Abstract
Mitochondrial mutations, an important cause of incurable human neuromuscular diseases, are mostly heteroplasmic: mutated mitochondrial DNA is present in cells simultaneously with wild-type genomes, the pathogenic threshold being generally >70% of mutant mtDNA. We studied whether heteroplasmy level could be decreased by specifically designed oligoribonucleotides, targeted into mitochondria by the pathway delivering RNA molecules in vivo. Using mitochondrially imported RNAs as vectors, we demonstrated that oligoribonucleotides complementary to mutant mtDNA region can specifically reduce the proportion of mtDNA bearing a large deletion associated with the Kearns Sayre Syndrome in cultured transmitochondrial cybrid cells. These findings may be relevant to developing of a new tool for therapy of mtDNA associated diseases.
INTRODUCTION
Mitochondria are responsible for many cellular processes ranging from respiration to apoptosis, and their dysfunctions have severe consequences. Mammalian mitochondrial DNA (mtDNA) is a closed circular molecule of 16.5 kb, encoding 13 core proteins that contribute to oxidative phosphorylation, 2 ribosomal RNAs (rRNAs) and 22 transfer RNAs (tRNAs) needed for the biosynthesis of these 13 peptides in the mitochondrial matrix (Figure 1A). More than 200 human neuromuscular incurable diseases were associated with mtDNA mutations (1). One of the striking particularities of the majority of these pathologies is the simultaneous presence of mutant and wild-type (WT) mitochondrial genomes in the same cell, the heteroplasmy (2). Two approaches were proposed to address mtDNA-associated pathologies by gene therapy. In some cases, defects may be rescued by targeting into mitochondria nuclear DNA-expressed counterparts of the affected molecules, approach called ‘allotopic’ strategy (3–7). Reducing the level of heteroplasmy, ‘antigenomic’ strategy, is another possibility. To this last goal, several approaches were tested. One example is mitochondrial targeting of restriction endonucleases cleaving mutated mtDNA molecules (8–10). Another one was to specifically design synthetic peptide nucleic acids (PNA) molecules inhibiting mutant mtDNA replication. They revealed to be efficient in vitro but not able to cross the inner mitochondrial membrane and, as a result, not capable of interacting with mtDNA in vivo (11–14).
Figure 1.
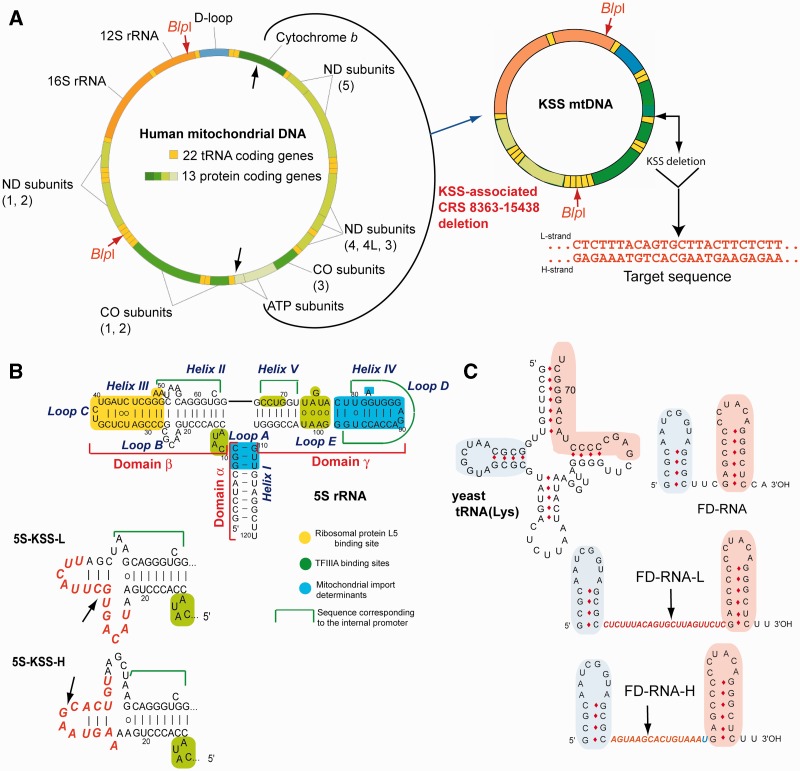
KSS deletion in human mtDNA and anti-genomic RNAs design. (A) Genetic maps of wild-type and mutant KSS mtDNAs. Target sequence, the sequence of deletion boundaries is shown in red. (B) Secondary structure of human 5S rRNA and its functional elements. Below, secondary structures of recombinant 5S rRNA β-domains (inserts in red). The arrows show the deletion point. (C) The structures of yeast tRNALys, short synthetic FD-RNA and two recombinant RNAs containing inserts corresponding to L- and H-strands of KSS-mtDNA (in red). Two domains derived from the tRNA (32) are shown in pink and blue. The arrows show the deletion boundaries point.
We have exploited RNA mitochondrial import pathway to target into human mitochondria RNAs able to hybridize with the mutated mtDNA and to inhibit selectively its replication. Import of nuclear DNA-encoded RNAs into mitochondria is a widely spread phenomenon; however, the types, number of imported RNAs and targeting mechanisms vary among species (15–17). Mammalian mitochondria were reported to import several types of small non-coding RNAs (18), including several microRNAs (19–21), some tRNAs (in natural or artificial manner) (22,23), RNA components of RNase P and MRP endonuclease (24,25) and 5S rRNA (26–28). Recently, we demonstrated that 5S rRNA import mechanism can rely on protein import factors identified as the mitochondrial enzyme rhodanese and the precursor of mitochondrial ribosomal protein MRP-L18 (29,30), interacting with two structural motifs of the 5S rRNA molecule (Figure 1B), whereas the third motif, so called β-domain, may be either deleted or replaced without loss of import capacity (31). Additionally, analysis of artificial import of yeast tRNAs derivatives into human mitochondria (26) permitted us to design short synthetic RNAs (referred to as FD-RNA) comprising two domains of the tRNA (Figure 1C) and characterized by a high efficiency of mitochondrial import (32). Here, we show that these two types of molecules can serve as vectors to deliver into mitochondria oligoribonucleotides with a therapeutic potential.
MATERIALS AND METHODS
Clinical case
Patient T was diagnosed with a Kearns Sayre Shy syndrome at the age of 15 years. He was the second child of non-consanguinous parents and had a healthy brother. He first came to medical attention at the age of 9 years for progressive left ptosis, ophthalmoplegia, mild deafness and walking difficulties. He was hospitalized at the age of 15 years. Physical examination disclosed mild ataxia and muscle weakness with amyotrophy. Brain imaging showed white matter hypersignals. Biological analyses showed high CK at 10 times normal values, high protein (2.65 g/l) and lactate (5.6 mM) levels in CSF and high lactate (2.7 mM) in blood. Histological examination of deltoid muscle biopsy showed numerous fibers with important mitochondrial proliferation (‘ragged red fibres’) and completely defective cytochrome c oxidase histochemical reaction. Spectrophotometric assays of muscle mitochondrial activities showed high values of both citrate synthase and succinate dehydrogenase activities (above the 95th centile of normal values) reflecting the muscle mitochondrial proliferation as well as low values (below or at the 5th centile of normal values) for cytochrome c oxidase (complex IV) activity and for the ratios of complex I, III, or IV activity to either citrate synthase or complex II. Southern blot analysis of total DNA from muscle showed the presence of a large size deletion involving 72% of the mtDNA molecules. Direct sequencing of the patient's mtDNA showed that the deletion spanned from nucleotide 8363 to 15 438, thus removing 7075 base pairs including 9 structural genes (MT-ATP8, MT-ATP6, MT-CO3, MT-ND3, MT-ND4L, MT-ND4, MT-ND5, MT-ND6 and part of MT-CYB) and 6 tRNA genes (MT-TG, MT-TR, MT-TH, MT-TS2, MT-TL2 and MT-TE).
Transmitochondrial cybrid cell line construction
Cultured skin fibroblasts were derived from the patient’s skin biopsy according to routine procedure. After their first passage, they were enucleated and fused to a human rho0 osteosarcoma cell line (143B) kindly provided by Prof. E. Shoubridge, Montreal Institute of Neurology, and cloned by limit dilution as reported (33). Of more than 80 screened clones, only two had the mtDNA deletion, involving 65% mtDNA molecules in one case and 20% in the second case. Cybrid cells containing 65% mutant mtDNA used in this study were characterized by the 10% ± 2% decrease of oxygen consumption comparing to control 143B cell line.
Plasmids and recombinant RNAs
For replication in vitro, two human mtDNA fragments were cloned in pUC119, 430 bp fragment (nucleotides 15 251–15 680 of mtDNA) referred to as WT-DNA and 512 bp fragment (nucleotides 8099–8365/15 438–15 680 of mutant mtDNA) comprising the fusion of the two boundaries of the Kearns Sayre Syndrome (KSS) deletion, referred to as KSS-DNA. Cloning was performed by polymerase chain reaction (PCR) with primers listed in the Supplementary Table S1. For the real-time PCR calibration curves, two plasmids containing the 12S rRNA region not touched by the KSS deletion (nucleotides 109–1714 of mtDNA) and the deleted region (nucleotides 10 739–11 839 of mtDNA) were constructed using pUC119 vector.
All recombinant 5S rRNA versions' genes were cloned into pUC119 as described (31). Recombinant RNAs were obtained by T7 transcription using the T7 RiboMAX Express Large Scale RNA Production System (Promega) on the plasmid DNA or the corresponding PCR products.
For synthesis of fluorescently labeled RNA transcript, Alexa Fluor 488-5-UTP (Molecular Probes) was incorporated in RNA during 2 h T7-transcription by T7-RNA polymerase (Promega). Reaction mixture of total volume 20 μl contained 0.5 μg of DNA template, 80 u of T7-RNA polymerase (2 additions of 40 u each), 0.5 mM of ATP, 0.5 mM CTP, 0.5 mM GTP, 0.37 mM UTP, 0.125 mM Alexa Fluor 488-5-UTP, 10 mM DTT and 40 u of RNaseOUT (Invitrogen). T7-transcript was purified by PAGE. To check the incorporation of the label in purified transcript, we compared the dye absorbance at 492 nm with the absorbance at 260 nm using NanoDrop Microarray Program. The efficiency of labeling obtained was approximately five labeled UTP per one RNA molecule.
For in vivo expression, 5S rRNA gene versions were PCR cloned either in pBK-CMV phagemid vector (Stratagene) or in pcDNA3neo/zeo vector (Invitrogen), as described (31).
Mitochondria isolation
Mitochondria from beef liver were isolated as described in (26,34). Mitochondria from cultured HepG2 or cybrid human cells were isolated as described in (34) by several rounds of high (20 000g) and low (4000g) centrifugations. After the second round of high-speed centrifugation of mitochondria, they were re-suspended in the buffer [0.44 M mannitol, 20 mM Tris–HCl (pH 7.0), 20 mM NaCl and 1 mM EDTA] and centrifuged through two layers of sucrose (0.5 M and 1.5 M). Mitochondria were collected on the top of the 1.5 M layer and harvested by high-speed centrifugation. The integrity of mitochondria was checked by a citrate synthase assay (35).
In vitro import
To isolate proteins directing import in vitro (IDP), HepG2 cells were harvested in phosphate-buffered saline (PBS) containing 1 mM EDTA, washed with PBS, suspended in NPMD buffer [20 mM Na-Phosphate buffer, pH 6.5 (alternatively replaced by Tris–HCl, pH 7.5), 150 mM NaCl, 1 mM MgCl2, 5 mM DTT] containing the cocktail of protease inhibitors (Boehringer-Mannheim) and disrupted by sonication. Cellular debris was removed by centrifugation, nucleic acids were removed by polyethylenimine treatment (36), proteins were precipitated by ammonium sulfate (80% of saturation) and dialyzed against the NPMD buffer. Import conditions were as described in (26,31) with several modifications. The standard assay (100 µl) contained isolated mitochondria (100 µg of mitochondrial protein), 3 pmoles of 5′-end 32P-labelled RNA and 10 µg of IDPs in the ‘import buffer’ [0.44M mannitol, 20 mM HEPES-KOH (pH 6.8), 50 mM KCl, 2.5 mM MgCl2, 1 mM ATP, 5mM DTT, 0.5 mM PMSF, 0.1 mM DIFP, 0.5 mM, phosphoenol pyruvate and 1 unit of pyruvate kinase]. The import assay was carried out at 30°C for 20 min (37). After treatment with a mixture of nucleases (20 u/ml of micrococcal nuclease, 15 µg/ml of RNase A and 25 u/ml of phosphodiesterase in 2 × concentrated stock), mitoplasts were generated by treatment with digitonin, at 100 µg per mg of mitochondrial protein. After incubation for 15 min at 25°C, mitoplasts were harvested by centrifugation and washed twice as described (38). Mitoplasts were then lysed in 1% SDS, 0.1 M sodium acetate (pH 4.8) and 0.05% diethyl pyrocarbonate at 100°C, and mtRNA was phenol extracted. RNAs were analyzed by denaturating gel electrophoresis, and import was quantified using PhosphorImager (Typhoon-Trio, GE Healthcare).
Isolation of protein fraction containing replication enzymes from beef liver mitochondria
Partial purification of mammalian mitochondrial replication enzymes was performed essentially as described in (39,40). One gram of mitochondria (mitochondrial protein) was lysed in 20 mM Tris–HCl, pH 8.0, 0.1 M NaCl, 20 mM EDTA, 10% Glycerol, 14 mM 2-mercaptoethanol, 1% Triton X-100 (v/v), 1 mM PMSF and 0.1 mM DIFP for 15 min at 4°C. After removing debris (40 000g, 1 h), the supernatant was diluted 1.5 times with 20 mM Tris–HCl, pH 8.0, 20 mM EDTA, 10% Glycerol, 14 mM 2-mercaptoethanol and 1 mM PMSF and loaded on DE52 cellulose column equilibrated with the same buffer with 70 mM NaCl. The main peak eluted at 0.3 M NaCl characterized by maximal activity was used in all analysis. Activity of the DNA polymerase and/or primase was checked by assaying 1–2 µl aliquots of the protein fraction in the presence of single-stranded DNA, [α-32P]-dCTP and either presence or absence of random primers (pd(N6) (Promega) in 20 mM Tris–HCl, pH 7.5, 20 mM MgCl2, 75 mM KCl, 0.1 mg/ml of BSA, 1 mM 2-mercaptoethanol, 100 µM of each dATP, dGTP and dTTP. The assay was performed at 37°C for 1 h, the 10% TCA-precipitated material fixed on a membrane and radioactivity measured in scintillation counter. Incorporation of [α-32P]-dCTP into nascent DNA was maximal (0.3 pmoles/μg) in the presence of random primers and 3-fold decreased in the absence of primers, indicating that eluted protein fraction contained mitochondrial enzymes capable of priming and elongating nascent DNA molecules.
In vitro replication assay
Double-stranded replication template was prepared by PCR on the cloned mtDNA fragment, to obtain 430 bp fragment (nucleotides 15 251–15 680 of mtDNA) referred to as WT-DNA and 512 bp fragment (nucleotides 8099–8365/15 438–15 680 of mutant mtDNA) referred to as KSS-DNA. To obtain single-stranded templates, we used the method described in (11). Mainly, the same PCRs were performed, but one of the primers was biotinylated (Supplementary Table S1). The PCR products were then fixed on Streptavidin-agarose beads (Fluka), the dsDNA denatured in NaOH (0.1 M) and the non-biotinylated strand was eluted and ethanol precipitated.
For the replication assay, 100 ng of dsDNA or ssDNA was mixed in final volume of 10 µl with [32P]-5′-labeled specific primer (designed in the way to generate full-size replication products of similar size for both WT and KSS mtDNA fragments) in 10 mM Tris–HCl, pH 7.5, 1 mM EDTA (TE), heated to 90°C and slowly cooled to 37°C. Then, the mixture was adjusted to 20 mM Tris–HCl, pH 7.5, 20 mM MgCl2, 75 mM KCl, 0.1 mg/ml of BSA, 1 mM 2-mercaptoethanol, 100 µM of each dNTP and 0.5 µl of mitochondrial protein fraction was added. The reaction was carried out in the presence of variable amounts of recombinant RNAs at 37°C for 1 h. Replication was stopped by addition of 10 µl of 0.2% SDS and 20 µl of TE-saturated phenol, nucleic acids were ethanol-precipitated and analyzed by 6.5% denaturing PAGE followed by phosphor imaging in Typhoon-Trio (GE Healthcare).
Transient and stable transfection of cybrid cells
Transient transfection with synthetic RNAs was performed as described in (31,41), with minor modification: for 2-cm2 well of 80% confluent cells, we used 0.25 µg RNA. Transfections were performed with Lipofectamin-2000 in OptiMEM medium (Invitrogen), as proposed in the supplier's protocol. OptiMEM was changed to a standard DMEM medium the next day after the transfection. The efficiency of transfection was estimated by fluorescence-activated cell sorting (FACS) analysis of transfected cells for Alexa Fluor 488 fluorescence 48 h after transfection (in collaboration with Fanny Monneaux and Frederic Gros, IBMC, Strasbourg). Transfection procedure did not lead to detectable decrease of viability of the cells or to significant change of the overall mtDNA amount, suggesting the absence of recombinant RNAs toxicity.
For stable transfection, 0.5–1.0 µg of corresponding linearized plasmid DNA was used for each 10-cm2 well of 85%-confluent cells. Transfection was performed with Lipofectamin, as described in the supplier's protocol. Selection (200–400 µg/ml of G418 for pBK-CMV-cloned versions and 150–200 µg/ml of Zeocine for pcDNA3-zeo) was applied after 48 h. Subsequent cloning was performed by serial dilution method.
RNA stability and mitochondrial import in vivo
Mitochondria were isolated from cybrid cells as described earlier (31,34), treated with digitonin to generate mitoplasts (mitochondria devoid of their outer membrane) and RNase A to get rid of non-specifically attached RNA. Such a treatment allows to obtain mitochondria completely free of cytosolic RNA contamination, including 5.8S rRNA, which can be strongly attached to the outer mitochondrial membrane upon cell disruption. However, during the RNase and digitonin treatment and multiple re-purification of organelles by centrifugation, an important part of mitochondria can be partially disrupted, and thus mitochondrial RNA can be partially degraded. In fact, we recover only 20–50% of intact mtRNA, and this yield varies in different experiments (Supplementary Figure S5B). Total and mitochondrial RNA were isolated with TRIZol reagent (Invitrogen).
Expression and mitochondrial import of recombinant 5S RNAs were detected by one-step reverse transcriptase (RT)-PCR (Qiagen) with primers listed in Supplementary Table S1. The PCR products were separated by gel electrophoresis, visualized by ethidium bromide staining and quantified using G-Box software. For quantification of recombinant 5S RNA molecules in cells and mitochondria, known amounts (0.5–500 fg) of corresponding T7-transcripts were used as a template for RT-PCR (Supplementary Figure S8). Linear dependence between the fluorescence of PCR fragment and the template RNA amount was obtained in the range from 0.5 to 50 fg RNA per assay, thus allowing the semi-quantitative estimation of recombinant 5S RNA molecules presence in cells and mitochondria.
Stability and mitochondrial import of small artificial FD-RNA molecules were analyzed by Northern hybridization of total and mitochondrial RNA with 32P-labeled oligonucleotide probes against the FD recombinant RNA or against control cytosolic and mitochondrial RNA (Supplementary Table S1). To compare the stability of different recombinant RNA, relative concentration of each RNA in various time period after transfection was calculated as a ratio between the specific probe signal and the signal for cytosolic 5.8S rRNA probe.
Relative amount of each imported RNA inside the mitochondria was estimated after quantification by Typhoon-Trio scanner as a ratio between the signal obtained after hybridization with FD probe and the signal obtained after hybridization with a probe against the mitochondrial tRNALeu in the same RNA preparation, as described previously (31). To compare import efficiencies of different RNA molecules, total level of each RNA molecules in transfected cells should be taken into account and normalized. For this, the amount of imported RNA inside the mitochondria was divided by the ratios calculated in the same way but in total RNA preparation, which indicate the total level of recombinant RNA in the cells.
To quantify the recombinant RNA molecules in the mitochondrial fraction after transfection, the signal obtained after mitochondrial RNA hybridization with specific probe was compared with the signals corresponding to 10–50 ng of corresponding T7-transcripts loaded on the same gel. The values were normalized to the amount of cells used for mitochondria isolation. MtDNA molecules were quantified by real-time qPCR (iCycler, MyiQ™ Real-Time Detection System, Biorad).
mtDNA heteroplasmy level analysis
To isolate total DNA from transfected cells, the cells were solubilized in 0.5 ml of buffer containing 10 mM Tris–HCl, pH 7.5, 10 mM NaCl, 25 mM Na-EDTA and 1% SDS, then 10 µl of proteinase K solution (20 mg/ml) was added and the mixture incubated for 2 h at 50°C; 50 µl of 5 M NaCl was added and DNA precipitated by isopropanol.
Heteroplasmy level was analyzed by real-time qPCR using SYBR Green (iCycler, MyiQ™ Real-Time Detection System, BioRad) with primers listed in the Supplementary Table S1. Two pairs of primers were used: 1) amplifying the 210 bp fragment of 12S rRNA gene region (nucleotides 1095–1305 in mtDNA) not touched by the KSS deletion as a value showing all mtDNA molecules, and 2) amplifying the 164 bp fragment of the deleted region (nucleotides 11 614–11 778) as a value showing WT mtDNA molecules. All reactions were performed in a 20 μl volume in triplicates. PCR using water instead of template was used as a negative control. PCR was performed by initial denaturation at 95°C for 10 min, followed by 40 cycles of 30 s at 95°C, 30 s at 60°C and 30 s at 72°C. Specificity was verified by melting curve analysis and gel electrophoresis. The threshold cycle (Ct) values of each sample were used in the post-PCR data analysis by MyiQ™ software, if only the standard deviation (SD) of triplicate samples was less than 0.2. In each experiment, the absolute amounts of the PCR product were determined by using in parallel the serial dilutions of known amounts of linearized plasmid DNA (104–107 copies per sample). The KSS heteroplasmy level was calculated using the formula: mutant mtDNA/total mtDNA = 1 − (WT mtDNA/total mtDNA). To evaluate mtDNA content upon transfections with recombinant RNAs, real-time PCR was performed using 12S rRNA gene specific probes for mtDNA and Actin gene-specific ones for nuclear DNA (for sequences of the primers, see Supplementary Table S1).
mtDNA isolation and replication intermediate analysis on 2D native agarose gel
Seven hundred square centimeters of 80%-confluent TSFR cybrids cells was transfected with 70 µg of FD-H RNA, and mitochondria and mtDNA were isolated in 4 days after transfection as described in (42) with small modifications. Cells were disrupted in hypotonic buffer [HEPES (pH 7.8) 20 mM, KCl 5 mM, MgCl2 1.5 mM, DTT 2 mM, BSA 1 mg/ml and PMSF 1 mM] using a tight-fitting Dounce homogenizer and immediately mixed with 2.5 × MSH buffer [Mannitol 525 mM, sucrose 175 mM, HEPES (pH 7.8) 20 mM, EDTA 5 mM, BSA 1 mg/ml, DTT 2 mM and PMSF 1 mM]. After several rounds of centrifugation, mitochondria were treated with DNase I (Boehringer Mannheim) at 4°C for 15 min. DNase I activity was inhibited by addition of 15 mM EDTA, mitochondria were washed two times with 1 × MSH buffer [Mannitol 210 mM, sucrose 70 mM, HEPES (pH 7.8) 20 mM, EDTA 2 mM, DTT 2 mM and PMSF 1 mM] and purified on sucrose gradient as described (42).
For 2D native agarose gel (2DNAGE), 2 µg of mtDNA was digested by FastDigest BlpI enzyme following manufacturer’s recommendations (Fermentas). Cleavage sites are referred in the following list (co-ordinates/fragment length, bp): 788–1093/306; 1094–1248/155; 1249–4533/3285; 4534–5646/1113; 5647–5699/53 and 5700–787/11 657 (WT mtDNA) or 4583 (KSS mtDNA). Neutral 2D-AGE was performed by the standard method and run on 0.5% agarose gel for the first dimension and on 1% agarose gel containing 0.5 µg/ml EtBr for the second dimension, as described (43,44). Blots were hybridized with radiolabeled PCR fragment (nucleotides 8099–8365/15 438–15 931 of mutant KSS mtDNA), exposed to PhosphorImager screen, revealed by Typhoon-Trio (GE Healthcare) and analyzed by ImageQuantTL software (GE Healthcare).
Recombinant RNA modeling
To predict secondary structures of recombinant RNA molecules and estimate their free energy (dG), the ‘Mfold’ program was used (45,46); 5S rRNA variants secondary structures were corrected in accordance with data published (47,48). To estimate melting temperatures for DNA–DNA and RNA–DNA duplexes, we used IDT Sci-Tools OligoAnalyser 3.1 software (49). Specific hybridization with target mtDNA was tested using Southern-blot hybridization of WT and KSS double-stranded replication templates (see earlier), corresponding plasmids and mtDNA fragments with 32P-labeled recombinant RNAs in 1 × PBS at 37°C.
In vivo mitochondrial translation
Analysis of mitochondrial protein synthesis was performed as described previously (22) with minor modifications. Briefly, 600 × 103 cells were incubated for 10 min in DMEM w/o methionine (Sigma) in the presence of 100 μg/ml of emetine to block cytoplasmic translation, followed by 30 min with 200 μCi/ml [35S]-methionine (Amersham) and, finally, 10 min chase in the normal growth medium. Cells were solubilized in a Laemmli’s buffer, sonicated for 5 s to break cellular DNA, incubated for 10 min at 37°C and 100 μg of protein was run on a 10–20% gradient SDS-PAGE. Protein amounts loaded were before normalized by anti-tubulin and anti-porin immunoblotting of the same preparations. Visualization and quantification were performed using Typhoon-Trio and ImageQuantTL software (GE Healthcare).
RESULTS
Design of RNA-based mitochondrial vectors
To test the anti-genomic strategy (Figure 2A), we used as a model mtDNA containing a large deletion (nucleotides 8363–15 438) underlying a case of a frequent mitochondrial pathology, the KSS (Figure 1A). The deletion in mtDNA generates a new sequence at the fusion of the deletion boundaries, further referred to as KSS-DNA, which can serve as the target for the anti-genomic RNAs. To use 5S rRNA and FD-RNA as mitochondrial vectors, the distal portion of the β-domain of 5S rRNA (Figure 1B) and the linker part between hairpin domains in FD-RNA (Figure 1C) were replaced by sequences corresponding to either H- or L-strand of KSS-DNA (referred to as 5S-H and 5S-L or FD-H and FD-L correspondingly). The choice of the insert length was driven by secondary structure and melting temperature prediction (Table 1 and Supplementary Figure S1). To avoid formation of stable alternative structures for FD-RNA-L, carrying the longest insertion of 22 bases, a C to G mismatch was introduced (Figure S1), resulting in comparable predicted melting temperature for hybrids between the recombinant RNAs and mutant KSS mtDNA (Table 1). According to Tm predictions, annealing of recombinant RNAs to WT mtDNA should be negligible at 37°. To validate the selected versions experimentally, we hybridized labeled recombinant RNAs with mtDNA fragments under physiological conditions, demonstrating specific annealing with the mutant (KSS) but not with WT mtDNA (Figure 2A and B and Supplementary Figure S2).
Figure 2.
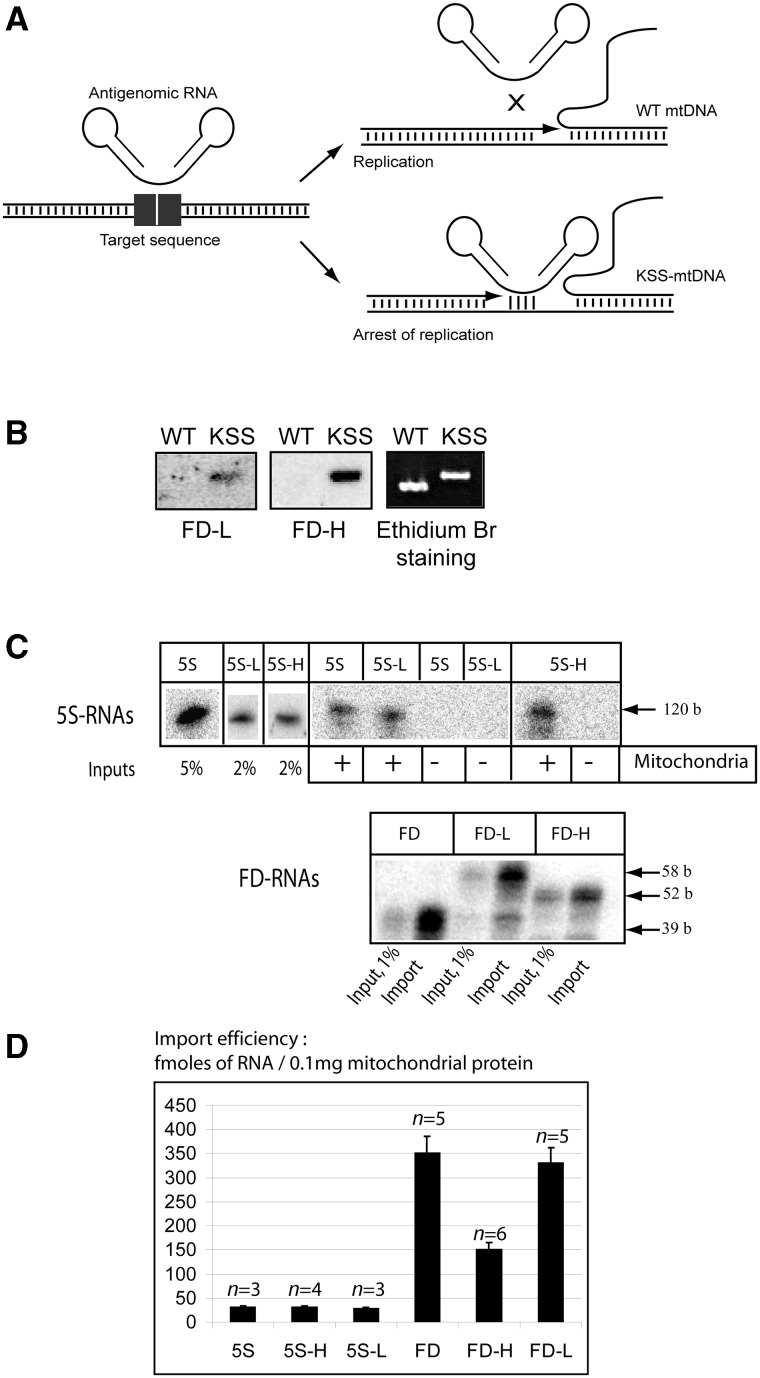
(A) Scheme of the anti-genomic strategy. (B) Southern hybridization of wild-type or KSS mtDNA fragments with labeled recombinant RNAs (FD-L and FD-H) at 37°C. (C) In vitro import of the recombinant RNAs. Autoradiography of isolated from purified mitochondria RNA separated in denaturing 10% PAAG is presented; 5S rRNA derived molecules above and FD-RNA derived ones below. The size of RNA molecules is shown at the right. ‘Input’, 1–5% of RNA used for each assay, representing 30–150 fmoles of labeled RNA. Mitochondria (+) corresponds to the import assay, Mitochondria (−) to the mock import assay without mitochondria, used as a control for non-specific protein–RNA aggregation. (D) The import efficiencies, expressed in fmoles of protected RNA to 0.1 mg of mitochondrial protein, as described elsewhere (32); n, number of independent experiments; n = 3–6, as detailed upon each graph.
Table 1.
Melting temperatures prediction for hybrids between the recombinant RNAs and mutant or wild-type mtDNA regions (SD = 1.3°C, “b”, bases)
| RNA | Homology with KSS mtDNA | Tm for KSS mtDNA | Homology with wt mtDNA (5′-deletion boundary) | Tm for 5′-deletion boundary | Homology with wt mtDNA (3′-deletion boundary) | Tm for 3′-deletion boundary |
|---|---|---|---|---|---|---|
| 5S-H | 15 b | 52.1°C | 8 b | 18.2°C | 8 b | 28.1°C |
| 5S-L | 14 b | 45.2°C | 7 b | 16.1°C | 8 b | 16.6°C |
| FD-H | 15 b | 52.1°C | 9 b | 24.2°C | 7 b | 21.9°C |
| FD-L | 21 b (1 mismatch) | 51.4°C | 12 b | 34.4°C | 10 b (1 mismatch) | 27.7°C |
All the recombinant RNAs were tested for their import into isolated human mitochondria as described in (26). It was observed that the insertion sequences did not affect the capacity of RNA molecules to be internalized by mitochondria (Figure 2B). Mitochondrial targeting of FD-H RNA revealed to be less important compared to FD-L and FD RNAs, however, still rather efficient. This can be explained by a possibility of an alternative secondary structure for this RNA, missing the D stem-loop domain and thus less efficient as an import substrate (Supplementary Figure S1).
To test whether recombinant RNAs imported into mitochondria are able to interfere with replication of the mutant mtDNA, an in vitro replication run-off assay was first established (Supplementary Figure S3), permitting to replicate both strands of mtDNA PCR-amplified fragments. Addition of recombinant RNAs containing KSS-H or KSS-L inserts inhibited replication of KSS-DNA in a dose-dependent manner. In contrast, the presence of the recombinant RNAs did not significantly inhibit replication of WT mtDNA, supporting the above data suggesting that recombinant RNAs can discriminate KSS-DNA from the WT one (Figure 2B). The anti-replicative effect of FD-RNAs was more pronounced than that of 5S-L and 5S-H RNAs and allowed up to 60% inhibition (Supplementary Figure S3). These values are comparable with those previously reported for PNA (11), which indicates that natural RNAs can be as efficient as synthetic PNAs for the anti-genomic strategy.
Transfection of cybrid cells with anti-replicative RNA
To validate the anti-replicative effect of recombinant RNAs in vivo, we constructed a transmitochondrial cybrid cell line containing mitochondria from fibroblasts of a patient diagnosed with KSS syndrome. Heteroplasmy level (proportion of mutant KSS mtDNA to all mtDNA molecules) has been analyzed by real-time PCR with primers listed in the Supplementary Table S1. The cybrid cell line obtained was characterized by presence of 65% ± 2% of KSS mutation in mtDNA, the level being stable upon more than 3 months of continuous cultivation. The sequence of deletion boundaries was shown to be the same in five independent clones and corresponded to deletion of nucleotides 8363–15 438 in mtDNA, as was shown by direct sequencing of the patient's mtDNA (Figure 1A). However, only a very weak (10% ± 2%) decrease of oxygen consumption can be detected in cybrid cells (not shown). The same situation was already reported for many other mutations in mtDNA, where the respiration defects detected in patients’ muscles and nervous tissues have been prominently decreased and almost not detectable in fibroblasts and cybrid cells (50,51). On the other hand, the absence of strong respiration phenotype facilitated the manipulations with the cultured cells and allowed us to use this cell line as a model to validate the anti-replicative effect of recombinant RNAs in vivo.
Because the in vitro anti-replicative effect of FD-RNAs was more pronounced, for the in vivo assay, we performed the transient transfection of KSS cybrid cells with FD-L and FD-H RNA. First, the efficiency of transfection was evaluated by FACS using fluorescently labeled FD-H RNA, and it was shown that >95% of cells were transfected (Supplementary Figure S4). Then, cells were transfected with non-labeled FD-RNAs, and amount of RNA internalized by cells was evaluated by Northern hybridization of total cellular RNA comparing with the signals of the known amounts of T7-transcripts loaded on the same gel. By this approach, we could estimate that approximately 15% of RNA added to cells were internalized and can be detected in full-size form 24 h after transfection (not shown). We also measured the degradation ratio of recombinant RNA in cybrid cells at different time points spanning a 10-day period after transfection. Surprisingly, recombinant RNAs were detectable in cybrid cells up to 6 days after transfection, stability of RNAs FD-H and FD-L being slightly more prominent than that of RNA FD (Figure 3A).
Figure 3.
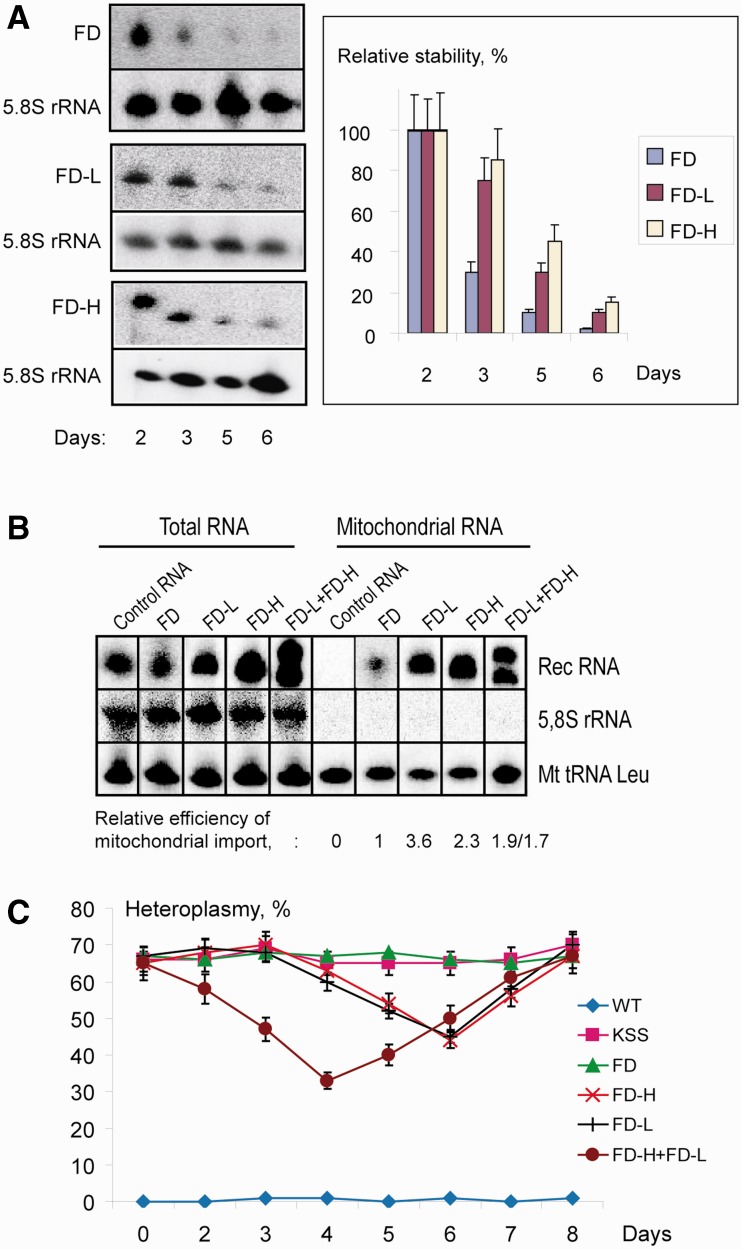
The effect of recombinant RNA mitochondrial targeting on heteroplasmy level in KSS cybrid cells. (A) Recombinant RNA stability in transiently transfected KSS cybrid cells. At the left, an example of Northern hybridization of total RNA isolated in different time after transfection (indicated below) with probes against FD-RNAs used for transfection (FD, FD-L or FD-H) and against 5.8S rRNA to quantify the level of recombinant RNA in the cells. At the right, time dependence of RNA decay, SD is calculated from at least four independent experiments. (B) Mitochondrial import of recombinant FD-RNAs in transiently transfected cybrid cells. Northern hybridizations of RNA extracted from cells or purified mitochondria 48 h after transfection. Above, RNAs used for transfection are indicated; control is an artificial RNA not imported into mitochondria (Supplementary Figure S5A). Relative efficiencies of mitochondrial import are shown below, import of FD-RNA is taken as 1 (±SD = 0.1, results of one from three independent experiments are presented). (C) Time dependence of KSS heteroplasmy level upon transient transfection of cybrid cells with various RNA (indicated at the right); KSS, mock-transfected cybrid cells. SD is calculated from three independent experiments.
Relative efficiency of RNA mitochondrial import was analyzed by Northern hybridization of RNA isolated from purified RNase-treated mitochondria 48 h after cybrid cells transfection with various RNAs (Figure 3B). As a negative control, cells were transfected with a short artificial RNA containing no import determinants (referred to as Control RNA, Figure 3B and Supplementary Figure S5A and B). Contrary to this RNA found not to be associated with mitochondrial fraction, recombinant FD-L and FD-H RNA were even more efficiently imported into mitochondria than FD-RNA without insert (Figure 3B and Supplementary Figure S5B). The quantitative discrepancy between in vitro (Figure 2B) and in vivo import data can be explained by different RNA folding in cells and, also, by an advantage of more flexible molecules containing linkers between the two structured domains interacting with the protein import factors.
The quantity of recombinant RNAs in the mitochondrial fraction 2 days after transfection (estimated by Northern hybridization comparing to known amounts of corresponding T7-transcripts used as quantitative reference) corresponded to a value of 3.2 ± 0.5 × 104 RNA molecules per cell. Comparing these values with mtDNA amount, measured by real-time PCR as 1.0 ± 0.1 × 103 molecules per cell, we can estimate the RNA:mtDNA molar ratio in organello as 30:1. This estimation validate our strategy, because the same order of recombinant RNA molar excess allowed us to decrease mtDNA replication in vitro up to 60% (Supplementary Figure S3).
Transient effect of imported RNAs on mutant mtDNA in vivo
Heteroplasmy levels (percentage of mutant KSS mtDNA to all mtDNA molecules) were measured in cybrid cells at different time points spanning a 10-day period after transfection (Figure 3C). We observed a decrease of the proportion of mutant mtDNA by 15–25% in cells transfected by recombinant RNA containing either KSS-DNA-H or KSS-DNA-L inserts but not in mock-transfected ones or cells transfected with FD-RNA (mitochondrially imported but having no homology to the mt-DNA), indicating that mitochondrially imported RNA molecules can function as anti-genomic agents in human cells. Because the effect of the inserts complementary to both mtDNA strands was similar, in the next experiment two recombinant RNA molecules, FD-L and FD-H, were used for transfection simultaneously. Both RNAs were detected in the mitochondria simultaneously in comparable amounts (Figure 3B and Supplementary Figure S5B) and provided the most pronounced decrease of heteroplasmy level by 35% ± 2% (Figure 3C).
To ascertain the absence of a non-specific effect on replication that reduced total mtDNA levels in cells, we estimated mtDNA amounts in WT and KSS cybrid cells after transfection with RNAs FD-L and FD-H (Supplementary Figure S6A). Using real-time PCR, we compared ratios between mtDNA (gene 12S rRNA) and nuclear DNA (gene of actin, see Supplementary Table S1). Variations of this ratio, taken as 1 24 h after transfection, did not correlate with the heteroplasmy shift. For WT cells, levels of mtDNA were stable in the range of 10%, comparable to natural fluctuation values, thus indicating that no depletion effect has been caused by adventitious reduction in replication of the WT mtDNA. In KSS cybrid cells, a slight decrease of 25% from initial mtDNA level has been observed, probably due to replication pausing caused by anti-replicative RNAs.
The shift of heteroplasmy in cybrid cells was reproducibly observed with a 4–6 days delay in respect to the cells transfection; however, the initial level was consistently restored within 7–8 days. The temporal character of the effect could be a consequence of the recombinant RNA degradation and/or of the novel mtDNA deletion formation and propagation. The latter suggestion was verified by direct sequencing of mtDNA isolated at different time points after cybrid cells transfection with recombinant RNAs. No heterogeneity of the deletion boundaries was found (Supplementary Figure S6B), proving the absence of other mutant mtDNA molecules that could not be affected by recombinant RNAs.
Taking into account the degradation ratio of recombinant RNA in the cells (Figure 3A), one can suggest that the restoration of heteroplasmy level in 7–8 days after transfection coincides perfectly with the time period when RNA/mtDNA molar ratio should be greatly decreased, being no more detectable by Northern hybridization. Thus, one can suggest that the stabilization of the heteroplasmy shift could be achieved by stable expression of anti-genomic RNA molecules in the cells.
RNA affects mutant mtDNA replication in vivo
To demonstrate that anti-replicative RNA molecules affect mutant mtDNA replication in vivo, we used 2DNAGE. This technique separates DNA molecules on the basis of size and shape, thus permitting to detect various types of replication intermediate (RI) in specific and predictable way (42,52). We compared RIs of mtDNA isolated from WT and KSS cybrid cells (Figure 4A), digested with restriction enzyme BlpI, chosen to separate fragments of different types of mtDNA, as indicated at the Figure 1A. Because of heteroplasmy, after hybridization of digested mtDNA with PCR fragment of Cyt b region, we can see one (for WT mtDNA) or two (for KSS mtDNA) main spots, corresponding to DNA fragments of WT and mutant KSS mitochondrial genomes and, therefore, to two sets of RIs (see cartoon interpretation on Figure 4B, panel d).
Figure 4.
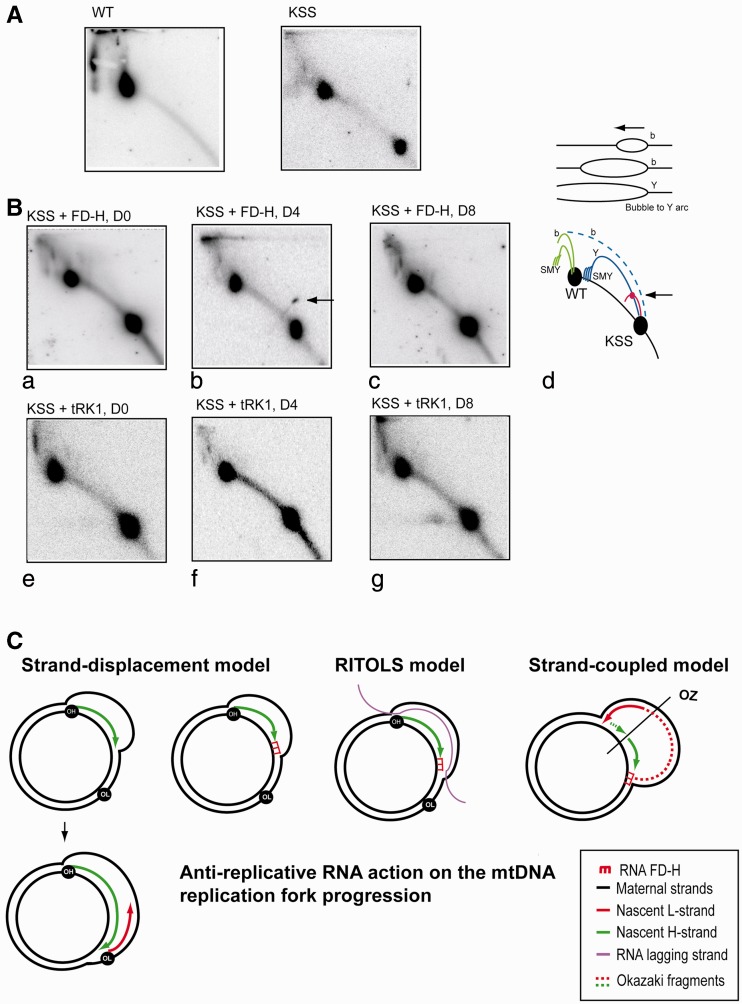
Recombinant RNA affects mutant mtDNA replication in vivo. (A) The 2DNAGE electrophoresis analysis of mtDNA isolated from wild-type (WT) and KSS cybrid cells. MtDNA was digested with BlpI, blotted and hybridized with P32 labeled fragment of the Cyt b gene (Figure 1A). Radio autographs detect DNA fragment of 11 645bp for wild-type mtDNA and two fragments of 11 645 and 4570 bp for heteroplasmic mtDNA, indicated as WT and KSS on the cartoon at the panel d. (B) The 2DNAGE analysis of mtDNA isolated from KSS cybrid cells in different periods of time after transfection with RNA FD-H (panels a–c) and as a control with yeast tRNA (tRK1, panels e, f and g) immediately after transfection (panels a and e), in 4 days (panels b and f) and in 8 days (panels c and g). Panel d, schematic representation of replication intermediates: in green, replication intermediates corresponding to wild-type mtDNA and in blue, corresponding to mutant mtDNA. b, bubble arcs; Y, y arc; SMY, slow migrating y arcs. The site of presumptive replication pausing is shown by arrow. (C) Schematic representation of three mtDNA replication models (see ‘Discussion’ for details and references) and possible effect of anti-replicative recombinant RNA FD-H. OH, OL and OZ, origins of H-strand, L-strand, and strand-coupled replication, respectively.
As it was demonstrated previously (53), mtDNA fragments containing the replication origin OH produce two types of RIs, bubble arc and simple Y arc, their relative abundance being altered markedly: when larger fragments of mtDNA were studied, the bubble arc signal increased, with a concomitant decrease in signal from the Y arc (illustrated on a scheme above panel d). Analysis of two DNA fragments on the same gel is a rather unusual situation for 2DNAGE approach, therefore RIs corresponding to WT mtDNA fragment are not well separated in our system; however, they correspond perfectly to the predicted pattern (54). We can detect bubble arc-type RI corresponding to WT mtDNA fragment, as well as slow-moving Y arcs, sensitive to S1 nuclease treatment (55,56) (Figure 4A and B). The intermediates of mutant KSS mtDNA replication can be partially masked by WT mtDNA fragment; however, we detected the descending part of a Y arc-type RI and a weak bubble arc, visible upon longer exposition of the blot (not shown).
We compared RIs of mtDNA isolated from KSS cybrid cells at different periods of time after transfection with RNA FD-H (Figure 4B, panels a–c) and, as a control, with a yeast tRNA (tRK1, panels e–g). The patterns of RIs corresponding to WT mtDNA fragment are rather similar in post-transfected cells (Figure 4B) indicating that the replication of WT mtDNA was not significantly altered by recombinant RNA. In contrast, we reproducibly observed a prominent spot on a small Y arc-resembling RI corresponding to the mutant mtDNA fragment 4 days after transfection with RNA FD-H but not immediately after transfection or 8 days later (Figure 4B, panel b, see also Supplementary Figure S7 for two other examples of similar experiments). This kind of spots, caused by a marked increase of RIs around a specific point, had been previously described as mtDNA replication pauses in the sites of specific protein binding (43,44), thus allowing us to suggest that the anti-replicative RNA FD-H enhances replication pausing at the site of its annealing to mutant mtDNA (Figure 4C). The origin of a new RI resembling a small Y arc is still not clear. We can hypothesize that it might be due to increased rate of lagging-strand initiation as proposed in (57). Indeed, a stalled leading H-strand may coincide with the alternative initiation of an L-strand through strand switching of the polymerase, resulting in a set of forks expected to form a Y arc upon 2DNAGE analysis (58).
In non-transfected cells (Figure 4A) or in cells transfected with yeast tRNALys in the same conditions, we cannot detect any RIs corresponding to replication pausing (Figure 4B, panels e–g). As mentioned before, yeast tRNALys is efficiently imported into human mitochondria (Supplementary Figure S5B) but does not contain any homology to human mtDNA (Figure 1C) and should not affect its replication. Thus, all our data clearly demonstrate that the recombinant RNA imported into mitochondria in vivo leads to a specific replication pause of the mutated mtDNA.
Stable effect of imported RNAs on mutant mtDNA in vivo
Because all our attempts to express FD-RNA in the cybrid cells in stable manner under the control of external promoters were unsuccessful, we used another set of recombinant RNA molecules based on 5S rRNA, one of which was previously shown to be expressed, correctly maturated and mitochondrially targeted in human cells (31). Recombinant 5S rRNA genes (Figure 1B) were cloned into a phagemid vector in the context optimal for correct transcript processing, as described previously (31), and used for a stable transfection of KSS cybrid cells. Transfectants selected for their resistance to appropriate antibiotic were cloned. Approximately 20–25 individual clones of each series—containing the empty vector, WT or recombinant 5S rRNA genes—were characterized by similar growth rates, suggesting the absence of toxic effect of the transgenes expression.
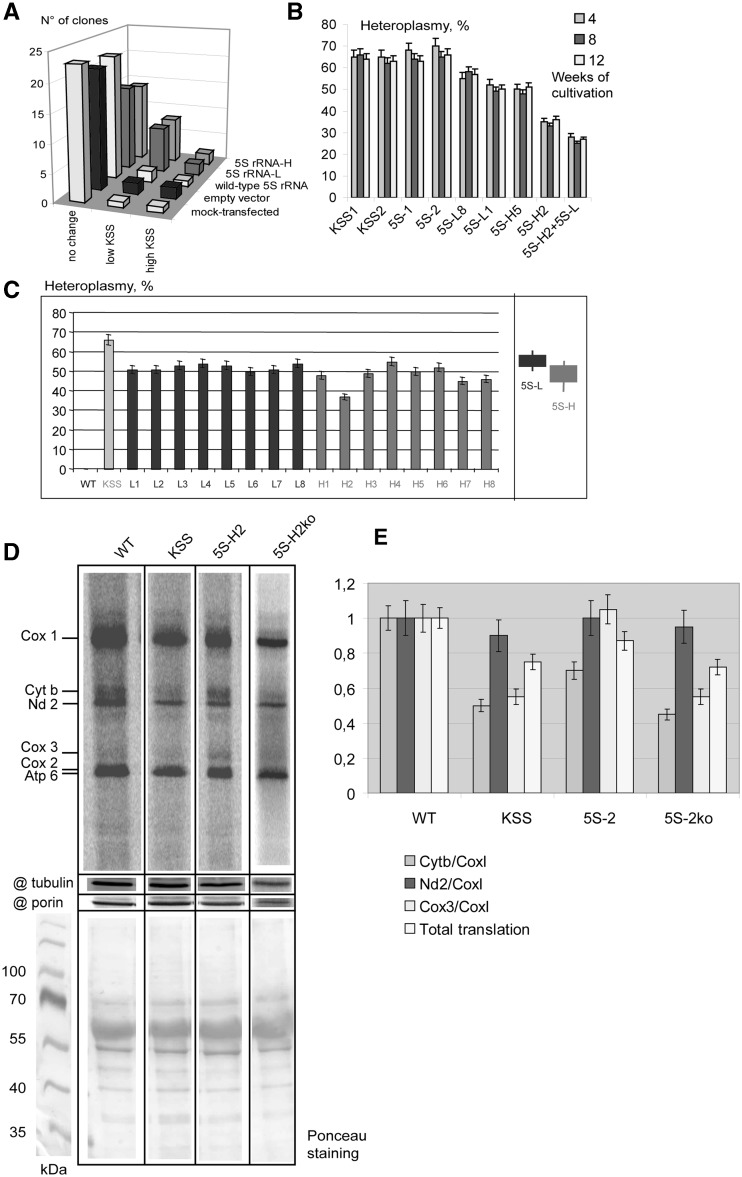
The KSS mutation heteroplasmy level was analyzed in each clone. Control cell lines (mock-transfection, transfection with the empty vector or with the vector containing the WT 5S rRNA gene) generated clones with little variations of heteroplasmy (Figure 5A), whereas for cells transfected with 5S-L and 5S-H RNA genes, the occurrence of 30% clones with decreased proportion of mutant mtDNA was observed (Figure 5A). The lower heteroplasmy levels found in these clones (51–54% for cells containing 5S-L gene and 37–52% for those containing 5S-H gene) were then stable during 3 months of continuous cultivation (Figure 5B and C).
Figure 5.
Analysis of clones obtained after stable transfection of KSS cybrid cells with 5S rRNA genes. (A) Heteroplasmy variations in series of clones of KSS cybrid cells after stable transfection with 5S rRNA genes (indicated at the right). ‘No change,’ number of clones where the heteroplasmy shifts were comparable to natural fluctuation values (0–5%); ‘low KSS,’ the proportion of KSS mtDNA reduced by >10% and ‘high KSS,’ the proportion of KSS deletion increased by >10%. (B) Stability of the heteroplasmy level in individual clones. Two clones of each series are presented: KSS, mock-transfected KSS cybrid cells, clones from ‘no change’ set; 5S, cells transfected with the wild-type 5S rRNA gene, clones from ‘no change’ set are presented; 5S-H and 5S-L, cells transfected with corresponding recombinant 5S RNA genes, clones from ‘low KSS’ series, see also (C); 5S-H2+5S-L, clone 5S-H2 transfected with 5S-RNA-L gene. (C) Analysis of heteroplasmy levels in individual clones. All clones resulting from transfection with 5S-L and 5S-R RNAs where the hetroplasmy shift was >10% are presented, number of independent qPCR measurements n = 3. An example of such an assay, demonstrating that the shifts observed were significant and cannot be explained by errors of qPCR, is given in the Supplementary Figure S9. At the right, the summary of heteroplasmy shifts induced by 5S-L and 5S-H RNA is presented. (D) Analysis of mitochondrial protein synthesis in wild type, KSS cybrid cells, a clone of KSS cells expressing recombinant 5S rRNA (5S-H2) and the derivant of the same cell line after the loss of recombinant 5S rRNA expression (5S-H2ko), as indicated above the panel. Autoradiography of SDS-PAGE is demonstrated, products of mitochondrial translation are indicated at the left. @ tubulin and @ porin, immunoblotting of the same preparations, using as a loading control. The lower panel represents Ponceau S staining of the corresponding membrane. (E) The graphs represent the total level of mitochondrial translation (normalized to the amount of mitochondrial porin) and the ratio of signals corresponding to individual proteins. Cytb and Cox3, cytochrome b and subunit 3 of cytochrome oxidase, proteins encoded by the portion of mtDNA deleted in KSS mutants; CoxI and Nd2, subunit 1 of Cytochrom oxidaze and subunit 2 of NADH-dehyrogenase complexes, the both proteins encoded by the mtDNA region not touched by the KSS deletion.
To test the effect of the simultaneous expression of both 5S-L and 5S-H RNAs, the cell line derived from the clone 5S-H2, characterized by the lowest heteroplasmy level (37% ± 2%), was transiently transfected with plasmid containing the 5S-L gene. The resulting cell line, referred to as 5S-H2+5S-L, was characterized by an additional shift of the heteroplasmy, from 37% ± 2% to 30% ± 2% of mutated mtDNA molecules (Figure 5B). Expression of both recombinant 5S rRNA molecules in this cell line was confirmed by RT-PCR analysis with insert-specific primers (Supplementary Figure S8A). Mitochondrial import of recombinant 5S rRNAs was detected in all the clones expressing 5S-L or 5S-H RNA by RT-PCR with RNA isolated from purified mitochondria devoid of cytosolic contaminants (Supplementary Figure S8B).
The level of recombinant 5S rRNA expression in the clone 5S-H2 was estimated as 30 ± 5 molecules per cell using semi-quantitative RT-PCR (Supplementary Figure S8C). This very low expression is not surprising taking into account the competition of mutant 5S rRNA with much more abundant WT 5S rRNA molecules for interaction with protein factors of transcription, processing and export from the nucleus. In the same cell line, 8 ± 2 molecules of recombinant 5S rRNA per cell were found inside the mitochondria (Supplementary Figure S8C), indicating the very high efficiency of import (25%), as it was shown previously. This value is certainly under-estimated, because a significant part of mitochondrial RNA has been usually degraded during the mitochondria isolation and purification (see Supplementary Figure S5B and ‘Materials and Methods’ section).
Because the respiration phenotype of KSS cells can be hardly detectable, we used the assay of mitochondrial protein synthesis to compare WT cells, KSS cybrid cells and a clone of KSS cells expressing recombinant 5S rRNA (5S-H2) (Figure 5D and E). Cybrid cells were characterized by the ∼25% decrease of total mitochondrial translation. As expected, the synthesis of proteins encoded by the portion of mtDNA deleted in KSS mutants (Figure 1A) was affected in a more prominent way, the ratio between Cyt b and Cox1 proteins being twice lower in cybrid cells comparing to control cell line. Remarkably, in the clone 5S-H2, the total level of translation and the Cyt b synthesis were partially restored (Figure 5D and E). For Cox3 protein, a prominent decrease in KSS cybrid cells and re-increase in the clone 5S-H2 (Figure 5E) have also been observed.
The results obtained should be considered as a promising indication on the possibility to obtain a curative effect on the mitochondrial dysfunction by a heteroplasmy shift. So far, one cannot exclude that selected individual clones can have a slightly varying nuclear background issued from the transfection procedure. To explore this possibility, we analyzed individual subclones of the 5S-H2 cell lines after prolonged cultivation and subsequent cloning. As mentioned earlier, this transgenic line was stable during 3-month-long cultivation; however, upon additional 6-week cultivation and subsequent recloning, one among >100 individual subclones retaining the resistance to G418 was characterized by a spontaneous loss of 5S-H expression (as demonstrated by PCR and RT-PCR, Supplementary Figure S8A and D). This derivant cell line, designed as 5S-H2ko, was analyzed for mitochondrial translation by pulse-chase assay (Figure 5D and E). It was indeed observed that the effect of improved mitochondrial translation found in the original 5S-H2 cell line was lost, and the ratio of mitochondrially synthesized polypeptides in 5S-H2ko line was nearly identical to that of the initial KSS cybrid cell line. This result indicates that the curative effect observed in 5S-H2 line was due to the presence and, most probably, mitochondrial import of the recombinant 5S rRNA.
In conclusion, a weak but stable expression and mitochondrial import of the recombinant 5S rRNAs in KSS cybrid cells allowed us to obtain cell lines characterized by a stable shift of mitochondrial heteroplasmy level accompanied, for the best expressor, by an improvement of mitochondrial translation.
DISCUSSION
RNA import pathway as a therapeutic tool
Mutations in mtDNA have been associated with a wide variety of human disorders (1). Many patients suffering from mtDNA defects harbor subpopulations of mutated and WT mtDNA molecules within the same cell and tissue, a state known as heteroplasmy (14,59). Because there is no effective treatment for these often devastating pathologies, one attractive approach would be to specifically target mutant mtDNA to prevent it from replicating, thereby allowing propagation of only WT genomes. Application of such an ‘anti-genomic,’ or ‘anti-replicative’, strategy is confronted to two main problems: facilitating the translocation of the anti-genomic oligomers through the double mitochondrial membrane and allowing their access and specific binding to mutated region of mtDNA. To resolve the first problem, the natural pathway of RNA import into yeast and human mitochondria can be useful. Our recent studies permitted to identify the import determinants in tRNA and 5S rRNA structures, and a set of small RNA molecules with significantly improved efficiency of import into human mitochondria was designed on this basis (31,32). In this study, to create a vector system able to target ‘therapeutic’ oligonucleotides into deficient human mitochondria, we inserted into these RNAs short (14–21 bases) sequences corresponding to the boundaries of a large deletion in mtDNA associated with a neuromuscular syndrome KSS. All the recombinant RNAs so obtained were proven to retain their capacity to be internalized by human mitochondria in vitro (in isolated human mitochondria) and in vivo (in cultured cybrid cells). These data indicate that the pathway of RNA import into mitochondria can serve for targeting anti-replicative oligoribonucleotides into mitochondrial matrix.
The second problem consists in specific binding of the anti-replicative oligonucleotides to mutated region of mtDNA. In a previously published study (11,60), synthetic PNAs were used to increase, comparing to oligodeoxynucleotides, the selective and stable binding to the region of mtDNA around the mutation site. However, RNA–DNA duplexes are considered to be, in general, more stable that DNA–DNA ones (49), so the stretch of ribonucleotides could target the mtDNA duplex, especially in the region of the replication machinery progression, where it is already at least partially destroyed by the helicases. Our in vitro replication assay showed that, indeed, specific oligoribonucleotides interfere with mtDNA replication, reducing it to the level comparable with that described for PNA. Therefore, RNA molecules having an important advantage as natural mitochondrial import substrates were also suitable for selective inhibition of mutant mtDNA replication in vitro.
Anti-replicative RNA action in vivo
The mechanism of human mtDNA replication is still a subject of intense debate (61–63). The idea of anti-replicative heteroplasmy shift as a therapeutic strategy (11) was initially based on the strand-displacement mtDNA replication model that suggested the existence long single-stranded RIs (64). According to this model, leading-strand (H, heavy strand) synthesis starts at a fixed point (OH) and advances approximately two-thirds of the way around the molecule before second-strand (L, light strand) synthesis is initiated (Figure 4C). More recently, another two classes of RIs were described in mammalian mitochondria. Detection of RNA–DNA hybrid intermediates of mtDNA replication gave rise to a model involving Ribonucleotide Incorporation Throughout the Lagging Strand (RITOLS) (55,56). This model shares many features with the strand-displacement one, the main difference consisting in the presence of displaced H-strand not in a single-stranded form covered by SSB protein but in the form of RNA–DNA hybrid, where the lagging strand RNA may be created by primase or by a nascent RNA transcript hybridizing with the displaced H-strand in the 3′–5′ direction (55).
Another type of RIs, DNA–DNA duplexes, especially predominant in cultured human cells recovering from mtDNA depletion, has been proposed as compatible with the products of conventional strand-coupled DNA synthesis, initiating from sites dispersed across the broad zone named ori Z (52,53). Alternatively, these DNA–DNA duplexes could be considered as a consequence of frequent initiation of lagging-strand DNA synthesis resulting in an increased rate of conversion of RITOLS RIs to dsDNA ones (57,58).
According to the scheme shown at Figure 4C, for all the three current models of mtDNA replication, mitochondrially imported anti-replicative RNA should cause a replication stalling at the site of RNA annealing to the mutant mtDNA due to impairing of the replication fork progression. The similar mechanism has been proposed to explain the replication forks arrest in the D-loop region, namely due to the inability of the replisome helicase to separate regions of short RNA–DNA hybrid (53). In full agreement with our expectations, analysis of mtDNA RIs on the 2DNAGE revealed a clear site of replication pausing detectable only on the fragment of mutant KSS mtDNA isolated from cells transfected with recombinant RNA FD-H (Figure 4B). This is a direct demonstration of recombinant RNA ability to stall the mutant mtDNA replication in vivo.
Effect of imported RNAs on mtDNA heteroplasmy
To test the anti-replicative RNA efficiency in human cells, the changes of heteroplasmy level in KSS transmitochondrial cybrid cells were measured upon the presence of recombinant RNAs in mitochondria. Two different strategies were used. First, cells were transiently transfected with small artificial RNA molecules containing structural determinants of mitochondrial import and 15–21 bases insertions complementary to the region of mtDNA around the KSS deletion site. Our data show that this approach allows targeting a rather high level of recombinant RNA inside the mitochondria, and this reproducibly coincides with a shift of KSS mtDNA heteroplasmy level from 65% to 40–50%, detected within the 4–6 days period after transfection. This change of heteroplasmy, if stable, should be enough to rescue phenotypic defects of oxidative phosphorylation (59,65). However, the restoration of the initial 65% of mutant mtDNA in 7–8 days was consistently observed, most probably being a consequence of the recombinant RNA degradation, followed by accelerated replication of shorter mutant mtDNA molecules due to the previously hypothesized effect of ‘replicative advantage of deleted mtDNA’ (65,66).
To check whether the permanent presence of recombinant RNA in human cybrid cells would lead to a more stable effect on the heteroplasmy, another strategy was used, consisting in a stable expression of 5S rRNA-based recombinant RNA genes. This approach appeared to have also some inconvenient features, because the expression of recombinant 5S rRNA genes was relatively weak, detectable by RT-PCR only, which can be explained by competition with more than 200 naturally present 5S rRNA genes for the transcription and nuclear export protein factors. A variation in the recombinant RNA expression in different clones led to various effects on the heteroplasmy level—no changes were detected in 70% of clones, and only in 8 from 25 clones of each series the shift of KSS mtDNA heteroplasmy level from 65% to 37–54% was observed. On the other hand, this strategy provided a heteroplasmy shift stable during several weeks of cells cultivation, which is a first successful attempt to change the proportion of mutant mtDNA in living cells by exploiting RNA import.
In our previous study, we have shown that 5S rRNA molecules with substitution of the helix III and loop C (Figure 1B) by a heterologic sequence can be imported into isolated human mitochondria (which was confirmed also in this study, Figure 2A), but had been imported in vivo 20 fold more efficiently than the WT 5S rRNA, probably due to the absence of competition between mitochondrial import factors and the ribosomal protein L5 for binding to a mutant 5S rRNA (31). This finding has been confirmed in this study, because at least 25% of recombinant 5S rRNA internalized by cells has been found in purified mitochondrial fraction (Supplementary Figure S8C). Nevertheless, the amount of stably expressed recombinant 5S rRNA was almost 1000-fold lower than in cells transiently transfected with recombinant FD-RNAs. Surprisingly, this weak but stable expression of the recombinant 5S rRNA was sufficient for a shift of mitochondrial heteroplasmy level (Figure 5). We can hypothesize that the persistent presence of even small amounts of anti-replicative 5S rRNA in mitochondria might have an effect on the mutant mtDNA comparable with that of more abundant but rapidly degradable small FD-RNA.
Most part of pathogenic mutations in mtDNA lead to the manifestation of biochemical and clinical defects only when a threshold level of heteroplasmy has been reached. This level may be different not only for different mutations but also for the same mutation in various tissues (2). Normally, phenotypic effect of mtDNA mutation is much more prominent in muscles and nervous tissues than in fibroblasts (50,67). This was exactly the case of KSS deletion, for which low values (below or at the 5% of normal) for cytochrome c oxidase (complex IV) activity and for the ratios of complex I, III or IV activity to either citrate synthase or complex II in patient’s muscles containing 72% mutant mtDNA has been shown. However, cybrid cells containing 65% mutant mtDNA, generated and used in this study, were characterized by a very low (10% ± 2%) decrease of oxygen consumption comparing to control 143B cell line. Thus, this is not surprising that the mitochondrial translation was only slightly affected (25% decrease in KSS cybrids). We can hypothesize that in cybrid cells, normal mtDNA (which is present in proportion 1:3 to mutant molecules) supports the level of mt tRNA expression sufficient for almost normal mitochondrial translation. Nevertheless, our data show that a stable shift of heteroplasmy can result in partial restoration of specific mitochondrial translation defects found in KSS cybrid cells (Figure 5E).
Both strategies described here for cultured human cells can be developed as a potential basis for therapy of mitochondrial diseases. Stable expression of 5S-based recombinant RNA in cells should be optimized to give more pronounced effects on the heteroplasmy. Transient transfection approach also appears to be an interesting perspective, because it represents not genetic but rather pharmacological intervention, which can give positive results if the way to stabilize the effect of anti-replicative agents could be found.
To summarize, we demonstrate here for the first time that the replication of mtDNA containing a known pathogenic mutation may be specifically affected by RNA molecules bearing oligonucleotide stretches complementary to the mutant region and that these molecules may be targeted into human mitochondria using artificially engineered RNA based on 5S rRNA or tRNA as vectors. Although the inhibitory effect was partial (15–35% decrease of mutated mtDNA level), it may have a long-term therapeutic interest, because only high levels of mutations in human mtDNA become pathogenic.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–9.
FUNDING
CNRS (Centre National de Recherche Scientifique); University of Strasbourg; AFM (Association Française contre les Myopathies); ANR (Agence Nationale de la Recherche); FRM (Fondation pour la Recherche Médicale); ARCUS/Suprachem collaboration program; LIA collaboration program (ARNmitocure); National Program ‘Investissement d'Avenir’ (Labex MitoCross); FRM and AFM PhD fellowships (to Y.T.). Funding for open access charge: Institutional budget (Strasbourg University & CNRS).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to E. Shoubridge (Montreal Institute of Neurology) for providing the rho0 143B cell line, to F. Monneaux and F. Gros (IBMC, Strasbourg) for FACS analysis, to M. Vysokikh (Strasbourg) for cell respiration measurement and to J. Holt and A. Reyes (Cambridge, UK) for detailed protocol of mtDNA isolation and helpful advices concerning 2DNAGE.
REFERENCES
- 1.Ruiz-Pesini E, Lott MT, Procaccio V, Poole JC, Brandon MC, Mishmar D, Yi C, Kreuziger J, Baldi P, Wallace DC. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35:D823–D828. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 3.Kolesnikova OA, Entelis NS, Mireau H, Fox TD, Martin RP, Tarassov IA. Suppression of mutations in mitochondrial DNA by tRNAs imported from the cytoplasm. Science. 2000;289:1931–1933. doi: 10.1126/science.289.5486.1931. [DOI] [PubMed] [Google Scholar]
- 4.Mahata B, Mukherjee S, Mishra S, Bandyopadhyay A, Adhya S. Functional delivery of a cytosolic tRNA into mutant mitochondria of human cells. Science. 2006;314:471–474. doi: 10.1126/science.1129754. [DOI] [PubMed] [Google Scholar]
- 5.Manfredi G, Fu J, Ojaimi J, Sadlock JE, Kwong JQ, Guy J, Schon EA. Rescue of a deficiency in ATP synthesis by transfer of MTATP6, a mitochondrial DNA-encoded gene, to the nucleus. Nat. Genet. 2002;30:394–399. doi: 10.1038/ng851. [DOI] [PubMed] [Google Scholar]
- 6.Karicheva OZ, Kolesnikova OA, Schirtz T, Vysokikh MY, Mager-Heckel AM, Lombes A, Boucheham A, Krasheninnikov IA, Martin RP, Entelis N, et al. Correction of the consequences of mitochondrial 3243A>G mutation in the MT-TL1 gene causing the MELAS syndrome by tRNA import into mitochondria. Nucleic Acids Res. 2011;39:8173–8186. doi: 10.1093/nar/gkr546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang G, Shimada E, Zhang J, Hong JS, Smith GM, Teitell MA, Koehler CM. Correcting human mitochondrial mutations with targeted RNA import. Proc. Natl Acad. Sci. USA. 2012;109:4840–4845. doi: 10.1073/pnas.1116792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bacman SR, Williams SL, Garcia S, Moraes CT. Organ-specific shifts in mtDNA heteroplasmy following systemic delivery of a mitochondria-targeted restriction endonuclease. Gene Ther. 2010;17:713–720. doi: 10.1038/gt.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bacman SR, Williams SL, Hernandez D, Moraes CT. Modulating mtDNA heteroplasmy by mitochondria-targeted restriction endonucleases in a ‘differential multiple cleavage-site' model. Gene Ther. 2007;14:1309–1318. doi: 10.1038/sj.gt.3302981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minczuk M, Papworth MA, Miller JC, Murphy MP, Klug A. Development of a single-chain, quasi-dimeric zinc-finger nuclease for the selective degradation of mutated human mitochondrial DNA. Nucleic Acids Res. 2008;36:3926–3938. doi: 10.1093/nar/gkn313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor RW, Chinnery PF, Turnbull DM, Lightowlers RN. Selective inhibition of mutant human mitochondrial DNA replication in vitro by peptide nucleic acids. Nat. Genet. 1997;15:212–215. doi: 10.1038/ng0297-212. [DOI] [PubMed] [Google Scholar]
- 12.Chinnery PF, Taylor RW, Diekert K, Lill R, Turnbull DM, Lightowlers RN. Peptide nucleic acid delivery to human mitochondria. Gene Ther. 1999;6:1919–1928. doi: 10.1038/sj.gt.3301061. [DOI] [PubMed] [Google Scholar]
- 13.Muratovska A, Lightowlers RN, Taylor RW, Turnbull DM, Smith RA, Wilce JA, Martin SW, Murphy MP. Targeting peptide nucleic acid (PNA) oligomers to mitochondria within cells by conjugation to lipophilic cations: implications for mitochondrial DNA replication, expression and disease. Nucleic Acids Res. 2001;29:1852–1863. doi: 10.1093/nar/29.9.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith PM, Lightowlers RN. Altering the balance between healthy and mutated mitochondrial DNA. J. Inherit. Metab. Dis. 2010;34:309–313. doi: 10.1007/s10545-010-9122-6. [DOI] [PubMed] [Google Scholar]
- 15.Salinas T, Duchene AM, Marechal-Drouard L. Recent advances in tRNA mitochondrial import. Trends Biochem. Sci. 2008;33:320–329. doi: 10.1016/j.tibs.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 16.Schneider A. Mitochondrial tRNA import and its consequences for mitochondrial translation. Annu. Rev. Biochem. 2011;80:1033–1053. doi: 10.1146/annurev-biochem-060109-092838. [DOI] [PubMed] [Google Scholar]
- 17.Sieber F, Duchene AM, Marechal-Drouard L. Mitochondrial RNA import: from diversity of natural mechanisms to potential applications. Int. Rev. Cell. Mol. Biol. 2011;287:145–190. doi: 10.1016/B978-0-12-386043-9.00004-9. [DOI] [PubMed] [Google Scholar]
- 18.Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, et al. The human mitochondrial transcriptome. Cell. 2011;146:645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kren BT, Wong PY, Sarver A, Zhang X, Zeng Y, Steer CJ. MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol. 2009;6:65–72. doi: 10.4161/rna.6.1.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrey E, Saint-Auret G, Bonnamy B, Damas D, Boyer O, Gidrol X. Pre-microRNA and mature microRNA in human mitochondria. PLoS One. 2011;6:e20220. doi: 10.1371/journal.pone.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bandiera S, Ruberg S, Girard M, Cagnard N, Hanein S, Chretien D, Munnich A, Lyonnet S, Henrion-Caude A. Nuclear outsourcing of RNA interference components to human mitochondria. PLoS One. 2011;6:e20746. doi: 10.1371/journal.pone.0020746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolesnikova OA, Entelis NS, Jacquin-Becker C, Goltzene F, Chrzanowska-Lightowlers ZM, Lightowlers RN, Martin RP, Tarassov I. Nuclear DNA-encoded tRNAs targeted into mitochondria can rescue a mitochondrial DNA mutation associated with the MERRF syndrome in cultured human cells. Hum. Mol. Genet. 2004;13:2519–2534. doi: 10.1093/hmg/ddh267. [DOI] [PubMed] [Google Scholar]
- 23.Rubio MA, Rinehart JJ, Krett B, Duvezin-Caubet S, Reichert AS, Soll D, Alfonzo JD. Mammalian mitochondria have the innate ability to import tRNAs by a mechanism distinct from protein import. Proc. Natl Acad. Sci. USA. 2008;105:9186–9191. doi: 10.1073/pnas.0804283105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puranam RS, Attardi G. The RNase P associated with HeLa cell mitochondria contains an essential RNA component identical in sequence to that of the nuclear RNase P. Mol. Cell. Biol. 2001;21:548–561. doi: 10.1128/MCB.21.2.548-561.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang G, Chen HW, Oktay Y, Zhang J, Allen EL, Smith GM, Fan KC, Hong JS, French SW, McCaffery JM, et al. PNPASE Regulates RNA Import into Mitochondria. Cell. 2010;142:456–467. doi: 10.1016/j.cell.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Entelis NS, Kolesnikova OA, Dogan S, Martin RP, Tarassov IA. 5 S rRNA and tRNA Import into Human Mitochondria. Comparison of in vitro requirements. J. Biol. Chem. 2001;276:45642–45653. doi: 10.1074/jbc.M103906200. [DOI] [PubMed] [Google Scholar]
- 27.Magalhaes PJ, Andreu AL, Schon EA. Evidence for the presence of 5S rRNA in mammalian mitochondria. Mol. Biol. Cell. 1998;9:2375–2382. doi: 10.1091/mbc.9.9.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshionari S, Koike T, Yokogawa T, Nishikawa K, Ueda T, Miura K, Watanabe K. Existence of nuclear-encoded 5S-rRNA in bovine mitochondria. FEBS Lett. 1994;338:137–142. doi: 10.1016/0014-5793(94)80351-x. [DOI] [PubMed] [Google Scholar]
- 29.Smirnov A, Comte C, Mager-Heckel AM, Addis V, Krasheninnikov IA, Martin RP, Entelis N, Tarassov I. Mitochondrial enzyme rhodanese is essential for 5 S ribosomal RNA import into human mitochondria. J. Biol. Chem. 2010;285:30792–30803. doi: 10.1074/jbc.M110.151183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smirnov A, Entelis N, Martin RP, Tarassov I. Biological significance of 5S rRNA import into human mitochondria: role of ribosomal protein MRP-L18. Genes Dev. 2011;25:1289–1305. doi: 10.1101/gad.624711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smirnov A, Tarassov I, Mager-Heckel AM, Letzelter M, Martin RP, Krasheninnikov IA, Entelis N. Two distinct structural elements of 5S rRNA are needed for its import into human mitochondria. RNA. 2008;14:749–759. doi: 10.1261/rna.952208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolesnikova O, Kazakova H, Comte C, Steinberg S, Kamenski P, Martin RP, Tarassov I, Entelis N. Selection of RNA aptamers imported into yeast and human mitochondria. RNA. 2010;16:926–941. doi: 10.1261/rna.1914110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King MP, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 34.Mager-Heckel AM, Entelis N, Brandina I, Kamenski P, Krasheninnikov IA, Martin RP, Tarassov I. The analysis of tRNA import into mammalian mitochondria. Methods Mol. Biol. 2007;372:235–253. doi: 10.1007/978-1-59745-365-3_17. [DOI] [PubMed] [Google Scholar]
- 35.Srere PA. Spectral evidence for complex formation between oxaloacetate and citrate-condensing enzyme. Biochim. Biophys. Acta. 1965;99:197–200. doi: 10.1016/s0926-6593(65)80028-5. [DOI] [PubMed] [Google Scholar]
- 36.Kern D, Dietrich A, Fasiolo F, Renaud M, Giege R, Ebel JP. The yeast aminoacyl-tRNA synthetases. Methodology for their complete or partial purification and comparison of their relative activities under various extraction conditions. Biochimie. 1977;59:453–462. doi: 10.1016/s0300-9084(77)80050-3. [DOI] [PubMed] [Google Scholar]
- 37.Entelis NS, Krasheninnikov IA, Martin RP, Tarassov IA. Mitochondrial import of a yeast cytoplasmic tRNA (Lys): possible roles of aminoacylation and modified nucleosides in subcellular partitioning. FEBS Lett. 1996;384:38–42. doi: 10.1016/0014-5793(96)00259-1. [DOI] [PubMed] [Google Scholar]
- 38.Kun E, Kirsten E, Piper WN. Stabilization of mitochondrial functions with digitonin. Methods Enzymol. 1979;55:115–118. doi: 10.1016/0076-6879(79)55016-2. [DOI] [PubMed] [Google Scholar]
- 39.Wong TW, Clayton DA. In vitro replication of human mitochondrial DNA: accurate initiation at the origin of light-strand synthesis. Cell. 1985;42:951–958. doi: 10.1016/0092-8674(85)90291-0. [DOI] [PubMed] [Google Scholar]
- 40.Wong TW, Clayton DA. Isolation and characterization of a DNA primase from human mitochondria. J. Biol. Chem. 1985;260:11530–11535. [PubMed] [Google Scholar]
- 41.Kohrer C, Xie L, Kellerer S, Varshney U, RajBhandary UL. Import of amber and ochre suppressor tRNAs into mammalian cells: a general approach to site-specific insertion of amino acid analogues into proteins. Proc. Natl Acad. Sci. USA. 2001;98:14310–14315. doi: 10.1073/pnas.251438898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reyes A, Yasukawa T, Holt IJ. Analysis of replicating mitochondrial DNA by two-dimensional agarose gel electrophoresis. Methods Mol. Biol. 2007;372:219–232. doi: 10.1007/978-1-59745-365-3_16. [DOI] [PubMed] [Google Scholar]
- 43.Hyvarinen AK, Pohjoismaki JL, Reyes A, Wanrooij S, Yasukawa T, Karhunen PJ, Spelbrink JN, Holt IJ, Jacobs HT. The mitochondrial transcription termination factor mTERF modulates replication pausing in human mitochondrial DNA. Nucleic Acids Res. 2007;35:6458–6474. doi: 10.1093/nar/gkm676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reyes A, Yasukawa T, Cluett TJ, Holt IJ. Analysis of mitochondrial DNA by two-dimensional agarose gel electrophoresis. Methods Mol. Biol. 2009;554:15–35. doi: 10.1007/978-1-59745-521-3_2. [DOI] [PubMed] [Google Scholar]
- 45.Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol. Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- 47.Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. 5S Ribosomal RNA Database. Nucleic Acids Res. 2002;30:176–178. doi: 10.1093/nar/30.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. 5 S rRNA: structure and interactions. Biochem. J. 2003;371:641–651. doi: 10.1042/BJ20020872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sugimoto N, Nakano S, Katoh M, Matsumura A, Nakamuta H, Ohmichi T, Yoneyama M, Sasaki M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- 50.Bornstein B, Mas JA, Fernandez-Moreno MA, Campos Y, Martin MA, del Hoyo P, Rubio JC, Arenas J, Garesse R. The A8296G mtDNA mutation associated with several mitochondrial diseases does not cause mitochondrial dysfunction in cybrid cell lines. Hum. Mutat. 2002;19:234–239. doi: 10.1002/humu.10050. [DOI] [PubMed] [Google Scholar]
- 51.DiMauro S, Schon EA. Mitochondrial DNA mutations in human disease. Am. J. Med. Genet. 2001;106:18–26. doi: 10.1002/ajmg.1392. [DOI] [PubMed] [Google Scholar]
- 52.Holt IJ, Lorimer HE, Jacobs HT. Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell. 2000;100:515–524. doi: 10.1016/s0092-8674(00)80688-1. [DOI] [PubMed] [Google Scholar]
- 53.Bowmaker M, Yang MY, Yasukawa T, Reyes A, Jacobs HT, Huberman JA, Holt IJ. Mammalian mitochondrial DNA replicates bidirectionally from an initiation zone. J. Biol. Chem. 2003;278:50961–50969. doi: 10.1074/jbc.M308028200. [DOI] [PubMed] [Google Scholar]
- 54.Hyvarinen AK, Pohjoismaki JL, Holt IJ, Jacobs HT. Overexpression of MTERFD1 or MTERFD3 impairs the completion of mitochondrial DNA replication. Mol. Biol. Rep. 2011;38:1321–1328. doi: 10.1007/s11033-010-0233-9. [DOI] [PubMed] [Google Scholar]
- 55.Yasukawa T, Reyes A, Cluett TJ, Yang MY, Bowmaker M, Jacobs HT, Holt IJ. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 2006;25:5358–5371. doi: 10.1038/sj.emboj.7601392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pohjoismaki JL, Holmes JB, Wood SR, Yang MY, Yasukawa T, Reyes A, Bailey LJ, Cluett TJ, Goffart S, Willcox S, et al. Mammalian mitochondrial DNA Replication intermediates are essentially duplex but contain extensive tracts of RNA/DNA hybrid. J. Mol. Biol. 2010;397:1144–1155. doi: 10.1016/j.jmb.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wanrooij S, Goffart S, Pohjoismaki JL, Yasukawa T, Spelbrink JN. Expression of catalytic mutants of the mtDNA helicase twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35:3238–3251. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brown TA, Cecconi C, Tkachuk AN, Bustamante C, Clayton DA. Replication of mitochondrial DNA occurs by strand displacement with alternative light-strand origins, not via a strand-coupled mechanism. Genes Dev. 2005;19:2466–2476. doi: 10.1101/gad.1352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith PM, Lightowlers RN. Altering the balance between healthy and mutated mitochondrial DNA. J. Inherit. Metab. Dis. 2011;34:309–313. doi: 10.1007/s10545-010-9122-6. [DOI] [PubMed] [Google Scholar]
- 60.Smith PM, Ross GF, Taylor RW, Turnbull DM, Lightowlers RN. Strategies for treating disorders of the mitochondrial genome. Biochim. Biophys. Acta. 2004;1659:232–239. doi: 10.1016/j.bbabio.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 61.Holt IJ. Mitochondrial DNA replication and repair: all a flap. Trends Biochem. Sci. 2009;34:358–365. doi: 10.1016/j.tibs.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 62.Clayton DA. Mitochondrial DNA replication: what we know. IUBMB Life. 2003;55:213–217. doi: 10.1080/1521654031000134824. [DOI] [PubMed] [Google Scholar]
- 63.Pohjoismaki JL, Goffart S. Of circles, forks and humanity: topological organisation and replication of mammalian mitochondrial DNA. Bioessays. 2011;33:290–299. doi: 10.1002/bies.201000137. [DOI] [PubMed] [Google Scholar]
- 64.Clayton DA. Replication of animal mitochondrial DNA. Cell. 1982;28:693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- 65.Moraes CT, Kenyon L, Hao H. Mechanisms of human mitochondrial DNA maintenance: the determining role of primary sequence and length over function. Mol. Biol. Cell. 1999;10:3345–3356. doi: 10.1091/mbc.10.10.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Diaz F, Bayona-Bafaluy MP, Rana M, Mora M, Hao H, Moraes CT. Human mitochondrial DNA with large deletions repopulates organelles faster than full-length genomes under relaxed copy number control. Nucleic Acids Res. 2002;30:4626–4633. doi: 10.1093/nar/gkf602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bornstein B, Mas JA, Patrono C, Fernandez-Moreno MA, Gonzalez-Vioque E, Campos Y, Carrozzo R, Martin MA, del Hoyo P, Santorelli FM, et al. Comparative analysis of the pathogenic mechanisms associated with the G8363A and A8296G mutations in the mitochondrial tRNA(Lys) gene. Biochem. J. 2005;387:773–778. doi: 10.1042/BJ20040949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.