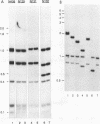
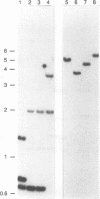
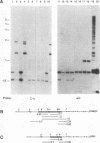
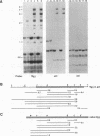
Abstract
Transformation of eukaryotic cells can be used to test potential signals for DNA methylation. This approach is not always reliable, however, because of chromosomal position effects and because integration of multiple and/or rearranged copies of transforming DNA can influence DNA methylation. We developed a robust system to evaluate the potential of DNA fragments to function as signals for de novo methylation in Neurospora crassa. The requirements of the system were (i) a location in the N. crassa genome that becomes methylated only in the presence of a bona fide methylation signal and (ii) an efficient gene replacement protocol. We report here that the am locus fulfills these requirements, and we demonstrate its utility with the identification of a 2.7-kb fragment from the psi 63 locus as a new portable signal for de novo methylation.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akins R. A., Lambowitz A. M. General method for cloning Neurospora crassa nuclear genes by complementation of mutants. Mol Cell Biol. 1985 Sep;5(9):2272–2278. doi: 10.1128/mcb.5.9.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson B. D., Lindgren K. M., Dunlap J. C., Loros J. J. An efficient method for gene disruption in Neurospora crassa. Mol Gen Genet. 1994 Feb;242(4):490–494. doi: 10.1007/BF00281802. [DOI] [PubMed] [Google Scholar]
- Asch D. K., Kinsey J. A. Relationship of vector insert size to homologous integration during transformation of Neurospora crassa with the cloned am (GDH) gene. Mol Gen Genet. 1990 Mar;221(1):37–43. doi: 10.1007/BF00280365. [DOI] [PubMed] [Google Scholar]
- Cambareri E. B., Jensen B. C., Schabtach E., Selker E. U. Repeat-induced G-C to A-T mutations in Neurospora. Science. 1989 Jun 30;244(4912):1571–1575. doi: 10.1126/science.2544994. [DOI] [PubMed] [Google Scholar]
- Case M. E., Geever R. F., Asch D. K. Use of gene replacement transformation to elucidate gene function in the qa gene cluster of Neurospora crassa. Genetics. 1992 Apr;130(4):729–736. doi: 10.1093/genetics/130.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case M. E. Genetical and molecular analyses of qa-2 transformants in Neurospora crassa. Genetics. 1986 Jul;113(3):569–587. doi: 10.1093/genetics/113.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case M. E., Schweizer M., Kushner S. R., Giles N. H. Efficient transformation of Neurospora crassa by utilizing hybrid plasmid DNA. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5259–5263. doi: 10.1073/pnas.76.10.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fincham J. R. Transformation in fungi. Microbiol Rev. 1989 Mar;53(1):148–170. doi: 10.1128/mr.53.1.148-170.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss H. M., Roberts C. J., Claeys K. M., Selker E. U. Abnormal chromosome behavior in Neurospora mutants defective in DNA methylation. Science. 1993 Dec 10;262(5140):1737–1741. doi: 10.1126/science.7505062. [DOI] [PubMed] [Google Scholar]
- Grayburn W. S., Selker E. U. A natural case of RIP: degeneration of the DNA sequence in an ancestral tandem duplication. Mol Cell Biol. 1989 Oct;9(10):4416–4421. doi: 10.1128/mcb.9.10.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasse A., Schulz W. A. Enhancement of reporter gene de novo methylation by DNA fragments from the alpha-fetoprotein control region. J Biol Chem. 1994 Jan 21;269(3):1821–1826. [PubMed] [Google Scholar]
- Jähner D., Jaenisch R. Retrovirus-induced de novo methylation of flanking host sequences correlates with gene inactivity. Nature. 1985 Jun 13;315(6020):594–597. doi: 10.1038/315594a0. [DOI] [PubMed] [Google Scholar]
- Kafri T., Ariel M., Brandeis M., Shemer R., Urven L., McCarrey J., Cedar H., Razin A. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 1992 May;6(5):705–714. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- Kim S. Y., Marzluf G. A. Transformation of Neurospora crassa with the trp-1 gene and the effect of host strain upon the fate of the transforming DNA. Curr Genet. 1988;13(1):65–70. doi: 10.1007/BF00365758. [DOI] [PubMed] [Google Scholar]
- Kinnaird J. H., Fincham J. R. The complete nucleotide sequence of the Neurospora crassa am (NADP-specific glutamate dehydrogenase) gene. Gene. 1983 Dec;26(2-3):253–260. doi: 10.1016/0378-1119(83)90195-6. [DOI] [PubMed] [Google Scholar]
- Kinnaird J. H., Keighren M. A., Kinsey J. A., Eaton M., Fincham J. R. Cloning of the am (glutamate dehydrogenase) gene of Neurospora crassa through the use of a synthetic DNA probe. Gene. 1982 Dec;20(3):387–396. doi: 10.1016/0378-1119(82)90207-4. [DOI] [PubMed] [Google Scholar]
- Kinsey J. A. Direct selective procedure for isolating Neurospora mutants defective in nicotinamide adenine dinucleotide phosphate-specific glutamate dehydrogenase. J Bacteriol. 1977 Dec;132(3):751–756. doi: 10.1128/jb.132.3.751-756.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey J. A., Hung B. S. Mutation at the am locus of Neurospora crassa. Genetics. 1981 Nov-Dec;99(3-4):405–414. doi: 10.1093/genetics/99.3-4.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E., Bestor T. H., Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992 Jun 12;69(6):915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Magewu A. N., Jones P. A. Ubiquitous and tenacious methylation of the CpG site in codon 248 of the p53 gene may explain its frequent appearance as a mutational hot spot in human cancer. Mol Cell Biol. 1994 Jun;14(6):4225–4232. doi: 10.1128/mcb.14.6.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead D. A., Szczesna-Skorupa E., Kemper B. Single-stranded DNA 'blue' T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1986 Oct-Nov;1(1):67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- Metzenberg R. L., Stevens J. N., Selker E. U., Morzycka-Wroblewska E. Identification and chromosomal distribution of 5S rRNA genes in Neurospora crassa. Proc Natl Acad Sci U S A. 1985 Apr;82(7):2067–2071. doi: 10.1073/pnas.82.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummaneni P., Bishop P. L., Turker M. S. A cis-acting element accounts for a conserved methylation pattern upstream of the mouse adenine phosphoribosyltransferase gene. J Biol Chem. 1993 Jan 5;268(1):552–558. [PubMed] [Google Scholar]
- Nehls U., Friedrich T., Schmiede A., Ohnishi T., Weiss H. Characterization of assembly intermediates of NADH:ubiquinone oxidoreductase (complex I) accumulated in Neurospora mitochondria by gene disruption. J Mol Biol. 1992 Oct 20;227(4):1032–1042. doi: 10.1016/0022-2836(92)90519-p. [DOI] [PubMed] [Google Scholar]
- Nelson M., McClelland M. Site-specific methylation: effect on DNA modification methyltransferases and restriction endonucleases. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2045–2071. doi: 10.1093/nar/19.suppl.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paietta J. V., Marzluf G. A. Gene disruption by transformation in Neurospora crassa. Mol Cell Biol. 1985 Jul;5(7):1554–1559. doi: 10.1128/mcb.5.7.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pröls F., Meyer P. The methylation patterns of chromosomal integration regions influence gene activity of transferred DNA in Petunia hybrida. Plant J. 1992 Jul;2(4):465–475. doi: 10.1046/j.1365-313x.1992.t01-20-00999.x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E. U., Cambareri E. B., Jensen B. C., Haack K. R. Rearrangement of duplicated DNA in specialized cells of Neurospora. Cell. 1987 Dec 4;51(5):741–752. doi: 10.1016/0092-8674(87)90097-3. [DOI] [PubMed] [Google Scholar]
- Selker E. U., Fritz D. Y., Singer M. J. Dense nonsymmetrical DNA methylation resulting from repeat-induced point mutation in Neurospora. Science. 1993 Dec 10;262(5140):1724–1728. doi: 10.1126/science.8259516. [DOI] [PubMed] [Google Scholar]
- Selker E. U., Garrett P. W. DNA sequence duplications trigger gene inactivation in Neurospora crassa. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6870–6874. doi: 10.1073/pnas.85.18.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E. U., Jensen B. C., Richardson G. A. A portable signal causing faithful DNA methylation de novo in Neurospora crassa. Science. 1987 Oct 2;238(4823):48–53. doi: 10.1126/science.2958937. [DOI] [PubMed] [Google Scholar]
- Selker E. U. Premeiotic instability of repeated sequences in Neurospora crassa. Annu Rev Genet. 1990;24:579–613. doi: 10.1146/annurev.ge.24.120190.003051. [DOI] [PubMed] [Google Scholar]
- Selker E. U., Richardson G. A., Garrett-Engele P. W., Singer M. J., Miao V. Dissection of the signal for DNA methylation in the zeta-eta region of Neurospora. Cold Spring Harb Symp Quant Biol. 1993;58:323–329. doi: 10.1101/sqb.1993.058.01.038. [DOI] [PubMed] [Google Scholar]
- Selker E. U., Stevens J. N. DNA methylation at asymmetric sites is associated with numerous transition mutations. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8114–8118. doi: 10.1073/pnas.82.23.8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E. U., Stevens J. N. Signal for DNA methylation associated with tandem duplication in Neurospora crassa. Mol Cell Biol. 1987 Mar;7(3):1032–1038. doi: 10.1128/mcb.7.3.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart W. D., Koo K., Vollmer S. J. Cloning of mtr, an amino acid transport gene of Neurospora crassa. Genome. 1988 Apr;30(2):198–203. doi: 10.1139/g88-034. [DOI] [PubMed] [Google Scholar]
- Szyf M. DNA methylation patterns: an additional level of information? Biochem Cell Biol. 1991 Dec;69(12):764–767. doi: 10.1139/o91-117. [DOI] [PubMed] [Google Scholar]
- Toth M., Lichtenberg U., Doerfler W. Genomic sequencing reveals a 5-methylcytosine-free domain in active promoters and the spreading of preimposed methylation patterns. Proc Natl Acad Sci U S A. 1989 May;86(10):3728–3732. doi: 10.1073/pnas.86.10.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth M., Müller U., Doerfler W. Establishment of de novo DNA methylation patterns. Transcription factor binding and deoxycytidine methylation at CpG and non-CpG sequences in an integrated adenovirus promoter. J Mol Biol. 1990 Aug 5;214(3):673–683. doi: 10.1016/0022-2836(90)90285-T. [DOI] [PubMed] [Google Scholar]
- Wu J., Issa J. P., Herman J., Bassett D. E., Jr, Nelkin B. D., Baylin S. B. Expression of an exogenous eukaryotic DNA methyltransferase gene induces transformation of NIH 3T3 cells. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):8891–8895. doi: 10.1073/pnas.90.19.8891. [DOI] [PMC free article] [PubMed] [Google Scholar]