Summary
Access to online repositories for genomic and associated “-omics” datasets is now an essential part of everyday research activity. It is important therefore that the Tuberculosis community is aware of the databases and tools available to them online, as well as for the database hosts to know what the needs of the research community are. One of the goals of the Tuberculosis Annotation Jamboree, held in Washington DC on March 7th–8th 2012, was therefore to provide an overview of the current status of three key Tuberculosis resources, TubercuList (tuberculist.epfl.ch), TB Database (www.tbdb.org), and Pathosystems Resource Integration Center (PATRIC, www.patricbrc.org). Here we summarize some key updates and upcoming features in TubercuList, and provide an overview of the PATRIC site and its online tools for pathogen RNA-Seq analysis.
1. Introduction
Bacterial genomes can now be sequenced in a matter of days for a few hundred dollars. Genomic, transcriptomic, and associated data-sets are becoming so large that the extent to which we provide user-friendly access will determine how much the Tuberculosis (TB) research community can learn from them. The TB community therefore needs to take stock of how we are placed to best exploit this data, and how we will deal with issues such as data analysis, curation and dissemination.
Web-accessible databases and analysis tools are an essential part of how we interpret and interact with genome data; as a community we need to be kept up to date with developments in these areas. This was one of the key aims of the TB Annotation Jamboree held in Washington on March 7th–8th 2012, where a session was devoted to databases and related issues. We focused our discussions on three of the key resources for TB genome data on the web, namely TubercuList (tuberculist.epfl.ch), TB Database (www.tbdb.org), and Pathosystems Resource Integration Center (PATRIC, www.patricbrc.org). The goal of this manuscript is to update and introduce the community to developments in TubercuList and PATRIC, as detailed below. Web-links to the databases and tools mentioned in this article can be found in supplemental table 1.
2. TubercuList
2. a. Overview
TubercuList (tuberculist.epfl.ch) is a relational database for the genome sequence annotation of Mycobacterium tuberculosis H37Rv, the reference strain commonly used in the study of TB. This infectious disease continues to be a serious global health issue, killing 1.4 million people in 20101. The database is a well-established resource, having been maintained since its inception in 19982. It is a gene-centric database, and in its current form provides information on annotated M. tuberculosis H37Rv genes and proteins, including functional annotation, orthologous genes in closely related species, gene ontology terms, structural information, and cross-references to several external resources including the TB Drug Resistance Mutation Database, a comprehensive list of polymorphisms associated with drug resistance3, and The TDR Targets Database, designed to facilitate the prioritization of drug targets4.
One of the greatest strengths of the TubercuList database lies in the fact that it has been subject to continuous manual annotation since the first release of the genome sequence and annotation5, 6. It is updated with experimental evidence from the scientific literature resulting in changes to gene boundaries, addition of new genes both protein- and RNA-encoding, improvements in functional annotation, and assignment or modification of gene names. This is enriched with data on the characterization of mutant strains, protein localization determined by proteomics studies, gene essentiality under different growth conditions, gene regulatory information, and operon structure. Citations are also provided for all such manually selected publications from which data has been extracted.
With advances in next-generation sequencing technologies and decreasing costs, the number of genome projects is increasing at a remarkable rate. According to the Genomes OnLine Database, there were 11472 genome sequencing projects as of September 2011, with 2907 complete7. Although these numbers are impressive, for the majority of newly sequenced genomes, the annotation will not go beyond computer-generated predictions8, 9. Moreover, vast amounts of empirical data are constantly being produced at the bench, particularly from high-throughput and genome-wide studies, and it is critical to extract key findings and apply them to the genome annotation so that it is readily accessible in a useful form for the entire research community. It is through this challenging task of manual annotation from the literature, collecting and organizing data from disparate sources, that we strive to extend the value of TubercuList for TB researchers as a current and reliable resource.
Through a partnership between the École Polytechnique Fédérale de Lausanne (EPFL) and the SIB Swiss Institute of Bioinformatics (SIB), updating of the TubercuList database is now carried out by the SIB who, together with the European Bioinformatics Institute (EBI) and the Protein Information Resource (PIR), form the UniProt consortium and produce the Swiss-Prot section of the UniProt Knowledgebase (UniProtKB/Swiss-Prot). UniProtKB/Swiss-Prot is an expertly curated database for protein information and M. tuberculosis is one of several model organisms on which the database focuses (www.uniprot.org)10. Annotation carried out by curators for UniProtKB/Swiss-Prot and for TubercuList is exchanged thereby maximizing the results of manual annotation efforts made by both groups.
As improvements, modifications, and the incorporation of new data to the M. tuberculosis H37Rv genome annotation are continually being made in TubercuList, we describe in the following sections some of the recent changes that have been made and the updates that will appear in the next release of the database (R26).
2. b. Updates to TubercuList annotation, Release 25
The TubercuList database is updated approximately every four months with information from the literature as well as with new or updated cross-references to external databases. The number of genes annotated in the current release of the database, version R25 completed in April 2012, has not changed significantly, now at 4095, and the coordinates of only four coding sequences (CDS) have been altered. New genes added since version R20 from June 20106 include four CDS, one of which is a replacement for rv0061, now annotated in the opposite orientation as rv0061c as indicated by RNA-Seq data11, 12 (Uplekar et al., in preparation). Continuing with the trend reported previously6, new CDS added to the annotation are typically small, being less than or close to 100 amino acids. Also added are two non-coding RNAs, regulatory molecules that are a topic of increasing interest13, 14. There are now a total of 23 such small RNA genes annotated in TubercuList.
Advances in mass spectrometry-based proteomic methods are providing the ability to identify wider ranges of proteins, reliably and accurately15–17. In TubercuList, 2828 proteins are annotated as having been identified in a proteomics study, 1114 more than in the R20 version of the database. This validates 70% of protein-coding features annotated in the genome, although a recent study, whose results await addition to the database, reports a higher ~80% coverage of the predicted genes18. Of the 2828 proteins, 23% are categorized as 8-Unknown or 10-Conserved hypothetical proteins, verifying that these predicted CDS are actual proteins produced by the bacterium.
The current distribution of all M. tuberculosis H37Rv genes across eleven functional categories is shown in Table 1. The function of one quarter (1048) of annotated CDS remains unknown, although this number is steadily being reduced as more proteins are characterized. Changes have been made to the functional category of 55 CDS (See Table 1) and approximately half of these changes move CDS from 10-Conserved hypothetical proteins to involvement in 1-Lipid metabolism. In addition to this functional annotation, 85 gene names have been added or modified, and more than half of these concern toxin-antitoxin genes, mainly for vapBC gene pairs.
Table 1.
Distribution of genes across functional categories (TubercuList version R25)
| Functional Category | Gene number | Change from R20 | |
|---|---|---|---|
| 0 | Virulence, detoxification, adaptations | 238 | 10 |
| 1 | Lipid metabolism | 272 | 25 |
| 2 | Information pathways | 242 | 1 |
| 3 | Cell wall and cell processes | 773 | - |
| 4 | Stable RNAs | 73 | 2 |
| 5 | Insertion sequences and phages | 147 | - |
| 6 | PE/PPE | 168 | - |
| 7 | Intermediary metabolism and respiration | 936 | 13 |
| 8 | Unknown | 16 | - |
| 9 | Regulatory proteins | 198 | 3 |
| 10 | Conserved hypothetical proteins | 1032 | −49 |
Structural biology plays an important role in understanding the mechanisms of protein function as well as in predicting functions for unknown proteins, and can also make a significant contribution to drug development19. TubercuList now links to 1019 structures in the Protein Data Bank (www.pdb.org), representing 365 unique proteins. This is a significant achievement in the field of TB research. However, as the protein structures of most of the CDS annotated in the genome remain unknown, protein structure prediction methods are a necessary tool to be used where experimental structure determination has not yet succeeded20 (see Mao et al., this issue).
In this period, we have also added information on gene regulation involving eight regulatory proteins21–29; 544 genes now have annotation on regulation. This includes data on predicted and confirmed regulons, identification of DNA-binding motifs as well as demonstration of DNA-binding by the regulatory proteins, and changes in expression levels in the absence or overexpression of the regulatory protein. Experimental evidence of operon structure has also been added for 38 genes.
TubercuList contains references to 1285 publications. In the last five updates of the database, 181 publications served as sources for the annotation changes described above and were selected through searches in PubMed (www.ncbi.nlm.nih.gov/pubmed) or were indicated by communication with the scientific community (email: tuberculist@epfl.ch).
2. c. Upcoming changes to TubercuList annotation (R26)
At the time of writing, we are in the process of preparing the next release of the TubercuList database. For the next version of the annotation, eleven new protein-coding genes have been added. All are proteins of unknown function with the exception of one which is a possible PemK-like mRNA interferase. Many of these are already included in the annotation of the CDC1551 strain of M. tuberculosis, however there is now experimental data indicating their presence in the H37Rv strain. Their transcription is supported by RNA-Seq gene expression data and peptides from these proteins have been identified in large-scale proteomics studies18, 30 (Uplekar et al., in preparation). Note that novel genes indicated by large-scale studies have been considered only when more than one source of evidence is available. Seven non-coding regulatory RNAs will also be added to the annotation31, 32 (Uplekar et al., in preparation). Currently, these non-coding RNAs are named in the way referred to in the publications reporting their discovery, however, the naming convention described by Lamichhane and colleagues in this issue, an outcome of the TB Annotation Jamboree, will be incorporated into the database when available. Likewise, new functional annotation that resulted from the jamboree will soon be included in TubercuList where appropriate.
Beyond contributing to the annotation of new genes, described above, proteomics data is a powerful tool that can be used to validate and correct existing annotation. For example rv0157A will no longer be described as a pseudogene since peptides from its protein product have been identified in cellular fractions of M. tuberculosis H37Rv30. In addition, expression of rv0157A has been measured by RNA-Seq (Uplekar et al., in preparation). There are also studies reporting hundreds of confirmed protein translation start sites, as well as the correction of gene coordinates for a small subset of these proteins18, 30 (Uplekar et al., in preparation). These changes are still to be addressed and incorporated into the TubercuList annotation. Furthermore, proteomic studies have already been used in identifying sites of post-translational modifications of mycobacterial proteins, and continued refinement and development of proteomic methodologies are expected to lead to greater use of proteomics in addressing increasingly complex issues, such as the molecular mechanisms of specific steps of the host infection process15, 16, 33, 34. It is evident that proteomic data will continue to have an important role in improving the M. tuberculosis genome annotation and this will be both completed and complemented by RNA-Seq analysis.
As mentioned above, approximately 25% of M. tuberculosis H37Rv CDS in TubercuList encode proteins of unknown function. As such, since experimental data for such proteins are lacking, predictions based on in-depth bioinformatic and computational analyses may be of immense value with regards to hypothesis generation and testing. In an effort to address this, the upcoming annotation will include the results of functional predictions for 652 such proteins, based on functional interaction networks35. From this analysis, 47 6-PE/PPE and 605 10-Conserved hypothetical proteins can be re-classified into a more informative functional category. Nearly half of these are predicted to be involved in 7-Intermediary metabolism and respiration, while another quarter may be involved in 3-Cell wall and cell processes.
As many advances have been made in DNA sequencing technologies, and as more M. tuberculosis genomes have been sequenced, errors in the H37Rv reference genome sequence have been detected36, 37 (Uplekar et al., in preparation). Thus far, thirty-three single nucleotide substitutions have been made to the genome sequence for database version R26, affecting the amino acid sequence for fourteen CDS. Only those changes common to the different datasets have been considered. These sequence corrections, in addition to the annotation in TubercuList, are in the process of being submitted to EMBL-EBI (www.ebi.ac.uk) as an update to the M. tuberculosis H37Rv genome sequence entry (AL123456).
3. Pathogen Portal and PATRIC
3. a. Introduction
The Pathosystems Resource Integration Center (PATRIC, www.patricbrc.org)38 is a web-based information system designed to support basic and applied biomedical research on bacterial infectious diseases. It integrates genome-scale data, metadata, and analysis tools for all bacterial pathogens and all bacteria. Together with the Virus Pathogen Resource (ViPR, www.viprbrc.org), the Eukaryotic Pathogen Database Resources (EuPathDB, www.eupathdb.org), VectorBase (www.vectorbase.org for invertebrate vectors of human pathogens), and the Influenza Research Database (IRD, www.fludb.org), it represents the five Bioinformatics Resource Centers (BRCs) funded by the National Institute of Allergy and Infectious Diseases (NIAID). All five BRCs are connected by a common informatics coordination center, the Pathogen Portal (www.pathogenportal.org), which provides tools that can be utilized by all BRCs (for example RNA-Seq analyses) and consolidates information across the individual BRCs and other NIAID-funded “big data” resource centers.
PATRIC focuses on NIAID Category A–C bacterial pathogens, which include Mycobacterium tuberculosis, but provides data, tools and analysis services for all publicly available bacterial genomes. As of October 2012, PATRIC has released the genomic data for total of 6642 bacterial genomes including 175 mycobacterial genomes (Table 2). To enable comprehensive comparative analyses, genome annotation in PATRIC is performed in a standardized manner using the RAST (Rapid Annotation using Subsystem Technology) system39. PATRIC also provides a free end-user genome annotation service through RAST to allow users to annotate their own genomes (www.patricbrc.org/portal/portal/patric/RAST). In addition to the RAST annotations, PATRIC includes other reference annotations (e.g., the annotation and nomenclature introduced by Cole et al. for M. tuberculosis H37Rv2) and an Identifier (ID) Mapping tool, which allows users to quickly map between PATRIC annotation identifiers and identifiers used by various resources, such as PDB, RefSeq, etc. PATRIC also supports comparative analysis across multiple genomes using protein families and metabolic pathways. Furthermore, PATRIC’s Disease View integrates infectious disease, host, pathogen and disease outbreak data, which enables infectious disease-centric access and analysis of host-pathogen interactions40 (enews.patricbrc.org/faqs/virulence-and-disease-faqs). Users of PATRIC can register and create their own user-accounts in PATRIC to save, manage and analyze the data groups gathered from the PATRIC site to their PATRIC personal workspace (enews.patricbrc.org/faqs/workspace-faqs). Specific workflows supporting various types of analyses have also been recently released in PATRIC and enable finding genomic islands, identifying proteins from outbreak strains, and comparing diverse biochemical pathways, for example. Increasing functionality and improved workflows are being developed through the private, personal PATRIC workspace, to support infectious disease researchers in the process of uploading their own data for analysis with collaborators, generating results and figures for publications, presentations and grant applications, and, ultimately, releasing the results to the broader scientific community. A detailed description of PATRIC has recently been published38, 40. Here, we provide an update on the specific data, tools and services that PATRIC provides for the analysis of bacterial gene expression, which have been released after the Gillespie et al. (2011) publication.
Table 2.
Number of genomes available at PATRIC (as of October 2012).
| All bacteria | Mycobacterium spp. | |
|---|---|---|
| Number of genomes | 6642 | 175 |
| Number of complete genomes | 1927 | 43 |
| Number of incomplete genomes | 4713 | 132 |
3. b. RNA-Seq analysis pipeline at Pathogen Portal
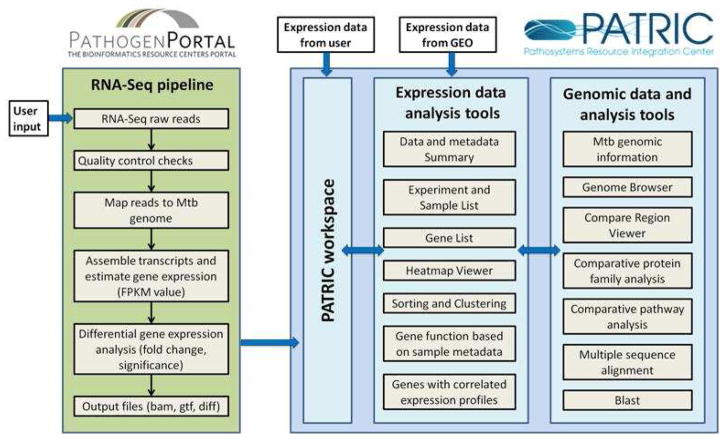
Pathogen Portal (www.pathogenportal.org) is focused scientifically on enabling comparative analysis of host response to pathogens, while using its infrastructure to provide common services across BRCs when appropriate. For example, the Pathogen Portal provides an RNA-Seq pipeline for processing and analyzing high throughput sequencing data to characterize the transcriptome of all BRC pathogens and their key hosts. As the Pathogen Portal is part of the BRC program, it can be used to analyze transcriptome data for the thousands of genomes stored at PATRIC, VectorBase, and EuPathDB. The pipeline is built on Galaxy, an open source bioinformatics workflow system infrastructure41. The Galaxy system has been modified by the Pathogen Portal team to help simplify the process of RNA-Seq analysis for routine use by informatics-naïve, biologically focused users and provide a guided experience to quality control of read data, read mapping, assembling transcripts, estimating gene expression values, and doing differential expression analysis (Figure 1). As of October 2012, the Portal’s RNA-Seq pipeline had 175 Mycobacterial genomes, of which 74 were M. tuberculosis. Genomes for use as references by the RNA-Seq pipeline are frequently updated as new sequences are published at PATRIC, VectorBase, or EuPathDB. Researchers can upload read data into their own private project space at the Pathogen Portal and use the system to contrast expression profiles for various states of Mycobacterium spp. The resulting data can also be used to discover new genes and alternative start transcription sites, for example. The pipeline records provenance information, including the tools and parameters used to process the data, supports batch analysis for multiple samples, and provides secure results sharing and publishing. The RNA-Seq Pipeline is available at rnaseq.pathogenportal.org and is free to use.
Figure 1.
RNA-Seq analysis workflow in Pathogen Portal and PATRIC.
3. c. Expression data at PATRIC
PATRIC provides a suite of integrated methods and tools to explore, visualize, analyze, and compare a large number of published gene expression data-sets available at PATRIC as well as upload and analyze their own unpublished gene expression data-sets, including RNA-Seq data. PATRIC has incorporated a large number of published gene expression data-sets related to bacterial pathogens from NCBI’s GEO database (www.ncbi.nlm.nih.gov/geo). Once a data-set is retrieved from GEO, it is curated by carefully reviewing and curating published experimental design and protocol. Organism and gene identifiers described in the expression array platform are mapped to corresponding genomes and genes in PATRIC. Data from replicates are merged, normalized and log-transformed using a manual curation process for quality control. Experimental procedures and sample metadata are also curated to accurately and consistently capture information such as sample strain, genetic modification, experimental condition, treatment, and time-point. Then, the expression data are combined with the other genomic data in PATRIC to provide integrated data analysis capabilities. As of October 2012, PATRIC has incorporated Mycobacterium-related gene expression data from 45 published data-sets, which correspond to 549 curated comparisons.
PATRIC also allows researchers to upload unpublished gene expression data into their free private workspace to explore them with various analysis and visualization tools (further described below) and to compare them with the public data-sets at PATRIC. Pre-processed gene expression data generated by using either microarray or high-throughput sequencing technologies can be uploaded to PATRIC as excel or tab-delimited files in the form of a gene list or a gene matrix. An additional file containing sample metadata is also provided to aid in the data analysis. These file formats are further described at enews.patricbrc.org/faqs/transcriptomics-data-faqs/. Transcriptomics data generated using high-throughput sequencing technologies can first be processed using the RNA-Seq analysis pipeline available at Pathogen Portal and then be imported to the PATRIC workspace for further analysis (Figure 1). Some of the expression data-related tools and functionality available on the PATRIC website are described below.
3. c.1 Experiment and sample list
For any taxonomy level or genome, all publicly available data-sets are displayed as experiment and sample lists. Metadata-based searching and progressive filtering allows researchers to quickly find data-sets of interest. For each of the samples, numbers of differentially expressed genes identified to be significant (|log ratio ≥ 1 or |Z-score| ≥ 2) are summarized and linked to the corresponding gene list. Multiple data-sets can be selected for further analysis across multiple experiments and/or can be saved as a group for repeated or future use.
3. c.2 Gene list based on expression data
PATRIC researchers can create a gene list based on the expression data-sets of their interest, which displays all the genes reported in the selected data-sets, their functions, and summarizes their expression levels. Gene lists can be dynamically filtered to find the most differentially expressed genes by applying different log ratio and/or Z-score thresholds. Lists can also be filtered by gene names or functions to analyze expression of any genes of interest. Researchers also can find genes that are up- or down-regulated in only a subset of samples of interest. Once a subset of genes of interest are found, they can be downloaded, along with their annotations and expression values, as a tab-delimited or Excel file or they can be saved as a group for further analysis. The pathway summary tool (enews.patricbrc.org/faqs/transcriptomics-data-faqs) allows researchers to quickly find top metabolic pathways corresponding to their genes of interest and visualize them in KEGG42 (Kyoto Encyclopedia of Genes and Genomes) maps along with all other genes annotated in a pathway.
3. c.3 Heatmap visualization and clustering
Complementary to the gene list is the 2-D visualization or “heatmap tool” that shows expression levels of all the genes in a gene list across all selected samples. Genes displayed in the heatmap can be filtered and researchers can switch between the gene list and the heatmap view at any point. Genes in the heatmap can be sorted based on their genomic locations to visually detect genomic regions that are similarly or differently expressed across multiple samples. Genes and samples can also be sorted by applying hierarchical clustering, which allows researchers to quickly identify group of genes that are similarly expressed across multiple samples. Any area of the heatmap can be selected to download corresponding expression data or to save the genes as a group.
3. c.4 Sample metadata analysis
For a given gene, top samples in which the gene expression passes the specified threshold are listed along with their metadata, such as sample strain, genetic modification, and experimental conditions. A visual summary of the metadata allows researchers to find top experimental conditions and gene manipulations that a gene responds to and confirm or infer potential function of the gene.
3. c.5 Genes with correlated expression
For a gene of interest, lists of genes with the most highly correlated expression profiles (positively or negatively) across all publicly available data-sets are displayed along with their function. This allows researchers to identify potentially co-regulated genes or genes that perform similar functions and, often, generate hypothesis about potential function of a hypothetical gene.
4. Conclusions
Access to high quality genome annotation and analysis tools is essential for a modern research community, and we are well served in this regard by the TubercuList and PATRIC resources outlined above. Just some of the information and tools that are available in TubercuList and PATRIC have been presented here, highlighting the increasing sophistication of resources that are available to users. To further tailor these resources to the needs of the community we look forward to getting feedback for TubercuList (email: tuberculist@epfl.ch) and PATRIC (email: patric@vbi.vt.edu).
Supplementary Material
Acknowledgments
Funding: TubercuList received funding from the Swiss-South Africa Joint Research Programme of the University of Basel (contract no. JRP09), and the TB Drug Accelerator program, part of the Bill and Melinda Gates Foundation (grant no. 42917_BMGF). The UniProt and Vital-IT activities at SIB are supported by the Swiss Confederation through the Secrétariat à l’Education et la Recherche (SER). UniProt activity is also supported through the National Institutes of Health (grant 1 U41 HG006104-02). PATRIC/Pathogen Portal has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN272200900040C, awarded to BWS Sobral.
Footnotes
Competing interest: None declared.
Ethical approval: None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.WHO. Tuberculosis Fact Sheet No 104. 2012. [Google Scholar]
- 2.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 3.Sandgren A, Strong M, Muthukrishnan P, Weiner BK, Church GM, Murray MB. Tuberculosis drug resistance mutation database. PLoS Medicine. 2009;6:e2. doi: 10.1371/journal.pmed.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magarinos MP, Carmona SJ, Crowther GJ, Ralph SA, Roos DS, Shanmugam D, Van Voorhis WC, Aguero F. TDR Targets: a chemogenomics resource for neglected diseases. Nucl Acid Res. 2012;40:D1118–1127. doi: 10.1093/nar/gkr1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camus JC, Pryor MJ, Medigue C, Cole ST. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology. 2002;148:2967–2973. doi: 10.1099/00221287-148-10-2967. [DOI] [PubMed] [Google Scholar]
- 6.Lew JM, Kapopoulou A, Jones LM, Cole ST. TubercuList--10 years after. Tuberculosis. 2011;91:1–7. doi: 10.1016/j.tube.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Pagani I, Liolios K, Jansson J, Chen IM, Smirnova T, Nosrat B, Markowitz VM, Kyrpides NC. The Genomes OnLine Database (GOLD) v.4: status of genomic and metagenomic projects and their associated metadata. Nucl Acid Res. 2012;40:D571–579. doi: 10.1093/nar/gkr1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madupu R, Brinkac LM, Harrow J, Wilming LG, Bohme U, Lamesch P, Hannick LI. Meeting report: a workshop on Best Practices in Genome Annotation. Database. 2010:baq001. doi: 10.1093/database/baq001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Medigue C, Moszer I. Annotation, comparison and databases for hundreds of bacterial genomes. Res Microbiol. 2007;158:724–736. doi: 10.1016/j.resmic.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Consortium U. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucl Acid Res. 2012;40:D71–75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abramovitch RB, Rohde KH, Hsu FF, Russell DG. aprABC: a Mycobacterium tuberculosis complex-specific locus that modulates pH-driven adaptation to the macrophage phagosome. Mol Microbiol. 2011;80:678–694. doi: 10.1111/j.1365-2958.2011.07601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haft DH. Bioinformatic evidence for a widely distributed, ribosomally produced electron carrier precursor, its maturation proteins, and its nicotinoprotein redox partners. BMC Genom. 2011;12:21. doi: 10.1186/1471-2164-12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnvig K, Young D. Non-coding RNA and its potential role in Mycobacterium tuberculosis pathogenesis. RNA Biol. 2012;9 doi: 10.4161/rna.20105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiChiara JM, Contreras-Martinez LM, Livny J, Smith D, McDonough KA, Belfort M. Multiple small RNAs identified in Mycobacterium bovis BCG are also expressed in Mycobacterium tuberculosis and Mycobacterium smegmatis. Nucl Acid Res. 2010;38:4067–4078. doi: 10.1093/nar/gkq101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Souza GA, Wiker HG. A proteomic view of mycobacteria. Proteomics. 2011;11:3118–3127. doi: 10.1002/pmic.201100043. [DOI] [PubMed] [Google Scholar]
- 16.Mehaffy MC, Kruh-Garcia NA, Dobos KM. Prospective on Mycobacterium tuberculosis proteomics. J Prot Res. 2012;11:17–25. doi: 10.1021/pr2008658. [DOI] [PubMed] [Google Scholar]
- 17.Renuse S, Chaerkady R, Pandey A. Proteogenomics. Proteomics. 2011;11:620–630. doi: 10.1002/pmic.201000615. [DOI] [PubMed] [Google Scholar]
- 18.Kelkar DS, Kumar D, Kumar P, Balakrishnan L, Muthusamy B, Yadav AK, Shrivastava P, Marimuthu A, Anand S, Sundaram H, Kingsbury R, Harsha HC, Nair B, Prasad TS, Chauhan DS, Katoch K, Katoch VM, Chaerkady R, Ramachandran S, Dash D, Pandey A. Proteogenomic analysis of Mycobacterium tuberculosis by high resolution mass spectrometry. Molec Cell Prot. 2011;10:M111 011627. doi: 10.1074/mcp.M111.011445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ehebauer MT, Wilmanns M. The progress made in determining the Mycobacterium tuberculosis structural proteome. Proteomics. 2011;11:3128–3133. doi: 10.1002/pmic.201000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daga PR, Patel RY, Doerksen RJ. Template-based protein modeling: recent methodological advances. Curr Top Med Chem. 2010;10:84–94. doi: 10.2174/156802610790232314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agarwal N, Raghunand TR, Bishai WR. Regulation of the expression of whiB1 in Mycobacterium tuberculosis: role of cAMP receptor protein. Microbiology. 2006;152:2749–2756. doi: 10.1099/mic.0.28924-0. [DOI] [PubMed] [Google Scholar]
- 22.Bai G, Gazdik MA, Schaak DD, McDonough KA. The Mycobacterium bovis BCG cyclic AMP receptor-like protein is a functional DNA binding protein in vitro and in vivo, but its activity differs from that of its M. tuberculosis ortholog, Rv3676. Infect Immun. 2007;75:5509–5517. doi: 10.1128/IAI.00658-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendoza Lopez P, Golby P, Wooff E, Nunez Garcia J, Garcia Pelayo MC, Conlon K, Gema Camacho A, Hewinson RG, Polaina J, Suarez Garcia A, Gordon SV. Characterization of the transcriptional regulator Rv3124 of Mycobacterium tuberculosis identifies it as a positive regulator of molybdopterin biosynthesis and defines the functional consequences of a non-synonymous SNP in the Mycobacterium bovis BCG orthologue. Microbiology. 2010;156:2112–2123. doi: 10.1099/mic.0.037200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pang X, Vu P, Byrd TF, Ghanny S, Soteropoulos P, Mukamolova GV, Wu S, Samten B, Howard ST. Evidence for complex interactions of stress-associated regulons in an mprAB deletion mutant of Mycobacterium tuberculosis. Microbiology. 2007;153:1229–1242. doi: 10.1099/mic.0.29281-0. [DOI] [PubMed] [Google Scholar]
- 25.Rodrigue S, Brodeur J, Jacques PE, Gervais AL, Brzezinski R, Gaudreau L. Identification of mycobacterial sigma factor binding sites by chromatin immunoprecipitation assays. J Bacteriol. 2007;189:1505–1513. doi: 10.1128/JB.01371-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veyrier F, Said-Salim B, Behr MA. Evolution of the mycobacterial SigK regulon. J Bacteriol. 2008;190:1891–1899. doi: 10.1128/JB.01452-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de la Paz Santangelo M, Klepp L, Nunez-Garcia J, Blanco FC, Soria M, Garcia-Pelayo MC, Bianco MV, Cataldi AA, Golby P, Jackson M, Gordon SV, Bigi F. Mce3R, a TetR-type transcriptional repressor, controls the expression of a regulon involved in lipid metabolism in Mycobacterium tuberculosis. Microbiology. 2009;155:2245–2255. doi: 10.1099/mic.0.027086-0. [DOI] [PubMed] [Google Scholar]
- 28.Fontan PA, Aris V, Alvarez ME, Ghanny S, Cheng J, Soteropoulos P, Trevani A, Pine R, Smith I. Mycobacterium tuberculosis sigma factor E regulon modulates the host inflammatory response. J Infect Dis. 2008;198:877–885. doi: 10.1086/591098. [DOI] [PubMed] [Google Scholar]
- 29.Golby P, Nunez J, Cockle PJ, Ewer K, Logan K, Hogarth P, Vordermeier HM, Hinds J, Hewinson RG, Gordon SV. Characterization of two in vivo-expressed methyltransferases of the Mycobacterium tuberculosis complex: antigenicity and genetic regulation. Microbiology. 2008;154:1059–1067. doi: 10.1099/mic.0.2007/014548-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Souza GA, Arntzen MO, Fortuin S, Schurch AC, Malen H, McEvoy CR, van Soolingen D, Thiede B, Warren RM, Wiker HG. Proteogenomic analysis of polymorphisms and gene annotation divergences in prokaryotes using a clustered mass spectrometry-friendly database. Molec Cell Prot. 2011;10:M110 002527. doi: 10.1074/mcp.M110.002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnvig KB, Comas I, Thomson NR, Houghton J, Boshoff HI, Croucher NJ, Rose G, Perkins TT, Parkhill J, Dougan G, Young DB. Sequence-based analysis uncovers an abundance of non-coding RNA in the total transcriptome of Mycobacterium tuberculosis. PLoS Pathog. 2011;7:e1002342. doi: 10.1371/journal.ppat.1002342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pelly S, Bishai WR, Lamichhane G. A screen for non-coding RNA in Mycobacterium tuberculosis reveals a cAMP-responsive RNA that is expressed during infection. Gene. 2012;500:85–92. doi: 10.1016/j.gene.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poulsen C, Akhter Y, Jeon AH, Schmitt-Ulms G, Meyer HE, Stefanski A, Stuhler K, Wilmanns M, Song YH. Proteome-wide identification of mycobacterial pupylation targets. Molec Sys Biol. 2010;6:386. doi: 10.1038/msb.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prisic S, Dankwa S, Schwartz D, Chou MF, Locasale JW, Kang CM, Bemis G, Church GM, Steen H, Husson RN. Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. Proc Natl Acad Sci USA. 2010;107:7521–7526. doi: 10.1073/pnas.0913482107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazandu GK, Mulder NJ. Function Prediction and Analysis of Mycobacterium tuberculosis Hypothetical Proteins. Int J Molec Sci. 2012;13:7283–7302. doi: 10.3390/ijms13067283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niemann S, Koser CU, Gagneux S, Plinke C, Homolka S, Bignell H, Carter RJ, Cheetham RK, Cox A, Gormley NA, Kokko-Gonzales P, Murray LJ, Rigatti R, Smith VP, Arends FP, Cox HS, Smith G, Archer JA. Genomic diversity among drug sensitive and multidrug resistant isolates of Mycobacterium tuberculosis with identical DNA fingerprints. PLoS One. 2009;4:e7407. doi: 10.1371/journal.pone.0007407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ioerger TR, Feng Y, Ganesula K, Chen X, Dobos KM, Fortune S, Jacobs WR, Jr, Mizrahi V, Parish T, Rubin E, Sassetti C, Sacchettini JC. Variation among genome sequences of H37Rv strains of Mycobacterium tuberculosis from multiple laboratories. J Bacteriol. 2010;192:3645–3653. doi: 10.1128/JB.00166-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gillespie JJ, Wattam AR, Cammer SA, Gabbard JL, Shukla MP, Dalay O, Driscoll T, Hix D, Mane SP, Mao C, Nordberg EK, Scott M, Schulman JR, Snyder EE, Sullivan DE, Wang C, Warren A, Williams KP, Xue T, Yoo HS, Zhang C, Zhang Y, Will R, Kenyon RW, Sobral BW. PATRIC: the comprehensive bacterial bioinformatics resource with a focus on human pathogenic species. Infect Immun. 2011;79:4286–4298. doi: 10.1128/IAI.00207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Driscoll T, Gabbard JL, Mao C, Dalay O, Shukla M, Freifeld CC, Hoen AG, Brownstein JS, Sobral BW. Integration and visualization of host-pathogen data related to infectious diseases. Bioinformatics. 2011;27:2279–2287. doi: 10.1093/bioinformatics/btr391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nuc Acid Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.