Abstract
RNA viruses are the causative agents for AIDS, influenza, SARS, and other serious health threats. Development of rapid and broadly applicable methods for complete viral genome sequencing is highly desirable to fully understand all aspects of these infectious agents as well as for surveillance of viral pandemic threats and emerging pathogens. However, traditional viral detection methods rely on prior sequence or antigen knowledge. In this study, we describe sequence-independent amplification for samples containing ultra-low amounts of viral RNA coupled with Illumina sequencing and de novo assembly optimized for viral genomes. With 5 million reads, we capture 96 to 100% of the viral protein coding region of HIV, respiratory syncytial and West Nile viral samples from as little as 100 copies of viral RNA. The methods presented here are scalable to large numbers of samples and capable of generating full or near full length viral genomes from clone and clinical samples with low amounts of viral RNA, without prior sequence information and in the presence of substantial host contamination.
INTRODUCTION
Massively parallel sequencing allows for rapid and low-cost deep sequencing of viral genomes and provides an opportunity to gain greater insight into viral evolution, fitness, emergence and transmission. Currently, reverse transcription followed by polymerase chain reaction (RT-PCR) with primers designed to amplify specific viral RNA sequences is the most common method for amplifying RNA viruses prior to sequencing and other downstream applications. Recent studies have used both 454 (1–11) (Newman et al., manuscript submitted) and Illumina (12–14) sequencing of RT-PCR amplicons for RNA viruses. However, standard RT-PCR methods are not adequate for the generation of templates suitable for sequencing low-copy viral RNA samples, where labor-intensive methods such as nested PCR (15) or single-genome amplification (SGA) (16,17) are typically required. Examples of low-copy viral RNA samples include the following: HIV controllers (capable of controlling the virus in the absence of antiretroviral therapy) (18), dengue virus (DENV) clinical samples collected after peak viremia (19), and West Nile virus (WNV) surveillance samples (20). Obtaining genomic sequence from such samples can provide valuable insights into viral attenuation, response to host immune pressure and drug treatment during infection, disease severity, transmission and epidemic spread.
Development of high-throughput methods for viral sequencing from low-copy viral samples presents challenges. First, the method should be robust in generating sufficient material for sequencing libraries. Second, it should be broadly applicable to a variety of sample types. Finally, it should consistently generate sequence coverage for the entire target region, typically the protein coding region, CDS, of a viral genome.
Sequence-independent methods (21–25) are attractive for sequencing viral genomes in that they do not rely on prior viral sequence knowledge. This allows sequencing of viral genomes with little or no genomic information and of highly divergent viruses for which robust primers targeting conserved regions are difficult to design. To date these methods have difficulty with sequencing complete genomes from clinical samples due to high levels of host contamination and low viral amounts. The SISPA method described by Djikeng et al. is capable of capturing complete genomes from viral RNA samples, but requires high viral amounts and removal of host RNA contamination with RNases prior to viral RNA extraction (22). Work by Victoria et al. demonstrated the ability to capture complete genomes from stool samples, but it appears these samples had high viral titers (25). Recent work by Ninomiya et al. has demonstrated success of capturing hepatitis C virus (HCV) total RNA sequencing from clinical samples (23), but this method required 200 ng of input RNA which exceeds the amount typically present in most clinical samples. Moore et al. evaluated the sensitivity of detection of infectious agents using RNA sequencing (24). Their method worked well for detecting low amounts of virus but was limited to input amounts of 30,000 copies of viral RNA per sample.
In this study, we evaluated the use of a sequence-independent RNA amplification method, NuGEN’s Ovation RNA-Seq system, for capturing complete target regions of viral genomes from low-copy HIV, respiratory syncytial virus (RSV) and WNV samples. Previously, this amplification method was used for cellular transcriptome analysis (26,27) with input amounts of 500 pg to 100 ng of total RNA. Here, using only femtograms to attograms of viral RNA with this amplification method in combination with Illumina sequencing and de novo assembly of viral reads, we successfully generated consensus sequence for the complete, or very nearly complete, CDS of viral genomes.
MATERIALS AND METHODS
HIV sample information
Plasma samples from HIV-1 chronically infected subjects were obtained from the Ragon Institute of MGH, MIT and Harvard in Boston, MA. All subjects gave written informed consent and the study was approved by the Massachusetts General Hospital Review Board. NL4-3 (28) virus stocks were produced by transfection of HEK293T cells (ATCC, Manassas, VA) with plasmid DNA encoding a full-length infectious HIV RNA using Polyfect transfection reagent (Qiagen, Valencia, CA) according to a modified manufacturer protocol. Briefly, 1 day prior to transfection, 2.8 × 106 cells were seeded in a T75 flask. For the transfection, 15 µg of plasmid DNA, at a minimum concentration of 1 µg/µl, was diluted to a 150-µl final volume in Dulbecco modified Eagle medium without supplements; 115 µl of Polyfect reagent was added, and the solution was mixed by gentle pipetting and incubated for 10 min at room temperature. During the incubation, medium was removed from the cells to be transfected followed by a single wash with cold phosphate-buffered saline (PBS), and then 7 ml of fresh medium was added. After this incubation, the transfection mixture was transferred to the flask, swirled gently to mix, and incubated for 3 hours at 37°C with 5% CO2. The medium was removed and discarded; the cells were washed once with cold PBS, and 7 ml of fresh medium was added before returning the cells to the incubator. Transfection supernatant was harvested after 72 hours, filtered through a 0.45-µm filter, and stored in aliquots at −80°C. NL4-3 viral titer was determined by HIV-1 p24 antigen enzyme-linked immunosorbent assay (Perkin-Elmer, Waltham, MA) per the manufacturer's protocol.
West Nile sample information
WNV was produced from an infectious cDNA clone according to methods described elsewhere (29). Briefly, hamster BHK-21 cells (ATCC) were transfected (transMessenger, Qiagen) with RNA transcribed in vitro from pFLWNV (29), using the mMessage mMachine T7 kit (Life Technologies, Carlsbad, CA). Cells were inspected daily and virus harvested after approximately 3 days of replication. Virus-containing tissue culture supernatant was supplemented with 20% FBS, stored at −80°C in 1 ml aliquots and used without subsequent passage.
RSV sample information
Nasal washes were collected by study team members using similarly quantified-collection method as previously described (30). The study was conducted with the approval of the University of Tennessee Institutional Review Board and included appropriate informed consent, complying with all relevant federal guidelines and institutional policies.
RNA isolation and quantification by quantitative RT-PCR
For HIV clinical samples, 1.5 ml of plasma was thawed and centrifuged at 1500g for 10 min at 4°C to remove cellular debris. For HIV and WNV clone samples, 140 µl of cell culture supernatant was used for extraction. All samples were brought up to volume with 1× PBS and centrifuged at 120 000g for 1.5 hours at 4°C after which the pellet was re-suspended in 140 µl of 1× PBS and used as input for viral RNA extraction. For RSV clinical samples, 140 µl of nasal aspirate was used for extraction (without the 120 000g centrifugation step). Viral RNA was isolated using QIAamp Viral RNA Mini kit (Qiagen) per manufacturer’s protocol with the exception that 5 µg of linear polyacrylamide (Life Technologies) was used as the carrier instead of the carrier RNA provided with the kit. Viral RNA was eluted with 60 µl of AVE buffer, aliquoted and stored at −80°C. All viral samples were treated with Turbo DNase (Life Technologies) using the manufacturer’s rigorous treatment to ensure removal of DNA. For each HIV sample, quantitative RT-PCR (qRT-PCR) was used to measure the number of copies of HIV. These experiments were performed with the ABI 7900HT (Life Technologies) using HIV-1 gag SK145 (AGTGGGGGGACATCAAGCAGCCATGCAAAT) and SK431 (TGCTATGTCACTTCCCCTTGGTTCTCT) primers (31) at 300 nM final concentration and the SuperScript III Platinum SYBR Green One-Step qRT-PCR Kit (Life Technologies) per manufacturer’s protocol. The quantification standards consisted of linear HIV-1 pNL4-3 plasmid. Similarly to HIV, the number of WNV copies in each sample was also quantified by qRT-PCR performed on the ABI 7900HT using forward primer (TCAGCGATCTCTCCACCAAAG), reverse primer (GGGTCAGCACGTTTGTCATTG), probe (FAM-TGCCCGACCATGGGAGAAGCTC-TAMRA) and the SuperScript III Platinum One-Step qRT-PCR Kit (Life Technologies) per manufacturer’s protocol and previously described methods (32). In addition, the number of RSV copies in each sample was also quantified by qRT-PCR performed on the ABI 7900HT using forward primer (CATCCAGCAAATACACCATCCA), reverse primer (TTCTGCACATCATAATTAGGAGTATCAA), probe (FAM-CGGAGCACAGGAGAT-TAMRA) and the SuperScript III Platinum One-Step qRT-PCR Kit (Life Technologies) per manufacturer’s protocol and previously described methods (33). Host 18S ribosomal RNA (rRNA) was quantified for all samples using the ABI 7900HT and the TaqMan ribosomal RNA primer and probes following the manufacturer’s protocol (Life Technologies).
RNA amplification
For both Ovation RNA-Seq version 1 and 2 systems (NuGEN, San Carlos, CA), viral RNA was amplified into dsDNA per manufacturer’s protocol. Briefly, RNA was reversed transcribed into cDNA using oligo-dT and random primers, dsDNA was generated and amplified using single-primer isothermal linear amplification (SPIA), as previously described (34). The following modifications were performed. First, we used 1.4 volumes of AMPure RNA clean beads prior to SPIA amplification. Second, the final amplified products were purified using 0.8 volumes of AMPure XP beads (Beckman Coulter Genomics, Danvers, MA). Amplified products were eluted in 30 µl TE buffer (Life Technologies). Technical replicates were performed for HIV NL4-3 clone, HIV clinical samples and WNV clone dilutions samples.
Illumina library construction and sequencing
We prepared paired-end libraries for Illumina sequencing according to previously published methods (35). For Ovation RNA-Seq version 1 reactions, no shearing or size selection was performed since the majority of the amplified products were between 200 and 600 bp. For Ovation RNA-Seq version 2 reactions, amplified products were sheared in a single tube format using an adaptive focused acoustic shear technology (S2 machine, Covaris, Woburn, MA). Shearing conditions were as follows: time = 180 seconds, duty cycle = 10, intensity = 5; cycles per burst = 100, mode = frequency sweeping. Sheared samples were purified using 2.2 volumes of AMPure XP beads resulting in fragments between 150 and 500 bp. Briefly, libraries were prepared by end-repair of the products of the NuGEN reactions (sheared for version 2) followed by A-base addition, adapter ligation, and PCR. The following modifications were made. The libraries were generated with forked adapters containing unique 8-base index sequences. The number of PCR cycles varied from 8 to 15 cycles per sample, using the lowest number of cycles needed for sequencing. The PCR reactions were purified using 2 rounds of 0.7 volumes of AMPure XP beads. We sequenced the HIV and WNV indexed libraries in pools of 12 to 36 samples on a HiSeq 2000 (Illumina, Hayward, CA; 1 lane; 101 base paired-end reads). We sequenced the RSV indexed libraries in a pool of two samples on a MiSeq (Illumina; 101 base paired-end reads). Sequence reads were binned by index read prior to further analysis.
Alignment to host and viral reference genomes
As the Illumina reads generated from RNA processed with the Ovation RNA-Seq kit start with a variable number of bases derived from the kit primers, the first 18 bases of each read were trimmed prior to alignment. To analyse input data composition, we first employed an efficient Illumina aligner BWA-v0.5.9 (26) by aligning reads to viral and host reference genomes. For HIV clone and clinical samples, the reference genomes used were the concatenation of the human genome assembly (NCBI36/hg18), human rRNA sequences (NR_003286.1, NR_003287.1, V00589.1, NR_003285.2, gi|251831106:648-1601, gi|251831106:1671-3229) and the HIV HXB2 sequence (K03455.1). For WNV clone samples, the reference genomes used were the concatenation of preliminary and unpublished sequencing data from the golden hamster, Mesocricetus auratus, (K. Lindblad-Toh, personal communication), hamster rRNA sequences (NR_045212.1, NR_045133.1, NR_045213.1, D89009.1, DQ334843.1) and WNV NY99 reference (NC_009942.1). For RSV clinical samples, the reference genomes used were the concatenation of the human genome assembly (NCBI36/hg18), human rRNA sequences (NR_003286.1, NR_003287.1, V00589.1, NR_003285.2, gi|251831106:648-1601, gi|251831106:1671-3229) and the RSV A sequence (M74568.1). The alignment was carried out by first aligning each read independently using command: bwa aln (-q 5 -l 32 -k 2 -t 4 -o 1), then the read pair information was used by invoking command: bwa sampe (-a 100 000). MergeBamAlignments, from the picard package (v1.59) [http://picard.sourceforge.net/], were used to return the unmapped reads to the aligned BAM file. A custom script was used to count the number of reads aligning to each of the concatenated references. Compared with the BWA aligner, the Mosaik aligner (version 1.1.0013) (http://bioinformatics.bc.edu/marthlab/Mosaik) is slower but more accurate when aligning relatively long Illumina reads like the ones used in the current studies. To determine more accurate coverage information for our viral samples only, we used Mosaik (MosaikAligner -st illumina -hs 10 -act 15 -bw 29 -mmp 0.25 -minp 0.25) to align reads to the target viral reference genome and the consensus assembly results. Primer sequences were not trimmed since soft-clipping is tolerated by Mosaik.
MEGAN analysis
To determine the composition of unmapped reads (from BWA alignments discussed earlier), we carried out taxonomic analysis on the unmapped reads using the MEGAN package (version 4.62.3) (36) (http://www-ab.informatik.uni-tuebingen.de/software/megan) for sequence from HIV (NL4-3) and WNV clones. A random subset of 50 000 unmapped reads was selected and aligned to the nt database (megablast -v 1 -b 1 -e 1 e-10 -a 8 -m 7) (37). The results were visualized with the MEGAN package (36).
De novo consensus assembly
We assembled Illumina data using the VICUNA assembly program (Yang et al., manuscript submitted). In short, VICUNA used the Ovation RNA-Seq primer sequence (inferred from several input datasets) for read trimming. Low complexity reads were discarded. Since some of the HIV, RSV and WNV datasets contained a large percentage of reads derived from the host, we invoked the optional target genome filter component of VICUNA. Based on sequence similarity, the remaining reads were clustered to form contigs, which were further validated such that each read in the contig had sufficient similarity compared with the consensus. Guided by paired reads, these contigs were compared using an efficient alignment algorithm. Any two contigs that shared a significant suffix–prefix overlap were merged. The assembly process terminated when no further merging could be applied. In several cases, a viral reference genome was used to further extend the assembly. Briefly, raw contigs of >350 bases obtained by the assembler were aligned to a reference using MUSCLE (version 3.8) (38). Each contig was broken down in segments when there was a deletion of 30 or more nucleotides compared with the reference. The contigs were added to form the final assembly in decreasing order of length until either the whole assembly was covered or no contigs remained. Overlapping segments were merged together by concatenating the nucleotide sequence of each segment at the central position of overlap.
Comparison of consensus assemblies
The comparison of the assemblies was performed by a custom script that first aligned them using MUSCLE (version 3.8) (38) and calculated their percent identity over the target region. The script then calculated the composition identity (i.e. the positions where the consensus bases either matched or were found as a non-dominant variant in the other sample) by comparing the nucleotide frequency tables using the alignment to match positions.
All consensus genome assemblies generated as part of this project were submitted to NCBI’s GenBank database (Accession numbers JX503071-JX503101). Illumina read data was submitted to NCBI’s Short Read Archive under Trace Identifiers SRR513075, SRR513078, SRR513080, SRR513086-87, SRR513092, and SRR527699-726.
RESULTS
Viral RNA amplification and Illumina library construction
We generated Illumina libraries for HIV, RSV and WNV samples using the Ovation RNA-Seq system with viral samples containing 500 ag to 240 fg (∼100 to ∼30 000 copies) of viral RNA. The concentration of extracted RNA could not be quantified using standard RNA quantification methods; therefore the quantity of viral genomes from each extraction was determined by viral-specific qRT-PCR assays (Supplementary Table S1). For all samples, the amount of host RNA was determined by 18S rRNA qRT-PCR assays (Supplementary Table S1). We used Illumina sequencing to ensure that we obtained sufficient coverage to compensate for the high levels of host contamination (discussed later) in the clinical samples.
We used both the Ovation RNA-Seq version 1 and 2 systems for HIV NL4-3 clone and clinical samples. As the input amounts for all these viral reactions was below the minimum recommended amount of 500 pg of RNA, we evaluated the success of the amplification reactions using HIV-specific qPCR assay (Supplementary Table S1). For WNV clone samples, Ovation RNA-Seq version 2 system reactions were performed using 100–10 000 copies of input RNA with technical duplicates. The success of each amplification reaction was determined using WNV-specific qPCR assay (Supplementary Table S1). For RSV clinical samples, Ovation RNA-Seq version 2 system reactions were performed using 1795 and 30 000 copies of input RNA. The success of each amplification reaction was determined using RSV-specific qPCR assay. All amplification reactions generated sufficient final products to make Illumina libraries. Illumina libraries were generated, pooled, and sequenced. We obtained 5.2–86.9 million reads per library (Supplementary Table S1).
Composition of sequence data
To determine the origin of the Illumina reads, we aligned reads to host, rRNA, and viral references. For the HIV clone samples, the majority of the reads (51–69%) aligned to the HIV reference and less than 6% of reads aligned to host (Table 1 and Supplementary Table S2). For the HIV clinical samples, the percent of HIV aligning reads was significantly lower (0.4–7.1%) while the majority of the reads (30–65%) aligned to host (Table 1 and Supplementary Table S2). The percent of reads aligning to HIV was higher in the version 1 than version 2 samples (Table 1 and Supplementary Table S2). This resulted in a higher average of coverage for the CDS for version 1 than version 2, 2830-fold versus 442-fold, respectively (Table 1 and Supplementary Table S2). For WNV clone samples, 13–31% of the reads align to WNV reference and 48–60% aligned to host (Table 1 and Supplementary Table S2). The average coverage of CDS was 7671-fold for WNV samples (Table 1 and Supplementary Table S2). For RSV clinical samples, the percent of RSV aligning reads were 10–17%, whereas the majority of the reads (63–69%) aligned to host (Table 1 and Supplementary Table S2). The average coverage of the CDS was 4492-fold for RSV samples.
Table 1.
Composition of sequence data and de novo assembly statistics
| Sample | Sample ID | Virus | Copies viral RNA used | Versiona | Reads aligning to viral referenceb (%) | rRNAc (%) | Hostd (%) | CDS covered by all contigse (%) | Average coverage in target region | Genes intactf |
|---|---|---|---|---|---|---|---|---|---|---|
| NL4-3 | D615 | HIV | 10 000 | 1 | 67.1 | 0.3 | 3.5 | 100 | 36 021 | 9 |
| Clinical sample A | D614 | HIV | 10 000 | 1 | 7.1 | 32.3 | 32.5 | 100 | 3869 | 9 |
| Clinical sample A | D613 | HIV | 1 000 | 1 | 6.5 | 25.9 | 28.8 | 96 | 3489 | 7 |
| Clinical sample B | D616 | HIV | 800 | 1 | 5.9 | 18.6 | 18.9 | 100 | 3109 | 9 |
| Clinical sample C | D617 | HIV | 200 | 1 | 2.2 | 17.5 | 12.8 | 100 | 965 | 9 |
| NL4-3 | D619 | HIV | 10 000 | 2 | 68.7 | 0.6 | 4.6 | 100 | 38 725 | 9 |
| Clinical sample B | D620 | HIV | 800 | 2 | 1.1 | 16.8 | 18.2 | 100 | 661 | 9 |
| Clinical sample C | D621 | HIV | 200 | 2 | 0.4 | 14.8 | 8.8 | 97 | 233 | 8 |
| Clinical sample B | G15482 | HIV | 200 | 2 | 1.3 | 18.2 | 27.9 | 100 | 647 | 9 |
| Clinical sample B | G15480 | HIV | 100 | 2 | 1.7 | 17.5 | 28.0 | 99 | 385 | 8 |
| WNV clone | G15493 | WNV | 10 000 | 2 | 31.1 | 0.11 | 47.6 | 100 | 14 822 | 10 |
| WNV clone | G15494 | WNV | 1500 | 2 | 14.5 | 0.09 | 59.1 | 100 | 6925 | 10 |
| WNV clone | G15495 | WNV | 1000 | 2 | 14.3 | 0.09 | 59.1 | 99 | 6800 | 9 |
| WNV clone | G15496 | WNV | 750 | 2 | 13.7 | 0.08 | 59.1 | 100 | 6594 | 10 |
| WNV clone | G15497 | WNV | 500 | 2 | 13.9 | 0.10 | 59.8 | 100 | 6681 | 10 |
| WNV clone | G15498 | WNV | 250 | 2 | 14.3 | 0.08 | 59.0 | 100 | 6786 | 10 |
| WNV clone | G15499 | WNV | 150 | 2 | 15.1 | 0.09 | 58.7 | 100 | 7253 | 10 |
| WNV clone | G15500 | WNV | 100 | 2 | 13.8 | 0.09 | 59.4 | 100 | 6576 | 10 |
| Clinical sample 1 | V6100 | RSV | 30 470 | 2 | 16.6 | 31.8 | 37.0 | 100 | 5599 | 10 |
| Clinical sample 2 | V6103 | RSV | 1795 | 2 | 10.1 | 36.7 | 26.5 | 100 | 3386 | 10 |
aNuGEN’s Ovation RNA-Seq version 1 or 2 system. bFor HIV, the viral reference genome used was HXB2. For WNV, the viral reference genome used was NY99. For RSV, the viral reference genome used was RSV A2. cPercent of reads aligning to both cytoplasmic and mitochondrial rRNA. For HIV and RSV, human rRNA sequences were used. For WNV, hamster rRNA sequences were used. dFor HIV and RSV, the host used was human. For WNV, the host used was hamster. eThose samples with 100% genome covered were in a single contig except V6103 which was covered in two contigs. Those with less than 100% were covered in two contigs except D613 which was covered in three contigs. fFor HIV, the total number of genes is 9. For WNV and RSV, the total number of genes is 10.
By further analysing the read data, we identified several types of artifacts of the Ovation RNA-Seq system. First, the majority of reads (>95%) contained a partial (>9 bp) Ovation RNA-Seq SPIA (single primer isothermal amplification) primer, which often appears in the start of the read. Second, a substantial fraction (up to 25%) of the reads did not align to the viral or host references. With a fraction of the unaligned reads (50 000), we used MEGAN to identify the composition of unaligned reads for the HIV and WNV clones (Supplementary Figure S3). For the HIV clone, the read composition was: 12% bacterial, 3% eukaryotic, 4% retrovirus, 33% not assigned and 48% no hit. The retrovirus hits were unaligned HIV sequences. The majority of the not assigned reads were bacterial rRNA and HIV hits that could not be classified to specific taxonomical group. The no hit reads were mostly simple sequence repeats and SPIA primer sequences. The high abundance of these reads might be due to the low input amounts, which led to the greater abundance of these non-specific by-products. For the WNV clone, the read composition was: 0.5% bacterial, 3% eukaryotic, 0.5% flavivirus, 2% not assigned and 95% no hit. The eukaryotic hits were mostly rat and mouse. The flavivirus hits were unaligned WNV sequences. Some of the no hits for WNV clone were simple sequence repeats and SPIA primer, but the majority of the no hit reads for WNV could not be identified as similar to known sequences. It is possible that these reads could be hamster-specific sequences that were not aligned to the preliminary golden hamster assembly.
Complete coverage of viral coding region
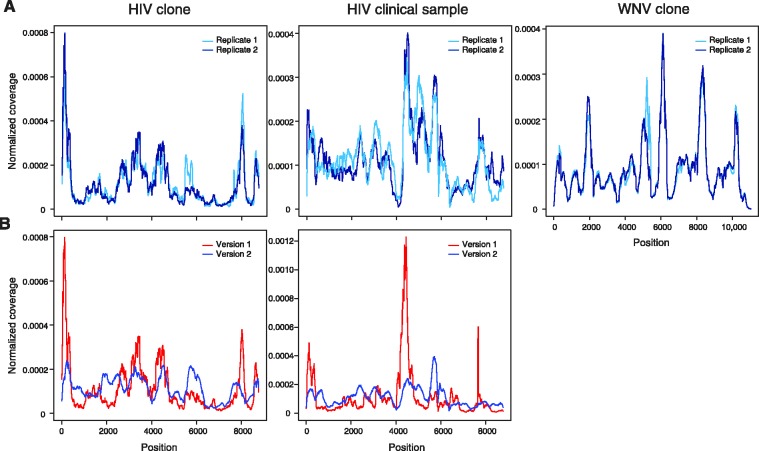
To determine whether our method captured the entire target region (CDS), reads were aligned to the CDS of the viral references using the Mosaik alignment tool. For both HIV clone and clinical samples, reproducible coverage of the entire CDS was obtained (Figure 1A). In all 13 cases, complete coverage was obtained (Supplementary Figure S1). As previous reports using this system utilized only the version 1 system (39), we compared the coverage pattern between version 1 and version 2 systems for HIV clone and clinical samples (Figure 1B). For both HIV clone and clinical samples, version 2 had more even coverage than version 1 (Figure 1B) based on a lower coefficient of variation (CV) (Supplementary Table S3). More even coverage is highly desirable for sequencing viral samples and other applications; therefore the version 2 system would be the preferred system.
Figure 1.
Complete sequence coverage of viral coding region. (A) Reproducibility of read coverage for technical replicates for HIV clone and clinical and WNV clone samples. (B) Comparison of read coverage for HIV clone and clinical samples between Ovation RNA-Seq version 1 (red) and version 2 (blue) systems. Reads were aligned to the CDS of the relevant viral reference using Mosaik. Coverage was computed as the total number of reads covering a given residue and was normalized by the total coverage summed across all residues; at each residue, the coverage was divided by the total coverage and sum of normalized coverage equals one.
Next, we assessed the read coverage for the serial dilution of the WNV clone relative to a WNV reference. Our method captured the entire WNV CDS with as little as 100 copies of input RNA (Figure 1A). In all 16 dilutions of WNV clone RNA, complete coverage was obtained (Supplementary Figure S1). In addition, the pattern of variable coverage was very reproducible for WNV clone with input amounts of ranging from 100 to 10 000 copies (Supplementary Figure S2).
Finally, we assessed the read coverage for two RSV clinical samples. Our method captured the entire RSV CDS for both samples (Supplementary Figure S1C). Similarly to HIV and WNV samples, the coverage is variable across the CDS.
De novo assembly of nearly complete HIV and WNV consensus genomes
Due to the challenges of large amounts of host contamination, highly variable coverage and the presence of SPIA primer found in our sequence data, we used a newly developed assembler, VICUNA, to generate de novo assemblies for HIV, RSV and WNV low input samples (Yang et al., manuscript submitted). VICUNA is designed to assemble genetically heterogeneous viral genomes in the presence of large amounts of host contamination. To enable a direct comparison among all our viral libraries, we randomly sampled 5 million reads for each sample. This choice reflects a realistic sequencing process in production and allowed us to obtain a high average coverage (e.g. over 1000-fold) of the target region as well as sufficient coverage across the genome, given that up to 99% of the reads are non-viral sequences and coverage is uneven. For each HIV clinical sample, we generated consensus sequence that covers over 96% of the target region (Table 1). In 7 out of 10 clinical samples, we generated consensus sequence that covers 100% of the target region (Supplementary Table S2). For WNV clone samples, we generated a consensus sequence that covers over 99% of the target region (Table 1). In 15 out of 16 WNV samples, we generated a consensus sequences that covers 100% of the target region (Supplementary Table S2). For both RSV clinical samples, we generated a consensus sequence that covers 100% of the target region (Table 1).
For selected samples from HIV, WNV and RSV, we compared the assembly statistics between three different assemblers: VICUNA, AV454 (10) and SOAPdenovo (40) (Supplementary Table S4). VICUNA outperformed the other assemblers by returning fewer and longer contigs that covered a larger part of the CDS. The percent of CDS covered by all contigs could not be calculated for SOAPdenovo due to the large number of short contigs which would require a second assembly in order to reconstitute a consensus sequence.
During the assembly process, we observed in several cases that by invoking the optional reference guided merging component in VICUNA after the initial assembly, we could improve the final consensus. The majority of the clinical samples were assembled in only one or two contigs prior to reference guided merging. A significant portion of the viral genome was assembled into a single large contig (Supplementary Table S4) without the need for reference guided merging, but the reference guided merging increased the coverage of the final CDS to >96% of the viral genome (Table 1 and Supplementary Table S2). In addition, upon further investigation, we discovered an artifact that produced chimeric fragments, which might be caused by stem loops of RNA secondary structure (Supplementary Figure S4). This artifact complicated the de novo assembly by increasing the number of contigs and required reference guided merging of contigs.
We next compared the consensus assembly between technical replicates for HIV clone and clinical samples (Table 2). First, we compared the nucleotide differences between the replicates (Assembly identity (%) in Table 2). For HIV clone replicates, the assemblies were 99.94% identical. For HIV clinical sample B, the assemblies were 99.26% and 98.55% identical for the 200 and 100 copy replicates, respectively. For WNV clone replicates, the assemblies were 99.98% and 100% identical for the 250 and 100 copy replicates, respectively. The discrepancies between assembled consensus sequences may be either caused by the differences in data generation or by the assembly process. To further evaluate the reproducibility of the method, we examined the nucleotide composition differences (Table 2) between any two assemblies. At each mismatch residue, the consensus base in the first assembly was considered to be consistent with the consensus base in the second assembly if the mismatch was supported by read alignments in the second assembly (Composition mismatches in Table 2). For HIV clone replicates, the assemblies were 99.98% identical when we include read support for any differences in the assembly. For HIV clinical sample B, the assemblies were 99.79% and 99.73% identical for the 200 and 100 copy replicates, respectively. For WNV clone replicates, the assemblies were 100% identical for both 250 and 100 copy replicates. Thus assemblies of both HIV and WNV consensus sequences are very reproducible.
Table 2.
Comparison of HIV and WNV technical replicate assemblies
| Samples | Aligned bases in CDS | Assembly identitya (%) | Composition mismatchesb | Indel events | Indel bases (composition)c | Composition identity (%) |
|---|---|---|---|---|---|---|
| HIV clinical sample B (100 copies) | 8 379 | 98.90 | 18 | 2 | 18 (12–6) | 99.76 |
| HIV clinical sample B (200 copies) | 8 523 | 99.26 | 15 | 3 | 9 (3–3–3) | 99.79 |
| HIV clone (NL4-3) | 8 478 | 99.94 | 0 | 2 | 5 (4–1) | 99.98 |
| WNV clone (100 copies) | 10 303 | 100.00 | 0 | 0 | 100.00 | |
| WNV clone (250 copies) | 10 303 | 99.98 | 0 | 0 | 100.00 |
aNucleotide % identity between assemblies of replicates. bNumber of mismatches between assemblies that were not supported by read data (see Materials and Methods). cNumber outside the parentheses is the total number of indel bases. Number within the parentheses refers to size of individual indels with a hyphen between each occurrence.
For point of comparison with our newly developed methods, we attempted to generate RT-PCR amplicons for HIV clone and clinical samples A and B and sequence them by 454 using previously described methods (10). For clinical sample B, we were not able to generate all four-overlapping PCR amplicons spanning the CDS after four attempts. With our newly developed methods, this sample was successful in all six attempts to generate consensus assembly covering >98% of the CDS (Supplementary Table S1). We did generate all four-overlapping PCR amplicons, sequence by 454 and built consensus assemblies for HIV clone and clinical sample A. For HIV clone, assemblies were 100% identical between the two methods (Table 3). For clinical sample A, the assemblies were 98.25 and 99.41 % identical at the nucleotide and composition levels, respectively (Table 3). Our newly developed methods generated assemblies for HIV clone and clinical samples that were highly similar to those generated using standard RT-PCR methods.
Table 3.
Comparison between Ovation RNA-Seq-Illumina and RT-PCR-454 assemblies
| Samples | Sample ID | Aligned bases | Assembly identitya (%) | Composition mismatchesb | Indel events | Indel bases (composition)c | Composition identity (%) |
|---|---|---|---|---|---|---|---|
| Clinical sample A | D614 | 8 642 | 98.25 | 45 | 6 | 24 (3–3–3–3–6–6) | 99.41 |
| HIV clone (NL4-3) | D618 | 8 627 | 100.00 | 0 | 0 | 0 | 100.00 |
aNucleotide % identity between Ovation RNA-Seq-Illumina and RT-PCR-454 assemblies. bNumber of mismatches between assemblies that were not supported by read data (see Materials and Methods). cNumber outside the parentheses was the total number of indel bases. Number within the parentheses refers to size of individual indels with a hyphen between each occurrence.
DISCUSSION
In this study, we used a sequence-independent amplification method, coupled with Illumina sequencing to generate full genome assemblies for HIV, RSV and WNV samples with as little as 100 viral RNA genomes. Previous studies using Ovation RNA-Seq with HIV viral RNA used ex vivo amplification, large amounts of viral RNA, and utilized a reference-based assembly approach (39). For several reasons, these conditions are not ideal for sequencing from clinical samples. First, most clinical samples contain a few picograms or less of viral RNA. In our study, we were able to study viral samples with femtogram and attogram amounts of viral RNA (over 1 million times less material than had been previously used) as input to the Ovation RNA-Seq system and generate sufficient dsDNA for Illumina sequencing. Second, ex vivo amplification is laborious and could lead to bias in sample generation during the 9 days in culture. We were able to capture complete sequence coverage of the CDS directly from clinical samples therefore generating sequence data more representative of the dominant viral genome within the infected individual. Third, we utilized a recently developed de novo genome assembly algorithm, VICUNA, to assemble a full-length consensus (Yang et al., manuscript submitted). Previous studies have shown (10,41) that de novo assembly can significantly improve the accuracy of assembly and utilization of data compared with reference-based assembly approaches. In our study, VICUNA outperformed alternate assemblers (Supplementary Table S4) by returning the fewest and longest contigs.
Our method, like other sequence-independent methods (25), generates highly variable coverage across the viral genome. RNA viruses can have a large number of functional RNA secondary structures (42,43) which can result in stalling of cDNA synthesis (44). This uneven coverage may be due to viral secondary structure. The VICUNA assembler is able to work with this highly variable coverage to generate full-length consensus assemblies for the HIV, RSV and WNV samples.
In our study, we were not able to use the Ovation RNA-Seq method to quantify the amount of viral RNA amounts in our clinical samples. For RSV and HIV clinical sample, the clinical samples with highest input viral RNA amounts had the highest percent of reads aligning to viral reference (Supplementary Table S1), but there was no correlation between input RNA and viral sequence coverage. This method may provide approximation of viral amounts in clinical sample, but it is not quantitative.
A major disadvantage for sequence-independent amplification of viral RNA samples is the large amount of host contamination that is amplified using these methods. We tested several different RNA sequence-independent amplification kits to evaluate their utility with viral clinical samples (unpublished results). The TransPlex Whole Transcriptome Amplification (WTA1—Sigma Aldrich, St. Louis, MO), Complete TransPlex Whole Transcriptome Amplification (WTA2—Sigma Aldrich), and SMARTer cDNA synthesis (Clontech, Mountain View, CA) kits produced 10-fold fewer reads aligning to the HIV genome for clinical samples compared with the Ovation RNA-Seq system (C. Malboeuf, unpublished data). The Ovation RNA-Seq system performed best, in part, due to it having the lowest percent of reads aligning to host contamination, particularly rRNA, which comprises >80% of total RNA. In our Ovation RNA-Seq experiments, <37% of the total reads from HIV and RSV clinical samples aligned to rRNA (Table 1). This significant reduction in amplification of rRNA may have led to increased amplification of viral RNA thus allowing us to utilize only 5 million reads to generate full-length consensus assembles with >400-fold average coverage.
Our method has several advantages over standard RT-PCR based methods. It does not require prior viral sequence knowledge for primer design enabling the study of viruses with limited sequence data. The lack of viral-specific primers allows for identification of viral recombinants that might not be found with standard RT-PCR methods. Our method is capable of amplifying viral RNA genomes that are not successful by traditional RT-PCR methods. For HIV clinical sample B, four RT-PCR attempts were not successful in capturing the CDS, but in all six attempts with our method 97–100% of the CDS was captured (Table 1). Furthermore, this method is easily applicable to multiple sample types. We applied the method to HIV, RSV and WNV samples without the need for optimization. Developing a robust RT-PCR process with viral specific primers can require a significant effort, up to several months (unpublished results), so that our method potentially speeds up viral genome sequencing projects. This method works with a highly variable virus such as HIV—capturing the entire CDS of the genome starting from only 100 viral genomes (Table 1). In addition, the process from viral RNA to dsDNA requires only 4.5 hours. Furthermore if this method were coupled with Nextera Illumina library construction (45), one could generate an Illumina library in less than a single day. Although the Nextera method is very fast, the transposase-based method of Illumina library construction may introduce GC-bias compared with standard methods (46). As only 5 million reads were necessary to assemble complete viral genomes, MiSeq or Ion Torrent sequencing (47) could be used. This would allow a researcher to go from sample to sequence data in less than a week. Finally, the Ovation RNA-Seq method is available in automated format allowing for high-throughput processing of clinical samples. As only 5 million reads are needed per sample, one could pool up to 96 samples per lane of HiSeq2000.
The methods described here have several promising applications. The sequence-independent amplification methods coupled with our de novo assembly of viral genomes provides a new methodology for sequencing viral genomes from many different sample types. These methods can generate next-generation sequencing data of pathogens allowing for greater insight into pathogen evolution, fitness, emergence and transmission as well as factors that are important for the development of new therapeutics and vaccines. In addition, these methods could be used to discover known or unknown pathogens in clinical samples, not limited to viruses. Likewise, these methods could also be used for surveillance to detect viral RNA from environmental sources, such as WNV in mosquito samples. This surveillance ability could provide valuable insight into what pathogens are circulating and provide an “early warning” of future outbreaks (48). Additionally, one would be able to identify and study multiple viruses from a single sample. This could be beneficial in understanding the dynamics of viral co-infection, such as HIV/HCV. Furthermore, these methods could be applied to parasite, bacterial and fungal pathogen discovery by sequencing total RNA that contains expressed pathogen RNA. Expressed pathogens could be discovered incidentally through RNA-Seq data. Additionally, it may be more cost effective for some projects to sequence RNA rather than DNA to detect pathogens because greater sequencing depth may be needed for DNA than RNA (24). Morin et al. have demonstrated the capability of detecting expressed pathogen RNA through RNA sequencing (49).
In summary, we demonstrate that our method is reproducible and robust for generating full length genomes from challenging ultra-low copy viral RNA samples. We demonstrate success with HIV, RSV and WNV samples, but these methods can be applied to other viral samples with various viral RNA amounts and host contamination.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4 and Supplementary Figures 1–4.
FUNDING
National Institute of Allergy and Infectious Disease, the National Institutes of Health and the Department of Health and Human Services [contracts HHSN272200900018C and HHSN272200900006C], and grant P01-AI074415 (to T.M.A.), and the Bill and Melinda Gates Foundation (to T.M.A. and M.R.H.). Funding for open access charge: NIAID [contract no. HHSN27220090018C].
Conflict of intertest statement. After the development of the lab method and initial sequencing results were obtained in April, 2011, Joshua Levin participated in a one day Medicinal/Clinical Sequencing Technology Advancement Summit at NuGEN and was compensated for his time and travel expenses.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Andreas Gnirke, Bruce Birren and Chad Nusbaum for support and advice; the Broad Institute Genomics Platform and Genome Sequencing and Analysis Program, Federica Di Palma and Kerstin Lindblad-Toh for making the data for Mesocricetus auratus genome available; Rachel Erlich, Niall Lennon, Ruchi Newman and Elizabeth Ryan for useful discussion; Ryan Murphy for help in providing the RSV clinical samples; Karen Power for help in providing HIV clinical samples; Fernando Viloria, Lizz Gottardi and Tiffany Poon for project sample management; the Broad Sample Repository and Sequencing platforms for assistance in sequencing all samples; and Leslie Gaffney for assistance with preparation of the figures.
REFERENCES
- 1.Bimber BN, Dudley DM, Lauck M, Becker EA, Chin EN, Lank SM, Grunenwald HL, Caruccio NC, Maffitt M, Wilson NA, et al. Whole-genome characterization of human and simian immunodeficiency virus intrahost diversity by ultradeep pyrosequencing. J. Virol. 2010;84:12087–12092. doi: 10.1128/JVI.01378-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fischer W, Ganusov VV, Giorgi EE, Hraber PT, Keele BF, Leitner T, Han CS, Gleasner CD, Green L, Lo CC, et al. Transmission of single HIV-1 genomes and dynamics of early immune escape revealed by ultra-deep sequencing. PLoS One. 2010;5:e12303. doi: 10.1371/journal.pone.0012303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann C, Minkah N, Leipzig J, Wang G, Arens MQ, Tebas P, Bushman FD. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 2007;35:e91. doi: 10.1093/nar/gkm435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simen BB, Simons JF, Hullsiek KH, Novak RM, Macarthur RD, Baxter JD, Huang C, Lubeski C, Turenchalk GS, Braverman MS, et al. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J. Infect. Dis. 2009;199:693–701. doi: 10.1086/596736. [DOI] [PubMed] [Google Scholar]
- 5.Tsibris AM, Korber B, Arnaout R, Russ C, Lo CC, Leitner T, Gaschen B, Theiler J, Paredes R, Su Z, et al. Quantitative deep sequencing reveals dynamic HIV-1 escape and large population shifts during CCR5 antagonist therapy in vivo. PLoS One. 2009;4:e5683. doi: 10.1371/journal.pone.0005683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang C, Mitsuya Y, Gharizadeh B, Ronaghi M, Shafer RW. Characterization of mutation spectra with ultra-deep pyrosequencing: application to HIV-1 drug resistance. Genome Res. 2007;17:1195–1201. doi: 10.1101/gr.6468307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang GP, Sherrill-Mix SA, Chang KM, Quince C, Bushman FD. Hepatitis C virus transmission bottlenecks analyzed by deep sequencing. J. Virol. 2010;84:6218–6228. doi: 10.1128/JVI.02271-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lauck M, Alvarado-Mora MV, Becker EA, Bhattacharya D, Striker R, Hughes AL, Carrilho FJ, O'Connor DH, Pinho JR. Analysis of hepatitis C virus intrahost diversity across the coding region by ultradeep pyrosequencing. J. Virol. 2012;86:3952–3960. doi: 10.1128/JVI.06627-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pesko KN, Fitzpatrick KA, Ryan EM, Shi PY, Zhang B, Lennon NJ, Newman RM, Henn MR, Ebel GD. Internally deleted WNV genomes isolated from exotic birds in New Mexico: function in cells, mosquitoes, and mice. Virology. 2012;427:10–17. doi: 10.1016/j.virol.2012.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henn MR, Boutwell CL, Charlebois P, Lennon NJ, Power KA, Macalalad AR, Berlin AM, Malboeuf CM, Ryan EM, Gnerre S, et al. Whole genome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathog. 2012;8:e1002529. doi: 10.1371/journal.ppat.1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parameswaran P, Charlebois P, Tellez Y, Nunez A, Ryan EM, Malboeuf CM, Levin JZ, Lennon NJ, Balmaseda A, Harris E, et al. Genome-wide patterns of intra-human dengue virus diversity reveal associations with viral phylogenetic clade and inter-host diversity. J Virol. 2012;86:8546–8558. doi: 10.1128/JVI.00736-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cordey S, Junier T, Gerlach D, Gobbini F, Farinelli L, Zdobnov EM, Winther B, Tapparel C, Kaiser L. Rhinovirus genome evolution during experimental human infection. PLoS One. 2010;5:e10588. doi: 10.1371/journal.pone.0010588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckerle LD, Becker MM, Halpin RA, Li K, Venter E, Lu X, Scherbakova S, Graham RL, Baric RS, Stockwell TB, et al. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6:e1000896. doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright CF, Morelli MJ, Thebaud G, Knowles NJ, Herzyk P, Paton DJ, Haydon DT, King DP. Beyond the consensus: dissecting within-host viral population diversity of foot-and-mouth disease virus using next-generation genome sequencing. J. Virol. 2010;85:2266–2275. doi: 10.1128/JVI.01396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miura T, Brockman MA, Brumme CJ, Brumme ZL, Carlson JM, Pereyra F, Trocha A, Addo MM, Block BL, Rothchild AC, et al. Genetic characterization of human immunodeficiency virus type 1 in elite controllers: lack of gross genetic defects or common amino acid changes. J. Virol. 2008;82:8422–8430. doi: 10.1128/JVI.00535-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmer S, Kearney M, Maldarelli F, Halvas EK, Bixby CJ, Bazmi H, Rock D, Falloon J, Davey RT, Jr, Dewar RL, et al. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 2005;43:406–413. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, Derdeyn CA, Farmer P, Hunter E, Allen S, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deeks SG, Walker BD. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity. 2007;27:406–416. doi: 10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 19.Tricou V, Minh NN, Farrar J, Tran HT, Simmons CP. Kinetics of viremia and NS1 antigenemia are shaped by immune status and virus serotype in adults with dengue. PLoS Negl. Trop. Dis. 2011;5:e1309. doi: 10.1371/journal.pntd.0001309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu W, Unnasch TR, Katholi CR, Lampman R, Novak RJ. Fundamental issues in mosquito surveillance for arboviral transmission. Trans. R. Soc. Trop. Med. Hyg. 2008;102:817–822. doi: 10.1016/j.trstmh.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheval J, Sauvage V, Frangeul L, Dacheux L, Guigon G, Dumey N, Pariente K, Rousseaux C, Dorange F, Berthet N, et al. Evaluation of high-throughput sequencing for identifying known and unknown viruses in biological samples. J. Clin. Microbiol. 2011;49:3268–3275. doi: 10.1128/JCM.00850-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Djikeng A, Halpin R, Kuzmickas R, Depasse J, Feldblyum J, Sengamalay N, Afonso C, Zhang X, Anderson NG, Ghedin E, et al. Viral genome sequencing by random priming methods. BMC Genomics. 2008;9:5. doi: 10.1186/1471-2164-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ninomiya M, Ueno Y, Funayama R, Nagashima T, Nishida Y, Kondo Y, Inoue J, Kakazu E, Kimura O, Nakayama K, et al. Use of illumina deep sequencing technology to differentiate hepatitis C virus variants. J. Clin. Microbiol. 2012;50:857–866. doi: 10.1128/JCM.05715-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore RA, Warren RL, Freeman JD, Gustavsen JA, Chenard C, Friedman JM, Suttle CA, Zhao Y, Holt RA. The sensitivity of massively parallel sequencing for detecting candidate infectious agents associated with human tissue. PLoS One. 2011;6:e19838. doi: 10.1371/journal.pone.0019838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Victoria JG, Kapoor A, Li L, Blinkova O, Slikas B, Wang C, Naeem A, Zaidi S, Delwart E. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J. Virol. 2009;83:4642–4651. doi: 10.1128/JVI.02301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beane J, Vick J, Schembri F, Anderlind C, Gower A, Campbell J, Luo L, Zhang XH, Xiao J, Alekseyev YO, et al. Characterizing the impact of smoking and lung cancer on the airway transcriptome using RNA-Seq. Cancer Prev. Res. (Phila) 2011;4:803–817. doi: 10.1158/1940-6207.CAPR-11-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tariq MA, Kim HJ, Jejelowo O, Pourmand N. Whole-transcriptome RNAseq analysis from minute amount of total RNA. Nucleic Acids Res. 2011;39:e120. doi: 10.1093/nar/gkr547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi PY, Tilgner M, Lo MK, Kent KA, Bernard KA. Infectious cDNA clone of the epidemic west nile virus from New York City. J. Virol. 2002;76:5847–5856. doi: 10.1128/JVI.76.12.5847-5856.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devincenzo JP. Natural infection of infants with respiratory syncytial virus subgroups A and B: a study of frequency, disease severity, and viral load. Pediatr. Res. 2004;56:914–917. doi: 10.1203/01.PDR.0000145255.86117.6A. [DOI] [PubMed] [Google Scholar]
- 31.Christopherson C, Sninsky J, Kwok S. The effects of internal primer-template mismatches on RT-PCR: HIV-1 model studies. Nucleic Acids Res. 1997;25:654–658. doi: 10.1093/nar/25.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kauffman EB, Jones SA, Dupuis AP, 2nd, Ngo KA, Bernard KA, Kramer LD. Virus detection protocols for west nile virus in vertebrate and mosquito specimens. J. Clin. Microbiol. 2003;41:3661–3667. doi: 10.1128/JCM.41.8.3661-3667.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perkins SM, Webb DL, Torrance SA, El Saleeby C, Harrison LM, Aitken JA, Patel A, DeVincenzo JP. Comparison of a real-time reverse transcriptase PCR assay and a culture technique for quantitative assessment of viral load in children naturally infected with respiratory syncytial virus. J. Clin. Microbiol. 2005;43:2356–2362. doi: 10.1128/JCM.43.5.2356-2362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurn N, Chen P, Heath JD, Kopf-Sill A, Stephens KM, Wang S. Novel isothermal, linear nucleic acid amplification systems for highly multiplexed applications. Clin. Chem. 2005;51:1973–1981. doi: 10.1373/clinchem.2005.053694. [DOI] [PubMed] [Google Scholar]
- 35.Levin JZ, Yassour M, Adiconis X, Nusbaum C, Thompson DA, Friedman N, Gnirke A, Regev A. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods. 2010;7:709–715. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 38.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willerth SM, Pedro HA, Pachter L, Humeau LM, Arkin AP, Schaffer DV. Development of a low bias method for characterizing viral populations using next generation sequencing technology. PLoS One. 2010;5:e13564. doi: 10.1371/journal.pone.0013564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Archer J, Rambaut A, Taillon BE, Harrigan PR, Lewis M, Robertson DL. The evolutionary analysis of emerging low frequency HIV-1 CXCR4 using variants through time—an ultra-deep approach. PLoS Comput. Biol. 2010;6:e1001022. doi: 10.1371/journal.pcbi.1001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hofacker IL, Stadler PF, Stocsits RR. Conserved RNA secondary structures in viral genomes: a survey. Bioinformatics. 2004;20:1495–1499. doi: 10.1093/bioinformatics/bth108. [DOI] [PubMed] [Google Scholar]
- 44.Harrison GP, Mayo MS, Hunter E, Lever AM. Pausing of reverse transcriptase on retroviral RNA templates is influenced by secondary structures both 5' and 3' of the catalytic site. Nucleic Acids Res. 1998;26:3433–3442. doi: 10.1093/nar/26.14.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adey A, Morrison HG, Asan, Xun X, Kitzman JO, Turner EH, Stackhouse B, MacKenzie AP, Caruccio NC, Zhang X, et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marine R, Polson SW, Ravel J, Hatfull G, Russell D, Sullivan M, Syed F, Dumas M, Wommack KE. Evaluation of a transposase protocol for rapid generation of shotgun high-throughput sequencing libraries from nanogram quantities of DNA. Appl. Environ. Microbiol. 2011;77:8071–8079. doi: 10.1128/AEM.05610-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.ten Bosch JR, Deignan JL, Grody WW. Next-generation Sequencing in Clinical Molecular Diagnostics. New York: John Wiley; 2011. [Google Scholar]
- 48.Dong J, Olano JP, McBride JW, Walker DH. Emerging pathogens: challenges and successes of molecular diagnostics. J. Mol. Diagn. 2008;10:185–197. doi: 10.2353/jmoldx.2008.070063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morin R, Bainbridge M, Fejes A, Hirst M, Krzywinski M, Pugh T, McDonald H, Varhol R, Jones S, Marra M. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Biotechniques. 2008;45:81–94. doi: 10.2144/000112900. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.