Abstract
The adenovirus repression domain of E1A 243R at the E1A N-terminus (E1A 1–80) transcriptionally represses genes involved in differentiation and cell cycle progression. E1A 1–80 represses transcription in vitro from naked DNA templates through its interaction with p300 and TFIID. E1A 1–80 can also interact with several chromatin remodeling factors and associates with chromatin in vivo. We show here that E1A 243R and E1A 1–80 can repress transcription from a reconstituted chromatin template in vitro. Temporal analysis reveals strong repression by E1A 1–80 when added at pre-activation, activation and early transcription stages. Interestingly, E1A 1–80 can greatly enhance transcription from chromatin templates, but not from naked DNA, when added at pre-initiation complex (PIC) formation and transcription-initiation stages. These data reveal a new dimension for E1A 1–80's interface with chromatin and may reflect its interaction with key players in PIC formation, p300 and TFIID, and/or possibly a role in chromatin remodeling.
Keywords: Adenovirus, Ad E1A, Reconstituted chromatin, N-Terminal transcription repression, domain
Introduction
E1A is the first viral gene expressed during productive infection with human adenoviruses (Ads). E1A encodes multifunctional proteins that are essential for viral replication, have many interesting properties and profoundly impact cell function. The Ad group C (types 2 and 5) E1A oncogene encodes two major proteins of 243 and 289 amino acid residues (243R and 289R) which contain multiple functional domains that interact with a number of important cellular regulatory proteins. The diverse functions of E1A include transcription-activation, induction of cellular DNA synthesis, cell immortalization, cell transformation, and of particular interest transcription repression. E1A 289R differs from E1A 243R by conserved region 3 (CR3), a 46 amino acid domain unique to 289R that is involved in transcription-activation of Ad early genes. The Ad E1A 243R oncoprotein encodes two conserved domains and a less well conserved N-terminus in exon 1 which is essential for cell immortalization and cell transformation (Rasti et al., 2005). This region of E1A 243R can induce S-phase DNA synthesis and cell cycle progression by two pathways: (i) the Rb-E2F pathway which involves sequences within CR1 (residues 41 to 80) and CR2 (residues 121 to 139) that interact with the Rb family member proteins; and (ii) the N-terminal pathway within the E1A N-terminal 80 amino acids (E1A 1–80) consisting of CR1 and less well conserved residues 1 to 40. It is significant that the growth regulatory functions of E1A require sequences that reside within E1A 1–80. An important biochemical function encoded in the E1A N-terminus is the ability to transcriptionally repress cellular genes involved in cell proliferation and differentiation. The E1A repression domain takes on added importance in that it can transcriptionally repress promoters of clinical significance such as the HER2 proto-oncogene (Loewenstein and Green, 2011).
A major focus of our laboratory is the mechanism of E1A transcription-repression. Our combined findings suggest a two-step model for E1A repression. Briefly, E1A gains access to repressible promoters by interaction with cellular partners such as p300 as molecular scaffolds, thereby facilitating contact with TBP where it can disrupt the basal transcription machinery. This model of E1A repression is based on a detailed functional-mutational analysis using several experimental approaches including in vivo microinjection, DNA footprinting, gel mobility shift, DNA transfection, ChIP (chromatin immunoprecipitation) and extensive in vitro transcription analysis (Song et al., 1995a,b,c, 2007; Boyd et al., 2002; Loewenstein et al., 2006, 2007; Green et al., 2008a,b). The assays used to develop this model of E1A transcription-repression utilized naked DNA templates to measure transcription and E1A induced repression of transcription. Transcription from a naked DNA template occurs at a high basal rate and it can be argued that E1A's access to a naked promoter does not accurately reflect the complexity of transcription from the natural chromatin state. Cellular chromatin is organized into compact repeating units (nucleosomes) composed of about 146 bp of DNA wrapped around an octamer of four core histones (Wolffe, 1998). Within the cell, the structure of chromatin is dynamic which is necessary for the regulation of gene transcription. We have previously shown that E1A interacts with chromatin in adenovirus infected and transformed cells (Green et al., 2008a). It is therefore important to study E1A repression of transcription from a chromatin template in order to completely understand its regulation and further define the role of p300/CBP and TBP in E1A repression. Further, the use of chromatin templates may elucidate the possible roles of histone acetyltransferases (HATs) other than p300/CBP that interact with the E1A N-terminus including PCAF, hGCN5, TRRAP, p400 and Tip60 (reviewed in Frisch and Mymryk, 2002). The interaction of E1A N-terminal sequences with these HATs suggests that the E1A repression domain may also regulate chromatin remodeling. The development of in vitro reconstituted chromatin transcription systems, as described by Thomas and Chiang (2005) to analyze HPV E6 oncoprotein repression of p53 dependent gene activation, provides an opportunity to further extend the model of E1A repression using a more natural chromatin template system.
Results
Assembly of reconstituted chromatin
Reconstituted chromatin assembly described by Thomas and Chiang (2005) requires purified HeLa core histones (CHs), Drosophila nucleosome assembly factor ACF which is assembled from recombinant Acf1 and FLAG-tagged ISWI, recombinant human nucleosome assembly protein-1 (NAP-1) histone chaperone and a DNA template. For E1A transcription-repression analysis, chromatin was assembled around the pG5MLT DNA template. This plasmid has five Gal4 DNA binding sites upstream of the E1A repressible Ad major-late core promoter which precedes a 380 bp G-less cassette. The Gal4 DNA sites facilitate the binding of the Gal4-VP16 activator used in these studies. The G-less cassette facilitates the measurement of correctly initiated transcription from the template.
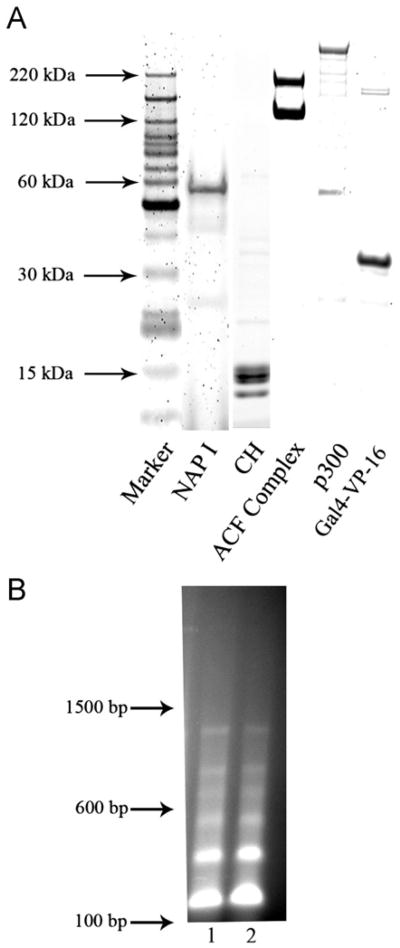
The assembly of in vitro reconstituted chromatin requires highly purified components. Aliquots of pG5MLT purified by double CsCl density gradient banding were analyzed by agarose gel electrophoresis (data not shown) and protein components purified as described in Materials and methods were analyzed by SDS-PAGE and quantified by SYPRO orange staining and laser fluoroscopy. Fig. 1A shows SDS-PAGE analysis of near homogeneous protein components used for chromatin assembly and for in vitro transcription from reconstituted chromatin. NAP-1 preparations show the expected ∼ 58 kDa band with greater than 90% purity. The purified core histone preparation shows the expected bands. The reconstituted ACF complex preparation shows the clean Acf1 band (∼ 200 kDa) and the FLAG-tagged ISWI band (∼130 kDa). The major band of the p300 preparation is the expected size. The Gal4-VP16 activator also runs at the expected size (about 30 kDa).
Fig. 1.
(A) SDS-PAGE gel of the purified protein components required for the assembly and in vitro transcription of chromatin templates. Proteins were purified as described in Materials and methods. Aliquots were subjected to SDS-PAGE electrophoresis and the protein bands stained with SYPRO orange and visualized by laser fluoroscopy as described. Proteins preparations were of the expected size. (B) Micrococal nuclease digests of a representative pG5MLT reconstituted chromatin template shows the expected regularly spaced nucleosome pattern. Lanes 1 and 2 are 150 ng of pG5MLT reconstituted chromatin digested with 1.8 units of micrococcal nuclease for 3 and 9 min respectively. Indicated sizes reflect the migration of a 100 bp DNA ladder.
The chromatin template was assembled as described (Thomas and Chiang, 2005) but with pG5MLT as template. Assembled chromatin was characterized on agarose gels after digestion with micrococcal nuclease to assure that the expected regularly spaced nucleosome pattern was formed (Fig. 1B). Analysis of micrococcal nuclease digested chromatin preparations using a Gel Doc XR imager (Bio Rad) running Quantity One software found that on average, 94% of pG5MLT DNA was associated with nucleosomes. Assembled chromatin was then used in in vitro transcription-repression assays as described in Materials and methods.
Transcription from a reconstituted chromatin template is authentic and is repressed by the E1A N-terminal repression domain
In order to facilitate analysis, transcription from reconstituted chromatin was performed by step-wise addition of activator, p300/acetyl-CoA, nuclear extract and ribonucleoside triphosphates (rNTPs). Transcription from a chromatin template can conveniently be divided into several stages. The first is the recruitment of the potent Gal4-VP16 activator to the Gal4 DNA binding sites of the chromatin pG5MLT promoter. The second stage is activation where p300 and acetyl-CoA are added and are recruited to the promoter-bound Gal4-VP16. During this step, acetylation of the nucleosomal core histones on reconstituted chromatin occurs. The third stage is the formation of a transcription pre-initiation complex (PIC). PIC formation begins with the addition of nuclear extract containing the general transcription factors including TFIID as well as Mediator and Pol II complex. The fourth stage is initiation of transcription which occurs upon the addition of rNTPs.
In the experiments described in Fig. 2, the first two stages were combined into a single activation step in which the pG5MLT reconstituted chromatin template was incubated with Gal4-VP16 activator, acetyl-CoA and p300 for 20 min. at 30 °C. The third stage (PIC formation) was accomplished by the addition of nuclear extract and incubation at 30 °C for an additional 20 min. The final stage (transcription initiation) begins with the addition of rNTPs and incubation for 60 min at 30 °C.
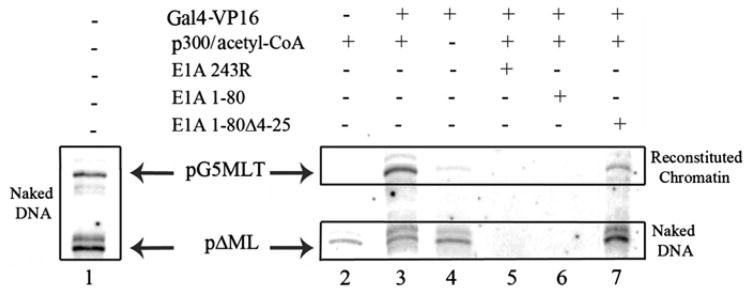
Fig. 2.
Transcription from pG5MLT is p300 and acetyl-CoA dependent and is repressed by E1A 243R and E1A 1–80 but not by E1A 1–80 Δ4–25. Lane 1 shows transcription from naked DNA of both pG5MLT and pΔMLP. Lanes 2 to 7 show transcription from pG5MLT reconstituted chromatin and the naked DNA control pΔMLP. Components of chromatin transcription (Gal4-VP16, p300 and acetyl-CoA) as well as E1A polypeptides were added as indicated. Radio-labeled transcripts were visualized by phosphor-image analysis.
Fig. 2, lane 1 shows in vitro transcription from two naked DNA templates, pG5MLT DNA and pΔMLP. pG5MLT DNA as reconstituted chromatin template is used in all other reactions. pΔMLP is added to all reactions as a naked DNA control at the initiation step of transcription. pΔMLP consists of the MLP core promoter alone with a G-less cassette of 210 bp in order to distinguish it from the transcript of the chromatin template of 380 bp (Wu and Chiang, 2001). Both naked DNA templates are efficiently transcribed in the absence of Gal4-VP16 activator or added p300/acetyl-CoA (lane 1).
Fig. 2, lane 2 shows that the pG5MLT template in reconstituted chromatin is silent in the absence of Gal4-VP16 activator, even in the presence of p300 and acetyl-CoA. As expected, transcription from the naked DNA control occurs in the absence of activator. However, when Gal4-VP16 activator and p300/acetyl-CoA are both present (lane 3), there is strong transcription from the chromatin template; in the presence of p300/acetyl Co-A, multiple transcription start sites within the naked pΔML DNA become more apparent and the authentic transcript is less distinct. Lane 4 shows transcription from a reaction containing the Gal4-VP16 activator but lacking p300/acetyl-CoA. In this case transcription from the chromatin template is severely limited whereas transcription from the naked DNA control is unaffected. These data show that transcription from the pG5MLT reconstituted chromatin template requires activation and is very largely p300/acetyl-CoA dependent as is expected for reconstituted chromatin. The small amount of transcription from pG5MLT occasionally seen in lane 4 is likely due to endogenous p300 and acetyl-CoA present in the nuclear extract (Thomas and Chiang, 2005).
We have shown in several studies that E1A 243R and E1A 1–80 efficiently inhibit transcription from several E1A repressible naked DNA templates both by in vitro transcription and by DNA transfection analysis. It is important to establish the ability of E1A 243R and the E1A repression domain alone to repress authentic transcription from a reconstituted chromatin template. Therefore, we tested the ability of E1A 243R and E1A 1–80 to block transcription from the pG5MLT chromatin template when added at the activation stage of chromatin transcription. When 125 ng of E1A 243R is added to a reaction mixture that contains both an activator and p300/acetyl-CoA, transcription from both chromatin and naked DNA templates is inhibited (see lane 5). Of significance, when 125 ng of the E1A repression domain alone (E1A 1–80) is added, transcription from both templates is also blocked (lane 6). Taken together these data (Fig. 2) indicate that transcription from the pG5MLT reconstituted chromatin template is authentic and is repressible by E1A 243R and the E1A repression domain alone. As shown in lane 7, this conclusion is further validated by the inability of E1A 1-80Δ4-25, a mutant that is deficient in the E1A repression function (Loewenstein et al., 2006), to effect transcription from either the chromatin or the naked DNA template.
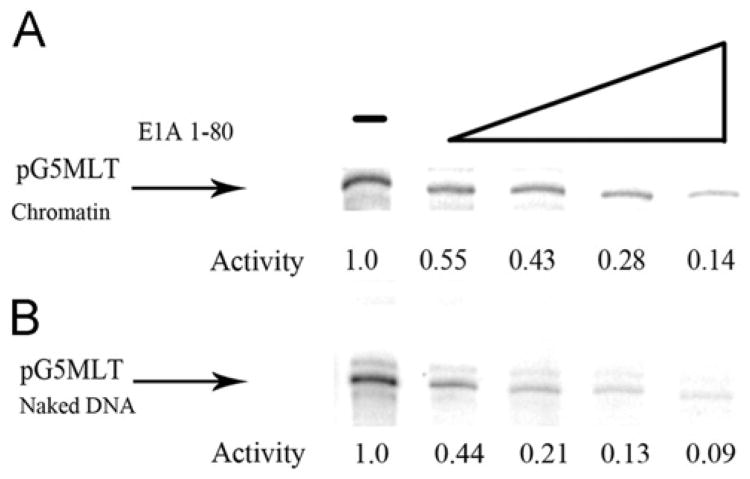
Our previous studies using the naked DNA template pHIV LTR, demonstrated that E1A 1–80 represses transcription in a dose-dependent manner (Loewenstein et al., 2006). Similar dose response transcription-repression assays were conducted to determine whether E1A 1–80 can repress transcription of a chromatin template as efficiently as it does a naked DNA template. As shown in Fig. 3, transcription from a pG5MLT chromatin template (Fig. 3A) and from a pG5MLT naked DNA template (Fig. 3B) is repressed by E1A 1–80 in a dose-dependent manner to approximately the same degree. For example 100 ng of E1A 1–80 inhibits transcription from pG5MLT as chromatin about 86% and inhibits transcription form pG5MLT as naked DNA about 91%. In these experiments pG5MLT as both chromatin and as naked DNA template are activated by Gal4 VP-16 (See Materials and methods).
Fig. 3.
Repression of transcription from a pG5MLT chromatin template (A) and a naked DNA template (B) is repressible by E1A 1–80 in a similar dose-dependent manner. Shown are transcripts from reactions containing increasing amounts of E1A 1–80 (12.5 ng, 25 ng, 50 ng and 100 ng). Indicated below are the relative amounts of transcription as determined by phosphor-image analysis.
Order-of-addition analysis suggests new activities for the E1A transcription-repression domain
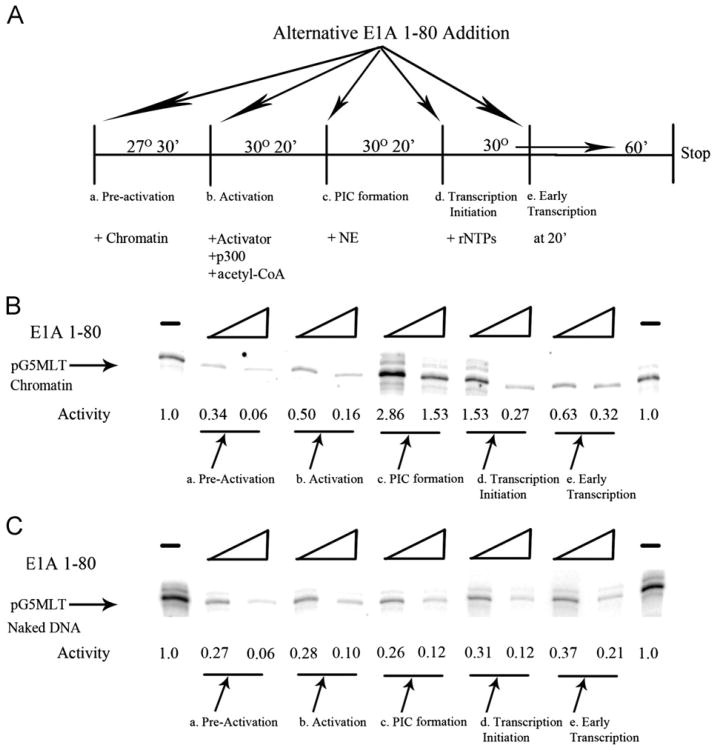
In addition to disrupting the basal transcription machinery as E1A does with naked DNA templates, it is entirely possible that the E1A repression domain can also modulate transcription from chromatin templates by other mechanisms. For instance, since transcription from chromatin templates requires acetylation of nucleosomal histones by p300, interference with this step during activation of the chromatin template could result in the blockage of transcription. In order to examine these possibilities, E1A 1–80 was added to chromatin transcription reactions at various stages depicted in Fig. 4A. E1A 1–80 at two levels was added to reactions (a) with the chromatin template prior to activation and incubated at 27° for 30 min (pre-activation); (b) at the stage when the Gal4-VP16 activator and p300/acetyl-CoA are added to the chromatin template (activation); (c) with the nuclear extract to the activated/acetylated chromatin template (PIC formation); (d) when the ribonucleoside triphosphates are added to the reaction (transcription initiation); and (e) at 20 min after the beginning of transcription (early transcription).
Fig. 4.
(A) Shown is a schematic of parallel in vitro transcription reactions using pG5MLT reconstituted chromatin as template in which two levels of E1A 1–80 (62.5 and 125 ng) were added at various stages. (B) Shown are transcripts from the reactions depicted above. The relative amounts of transcription are indicated. At pre-activation, strong, dose-dependent repression of transcription from the pG5MLT chromatin template by E1A 1–80 is seen, an average of 49% of the control activity for 62.5 ng and 12% for 125 ng of E1A 1–80 with a standard deviation (SD) of 6% and 9% respectively from 8 experiments. At activation, strong dose-dependent repression is also seen, an average of 56% of control for 62.5 ng and 22% for 125 ng of E1A 1–80 with a SD of 8% and 9% respectively from 8 experiments. At PIC formation, significantly enhanced activity is seen when of E1A 1–80 is added; 277% of control activity for 62.5 ng and 148% for 125 ng with a SD of 17% and 19% respectively from 5 experiments. At transcription-initiation, slightly enhanced activity is again seen when 62.5 ng of E1A 1–80 is added; an average of 116% control activity with a SD of 27% from 5 experiments. However, when 125 ng of E1A 1–80 is added, significant repression is seen with an average of 37% of the control activity with a SD of 16% for 5 experiments. At early transcription, strong dose-dependent repression is again seen, 72% of control activity for 62.5 ng and 41% with 125 ng of E1A 1–80 with an SD of 6% and 9% respectively from 8 experiments. (C) Shown is a parallel temporal analysis using pG5MLT as naked DNA template. In all stages, E1A 1–80 represses transcription in a dose-dependent manner.
As shown in Fig. 4B, when E1A 1–80 at 62.5 or 125 ng/reaction is added with the pG5MLT chromatin template at the pre-activation step, transcription-repression occurs in a dose-dependent manner. A similar dose-dependent transcription-repression occurs when E1A 1–80 is added at the beginning of the activation stage. Surprisingly, when E1A 1–80 (62.5 ng/reaction) is added immediately before nuclear extract during the PIC formation step, transcription from the reconstituted chromatin template is not repressed, but instead in stimulated greatly (about three-fold). Yet when E1A 1–80 is added at even higher levels (250 ng/reaction) the activated level of transcription is diminished about 50%. When E1A 1–80 is added immediately before transcription-initiation, lower levels of E1A 1–80 stimulate transcription whereas the higher levels of E1A 1–80 repress transcription significantly (i.e. over 5 fold from the activated level). Finally, when E1A 1–80 is added at 20 min after initiation of transcription, transcription is once again significantly repressed in a dose-dependent manner.
These data described above reveal an intriguing complexity that may be specific for the E1A transcription-repression domain in the chromatin environment. To further test this possibility, comparable experiments were carried out using Gal4 VP-16 activated naked pG5MLT DNA as template. Fig. 4C shows that E1A 1–80 transcription-repression occurs in a dose-dependent manner at the pre-activation, activation, PIC formation, transcription initiation and early transcription stages when added to naked DNA pG5MLT template using the same temporal scheme as for pG5MLT chromatin template (Fig. 4B). With pG5MLT as naked DNA template there is no stimulation of transcription when E1A 1–80 is added immediately before nuclear extract during the PIC formation stage or when low levels of E1A 1–80 is added at the beginning of the transcription initiation stage. This is in contrast to E1A 1–80 enhancement of transcription from pG5MLT as reconstituted chromatin at the PIC formation and transcription initiation stages (Fig. 4B).
Discussion
The E1A transcription-repression domain can repress transcription from a chromatin template
Transcription from silent chromatin templates requires activation by sequence specific activators and co-activators. Co-activators are generally divided into two groups: those that are involved in chromatin remodeling and those that interact directly with the general transcription factors and Pol II (Black et al., 2006). Although chromatin remodeling factors and other transcription factors needed to assemble a functional PIC are widely studied, the ordered recruitment of these factors necessary to activate chromatin transcription is incompletely understood. It would not be surprising therefore that the mechanism by which E1A represses transcription from chromatin, although intriguing, is even more difficult to elucidate.
Based on previous extensive mutational and functional analysis, we have proposed a model for E1A repression. To restate briefly: E1A gains access to repressible promoters by interaction with cellular partners such as p300 as molecular scaffolds and thereby contacts TFIID where it can disrupt the TBP/TATA interaction. It is clear from the data presented here that E1A 243R and the E1A repression domain alone can repress transcription from a chromatin template in vitro and does so in a dose-dependent manner and at about the same level as from a naked DNA template (Figs. 2 and 3). However, it is entirely possible that E1A modulates transcription from E1A repressible chromatin promoters at multiple stages during transcription. It is significant that the E1A N-terminal repression domain interacts with both p300 and with TFIID, members of both groups of transcription factors needed for activation of a chromatin template (Boyd et al., 2002; Loewenstein et al., 2006). Further, the E1A transcription-repression domain is known to interact with several HATs in addition to p300 that could also play a role in E1A repression. For instance, the E1A N-terminus can interact with PCAF, hGCN5, TRRAP, p400 and Tip60 (reviewed in Frisch and Mymryk, 2002). This suggests that E1A could possibly modulate chromatin remodeling needed for transcription from chromatin templates as well as interfere with the basal transcription machinery.
As a first attempt to investigate the possibility of multiple mechanisms of repression, we added E1A 1–80 at various stages of transcription from chromatin (Fig. 4). E1A 1–80, when added at the pre-activation step in the presence of only the reconstituted chromatin template and various buffers (no activator, p300 or acetyl-CoA), efficiently inhibits transcription. It seems unlikely that E1A 1–80, by itself, alters the nature of chromatin to prevent transcription. It seems more likely that E1A 1–80 is sufficiently stable to interfere with transcription at stages after pre-activation. E1A 1–80 also efficiently inhibits transcription from the chromatin promoter when added at the activation step where chromatin, Gal4-VP16 activator, p300 and acetyl-CoA are present. Again it is possible that E1A 1–80 does not modulate transcription at this step, but at a later step. However, from these data, we do not exclude the distinct possibility that the E1A repression domain can alter normal acetylation of nucleosomal histones through its interaction with p300. For example, E1A 243R has been reported to inhibit p300 HAT activity, but this activity does not require E1A N-terminal sequences and thus its relevance to E1A repression is unclear (Chakravarti et al., 1999; Hamamori et al. 1999). Of interest, E1A 243R can be acetylated by p300 which appears to regulate binding of the co-repressor CtBP to E1A CR4 (Zhang et al., 2000). Of particular interest, the human papillomavirus E6 oncoprotein inhibits the acetylation of both p53 and nucleosomal histones to repress p53-dependent transcription from a chromatin template (Thomas and Chiang, 2005).
Is an E1A N-terminal activation function revealed by temporal analysis of transcription from a chromatin template?
An unanticipated finding was that E1A-1–80 (62.5 ng) stimulates transcription about three fold when added with the nuclear extract at the PIC formation step of a chromatin transcription reaction whereas higher levels (125 ng) resulted in a reduced stimulation (about 50%). When E1A 1–80 (62.5 ng) is added at the end of the PIC formation but just prior to transcription-initiation, transcription is again stimulated but higher levels of E1A 1–80 (125 ng) clearly repress transcription. These phenomena appear to reflect the chromatin nature of the DNA template since transcription from a naked pG5MLT DNA template in analogous reactions is efficiently repressed rather that activated. Finally, when E1A 1–80 is added after transcription initiation of chromatin templates (early transcription), repression occurs at both levels as it does when added at the pre-activation and activation stages consistent with our model that E1A disrupts the basal transcription machinery at the PIC level.
Some studies have described an activation activity for E1A 243R. Several cellular genes that possess E2F DNA binding sites have been shown to be activated by E1A relief from repression by disrupting Rb-family-E2F complexes; however, E1A sequences required for this function are included within CR2, which is dispensable for E1A repression (Berk, 2007). The activation activity seen for E1A 1–80 in the temporal assay (Fig. 4B) is therefore not likely due to this pathway. However, other gene promoters may be activated by sequences found within E1A 1–80. For example hsp70 has been reported to be activated by sequences within the E1A repression domain by a mechanism which involves the disruption of binding of the Dr1 transcriptional repressor to the TATA sequence by E1A (Kraus et al., 1994). However, it seems unlikely that this mechanism is responsible for the activation seen in this temporal assay.
It seems more reasonable that the dual transcription activity of the E1A N-terminus seen in the temporal assay with a chromatin template (Fig. 4B) reflects its interaction with its major cellular partner's p300 and TFIID. In this context, it is of considerable interest that in a reconstituted chromatin transcription system, Mediator facilitates PIC formation by interacting first with p300 and then recruiting TFIID to the nascent PIC as p300 autoacetylates and falls from the complex (Black et al., 2006). It may be that E1A 1–80 mimics the role of Mediator in activating transcription from chromatin templates. It is interesting that when E1A 1–80 is added to a chromatin transcription reaction at the pre-PIC and PIC formation stages, it can stimulate transcription but that this stimulation is in inversely proportional to the amount of E1A 1–80 present. This might suggest that E1A at these stages could serve to recruit TFIID in the nuclear extract to the promoter through the ability of E1A 1–80 to interact with both p300 (bound to the Gal4-VP16 activator) and to TFIID. For example it has been reported than artificial tethering of TBP to a promoter results in transcriptional activation by assisting in PIC formation (He and Weintraub, 1998). We speculate that when the levels of E1A 1–80 and PIC formation are both high, E1A can then disrupt PICs resulting in transcription-repression as predicted by our model of E1A repression.
We note one caveat. Although chromatin is the natural template for the study of transcriptional activation and repression, any study which of necessity requires the temporal addition of modulating factors introduces additional challenges of interpretation. Within the cell nuclear environment, transcription from chromatin takes place in the simultaneous presence of template, sequence specific activators, co-activators including p300, TFIID and Mediator, as well as general transcription factors. Continued study will be required to sort out the details of transcriptional modulation by the E1A repression domain and its role in cell cycle regulation and virus replication.
Materials and methods
Constituents of chromatin assembly and for activated transcription from chromatin
All plasmids used were purified by double CsCl density gradient centrifugation and analyzed by agarose gel electrophoresis. Plasmids expressing FLAG-tagged Nap1 (pF:hNAP1-11d) and a FLAG-tagged Gal4 DNA binding domain fused to VP16, (pf:Gal4-VP16) (Chiang and Roeder, 1995), were transfected into BL21-Lys bacterial cells (Invitrogen). Bacterial cell pellets from IPTG-induced cultures were harvested and subjected to anti-FLAG agarose affinity chromatography and elution with FLAG peptide (Sigma) as previously described (Chiang and Roeder, 1993). Baculovirus vector stocks expressing Acf-1 and FLAG-tagged ISWI or FLAG-tagged p300 (Thomas and Chiang, 2005) were used to serially infect Sf-9 insect cells in order to produce sufficiently high titers to infect liter quantities of Sf-9 cells. FLAG-tagged ACF complex and FLAG-tagged p300 were purified by anti-FLAG agarose affinity chromatography.
HeLa cell core histones were purified from 20 l of cells grown to a density of 0.6 × 106 cells/ml (National Cell Culture Center). A nuclear transcription extract was first extracted from the HeLa cell nuclei (Dignam et al., 1983). The pellet remaining after extraction was then suspended by Dounce homogenization using an A pestle (Kontes) in 50 ml HB buffer (20 mM Tris pH 7.5, 1 mM EDTA, 10% glycerol, 400 mM NaCl, 1 mM β-mercaptoethanol, 0.5 mM PMSF, 2 μg/ml pepstatin A 2 μg/ml leupeptin and 5 μg/ml aprotinin). The nuclei were centrifuged again, resuspended and washed twice in 50 ml of the same buffer containing 0.2% NP-40. The pellet was washed twice in 50 ml of HAP buffer (50 mM Na2PO4 pH 6.8, 1 mM β-mercaptoethanol, 0.5 mM PMSF, 2 μg/ml pepstatin A, 2 μg/ml leupeptin and 5 μg/ml aprotinin) containing 400 mM NaCl. The pellet was then suspended in 40 ml of HAP buffer containing 600 mM NaCl and subjected to sonication until the average DNA fragment size was about 1.5 kb as determined by agarose gel electrophoresis of proteinase K treated aliquots. The sonicated nuclear preparation was added to 15 ml (bed volume) of hydroxylapatite resin (Bio Gel HTP—BioRad) and rotated overnight at 4 °C in a 50 ml centrifuge tube. The resin was washed with 5 volumes of the HAP buffer. Core histones were eluted batch-wise from the resin in 2 volumes of HAP buffer containing 2.5 M NaCl and subjected to dialysis against 10 mM HEPES pH 7.9, 1 mM EDTA 10 mM KCl, 10% glycerol, 0.5 mM PMSF, and 1mM dithiothreitol.
All of the purified proteins needed for the ordered assembly of chromatin and activated transcription were subjected to SDS-PAGE, staining with SYPRO orange followed by analysis and quantification by laser fluoroscopy using a Typhoon Trio scanner (GE Health Sciences).
Chromatin assembly and chromatin transcription-repression assays
Chromatin assembly using pG5MLT as template was performed and aliquots of assembled chromatin were subjected to analysis by digestion with micrococcal nuclease and agarose gel electrophoresis as described (Thomas and Chiang, 2005). Chromatin transcription reactions were conducted as described (Thomas and Chiang, 2005) except that E1A 243R, E1A 1-80Δ4-25 or E1A 1–80 polypeptides, produced and purified as described (Boyd et al., 2002), were added at various times during the reaction. Radio-labeled transcripts from the G-less cassette of the pG5MLT or pΔMLP templates was analyzed as described (Thomas and Chiang, 2005) and resolved by electrophoresis on 6% polyacrylamide gels containing 6 M urea. Transcription-repression reactions from a pG5MLT naked DNA template were conducted as described except that there was no added p300/Acetyl-CoA and that transcription was activated by the addition of 14 ng of Gal4 VP-16. Dried gels were exposed for 12 h to phosphor-storage screens and transcripts visualized and quantified on a Typhoon Trio scanner running ImageQuant software (GE Health Sciences).
Acknowledgments
We thank CE. Mulhall for insightful editorial assistance. This work was supported by Research Career Award AI-04739 and National Institutes of Health grants CA29561 to M.G and CA103867 and CA124760 to C.-M.C.
References
- Berk AJ. Fields Virology. Adenoviridae: The Viruses and their Replication. (5th) 2007;Chapter 63:2356–2394. [Google Scholar]
- Black JC, Choi JE, Lombardo SR, Carey M. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol Cell. 2006;23:809–818. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- Boyd J, Loewenstein PM, Tang Q, Yu L, Green M. The adenovirus 2 E1A N-terminal domain sequence requirements for repression of transcription in vitro and in vivo correlate with those required for E1A interference with TBP/TATA complex formation. J Virol. 2002;76:1461–1474. doi: 10.1128/JVI.76.3.1461-1474.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti D, Ogryzko V, Kao HY, Nash A, Chen H, Nakatani Y, Evans RM. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- Chiang CM, Roeder RG. Expression and purification of general transcription factors by FLAG epitope-tagging and peptide elution. Pept Res. 1993;6:62–64. [PubMed] [Google Scholar]
- Chiang CM, Roeder RG. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science. 1995;267:531–536. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch SM, Mymryk JS. Adenovirus-5 E1A: paradox and paradigm. Nat Rev, Mol Cell Biol. 2002;3:441–452. doi: 10.1038/nrm827. [DOI] [PubMed] [Google Scholar]
- Green M, Panesar N, Loewenstein PM. Adenovirus E1A proteins are closely associated with chromatin in productively infected and transformed cells. Virology. 2008a;371:1–7. doi: 10.1016/j.virol.2007.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M, Panesar N, Loewenstein PM. The transcription repression domain of the adenovirus E1A oncoprotein targets p300 at the promoter level. Oncogene. 2008b;27:4446–4455. doi: 10.1038/onc.2008.85. [DOI] [PubMed] [Google Scholar]
- Hamamori Y, Sartorelli V, Ogryzko V, Puri PM, Wu HY, Nakatani Y, Kedes L. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]
- He S, Weintraub SJ. Stepwise recruitment of components of the preinitiation complex by upstream activators in vivo. Mol Cell Biol. 1998;18:2876–2883. doi: 10.1128/mcb.18.5.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus VB, Inostroza JA, Yeung K, Reinberg D, Nevins JR. Interaction of the Dr1 inhibitory factor with the TATA binding protein is disrupted by adenovirus E1A. Proc Nat Acad Sci USA. 1994;91:6279–6282. doi: 10.1073/pnas.91.14.6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewenstein PM, Arackal S, Green M. Mutational and functional analysis of an essential subdomain of the adenovirus E1A N-terminal transcription repression domain. Virology. 2006;351:312–321. doi: 10.1016/j.virol.2006.03.041. [DOI] [PubMed] [Google Scholar]
- Loewenstein PM, Song CZ, Green M. The use of in vitro transcription to probe regulatory functions of viral protein domains. In: Totowa NJ, editor. Methods in Molecular Medicine, vol 131: Adenovirus Methods and Protocols, vol 2. second. Chapter 2. Humana Press, Inc., W.S.M. Wold and Ann Tollefson; 2007. [DOI] [PubMed] [Google Scholar]
- Loewenstein PM, Green M. Expression of the adenovirus early gene 1A transcription repression domain alone results in the death of human breast cancer cells up-regulated for the HER2 proto-oncogene. Genes & Cancer. 2011;2:737–744. doi: 10.1177/1947601911426570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasti M, Grand RJ, Mymryk JS, Gallimore PH, Turnell AS. Recruitment of CBP/p300, TATA-binding protein and S8 to distinct regions at the N-terminus of adenovirus E1A. J Virol. 2005;79:5594–5605. doi: 10.1128/JVI.79.9.5594-5605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CZ, Tierney CJ, Loewenstein PM, Pusztai R, Symington JS, Tang Q, Toth K, Nishikawa A, Bayley ST, Green M. Transcriptional repression by adenovirus E1A N-terminus/conserved domain 1 polypeptides in vivo and in vitro in the absence of protein synthesis. J Biol Chem. 1995a;270:23262–23267. doi: 10.1074/jbc.270.40.23263. [DOI] [PubMed] [Google Scholar]
- Song CZ, Loewenstein PM, Green M. Repression in vitro, by human adenovirus E1A protein domains, of basal and Tat-activated transcription of the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1995b;69:2907–2911. doi: 10.1128/jvi.69.5.2907-2911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CZ, Loewenstein PM, Toth K, Green M. TFIID is a direct functional target of the adenovirus E1A transcription repression domain. Proc Nat Acad Sci USA. 1995c;92:10330–10333. doi: 10.1073/pnas.92.22.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CZ, Loewenstein PM, Toth K, Tang Q, Nishikawa A, Green M. The adenovirus E1A repression domain disrupts the interaction between the TATA-binding protein (TBP) and the TATA box in a manner reversible by TFIIB. Mol Cell Biol. 2007;17:2186–2193. doi: 10.1128/mcb.17.4.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM. E6 oncoprotein represses p53-dependent gene activation via inhibition of protein acetylation independently of inducing p53 degradation. Mol Cell. 2005;17:251–264. doi: 10.1016/j.molcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Wolffe A. Chromatin: structure and function. 3rd. Academic Press; San Diego, CA: 1998. [Google Scholar]
- Wu SY, Chiang CM. TATA-binding protein-associated factors enhance the recruitment of RNA polymerase II by transcriptional activators. J Biol Chem. 2001;276:34235–34243. doi: 10.1074/jbc.M102463200. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Yao H, Vo N, Goodman RH. Acetylation of adenovirus E1A regulates binding of the transcriptional corepressor CtBP. Proc Nat Acad Sci USA. 2000;97:14323–14328. doi: 10.1073/pnas.011283598. [DOI] [PMC free article] [PubMed] [Google Scholar]