the binding solution and three times with 100 µl 50% ACN / 0.1% TFA, and eluted twice with 20 µl NH4PO4 (adjusted to pH 10 with ammonium hydroxide in ethanol). Peptide elutions were combined, quenched with 20 µl 50% ACN / 5% formic acid, dried and desalted on a µHLB OASIS C18 desalting plate. The liquid eluate from the µHLB OASIS plate (~60 µl) was transferred to deactivated glass micro inserts (Agilent), dried by vacuum centrifugation directly in inserts and analyzed by LC-MS/MS. Single-stage purifications were performed exactly as described [23].
LC-MS/MS Analysis
LC-MS/MS analysis was performed on a LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) equipped with an Agilent 1100 capillary HPLC, FAMOS autosampler (LC Packings, San Francisco, CA) and nanospray source (Thermo Fisher Scientific). Peptides were redissolved in 6% ACN / 1% formic acid and loaded onto an in-house packed polymer-fritted[24] trap column at 2.5 µl/min (1.5 cm length, 100 µm inner diameter, ReproSil, C18 AQ 5 µm 200 Å pore (Dr. Maisch, Ammerbuch, Germany)) vented to waste via a micro-tee. The peptides were eluted by split-flow at ~800 – 1000 psi head pressure from the trap and across a fritless analytical resolving column (18 cm length, 125 µm inner diameter, ReproSil, C18 AQ 3 µm 200 Å pore) pulled in-house (Sutter P-2000, Sutter Instruments, San Francisco, CA) with a 50 min gradient of 5–30% LC-MS buffer B (LC-MS buffer A: 0.0625% formic acid, 3% ACN; LC-MS buffer B: 0.0625% formic acid, 95% ACN). An LTQ-Orbitrap (LTQ Orbitrap MS control software v. 2.5.5, build 4 (06/20/08); previously tuned and calibrated per instrument manufacturer’s guidelines using caffeine, MRFA, and UltraMark “CalMix”) method consisting of one Orbitrap survey scan (AGC Orbitrap target value: 700K; R = 60K; maximum ion time: 800 milliseconds; mass range: 400 to 1400 m/z; Orbitrap “preview” mode enabled; lock mass[25] set to background ion 445.120029) was collected, followed by ten data-dependent tandem mass spectra on the top ten most abundant precursor ions (isolation width: 1.6 m/z; CID relative collision energy (RCE): 35%; MS1 signal threshold: 12,500; AGC LTQ target value: 3,500; maximum MS/MS ion time: 125 milliseconds; dynamic exclusion: repeat count of 1, exclusion list size of 500 (max), 24 seconds wide in time, +/− 20 ppm wide in m/z; for trypsin-only and Glu-C/trypsin double-digests doubly- and triply-charged precursors were selected for MS/MS; for Glu-C only precursor charge states greater than 1 were selected for MS/MS; no neutral-loss dependent or multi-stage activation methods were employed[26]).
Peptide spectral matching and bioinformatics
Raw data were searched using SEQUEST[10,11] (Thermo Fisher Scientific, San Jose, CA) against a target-decoy (reversed)[27] version of the human proteome sequence database (UniProt; downloaded 9/2010; 74,338 total (forward & reverse) proteins) with a precursor mass tolerance of +/− 1 Da and requiring either fully tryptic peptides (trypsin-only digests), or fully [D, E, K, R]-specific termini (sequential Glu-C / trypsin digests) with up to two mis-cleavages, carbamidomethylcysteine as a fixed modification and oxidized methionine and phosphorylated serine, threonine and tyrosine as variable modifications. The resulting peptide spectral matches were filtered to < 1% false discovery rate (FDR), based on reverse-hit counting (typically but not always using cutoffs of mass measurement accuracy (MMA) within −/+ 2.5ppm, a delta-XCorr (dCn) of greater than 0.08, and XCorr values of greater than 2 for +2-charge state peptides and greater than 2.6 for +3-charge state peptides). No additional criteria were used for assessing confidence of peptide spectral matches. Data filtering and comparative analyses were performed using the R statistical programming language (http://www.R-project.org). Summary information for all peptide assignments can be found in Electronic Supplementary Material Tables S1 – S6; Glu-C only, GSPT, GPST, GPTS, and trypsin-only replicates, respectively).
Results and Discussion
Assessment of Glu-C based LC-MS/MS workflow and introduction of double-digestion with Glu-C and trypsin
We began by first considering Glu-C as a stand-alone alternative protease to trypsin, and sought to characterize Glu-C digested, SCX separated peptide fractions. In contrast to trypsin-based SCX separations (Fig. 1b), Glu-C digestion produces peptides that are much more evenly distributed across the SCX chromatogram (Fig. 1c), likely due to the pseudo-random distribution of basic residues contained within these peptides. When combined with LC-MS/MS, we noted that the Glu-C based workflow produced a lower number of confident phosphopeptide identifications in later fractions, which consisted of longer and more highly-basic peptides (Electronic Supplementary Material Table S1). Significantly, the average gas-phase peptide charge state of these fractions rose outside the ideal range for CID-based peptide identification (optimally (M+2H+)2+ or (M+3H+)3+ charge states). Comparison of phosphopeptides from early and late fractions (Fig. 1e) revealed that later fractions contained many basic, trypsin-cleavable residues flanking phosphorylation events. We therefore reasoned that a secondary trypsin digestion would produce more analytically favorable species for peptides in these later-eluting fractions. To test this hypothesis, we performed a secondary digestion of fractions 13, 16, 19, 22 and 24 post-TiO2 enrichment with trypsin, and reanalyzed these fractions by LC-MS/MS. This resulted in a marked increase in phosphopeptide identifications for all of these fractions; for the most basic of these fractions (#24), we observed a 14-fold increase in the number of confident phosphopeptide identifications (Fig. 1f). In contrast to previous sequential digestion studies, where double-digestion reduced peptide length to the point of reduced capture efficiency on peptide trap and subsequent MS detection [22], we found a dramatic increase in the number of detected species. We hypothesize that this is due both to the proteases used and the differences in peptide pool that results from the SCX separation. Taken together, these data prompted the further development and refinement of three sequential digestion approaches.
Optimization of the sequential digestion workflow
To take advantage of non-tryptic digestion while maintaining the favorable peptide length and charge distributions of tryptically-digested peptides, we tested three sequential protease cleavage methods and evaluated the unique phosphorylation sites identified by each. Concurrently, we also tested if a single TiO2 phosphopeptide enrichment step [23], could be performed before SCX separation, precluding the need for laborious enrichment of individual SCX fractions. This single-stage TiO2 protocol has been shown to minimize the current rate-limiting step of phosphoproteomic sample preparation and to allow for higher throughput for phosphoproteomic workflows.
The three experimental workflows we tested differed only in the order of sample preparation steps (Fig 2a). Briefly, a single, large pool of NCI-H23 cells were lysed in 8M urea, and their proteins were digested using Glu-C protease, desalted, and lyophilized in three equal (5mg) aliquots. For method one, hereafter “GSPT” (Glu-C digestion, SCX, Phosphopeptide enrichment (TiO2), and trypsin digestion), one 5mg aliquot of the Glu-C digested peptides was separated by SCX chromatography using a 9.4mm inner diameter (I.D.) column, 24 fractions were collected and individually enriched for phosphopeptides using TiO2, and each fraction was individually digested with trypsin before analysis by LC-MS/MS. The second method, hereafter “GPST” (Glu-C digestion, Phosphopeptide enrichment (TiO2), SCX, and trypsin digestion), took advantage of a single-stage phosphopeptide enrichment step directly on an unfractionated 5mg aliquot of NCI-H23 peptide digest, followed by SCX chromatography using a 2.1 mm I.D. column into 24 fractions, and finally individual trypsin digestion of those fractions. The third method, hereafter “GPTS” (Glu-C digestion, Phosphopeptide enrichment (TiO2), trypsin digestion, and SCX), used the same single-stage phosphopeptide enrichment step on 5mg of Glu-C digested peptides, but then utilized a single trypsin digestion of the resulting phosphopeptides before separation by SCX chromatography on a 2.1 mm column, fraction collection and LC-MS/MS analysis (Electronic Supplementary Material Table S2 – S4).
In a typical trypsin-based workflow, phosphopeptides are unevenly distributed across SCX-separated fractions due to charge reduction of phosphorylated residues. Conversely, Glu-C digested peptides separate more uniformly across this separation space. As expected, this uniform distribution was also observed for the GSPT and GPST workflows, where SCX separation precedes trypsin digestion, while the GPTS workflow showed a post-SCX distribution of phosphopeptides across fractions similar to a trypsin-only workflow (Fig. 2b). No striking differences were observed for the number of phosphorylation sites per peptide as a function of SCX fraction number for any of the three workflows. In general, the number of phosphate groups per peptide was more evenly distributed across the SCX separation space for the two schemes that separated Glu-C digested peptides relative to the trypsin-separated approach. We surmised that the uniformity of peptide distribution in SCX chromatography from Glu-C digestions might allow us access to rare species that could be lost in the few phosphopeptide-heavy fractions via LC-MS/MS undersampling in a traditional workflow.
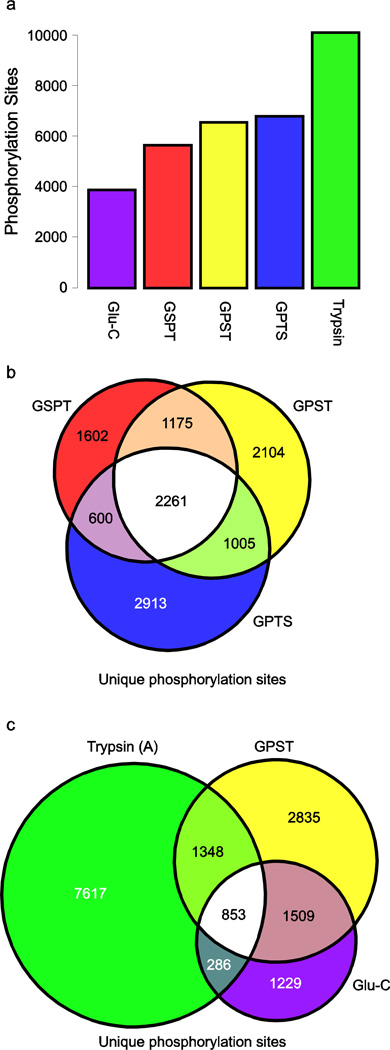
To test this hypothesis, we compared both the unique phosphopeptides and phosphorylation sites identified in each workflow against a representative trypsin-based dataset (Electronic Supplementary Material Table S5). First, we confirmed the sensitivity of our TiO2 enrichment steps by evaluating the number of unique phosphopeptides identified in each workflow. We identified 6808, 7845, and 7572 unique phosphopeptides across the GSPT, GPST, and GPTS workflows, respectively. This demonstrates the productivity of a single-stage TiO2 enrichment step which also requires substantially less effort to perform than individual enrichment of individual fractions. We note that the single-stage TiO2 enrichment process was slightly less productive in our trypsin-based workflow than performing 24 individual purifications, and speculate that the relatively high basic content of many of these Glu-C digested SCX fractions may have either impacted the SCX separation efficiency of later-eluting peptides, or the relative purification efficiency of these more basic peptides, or both. We also assessed phosphoproteomic coverage of the sequential digestion workflows by comparing the number of unique phosphorylation sites identified by each. Here, we find 5638, 6545, and 6779 unique phosphorylation sites for GSPT, GPST, and GPTS, respectively, with 2261 sites commonly observed in all methods (Fig. 3a). Notably, each of these combinations of Glu-C and trypsin approaches yielded a larger number of unique phosphorylation sites than when Glu-C was the only protease used for digestion (3877 unique phosphorylation sites), suggesting that we are able to recover a significant portion of the previously intractable, high charge state phosphopeptides characteristic of Glu-C digestion. Additionally, the GPTS and GPST methods produced the largest sets of unique phosphorylation events present exclusively in that workflow (2913 and 2104, respectively), compared to only 1602 for GSPT.
Fig. 3.
Phosphorylation sites identified by each approach. (a) Total number of distinct phosphorylation sites by method. Each experimental approach identified a larger number of phosphorylation sites than Glu-C only workflows. (b) Comparison of unique phosphorylation loci identified in common between the sequential digestion methods. (c) Comparison of the Glu-C only and the GPST workflows to a traditional trypsin only workflow. Notably, GPST is shown because this workflow yields the largest number of phosphorylation sites distinct from a trypsin only workflow despite a slightly lower absolute number of phosphorylation sites detected compared to GPTS
Comparison to a trypsin-only workflow
We next sought to establish whether our new methods were capable of producing novel biological information relative to the established trypsin-based workflow. To do this, we compared the sites found by sequential digestion methods to those found in a trypsin-only approach. Here, we find 36, 34, and 44% phosphorylation site overlap between trypsin and GSPT, GPST, and GPTS, respectively. Additionally, we considered the unique phosphorylation sites identified in the Glu-C only workflow and compared these results to both a trypsin-based approach and to the GPST method (Fig 3b). Given that data-dependent shotgun sequencing by standard LC-MS/MS approaches typically undersamples highly complex peptide mixtures [28]; we repeated the analysis of our trypsin-only sample by performing a technical replicate of all of the steps in the workflow. When comparing the trypsin sample to its technical replicate, 62% of the sites are common to both replicates, which suggests that the phosphorylation site information unique to any of the three sequential digestion methods cannot be accounted for solely by undersampling. Additionally, due to the greater overlap between GPTS and trypsin, we find that GPST identifies 14% more distinct phosphorylation events relative to trypsin than does GPTS (4344 and 3828 unique phosphorylation events respectively), despite detecting a lower absolute number of unique phosphorylation sites. We surmise that this arises primarily due to the fact that the peptide digest separated in the GPTS workflow is much more “trypsin-like”, in that the double-digest is performed prior to SCX (Fig. 2a; Electronic Supplementary Material Figure S1). We concluded from these analyses that the GPST format provided the most unique information relative to the standard trypsin-based workflow.
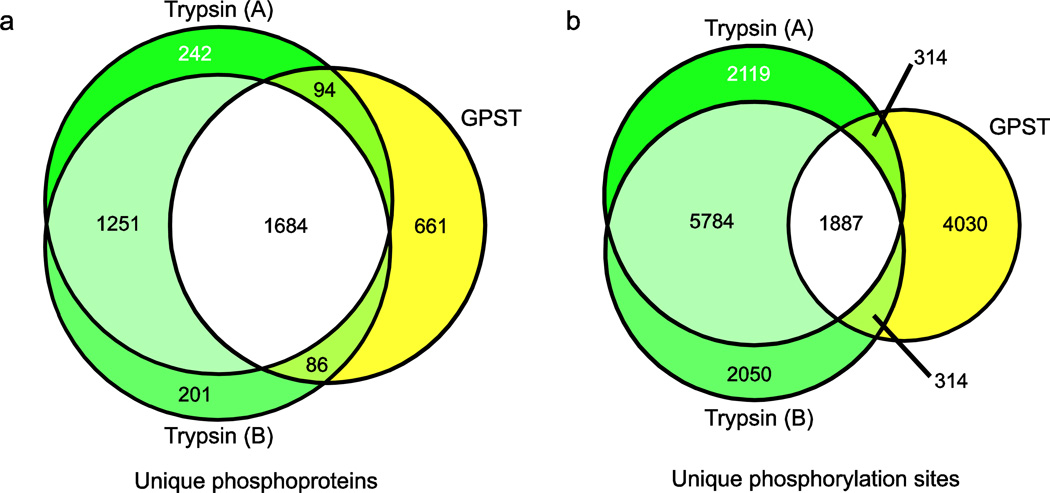
Given the high degree of similarity between sequential digestion methods we selected GPST as a representative workflow and compared the sets of unique phosphoproteins and phosphorylation sites between a pair of trypsin replicates (Electronic Supplementary Material Tables S5 & S6) and the GPST workflow (Fig. 4). In both cases, we observe substantially greater overlap between trypsin replicates than between either trypsin replicate and GPST. Furthermore, the relative protein overlap was greater than the phosphorylation site overlap, suggesting that gains in coverage come primarily from novel phosphorylation site localization on phosphoproteins already identified by trypsin workflows, rather than predominantly from sites on novel proteins. This reinforces the notion that the peptide selection bias of trypsin is preventing full characterization of the phosphoproteome, and underscores the necessity of developing workflows that complement traditional techniques.
Fig. 4.
Sequential digestion (GPST) versus trypsin-only workflow. Comparison of (a) phosphoprotein and (b) phosphorylation site identifications between GPST and a pair of trypsin-based replicates. GPST allows orthogonal access to the phosphoproteome, and produces phosphorylation site information inaccessible by traditional methods and which cannot be accounted for by undersampling
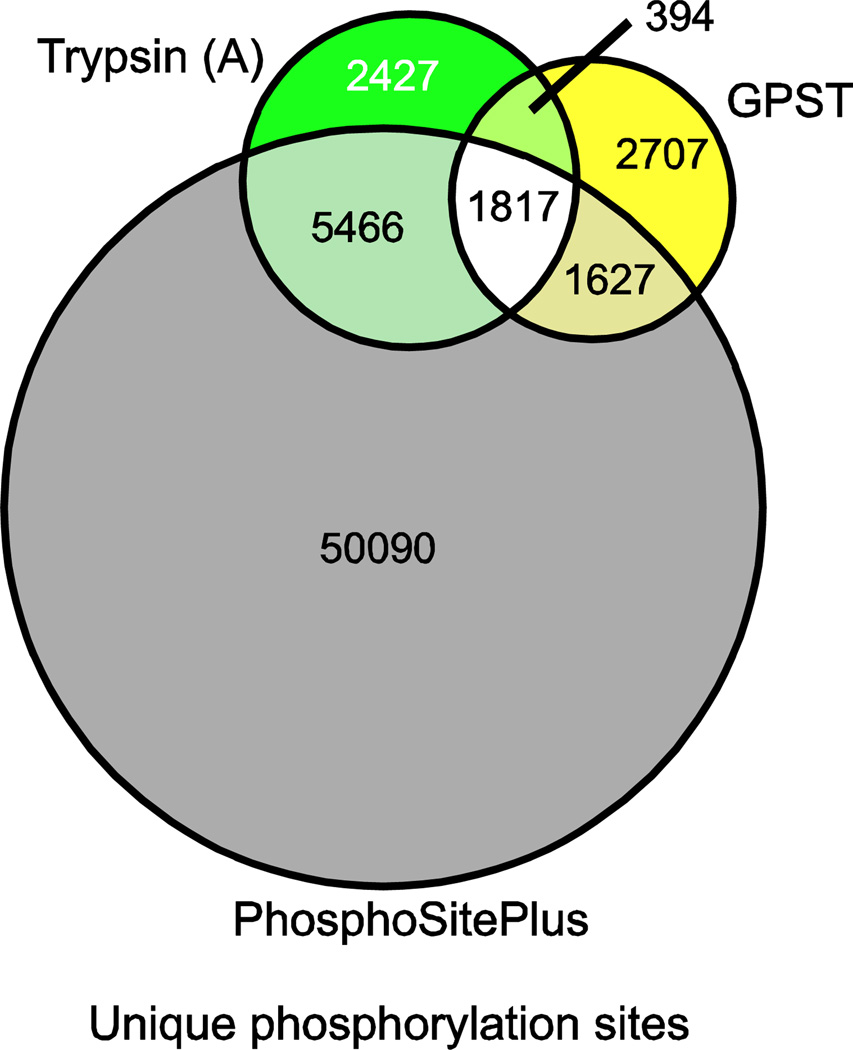
Finally, to place the contribution of sequential digestion workflows into a broader phosphoproteomic perspective, we compared the unique phosphorylation sites detected using the GPST method to the existing public phosphorylation database PhosphoSitePlus [29] (Fig. 5). Here, we find that a single sequential digestion workflow sample is able to confidently identify 3101 novel phosphorylation sites. Interestingly, our trypsin only workflow also yielded a large number of novel sites (2821), suggesting that characterization of NCI-H23 cell phosphorylation status is currently under represented in the greater phosphoproteomic collective. However, despite the incompleteness of PhosphoSitePlus, it is significant to note that the GPST workflow was able to identify both a larger absolute number of novel phosphorylation sites and a larger percentage of new sites, despite containing only two thirds as many unique phosphorylation sites as were present in the trypsin analysis.
Fig. 5.
Comparison of unique phosphorylation sites detected in sequential digestion GPST and a traditional, trypsin-only, workflow to human phosphorylation sites previously identified in the PhosphoSitePlus database
Conclusions
Despite rapid technical innovation in proteomics technology, probing the phosphoproteome remains a difficult problem. Certain phosphorylation events remain resistant to detection by classical approaches due to the chemical nature of the amino acids that surround them. Trypsin digestion has proven to be a valuable technique due to many analytically favorable characteristics of its resultant peptide pools, but it also suffers from a selection bias that may preclude the discovery of biologically significant phosphorylation events. Here, we present data demonstrating that a sequential digestion strategy allows access to some of these elusive phosphorylation sites. Unsurprisingly, we find that SCX separation of phosphopeptides is protease dependant. We also find, however, that the number and distribution of phosphorylation events observed using an optimized sequential digestion method cannot be accounted for by statistical undersampling alone. Furthermore, we show that many modification sites are located in peptides that would be poorly identifiable using either Glu-C or trypsin in isolation.
Ultimately, the sequential digestion strategy represents a compliment, rather than a replacement, to the traditional trypsin-only workflow, and is a more robust alternative than simple Glu-C digests or reliance on technical replicates. Furthermore, we find evidence to support previous work [23] that describes a single-stage phosphopeptide enrichment step designed to reduce sample manipulation and effort, and improve robustness of the workflow overall. This method minimizes the rate-limiting step of phosphopeptide sample preparation without loss of sensitivity and enables the rapid creation of biological replicates, which is likely to be a requirement for future experiments in the field. Ultimately, the combination of a sequential digestion strategy and single-stage phosphopeptide purification allows for rapid phosphoproteomic profiling that is essential for the accurate and robust analysis of cancer cells that exhibit dysregulated cellular signaling.
Supplementary Material
Acknowledgements
The authors would like to acknowledge funding from the American Cancer Society (IRG-82-003-24) and the National Institutes of Health (P20-RR018787) for the IDeA Program of the National Center for Research Resources (to S.A.G.).
References
- 1.Nita-Lazar A, Saito-Benz H, White FM. Quantitative phosphoproteomics by mass spectrometry: past, present, and future. Proteomics. 2008;8(21):4433–4443. doi: 10.1002/pmic.200800231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies H, Hunter C, Smith R, Stephens P, Greenman C, Bignell G, Teague J, Butler A, Edkins S, Stevens C, Parker A, O'Meara S, Avis T, Barthorpe S, Brackenbury L, Buck G, Clements J, Cole J, Dicks E, Edwards K, Forbes S, Gorton M, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jones D, Kosmidou V, Laman R, Lugg R, Menzies A, Perry J, Petty R, Raine K, Shepherd R, Small A, Solomon H, Stephens Y, Tofts C, Varian J, Webb A, West S, Widaa S, Yates A, Brasseur F, Cooper CS, Flanagan AM, Green A, Knowles M, Leung SY, Looijenga LH, Malkowicz B, Pierotti MA, Teh BT, Yuen ST, Lakhani SR, Easton DF, Weber BL, Goldstraw P, Nicholson AG, Wooster R, Stratton MR, Futreal PA. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65(17):7591–7595. doi: 10.1158/0008-5472.CAN-05-1855. [DOI] [PubMed] [Google Scholar]
- 3.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O'Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Cahill DP, Louis DN, Goldstraw P, Nicholson AG, Brasseur F, Looijenga L, Weber BL, Chiew YE, DeFazio A, Greaves MF, Green AR, Campbell P, Birney E, Easton DF, Chenevix-Trench G, Tan MH, Khoo SK, Teh BT, Yuen ST, Leung SY, Wooster R, Futreal PA, Stratton MR. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grimsrud PA, Swaney DL, Wenger CD, Beauchene NA, Coon JJ. Phosphoproteomics for the masses. ACS Chem Biol. 2010;5(1):105–119. doi: 10.1021/cb900277e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Villen J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(5):1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villen J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc. 2008;3(10):1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics. 2005;4(7):873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Pinkse MWH, Uitto PM, Hilhorst MJ, Ooms B, Heck AJR. Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-nanoLC-ESI-MS/MS and titanium oxide precolumns. Analytical Chemistry. 2004;76(14):3935–3943. doi: 10.1021/ac0498617. [DOI] [PubMed] [Google Scholar]
- 9.Sugiyama N, Masuda T, Shinoda K, Nakamura A, Tomita M, Ishihama Y. Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol Cell Proteomics. 2007;6(6):1103–1109. doi: 10.1074/mcp.T600060-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Eng JK, Mccormack AL, Yates JR. An Approach to Correlate Tandem Mass-Spectral Data of Peptides with Amino-Acid-Sequences in a Protein Database. Journal of the American Society for Mass Spectrometry. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 11.Faherty BK, Gerber SA. MacroSEQUEST: efficient candidate-centric searching and high-resolution correlation analysis for large-scale proteomics data sets. Anal Chem. 2010;82(16):6821–6829. doi: 10.1021/ac100783x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105(31):10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimsrud PA, den Os D, Wenger CD, Swaney DL, Schwartz D, Sussman MR, Ane JM, Coon JJ. Large-scale phosphoprotein analysis in Medicago truncatula roots provides insight into in vivo kinase activity in legumes. Plant Physiol. 2010;152(1):19–28. doi: 10.1104/pp.109.149625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, Mann M. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3(104):ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 15.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 16.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A. 2004;101(33):12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gauci S, Helbig AO, Slijper M, Krijgsveld J, Heck AJ, Mohammed S. Lys-N and trypsin cover complementary parts of the phosphoproteome in a refined SCX-based approach. Anal Chem. 2009;81(11):4493–4501. doi: 10.1021/ac9004309. [DOI] [PubMed] [Google Scholar]
- 18.Taouatas N, Altelaar AF, Drugan MM, Helbig AO, Mohammed S, Heck AJ. Strong cation exchange-based fractionation of Lys-N-generated peptides facilitates the targeted analysis of post-translational modifications. Mol Cell Proteomics. 2009;8(1):190–200. doi: 10.1074/mcp.M800285-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Chen R, Jiang X, Sun D, Han G, Wang F, Ye M, Wang L, Zou H. Glycoproteomics analysis of human liver tissue by combination of multiple enzyme digestion and hydrazide chemistry. J Proteome Res. 2009;8(2):651–661. doi: 10.1021/pr8008012. [DOI] [PubMed] [Google Scholar]
- 20.Schlosser A, Vanselow JT, Kramer A. Mapping of phosphorylation sites by a multi-protease approach with specific phosphopeptide enrichment and NanoLC-MS/MS analysis. Anal Chem. 2005;77(16):5243–5250. doi: 10.1021/ac050232m. [DOI] [PubMed] [Google Scholar]
- 21.Choudhary G, Wu SL, Shieh P, Hancock WS. Multiple enzymatic digestion for enhanced sequence coverage of proteins in complex proteomic mixtures using capillary LC with ion trap MS/MS. J Proteome Res. 2003;2(1):59–67. doi: 10.1021/pr025557n. [DOI] [PubMed] [Google Scholar]
- 22.Biringer RG, Amato H, Harrington MG, Fonteh AN, Riggins JN, Huhmer AF. Enhanced sequence coverage of proteins in human cerebrospinal fluid using multiple enzymatic digestion and linear ion trap LC-MS/MS. Brief Funct Genomic Proteomic. 2006;5(2):144–153. doi: 10.1093/bfgp/ell026. [DOI] [PubMed] [Google Scholar]
- 23.Kettenbach AN, Gerber SA. Rapid and reproducible single-stage phosphopeptide enrichment of complex peptide mixtures: Application to general and phosphotyrosine-specific phosphoproteomics experiments. Anal Chem. 2011 doi: 10.1021/ac201894j. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie R, Oleschuk R. Photoinduced polymerization for entrapping of octadecylsilane microsphere columns for capillary electrochromatography. Anal Chem. 2007;79(4):1529–1535. doi: 10.1021/ac061349t. [DOI] [PubMed] [Google Scholar]
- 25.Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics. 2005;4(12):2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Villen J, Beausoleil SA, Gygi SP. Evaluation of the utility of neutral-loss-dependent MS3 strategies in large-scale phosphorylation analysis. Proteomics. 2008;8(21):4444–4452. doi: 10.1002/pmic.200800283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4(3):207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 28.Hoopmann MR, Finney GL, MacCoss MJ. High-speed data reduction, feature detection, and MS/MS spectrum quality assessment of shotgun proteomics data sets using high-resolution mass spectrometry. Anal Chem. 2007;79(15):5620–5632. doi: 10.1021/ac0700833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4(6):1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.