Abstract
RNA trafficking to dendrites and local translation are crucial processes for superior neuronal functions. To date, several α-amino-3-hydroxy-5-methyl-4-isoxazolepropionate receptor (AMPAR) mRNAs have been detected in dendrites and are subject to local protein synthesis. Here, we report the presence of all AMPAR GluA1-4 mRNAs in hippocampal and cortical rat synaptic spines by synaptoneurosomes analysis. In particular, we showed that dendritic AMPAR mRNAs are present in the Flip versions in the cortex and hippocampus. To further confirm these data, we demonstrate, using in situ hybridization, the dendritic localization of the GluA2 Flip isoform in vitro and in vivo, whereas the Flop variant is restricted mainly to the soma. In addition, we report that dendritic AMPA mRNAs are edited at low levels at their R/G sites; this result was also supported with transfection experiments using chimeric GluA2 DNA vectors, showing that transcripts carrying an unedited nucleotide at the R/G site, in combination with the Flip exon, are more efficiently targeted to dendrites when compared with the edited-Flip versions. Our data show that post-transcriptional regulations such as RNA splicing, editing and trafficking might be mutually coordinated and that the localization of different AMPAR isoforms in dendrites might play a functional role in the regulation of neuronal transmission.
INTRODUCTION
The α-amino-3-hydroxy-5-methyl-4-isoxazolepropionate (AMPA) subtypes of glutamate receptors are among the primary mediators of fast excitatory neurotransmission in the brain, and the proper regulation of their functions plays a key role in superior learning and memory phenomena (1).
AMPA receptors (AMPAR) are heterotetramers consisting of combinations of four different subunits, GluA1 through GluA4. In the adult hippocampus (Hc), GluA1, GluA2 and GluA3 combine to form two distinct populations, GluA1/GluA2 and GluA2/GluA3, which play different roles and are delivered to synapses through distinct mechanisms (2).
Moreover, post-transcriptional modifications such as RNA editing and alternative splicing are functionally relevant for the regulation of AMPAR-mediated neurotransmission. Indeed, these modifications represent sophisticated mechanisms that mediate changes in each receptor subunit that determine key features of the channel, such as permeability, pore-opening kinetics and desensitization and recovery periods (3,4).
Each of the four AMPAR subunits is alternatively spliced in the extracellular region, generating so-called ‘Flip’ and ‘Flop’ variants (5). The splicing event consists of the insertion of a mutually exclusive exon into the mature mRNA, which results in the insertion of 38 amino acids into the extracellular loop in the region located before the fourth transmembrane domain (5). This modification generates channels that differ in their kinetic properties, including the desensitization and resensitization periods (6–10).
In addition to splice variants, the physiological properties of AMPARs are controlled through post-transcriptional RNA editing (11,12), which modifies one or more translation codons, resulting in the generation of functionally distinct proteins from a single gene. The predominant RNA editing mechanism in mammals is adenosine-to-inosine (A–I), which is catalyzed by ADAR (adenosine deaminase acting on RNA) enzymes 1 and 2 (13). GluA2, GluA3 and GluA4 are edited at different sites that are characterized by the type of amino acid substitution that occurs: the Q/R site in AMPAR GluA2 and the R/G site in GluA2, GluA3 and GluA4 (11,12). The GluA2 Q/R-edited subunits confer the capacity for linear current–voltage relationships (14) and impermeability to Ca2+. On the other hand, the R/G editing modulates the kinetic properties of the AMPAR channels (4), thus determining the time course of desensitization and resensitization (4,15). Interestingly, the R/G editing site is located 2 nt upstream of the next 5′-splice site, which generates the Flip/Flop isoforms, and editing affects both the desensitization properties of the AMPAR channels (4,15) and splicing (4,16), even though a functional correlation between the two post-transcriptional events has not yet been elucidated (17,18).
Recently, it has been shown that AMPAR mRNAs are actively transported in dendrites and are subjected to local protein synthesis (19). Growing evidence shows that these mechanisms are involved in a wide range of synaptic plasticity phenomena (20,21). The spatial restriction of gene expression within specific cell compartments, such as synaptic spines, localizes mRNAs and confers the capacity to regulate morphology and neurotransmission at the subcellular level in response to specific stimuli in neurons (21). The stimulation of the metabotropic glutamate receptors increases the dendritic localization of AMPAR mRNAs, whereas the activation of the NMDA receptors selectively decreases their dendritic abundance (19). These data suggest that neuronal activity can control the presence of AMPAR mRNAs within dendrites and synapses, contributing to synaptic plasticity.
Currently, however, there is no information concerning the differential subcellular localization of alternatively spliced and edited/unedited AMPAR transcripts. Using several biochemical and molecular approaches and in vitro and in vivo systems, we report here that RNA splicing, editing and transport might be correlated, thus creating a complex modulation of AMPAR properties.
MATERIALS AND METHODS
Perfusion and tissue processing
All animal procedures were performed in accordance with the guidelines of the European Community Council Directive 86/609/EEC and approved through the Italian Ministry of Health (Project ID: 320/2010). Adult Sprague–Dawley rats were deeply anesthetized following the intraperitoneal injection of Zoletil (60 mg/kg, Virbac, France) and transcardially perfused with 4% paraformaldehyde (PFA), pH 7.4, which was freshly prepared in 0.1% diethylpyrocarbonate-treated (DEPC, Sigma-Aldrich; Milan Italy) H2O containing 4% PFA (Sigma, Milan Italy), 0.4% NaOH (Sigma), 2.5% Sodium dihydrogen phosphate–monohydrate (Sigma, Milan Italy) and 1 mM MgCl2. The brains were post-fixed in the 4% PFA solution for 24 h and subsequently placed in 30% sucrose before sectioning. Coronal sections (40 µm) were stored in cryo-protectant sectioning buffer (30% ethylene glycol, 30% glycerol and 0.05 M phosphate buffer) at −20°C until further processing.
Primary cortical cultures
Rat cortical cultures were prepared as described previously (22). Briefly, cerebral cortices from Day 18 Sprague–Dawley rat embryos (E18, Charles River Laboratories Inc., Wilmington, MA, USA) were mechanically dissociated, and the neurons were resuspended in serum-free neurobasal medium (Life Technologies, Invitrogen Division, Milan, Italy) supplemented with B27 (Life Technologies, Invitrogen Division), 30 U/ml penicillin (Sigma-Aldrich, St. Louis, MO, USA), 30 mg/ml streptomycin (Sigma-Aldrich) and 0.5 mM Glutamax (Life Technologies, Invitrogen Division). The neurons were plated at a density of 32 000 cells/cm2 on 12-mm poly-d-lysine (Sigma-Aldrich) coated glass coverslips and grown at 37°C in 5% CO2. At 3 days after plating, 50% of the medium was replaced with fresh medium. Subsequently, half of the medium was replaced every 6 days for a maximum of 4 weeks.
Riboprobe preparation
Digoxigenin (DIG)-labeled riboprobes detecting β3-Tub, GluA1, pan-GluA2, GluA2 Flip and Flop isoforms and EGFP mRNAs were generated from PCR templates adapted with Sp6 and T7 RNA polymerase sites (see Supplementary Table S1 for primer sequences). Sense and antisense riboprobes were transcribed using a DIG RNA-Labeling Kit (Life Technologies, Ambion, Milan, Italy) according to the manufacturer’s instructions. Briefly, 20 μl of transcription mixture containing 200 ng of template purified cDNA, 1× Transcription Buffer, 0.5 mM ATP, 0.5 mM CTP, 0.5 mM GTP, 0.17 mM UTP, 0.33 mM DIG-UTP (Life Technologies, Ambion) and 40 U of either T7 or SP6 RNA polymerase was used. The reaction was incubated for 2 h at 37°C and terminated upon the addition of 0.5 M EDTA. The riboprobes were subsequently purified in NucAway™ spin columns (Life Technologies, Ambion) and quantified using Nanodrop 1000 (Thermo Scientific, Milan, Italy).
In situ hybridization on primary cortical cells
Fluorescent in situ hybridization was performed according to Muddashetty et al. (23) with minor modifications. Briefly, the neuronal cells were fixed for 20 min in 4% PFA, pH 10.0, at room temperature and permeabilized for 10 min in PBS containing 0.3% Triton X-100 (Sigma-Aldrich). After three washes with PBS/MgCl2, the cells were equilibrated in 1× saline sodium citrate (SSC) buffer. Escherichia coli tRNA (1 μg) and single-stranded salmon sperm DNA (1 μg) were dried and resuspended in 15 μl of 80% formamide/1× SSC, pH 7.0. The mixture was heat denatured and added to 15 μl of hybridization buffer [10% dextran sulfate; 2× SSC; 4 mg/ml BSA (bovine serum albumin); 10 mM sodium phosphate buffer, pH 7.0]. The coverslips were incubated cell-side-down in 30 μl of pre-hybridization mix on parafilm at 37°C in humidified chamber for 1 h. The riboprobes (10–100 ng) were dried with the E. coli tRNA (10 μg) and single-stranded salmon sperm DNA (10 μg) and resuspended in 15 μl of 80% formamide/1× SSC, pH 7.0. The mixture was heat denatured and subsequently mixed with 15 μl of hybridization buffer. The coverslips were incubated cell-side-down in 30 μl of a riboprobe-containing mixture overnight (ON) at 40°C in a humidified chamber. After hybridization, the coverslips were washed for 20 min in 40% formamide/1× SSC at 40°C, followed by three additional washes with 1× SSC for 10 min on a rotary shaker at room temperature.
The cells were washed in TBS50 buffer (50 mM Tris–HCl, pH 7.4; 150 mM NaCl) for 5 min and permeabilized with TBS50 containing 0.3% Triton X-100 for 5 min. The cells were further incubated in IF buffer (2% BSA; 0.1% Triton X-100 in TBS50) for 5 min. The coverslips were incubated cell-side-down in 30 μl of blocking buffer (2% BSA; 2% fetal bovine serum; 0.1% Triton X-100). Subsequently, the cells were incubated with a primary mouse anti-DIG antibody (1:100; Roche Applied Science, Mannheim, Germany) for 1 h in a humidified chamber and washed with IF buffer for 10 min on a rotary shaker at room temperature. AlexaFluor 555-conjugated donkey-anti-mouse (Life Technologies, Invitrogen) was used as the secondary antibody, and the cells were incubated for 1 h. Coverslips were mounted with Slow Fade Gold antifade reagent (Life Technologies, Invitrogen). Samples were imaged on a Zeiss LSM 510 Meta confocal microscope (Carl Zeiss, Milan, Italy), using identical settings for sense or antisense probes.
Immunofluorescence staining
For immunofluorescence analysis, the cells were fixed in 4% PFA in phosphate-buffered saline (4% PFA–PBS, Life Technologies, Invitrogen) for 15 min at room temperature and permeabilized in PBS containing 0.2% Triton X-100 (Sigma-Aldrich) for 5 min. Subsequently, the cells were rinsed in PBS and washed twice with 0.15 M glycine in PBS for 5 min. The cells were pre-incubated in blocking solution (Roche Applied Science) at room temperature (RT) for 30 min and then further incubated with primary antibodies diluted in the blocking solution at RT for 1 h. The primary antibodies used were rabbit anti-MAP2 (1:800; PRB-547C, Covance, Harrogate, UK), rabbit anti-FMRP (Fragile X Mental Retardation Protein) (1:300, ab69815 Abcam, Cambridge, UK) and rabbit anti-CPEB3 (Cytoplasmatic Polyadenilation Element-Binding Protein 3) (1:100, ab10883 Abcam). Unconjugated primary antibodies were recognized with secondary antibodies conjugated with Alexa-Fluor 488 or Alexa-Fluor 555 dyes (Life Technologies, Invitrogen). Following incubation with each antibody, the cells were washed three times for at least 20 min with PBS and mounted on coverslips in SlowFade Gold antifade reagent (Life Technologies, Invitrogen). The images were acquired on Zeiss LSM 510 Meta confocal microscope (Carl Zeiss, Milan, Italy). Colocalization analysis was performed using the colocalization function in the LSM software, following manufacturer’s instruction. As quantitative parameters the ‘weighted colocalization coefficients’ were used [sum of intensities of co-localizing pixels in the 555 channel (red-RNA hybridization signal) when compared with the overall sum of pixel intensities above threshold in the same channel]; the analyses were conducted referring to an area of 30 µm in length of the distal dendrites (n≥ 20).
In situ hybridization in vivo
In situ hybridization experiments on brain coronal slices were performed under RNAse-free conditions. All solutions were prepared with DEPC-treated H2O. The in situ hybridization was developed using the peroxidase method (Vectastain Elite, Vector laboratories, Burlingame, CA, USA). Free-floating sections were post-fixed for 3 h in 4% PFA at RT, washed twice in 0.1 Tween-20 in 1× PBS (PBST) at RT for 5 min and then treated with 0.3% H2O2 (Sigma-Aldrich) in H2O for 20 min to block endogenous peroxidase. Following three washes for 5 min in PBST, the slices were quickly washed in H2O and permeabilized with 2.3% sodium meta-periodate (Sigma-Aldrich) in H2O at RT for 5 min. Subsequently, the section was quickly washed in H2O and incubated in 1% sodium borohydride (Sigma-Aldrich) in 0.1 M Tris–HCl buffer, pH 7.5 at RT for 10 min followed by two washes in PBST at RT for 3 min. The slices were digested with 8 µg/ml proteinase K (Sigma-Aldrich) in PBST at RT for 20 min and washed twice in PBST at RT for 5 min. After digestion, the tissue slices were fixed in 4% PFA at RT for 5 min and washed three times in PBST at RT for 10 min. The slices were incubated at 55°C for 90 min in pre-hybridization solution containing 20 mM Tris–HCl (pH 7.5) (Sigma-Aldrich), 1 mM EDTA (Life Technologies, Gibco), 1× Denhardt’s solution (Life Technologies, Invitrogen), 300 mM NaCl, 100 mM DTT (Sigma-Aldrich), 0.5 mg/ml salmon sperm DNA (Life Technologies, Gibco), 0.5 mg/ml polyadenylic acid (Sigma-Aldrich) and 50% formamide (Sigma-Aldrich). The slices were subsequently incubated ON at 55°C in hybridization solution containing 10% dextransulfate and 100–200 ng/ml DIG-labeled riboprobes. The next day, the slices were washed twice in 2× SSC containing 0.1% Tween 20 (SSCT) and 50% deionized formamide at 55°C for 30 min, transferred to 2× SSCT at 55°C for 20 min and subjected to two final washes in 0.2× SSCT at 60°C for 30 min. Subsequently, the slices were blocked in PBS containing 0.1% Triton X-100 (Sigma) with serum contained in the kit (Vectastain Elite, Vector laboratories, Burlingame, CA, USA). The cells were then incubated for 1 h at RT followed by ON incubation with anti-DIG (1:50, Roche Applied Science). After rinsing with PBST, the slices were incubated for 1 h with a biotinylated secondary antibody (Vector laboratories, Vectastain Elite, Burlingame, CA, USA) at RT. Following intermittent rinses in PBST, the avidin–biotin–peroxidase complex was applied for 30 min, followed by peroxidase detection for 1 min (DAB, 3,3′-diaminobenzidine tetrahydrochloride; enhanced liquid substrates system, 1:30; Sigma Aldrich). The sections were mounted on gelatin-coated slides, completely dried, rinsed for 1 min in distilled water, dehydrated with graded concentrations of alcohol (50, 70, 90 and 100%; 1 min each), immersed in xylene and mounted on coverslips. The images were acquired using an optical microscope (Olympus BX50).
Padlock probe hybridization technique
For the in situ detection of individual transcripts with padlock probes and target-primed Rolling Cycle Amplification (RCA) (24), gene-specific cDNA was generated using locked nucleic acid (LNA)-modified primers. After RNA degradation, the cDNA was hybridized to the padlock probe. The probes were ligated and subjected to RCA. Rolling Circle Particles (RCPs) were identified through the hybridization of fluorescent detection oligoprobes. This procedure was performed as in Larsson et al. (24) with minor modifications.
DIV14 cortical neuron cells were fixed with 3% PFA, pH 10, for 30 min at room temperature. Permeabilization was performed using a series of 70, 85 and 99.5% ethanol washes for 3 min each for dehydration followed by a 10-min incubation in 0.15 M HCl and an additional 10-min incubation in PBS containing 0.3% Triton X-100. The cells were pre-incubated in a M-MuLV reaction buffer for in situ cDNA synthesis. A total of 1 μM of LNA primer (Eurogentec, Biosense, Milan Italy; sequences in Supplementary Table S2) was added to the slides with 20 U/μl of RevertAid H minus M-MuLV reverse transcriptase (Thermo Scientific, Fermentas, Milan, Italy), 100 µM dNTPs (Thermo Scientific, Fermentas), 0.2 μg/μl BSA (Thermo Scientific, Fermentas) and 1 U/μl RiboLock RNase Inhibitor (Thermo Scientific, Fermentas) in the M-MuLV reaction buffer. The slides were incubated ON at 37°C in a humidified chamber. After incubation, the slides were washed briefly in PBS-T (DEPC–PBS with 0.05% Tween-20; Sigma- Aldrich), followed by post-fixation in 2% PFA in DEPC–PBS for 30 min at room temperature. After post-fixation, the samples were washed twice in PBS-T.
To generate the target cDNA strands for padlock probe hybridization, the RNA portion of the RNA–DNA hybrids was degraded with ribonuclease H in the same step as the padlock probe hybridization and ligation. The samples were then pre-incubated in T4 DNA ligase buffer (Thermo Scientific, Fermentas). In parallel, the padlock probes (sequences in Supplementary Table S2) were phosphorylated using polynucleotide kinase (Thermo Scientific, Fermentas) according to the manufacturer’s instructions. Then, 100 nM of each padlock probe was added to T4 DNA ligase buffer supplemented with 1 mM ATP, 50 mM NaCl, 0.1 U/μl T4 DNA ligase, 0.4 U/μl RNase H and 0.2 μg/μl BSA. After ligation with T4 DNA ligase, the slides were washed in DEPC-treated 2× SSC with 0.05% Tween-20 at 37°C for 5 min and rinsed in PBS-T.
The slides were pre-incubated briefly in Φ29 DNA polymerase buffer (Thermo Scientific, Fermentas). The RCA was performed with 1 U/μl Φ29 DNA polymerase (Thermo Scientific, Fermentas) in the supplied reaction buffer containing 1 U/μl RiboLock RNase Inhibitor, 250 μM dNTPs, 0.2 μg/μl BSA and 5% glycerol. The incubation was conducted for 60 min at 37°C followed by a wash in PBS-T.
The RCPs were visualized using 100 nM of the Flip or Flop detection probe (Flip: Alexa-Fluor 555; Flop: Alexa-Fluor 488) in 2× SSC and 20% formamide at 37°C for 30 min. The cells were washed three times with PBST (0.05% Tween-20). The coverslips were mounted with Slow Fade Gold antifade reagent (Life Technologies, Invitrogen). The images were acquired on a Zeiss LSM 510 Meta confocal microscope. The accuracy of the protocol used and the specificity of the padlock probes were tested on PC12 cells transfected with DNA construct expressing GluA2 Flip and Flop mRNA (Supplementary Materials and Supplementary Figure S7).
Synaptoneurosomal preparation
Synaptoneurosomes were prepared from Cx and Hc by sequential centrifugation through Percoll and Optiprep gradients (25). The dissected tissues were homogenized in isotonic buffer [0.32 M sucrose, 1 mM EDTA pH 7.4, 1 mM dithiothreitol, 30 U/ml RNase OUT (Life Technologies, Invitrogen) and Protease Inhibitors (Roche, Applied Science)].
The homogenate was centrifuged at 1000g for 10 min, and the supernatant was loaded on a Sucrose–Percoll discontinuous gradient (3, 10, 15 and 23%) and centrifuged at 32 000g for 5 min (26). Crude synaptosomes were recovered from 15% to 23% interface, washed in PBS and centrifuged at 12 000g for 4 min. The synaptoneurosome-containing pellet was quickly resuspended in Optiprep solution (50% Optiprep, HEPES 10 mM pH 7.4, 65 mM sucrose, 1 mM MgSO4, 30 U/ml RNase OUT and Protease Inhibitor), loaded on an Optiprep discontinuous gradient (9, 12.5, 15, 25 and 35%) and centrifuged at 10 000g for 20 min (27). The synaptoneurosome fraction (15–25% interface) was removed and centrifuged at 30 000g for 5 min. The synaptoneurosome pellet was resuspended in the appropriate lysis buffer for subsequent analysis.
Ribonucleoprotein immunoprecipitation
A total of 100 mg of rat Cx was homogenized in 50 mM Tris–HCl, pH 8, 150 mM NaCl, 1% Triton X-100, 1 mM DTT, complete protease inhibitors (Roche, Applied Science) and 80 U/ml RNase Out. The RNA immunoprecipitation (RIP) was performed using an Immunoprecipitation Kit with Protein G Dynabeads® (Life Technologies, Invitrogen). The beads were initially saturated in buffer NT2 supplemented with 5% BSA for 1 h at 4°C.
FMRP and CPEB3 antibodies and rabbit IgG (10 µg) were bound to 50 µl of beads. The homogenate was incubated with the beads for 1 h at 4°C, and the beads were then collected using a magnet. After five washes with washing buffer, the immune complex was eluted with elution buffer and the RNAs co-immunoprecipitated with target RBPs were extracted for RT–PCR.
RNA extraction and retro-transcription
The RNA obtained from the synaptoneurosomal (SNS) preparation and the RIP samples was extracted using a Qiagen Micro Kit according to the manufacturer’s instructions (Qiagen, Milan, Italy). RNA quantification and quality controls were spectrophotometrically determined using AGILENT Bioanalyzer 2100 lab-on-a-chip technology (AGILENT Technologies, Santa Clara, CA, USA).
Reverse transcription was performed using the Moloney murine leukemia virus-reverse transcriptase (MMLV–RT; Invitrogen). Briefly, the 2 µg of RNA was mixed with 2.2 μl of 0.2 ng/μl random hexamer (Life Technologies, Invitrogen), 10 μl of 5× buffer (Life Technologies, Invitrogen), 10 μl of 2 mM dNTPs, 1 μl of 1 mM DTT (Life Technologies, Invitrogen), 0.4 μl of 33 U/μl RNasin (Promega, Madison, WI, USA) and 2 μl MMLV-RT (200 U/μl) in a final volume of 50 μl. The reaction mixture was incubated at 37°C for 2 h, and the enzyme was then inactivated at 95°C for 10 min. To perform PCR reactions, 1 µl of the reverse transcription product was mixed with 12.5 µl of 2.5× Master Mix (Promega, containing 3 mM MgCl2, 400 µM dNTP, 50 U/ml of Taq DNA polymerase) and 0.7 µl of each forward and reverse primer 20 µM, in a final volume of 25 µl. The primer sequences to amplify the target transcript are reported in Supplementary Table S3.
RNA editing level quantification
The quantification of AMPA GluA2 Q/R site and GluA2, GluA3 and GluA4 R/G sites editing levels was performed using sequence analysis as described previously (28). Briefly, in the electropherogram resulting after sequencing analysis of the PCR product obtained from a pool of transcripts that may or may not get edited, the nucleotide that undergoes the editing reaction appeared as two overlapping peaks: A for unedited nucleotides and G for the edited ones. We previously reported that the editing level might be reliably calculated as a function of the ratio between the G peak area and the A plus G peak areas (28–31). The areas representing the amount of each nucleotide were quantified using Discovery Studio (DS) Gene 1.5 (Accelrys Inc., San Diego, CA, USA) and the means and standard errors for each experimental group were calculated and used for subsequent statistical analyses. Regarding the R/G sites, the editing levels were determined independently for the Flip and Flop splicing variants of GluA2, GluA3 and GluA4 after specific and separate RT–PCR reactions (see primer sequence in Supplementary Table S3). Supplementary Figure S8 reported representative electropherograms used to calculate the editing levels.
Flip/Flop alternative splicing quantification
The levels of the Flip/Flop splicing variants were evaluated using sequence analysis as described previously (22,30). Briefly, for each of the AMPAR subunits, both isoforms were amplified using a pair of common primers located in the previous and following exons of Flip/Flop cassette (see primer sequence in Supplementary Table S3). The amplification products, consisting of variable level of Flip and Flop isoforms, were sequenced. Flip and Flop exons exhibited mismatches of A/G in specific locations. The relative expression of the Flip exon can be reliably calculated as the ratio of the peak area of G (Flip) and the sum of the peak areas A + G (Flip + Flop). The areas representing the amount of each nucleotide were quantified using DS Gene 1.5 (Accelrys Inc., San Diego, CA, USA), and the means and standard errors for each experimental group were calculated and used for subsequent statistical analyses.
Statistical analysis
The statistical analysis of the editing and splicing data was performed using one-way ANOVA followed by specific post hoc tests, as indicated in the figure legends.
DNA constructs
The DNA constructs used for transfection were derived from the plasmid vector pEGFP-c1 (Takara Bio Europe/Clontech, Saint-Germain-en-Laye France). The GluA2 mRNA sequence from nucleotides 2540 to 5607 (ref seq: NM_017261) was inserted downstream the EGFP-coding sequence into the multi-cloning site using a PstI/SacI restriction digestion. The inserted sequence included the last part of GluA2 cds (including Flip or Flop exon) and the complete 3′-UTR of 2524 nt. The 5′-UTR of GluA2 (nucleotides 1–433) was inserted upstream of the EGFP cds, using NheI/AgeI digestion. The DNA constructs would generate chimeric mRNA molecules consisting of 5′-UTR of GluA2 + EGFP cds + GluA2-Flip or Flop exon + 3′-UTR GluA2 (see the scheme reported in Supplementary Figure S9). Because of the insertion of a stop codon after the EGFP cds, the C-terminal domain of GluA2 is not translated. Therefore, every DNA construct, while producing different chimeric RNAs, would only express the EGFP protein that is use to detect transfected neurons. As a positive control for mRNA dendritic transport, the DTE (Dendritic Targeting Element) sequence of MAP2 was cloned (640 nt from 3′-UTR region) downstream of the EGFP sequence (32). To test whether the DNA constructs used might have different transcriptional and translational efficiency, RNA expression and western blot analysis were performed on transfected PC12 cells (Supplementary Materials and Supplementary Figure S9)
Magnetofection of primary cortical neurons
Primary cortical cultures were plated at a density of 44 000 cells/cm2 and transfected (DIV2) using paramagnetic nanobeads (NeuroMag, Oz Biosciences, Marseille, France). The complete medium was replaced before magnetofection with serum-free neurobasal medium. In general, each plasmid was incubated with NeuroMag beads at a ratio of 1 μg:3.5 μl in 100 μl of Neurobasal (Life Technologies) for 20 min and added drop-wise to the cultures. The cells were incubated on top of a magnetic plate (Oz Biosciences) for 20 min and the complete medium was restored after 1 h. Neurons were maintained in culture until DIV14. The pEGFP CMV promoter was induced (33) with 1 μM forskolin for 3 h to activate transcription. The presence of the chimeric transcript in the cell soma or in dendrites was assessed using fluorescence in situ hybridization with DIG-labeled riboprobes complementary to the EGFP sequence. Images of transfected neurons were acquired with a ZEISS LSM 510 confocal microscope. Using the integrated analysis software (Zeiss LSM image examiner), the fluorescence intensity from the cell soma up to 100 µm of the apical dendrites was calculated. For each experimental condition, a minimum of 20 dendrites were analyzed, and the averages and standard errors were calculated. The method was validated using DTE-MAP2 DNA construct as a positive control.
RESULTS
Dendritic localization of AMPAR mRNAs
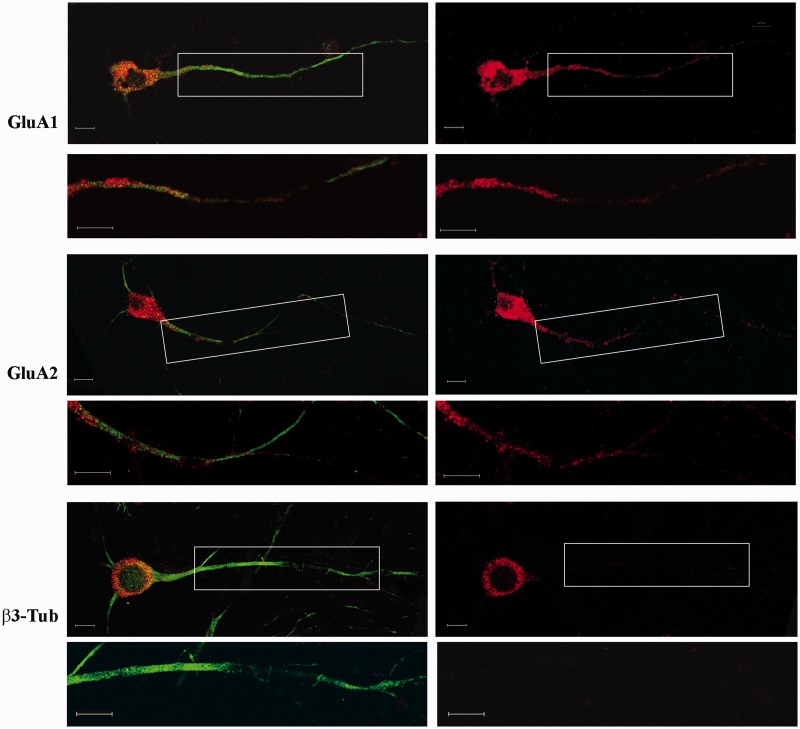
The subcellular localization of AMPAR-coding mRNAs was determined using in situ hybridization in cultured cortical neurons. The hybridization signals of GluA1 and GluA2 mRNAs were present in both the cell soma and dendrites of cultured cortical cells immunolabeled with a specific marker for Microtubule-Associated Protein-2 a (MAP2a). The Tubulin β-3 (β3-Tub) mRNA, which presented cell soma-restricted localization, was used as a negative control for dendritic localization (Figure 1). Sense probes did not show any hybridization signal (Supplementary Figure S1).
Figure 1.
Fluorescence in situ hybridization of GluA1, GluA2 and β3-Tub mRNAs in primary cortical neurons. The right panel shows the hybridization signal (red) of the different mRNAs. In the left panel, the figures are merged with the immunostaining for MAP2. Under each panel, high magnification of the boxed dendritic regions are reported. Scale bar is 10 µm.
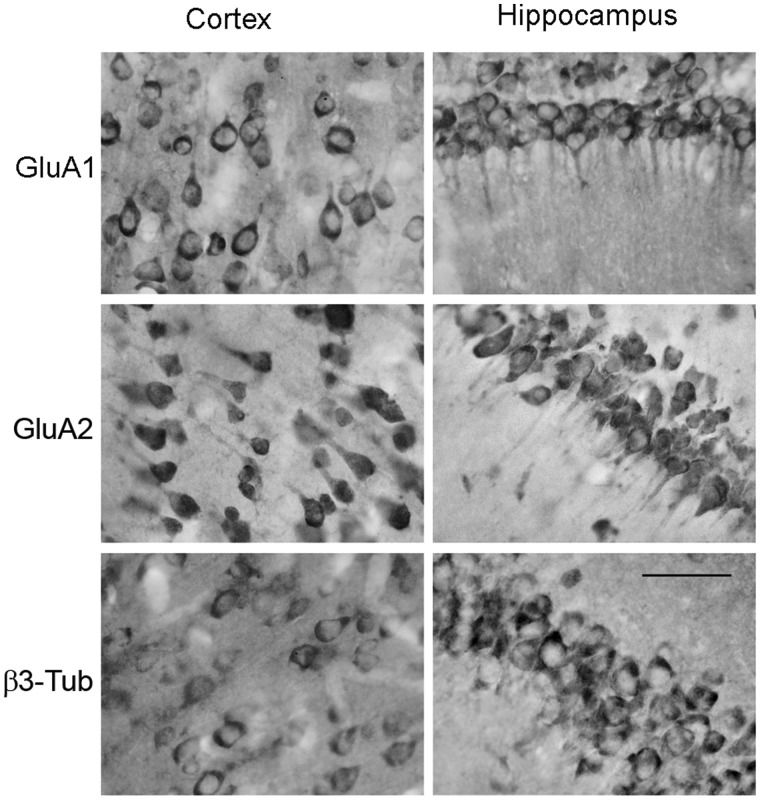
The distribution of GluA1 and GluA2 mRNAs was also analyzed in sections of the rat Hc and cerebral cortex (Cx) of adult naïve animals. For these studies, DIG-labeled antisense probes combined with alkaline phosphatase-conjugated anti-DIG antibodies were used. In vivo, GluA1 and GluA2 mRNAs showed a strong signal in the cell bodies across hippocampal and cortical regions, together with the presence of a clear signal in the proximal dendrites. The β3-Tub mRNA hybridization showed only a somatic-restricted signal (Figure 2). Sense probes did not show hybridization signal (Supplementary Figure S2). As a further control, the specificity of GluA1 and GluA2 antisense probes was tested in reverse dot blot experiments (Supplementary Materials and Supplementary Figure S3).
Figure 2.
Subcellular distribution of GluA1, GluA2 and β3-Tub mRNAs in the Cx (left panel) and rat Hc (right panel). In situ hybridization with alkaline phosphatase detection was used to visualize target mRNAs in coronal sections of the Cx and CA1 region of the Hc. Scale bar is 50 µm.
Colocalization of GluA1 and GluA2 mRNAs with RNA-binding proteins
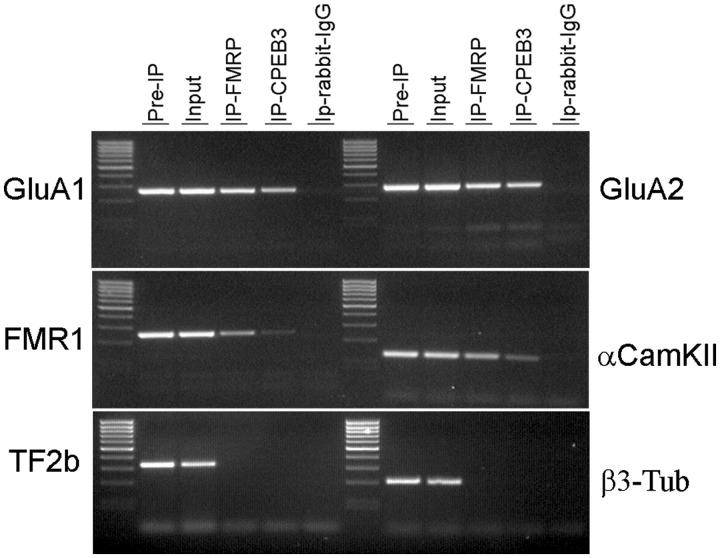
To test the possible interaction of GluA1 and GluA2 mRNAs with the proteins of neuronal RNA transport granules, the RNA–protein complex mediating cytoplasmic translation inhibition and dendritic transport, we performed RIP from rat Cx using antibodies specific for FMRP, a standard component of RNA granules (21) and CPEB3 that has recently been shown to bind directly GluA1 and GluA2 mRNAs (34,35).
The results of the RIP analysis show an enrichment of GluA1 and GluA2 mRNAs in both FMRP and CPEB3 IPs (Figure 3). As positive controls, fragile X mental retardation 1 (FMR1), Ca2+/calmodulin-dependent protein kinase (αCamKII) mRNAs were amplified from both FMRP and CPEB3 IPs; in contrast, transcription factor IIB (TF2b) mRNA and β3-Tub mRNAs, whose localizations are known to be restricted to the cell soma, were not detected in FMRP and CPEB3 IPs.
Figure 3.
RIP analysis of FMRP and CPEB3 protein from the rat Cx. The RT–PCR products for FMR1, αCamKII, GluA1, GluA2, TF2b and β3-Tub mRNAs are shown.
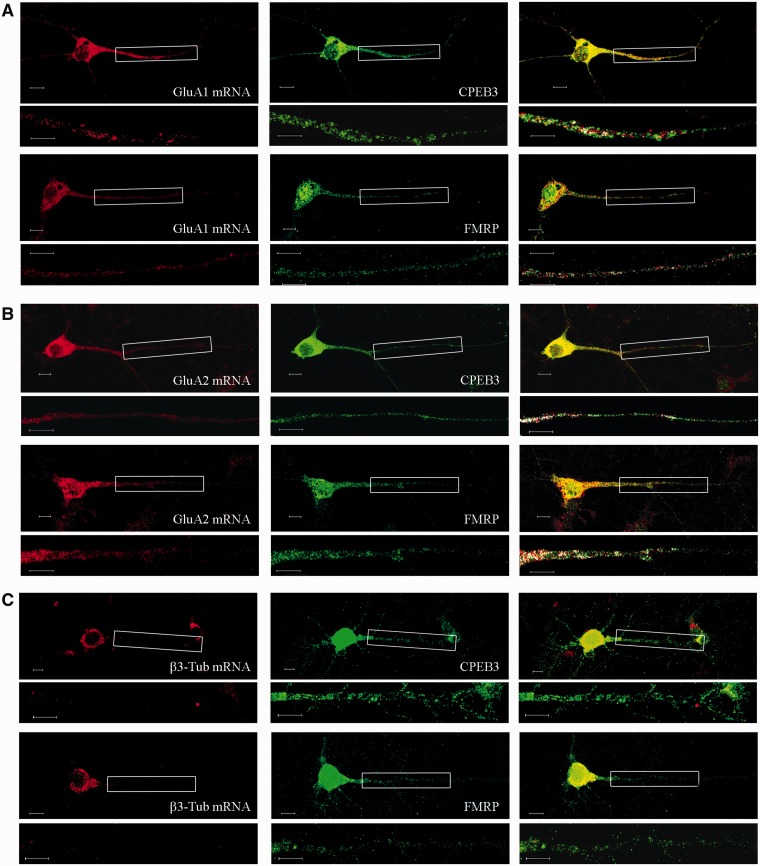
In primary cortical cultures, co-localization analysis of FMRP and CPEB3 immunofluorescence signals, together with GluA1 and GluA2 mRNA hybridization signals, showed a partial degree of co-localization (Figure 4). The percentages of co-localization calculated in a distal dendritic region are reported in Table 1. As a negative control, no co-localization can be detected for β3-Tub mRNA neither with FMRP or CPEB3. Taken together these data support the hypothesis that GluA1–2 mRNAs are actively transported through RNA granules to distal dendrites.
Figure 4.
Fluorescence in situ analysis of GluA1 (A), GluA2 (B) and β3-Tub (C) mRNA (red—left panel) is reported together with immunostaining with FMRP and CPEB3 (green—middle panel). The merged images are reported in the right panel. Magnifications of boxed dendritic regions are reported under each figure; the artificial white color shows co-localization of the mRNA and protein signals. Scale bar is 10 µm.
Table 1.
Percentage of co-localization for GluA1, GluA2 and beta3-Tub mRNAs with FMRP and CPEB3 proteins
| mRNA GluA1 | mRNA GluA2 | mRNA β3-Tub | |
|---|---|---|---|
| FMRP | 0.16 ± 0.07 | 0.28 ± 0.08 | – |
| CPEB3 | 0.14 ± 0.04 | 0.18 ± 0.06 | – |
SNS analysis of AMPAR mRNAs
To further examine the presence of the AMPAR mRNAs in dendrites, we extracted total RNA from soma and SNS preparations obtained from the Cx and the Hc of naïve rats. As reported in Figure 5A, all AMPAR mRNAs (GluA1–4) were detected in the SNS preparations by RT–PCR. To verify the quality of the SNS preparations, we used β3-Tub mRNA as negative control for dendritic transport and found that it was clearly detected in the soma but absent in SNS preparations. Although AMPAR might also be present in cells of glial origin (36–38), SNS preparations were tested for several glial markers (myelin basic protein, nerve glia antigen 2, and glial fibrillary acidic protein; S100 calcium-binding protein) confirming that the expression of AMPAR mRNAs derived mainly from neuronal spine (Supplementary Figure S4).
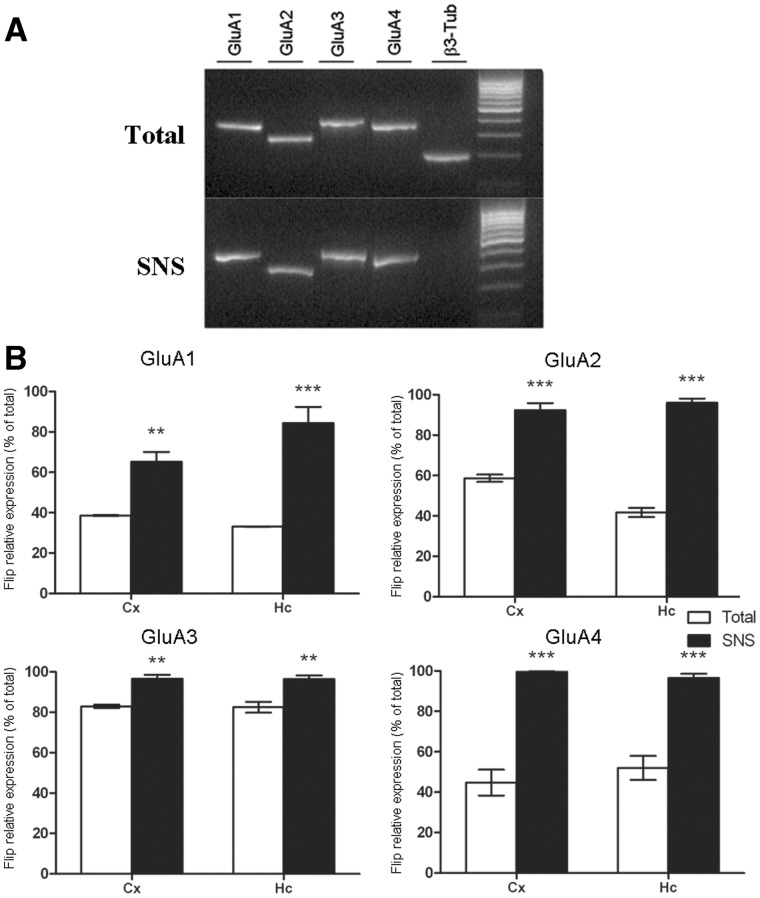
Figure 5.
(A) RT–PCR amplification products of GluA1 (lane 1), GluA2 (lane 2), GluA3 (lane 3) and GluA4 (lane 4) mRNAs in hippocampal total and SNS preparations. β3-Tub mRNA (lane 5), a soma-localized mRNA, was used as negative control and was not detected in SNS samples. (B) The relative expression of Flip isoform for GluA1–4 mRNAs in total and SNS preparations of Cx and Hc are reported. Bars represent Flip isoform expression (reported as a percentage of total transcript expression) in the different preparations. Data are presented as means ± SEM (n ≥ 5). Statistical analysis was performed using one-way ANOVA followed by Bonferroni post hoc test. SNS versus Total *P < 0.05, **P < 0.01, ***P < 0.001.
We next investigated whether AMPAR splicing isoforms could be differentially targeted to the dendrites. To do so, we used direct sequence analysis of specific PCR products, which evaluates the relative amount of the Flip and Flop isoforms for all AMPAR subunits. The results obtained (Figure 5B) showed that the GluA1 Flip isoform represented <40% in the two areas analyzed (Cx: 38.63 ± 0.13%; Hc: 33.1 ± 0.14%); however, in the SNS fractions, the Flip form was >60% in the Cx (65.25 ± 4.8% P< 0.01) and >80% in the Hc (84.37 ± 8.04% P< 0.001). Concerning the GluA2 mRNA, the Flip form represented ∼60% in the Cx (58.64 ± 1.8%) and 40% in the Hc (41.66 ± 2.28%); in the SNS fractions, strong splicing selection was observed, as >90% of the GluA2 mRNA was in the Flip form in both Cx and Hc neurons (Cx: 92.35 ± 3.4% P< 0.001; Hc: 96.00 ± 2.0% P< 0.001). The Flip form of GluA3 mRNA was primarily expressed in the cell soma of Cx and Hc neurons (Cx: 82.94 ± 3.4%; Hc: 96.00 ± 2.0%); however, the selective expression of this splicing variant could still be observed in the SNS preparation (Cx: 96.60 ± 1.9% P< 0.01; Hc: 96.43 ± 1.7% P< 0.01). The highest level of selective expression between soma and dendrites could be evidenced for the GluA4 mRNAs. In the Cx, the Flip isoform was expressed at ∼40% (Cx: 44.76 ± 6.37%), and Flip expression was ∼100% in the SNS preparations (99.6 ± 0.23% P< 0.01); a similar result was obtained also for the Hc (total Hc: 52.02 ± 5.9; SNS: 96.63 ± 2.02 P< 0.001).
Differential mRNA transport of GluA2 Flip and Flop isoforms
To confirm the splicing selectivity of the AMPAR mRNA dendritic transport, we focused on the GluA2 splicing variants. The choice to examine the Flip/Flop splicing variants of GluA2 was due to its high expression level, when compared with GluA3 and GluA4 mRNA, and the clear kinetic differences between the GluA2 Flip and Flop forms when compared with those of GluA1.
We performed fluorescent in situ hybridization in vivo using DIG-labeled riboprobes complementary to GluA2 Flip and Flop exons. Because of the high level of homology between Flip and Flop exons, the specificity of the two riboprobes was assessed using membrane reverse dot-blot analysis (Supplementary Figure S3) and in situ hybridization in transfected PC12 cells (Supplementary Figures S5 and S6). These control experiments demonstrated the specificity of the riboprobes used (Supplementary Materials and Supplementary Results section)
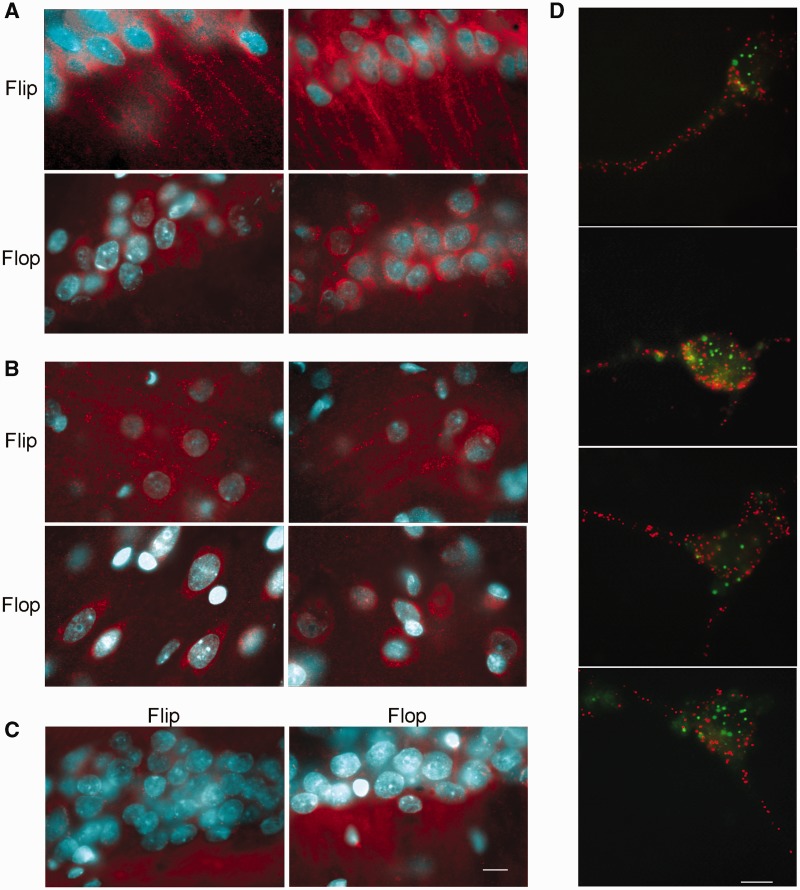
In the hippocampal region, a positive signal for the GluA2 Flip sequence was clearly detected in the CA1 neurons in the cell soma (stratum pyramidale) and the dendrites (stratum radiatum); in contrast, the signal for the GluA2 Flop form was restricted to the cell soma (Figure 6A). Although the neurites of pyramidal cells in the cerebral Cx are not organized in the structures observed in the Hc, a dendritic signal for GluA2 Flip mRNA was also observed, and GluA2 Flop mRNA was restricted to the cell soma (Figure 6B). The sense probes, used as negative controls, did not report any hybridization signal (Figure 6C)
Figure 6.
Fluorescent in situ hybridization of GluA2 mRNA in Flip and Flop splicing variants in (A) CA1 hippocampal and (B) cortical coronal sections. (C) The sense probe was used as negative control. Flip Flop hybridizations were performed on consecutive sections. (D) In situ detection of individual transcripts with padlock probes and target-primed RCA. After in situ reverse transcription and RNA degradation, the cDNA was probed using a Padlock probe. The probes were ligated and subject to RCA. RCPs were identified through hybridization with fluorescent detection probes. Figure reports the double in situ detection of GluA2 Flip mRNA (red) and GluA2 Flop mRNA (green). Scale bar is 10 µm.
The differential localization of GluA2 Flip and Flop was also verified using the in situ detection of individual transcripts with padlock probes and target-primed RCA (24) on neuronal cortical cultures (Figure 6D). This methodology is characterized by the high-resolution detection of mRNAs and allows an accurate discrimination of different transcripts within the same cell. As reported in the images, the Flip mRNA (red signal) was detected in both the cell soma and dendrites, whereas the Flop mRNA (green signal) was restricted to the soma. These data confirm the contribution of RNA splicing in determining the transport of GluA2 mRNA isoforms. The accuracy of the system and the specificity of the padlock probes were assessed in transfected PC12 as showed in Supplementary Figure S7.
RNA editing level of dendritic AMPAR mRNAs
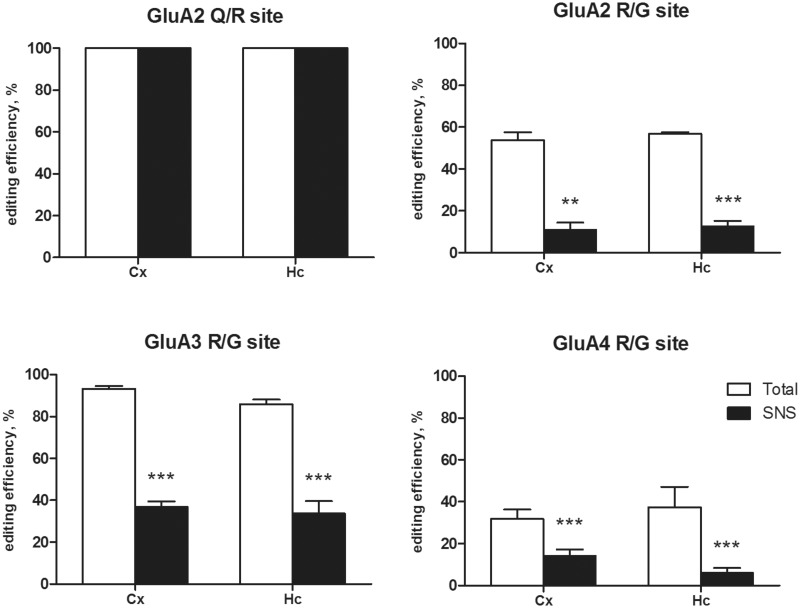
The next step was to investigate the involvement of RNA editing in the dendritic transport of AMPAR mRNAs. The Q/R and R/G editing levels of the different AMPAR mRNAs in both total and SNS preparations from Cx and Hc were analyzed (Figure 7). All the samples tested showed a fully edited GluA2 Q/R site. The R/G site was analyzed only in the Flip form, considering its predominant expression in the dendrites. The R/G site was partially edited and expressed at different levels for GluA2–4 in total Hc (GluA2 R/G 56.80 ± 0.80%; GluA3: R/G 85.90 ± 2.21%; GluA4 R/G: 37.39 ± 9.76%) and Cx preparations (Cx GluA2 R/G 53.68 ± 3.84%; GluA3 R/G 93.26 ± 1.34%; GluA4 R/G: 31.86 ± 4.48%). Extremely low levels of R/G editing were observed in the SNS preparations for all of the three AMPAR mRNAs, the Hc (GluA2 R/G 12.72 ± 2.56% P< 0.001; GluA3: R/G 33.78 ± 5.85% P< 0.001; GluA4 R/G: 6.05 ± 2.34% P< 0.001) and the Cx (GluA2 R/G 11.05 ± 3.38% P< 0.001; GluA3: R/G 36.90 ± 2.56 P< 0.001; GluA4 R/G: 14.18.39 ± 3.03 P< 0.001). Supplementary Figure S8 shows representative electropherogram used to calculate editing levels, showing the clearness and accuracy of the data.
Figure 7.
RNA editing analysis of the GluA2 Q/R and GluA2-4 R/G sites in total and SNS preparations derived from the rat Cx and Hc. Bars represent the editing efficiency (percentage of edited transcripts) in the different preparations for the Q/R and R/G sites. Data are presented as means ± SEM (n ≥ 5). Statistical analysis was performed using one-way ANOVA followed by Bonferroni post hoc test. SNS versus Total *P < 0.05, **P < 0.01, ***P < 0.001.
These data clearly show a difference in the presence of edited and unedited mRNA variants between the cell soma and the synaptic terminal, suggesting the preferential dendritic RNA transport of the unedited isoforms at the R/G site.
RNA localization of AMPA edited and unedited isoforms
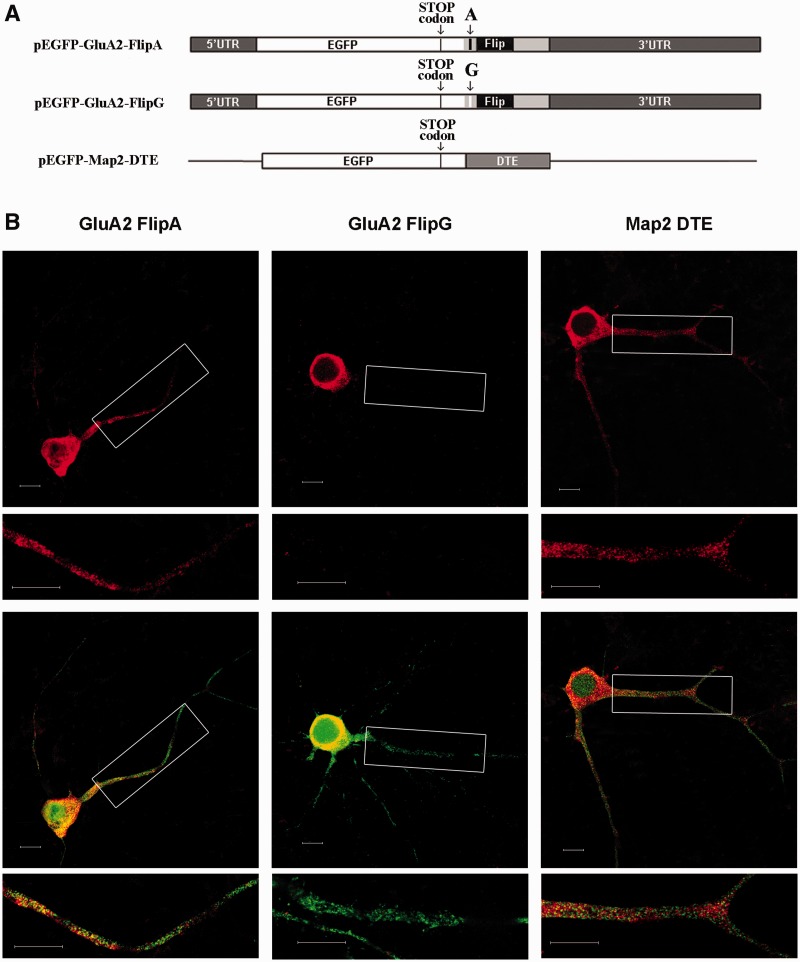
To explore whether the Flip/Flop sequences, in combination with the nucleotide subject to editing, could be implicated in a differential RNA transport, we transfected cortical neurons with different GluA2-based DNA vectors. As a positive control for dendritic transport, the MAP2 DTE was cloned and used for transfection as in (32). The scheme reported in Supplementary Figure S9 summarized the characteristics of the constructs used.
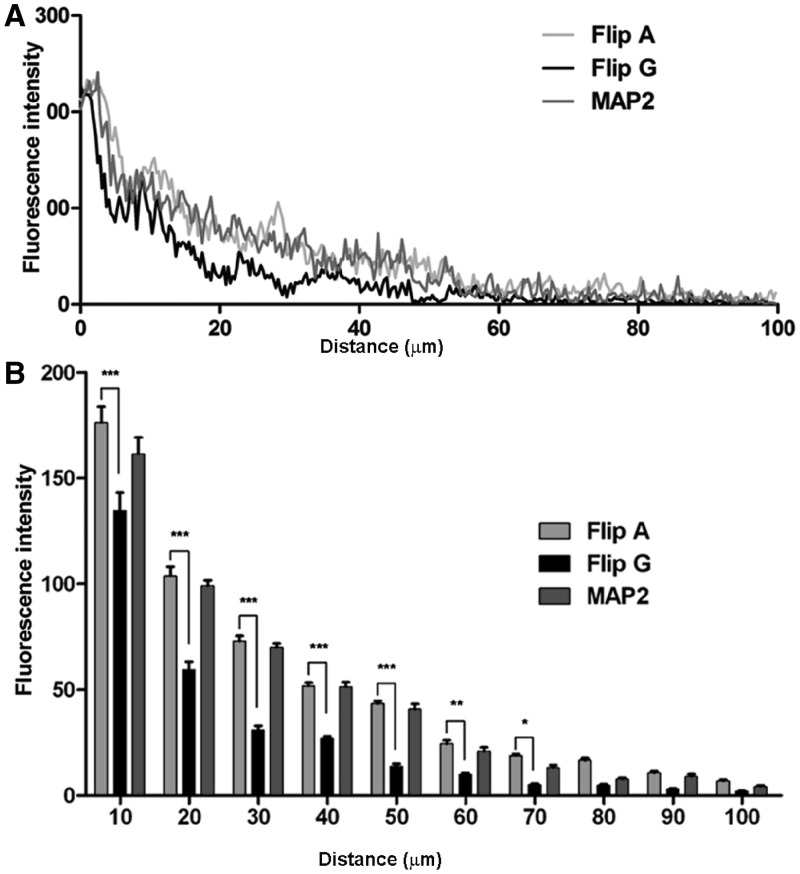
The presence of the chimeric transcripts in the cell soma or dendrites of primary neuronal cortical cells was assessed using fluorescent in situ hybridization with DIG-labeled riboprobes complementary to the EGFP sequence. Considering the Flip variant in the unedited (A) or edited (G) version (Figure 8A), the fluorescence intensity of the unedited Flip chimeric transcript (Figure 8B) was higher than that of the edited Flip chimeric transcript, in both the proximal (between 10 and 50 µm from the soma; P< 0.001) and distal dendrites (between 50 and 90 µm; P< 0.001–0.05) (Figure 9A and B). These data confirm that the unedited Flip sequence is more efficiently transported in the dendrites than the edited version. As expected, the MAP2-DTE chimeric transcript presented a higher degree of transport (Figure 8). The transcription and translation efficiency of the different DNA constructs were tested by RNA expression and western blot analysis in transfected PC12 cells (Supplementary Materials and Supplementary Figure S10) showing an overall similarity of expression for the two GluA2-Flip constructs.
Figure 8.
(A) Scheme of the chimeric mRNAs (GluA2-A-Flip; GluA2-G-Flip, MAP2-DTE) used for cortical neurons (DIV12) transfection. (B) In situ hybridization of chimeric mRNAs; mRNAs were detected using a DIG-labeled antisense EGFP riboprobe (red signal in top panels) in tranfected neurons (green signal in bottom panels). Magnifications of the boxed dendritic regions are reported above each image. Scale bar is 10 µm.
Figure 9.
(A) Histogram showing the quantification of chimeric transcripts dendritic localization. The mean fluorescence intensity in dendrites up to 100 µm from the cell soma has been reported for the different chimeric mRNAs (n ≥ 20). (B) Histogram showing the mean fluorescence intensity in dendritic regions of 10 µm in length, up to 100 µm from the cell soma (n ≥ 20).
Considering GluA2-Flop chimeric transcripts, in both edited and unedited versions, the transfection experiments confirmed that they are less efficiently transported in dendrites in comparison to GluA2-Flip-A version, showing a preferential localization in the cell soma or proximal dendrites (Supplementary Figure S11).
DISCUSSION
This study provides the first evidence that AMPAR mRNA transport to the dendrites might be coupled with AMPAR RNA splicing and RNA editing. An analysis of SNS preparations revealed that the four AMPAR subunit transcripts were localized to synapses, and we confirmed the dendritic localization of GluA1 and GluA2 mRNAs by in situ hybridization analysis in vitro and in vivo. In addition, we verified the co-localization and interaction of GluA1 and GluA2 with FMRP and CPEB3 proteins, which are known to be components of neuronal RNA transport granules (20). Furthermore, using GluA2 as a model, we provided evidence for a ‘splicing code’ for dendritic localization, where the Flip variant is dendritically transported while the Flop variant is restricted to the cell soma. Finally, we identified a previously unknown function for RNA editing, as the unedited Flip variant at the R/G site, was transported to distal compartments, whereas the edited Flip variant was localized primarily to the cell soma. These data suggest a central role for the RNA editing process in the regulation of the dendritic localization of AMPAR mRNAs.
mRNA trafficking and the subsequent local protein synthesis in dendrites have currently gained widespread acceptance as a fundamental mechanism to enable synaptic plasticity [for review see (20)]. By spatially restricting the gene expression in specific neuronal sites, localized mRNAs endow synapses with the capacity to regulate their morphology and activity in response to specific stimuli. A subset of mRNAs is transported in a dormant state and translated locally in an activity-dependent manner, conferring synapses with the capability to autonomously adjust their individual protein composition (39). These transcripts include mRNAs that encode cytosolic, cytoskeletal and transmembrane proteins (21). Among the latter proteins, special attention has been given to ionotropic AMPARs, which mediate fast excitatory neurotransmission; the proper regulation of their functions plays a key role in superior functions, such as learning and memory (40). In particular, it has been previously shown that GluA1 and GluA2 mRNAs are localized to dendrites (19,41), and their presence and local protein synthesis are tightly controlled through synaptic activity (19,42–45), Our analysis using RNA in situ hybridization confirmed the presence of GluA1 and GluA2 mRNAs both in vivo and in vitro. In addition, the SNS analysis showed the presence also of GluA3 and GluA4 mRNA in synaptic preparations, as reported recently by (46). These data add complexity to glutamate receptor function and plasticity at the synaptic sites.
The molecular mechanism that underlies mRNA localization and translational regulation involves several RNA-binding proteins (RBPs) (20). Two RBPs have been broadly accepted as AMPAR mRNA translational repressors: FMRP and CPEB3. FMRP has been reported to regulate AMPAR translation (23) (47,48), and GluA1 and GluA2 mRNAs have been recently reported as direct targets of the repressing activity of CPEB3 (34,35). Using RIP analysis, we confirmed the in vivo relation of GluA1 and GluA2 mRNAs with both FMRP and CPEB3 proteins. In addition, the co-immunostaining of FMRP and CPEB3 with GluA1 and GluA2 mRNA hybridization signals in primary cortical cultures supports a partial degree of co-localization in the dendrites. Taken together, these data confirm the idea that GluA1 and GluA2 mRNA might interact with FMRP and CPEB3 in the formation of transport neuronal RNA granules.
Although the presence of AMPAR transcripts at the synapses has been confirmed in numerous reports, no data are available on the analysis of the possible differential targeting of AMPAR splicing isoforms to the dendrites. The primary AMPAR splicing isoforms are generated through the alternative splicing of the Flip/Flop cassette. This splicing event consists of the insertion of one of two mutually exclusive exons of 115 nt each into the mature mRNA, a feature that is common to all four AMPAR subunits. At the protein level, this splicing event results in the insertion of two alternative strands of 38 amino acids in the extracellular loop in a region located before the fourth transmembrane domain. The two different AMPAR subunits generated are involved in determining the different kinetic properties of the channels by affecting the time course of desensitization (4).
Interestingly, the analysis of the SNS preparations showed that the Flip variants of AMPAR transcripts are enriched at the synapses. Indeed, almost 100% of the GluA2, GluA3 and GluA4 transcripts were present at the synapse in their Flip form; for GluA1, however, the Flip enrichment was clearly detectable but never reached 100%. To deepen the analysis of this peculiar splicing code affecting AMPAR RNA trafficking, we further examined the Flip/Flop splicing variants of GluA2. GluA2 was chosen for its high mRNA expression levels, when compared with GluA3 and GluA4, and for the clear kinetic differences between the GluA2 Flip and Flop forms, when compared with those of the GluA1 variants (1,6). We confirmed the differential mRNA transport of GluA2 Flip and Flop isoforms in vivo using a standardized fluorescent in situ hybridization assay on hippocampal and cortical slices and in vitro using the in situ detection of individual transcripts with padlock probes and target-primed RCA (24) on neuronal cortical cultures. This methodology is characterized by the high-resolution detection of mRNAs and allows the accurate discrimination of the two transcripts within the same cell.
Although three different experimental approaches clearly showed that the GluA2-Flip variant was dendritically transported and the GluA2-Flop variant was restricted to the cell soma, the mechanism by which this occurs is still unclear. Although bioinformatics analysis of the two exon sequences did not reveal clear evidence of binding sites for described RBPs, we might hypothesize that one or more still unknown RBPs might preferentially bind a hypothetic DTE in the Flip sequence and modulate the dendritic transport of the transcripts. Notably, this is one of the rare cases in which a DTE seems to be present in the coding sequence; indeed, other transcripts transported in dendrites, such as αCAMKII (49,50), ARC (51) and MAP2 (32), have a DTE in the 3′-UTR (52). However, only the mRNAs of vasopressin (53) and BDNF (54) have localization signals in the coding sequence acting in association with the UTRs. To date, only BDNF transcripts, which differ for the alternative splicing of the untranslated 5′-exons, differ in their subcellular localization (54,55), showing a ‘spatial code’ that directs protein expression either to the soma or proximal or distal dendrites (55). In the case of GluA2 mRNA, the ‘spatial code’ seems to be performed by a coding exon, adding a new function to the Flip/Flop cassette. Further analyses are needed to understand the molecular mechanism that underlies Flip exon recognition and its preferential trafficking to dendrites.
AMPAR mRNAs are also subjected to RNA editing, a sophisticated mechanism that modifies the RNA sequence at the post-transcriptional level and allows the modification of the receptor subunits in positions that are key in determining the permeability of the channel, the kinetics of pore opening, the time of desensitization of the receptor and the time of recovery from desensitization (11,12). We hypothesized that RNA editing might be involved and mutually coordinated in RNA trafficking. The analysis of the editing pattern of AMPAR RNAs in SNS preparation showed that the GluA2 Q/R site is fully edited, whereas the R/G site of GluA2, GluA3 and GluA4 mRNAs is barely edited in the SNS preparation. These data clearly indicate that the unedited transcripts at the R/G site (mainly in the Flip-form) are selectivity transported to the dendrites, whereas the edited form is restricted to the cell soma. These results were confirmed through transfection experiments in primary cortical neurons, indicating that the presence of the unedited nucleotide is required for efficient dendritic transport.
Intriguingly, the R/G editing site is located 2-nt upstream of the sequence involved in the splicing events determining the Flip/Flop isoforms, and therefore, a functional correlation might exist between these two post-transcriptional events, although it has not yet been completely elucidated (18). We can speculate that the modification of the single nucleotide subject to editing at the R/G site might vary the capability of RBPs to bind the target RNA, thus interfering with the RNA trafficking process. In support of this possibility are the results reported for the G196A (Val66Met) mutation in the BDNF gene, which blocks RNA trafficking by disrupting BDNF transcript binding with Translin (54). Clearly, it is not possible at present to rule out the possible existence of a direct interaction among ADAR proteins, the enzymes that catalyze the editing reaction and specific RBPs at the nuclear levels. Both classes of proteins might compete for RNA binding, recognizing similar secondary structures (13,20). It has been recently reported in Drosophila (56) that dADAR and dFMR1 might act in coordination for the proper formation and function of the fly neuromuscular junctions. In particular, the loss and overexpression of dFMR1 results in changes in the editing efficiency at several dADAR-dependent editing sites. The authors predict that dFMR1 and dADAR could associate in a common complex and converge on similar RNA substrates. It is hypothesized that dFRM1 could promote editing by either recruiting dADAR to the site via its own RNA-binding activity or forming and/or stabilizing RNA structures recognized by dADAR (56). Here, we report for the first time a possible link between the RNA editing pathway and RNA trafficking in mammals. Further work is needed to identify specific RBPs, if any, that interact or cooperate with ADAR enzymes in the choice of the different transcripts that should be subsequently transported to the synapse. The analysis of the editing levels in several editing sites in RBP knockout mice could help to characterize the possible link between editing and trafficking (57).
Another open question concerns the specific and constitutive localization of GluA2 R/G-unedited Flip variants at synaptic sites. Specific functional analyses, i.e. synaptic stimulation or inhibition, could be performed to understand the functional role of the preferential trafficking of the R/G-unedited Flip form, which modifies channel subunits with a slower desensitization time and a slower recovery from desensitization.
In conclusion, our in vivo and in vitro analyses revealed a previously unidentified function of RNA editing that in association with alternative splicing may influence the differential transport of AMPAR mRNAs to dendrites.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–11, Supplementary Methods and Supplementary References [28,30,58–60].
FUNDING
The Regione Lombardia ‘Network Enable Drug Design’ (NEDD) grant [14546 A SAL7]. Funding for open access charge: Ministero del'istruzione, dell'Università e della Ricerca (MIUR) grant [2009BRMW4W].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Nature Publishing Group Language Editing’ for assistance with the manuscript.
REFERENCES
- 1.Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol. Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- 2.Wenthold RJ, Petralia RS, Blahos J, II, Niedzielski AS. Evidence for multiple AMPA receptor complexes in hippocampal CA1/CA2 neurons. J. Neurosci. 1996;16:1982–1989. doi: 10.1523/JNEUROSCI.16-06-01982.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seeburg PH, Hartner J. Regulation of ion channel/neurotransmitter receptor function by RNA editing. Curr. Opin. Neurobiol. 2003;13:279–283. doi: 10.1016/s0959-4388(03)00062-x. [DOI] [PubMed] [Google Scholar]
- 4.Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger JR, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg PH. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 5.Sommer B, Keinanen K, Verdoorn TA, Wisden W, Burnashev N, Herb A, Kohler M, Takagi T, Sakmann B, Seeburg PH. Flip and flop: a cell-specific functional switch in glutamate-operated channels of the CNS. Science. 1990;249:1580–1585. doi: 10.1126/science.1699275. [DOI] [PubMed] [Google Scholar]
- 6.Grosskreutz J, Zoerner A, Schlesinger F, Krampfl K, Dengler R, Bufler J. Kinetic properties of human AMPA-type glutamate receptors expressed in HEK293 cells. Eur. J. Neurosci. 2003;17:1173–1178. doi: 10.1046/j.1460-9568.2003.02531.x. [DOI] [PubMed] [Google Scholar]
- 7.Koike M, Tsukada S, Tsuzuki K, Kijima H, Ozawa S. Regulation of kinetic properties of GluR2 AMPA receptor channels by alternative splicing. J. Neurosci. 2000;20:2166–2174. doi: 10.1523/JNEUROSCI.20-06-02166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosbacher J, Schoepfer R, Monyer H, Burnashev N, Seeburg PH, Ruppersberg JP. A molecular determinant for submillisecond desensitization in glutamate receptors. Science. 1994;266:1059–1062. doi: 10.1126/science.7973663. [DOI] [PubMed] [Google Scholar]
- 9.Hollmann M, Heinemann S. Cloned glutamate receptors. Ann. Rev. Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- 10.Monyer H, Seeburg PH, Wisden W. Glutamate-operated channels: developmentally early and mature forms arise by alternative splicing. Neuron. 1991;6:799–810. doi: 10.1016/0896-6273(91)90176-z. [DOI] [PubMed] [Google Scholar]
- 11.Barbon A, Barlati S. Glutamate receptor RNA editing in health and disease. Biochemistry. 2011;76:882–889. doi: 10.1134/S0006297911080037. [DOI] [PubMed] [Google Scholar]
- 12.Seeburg PH, Higuchi M, Sprengel R. RNA editing of brain glutamate receptor channels: mechanism and physiology. Brain Res. Brain Res. Rev. 1998;26:217–229. doi: 10.1016/s0165-0173(97)00062-3. [DOI] [PubMed] [Google Scholar]
- 13.Orlandi C, Barbon A, Barlati S. Activity regulation of adenosine deaminases acting on RNA (ADARs) Mol. Neurobiol. 2012;45:61–75. doi: 10.1007/s12035-011-8220-2. [DOI] [PubMed] [Google Scholar]
- 14.Boulter J, Hollmann M, O'Shea-Greenfield A, Hartley M, Deneris E, Maron C, Heinemann S. Molecular cloning and functional expression of glutamate receptor subunit genes. Science. 1990;249:1033–1037. doi: 10.1126/science.2168579. [DOI] [PubMed] [Google Scholar]
- 15.Krampfl K, Schlesinger F, Zorner A, Kappler M, Dengler R, Bufler J. Control of kinetic properties of GluR2 flop AMPA-type channels: impact of R/G nuclear editing. Eur. J. Neurosci. 2002;15:51–62. doi: 10.1046/j.0953-816x.2001.01841.x. [DOI] [PubMed] [Google Scholar]
- 16.Ryman K, Fong N, Bratt E, Bentley DL, Ohman M. The C-terminal domain of RNA Pol II helps ensure that editing precedes splicing of the GluR-B transcript. RNA. 2007;13:1071–1078. doi: 10.1261/rna.404407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schoft VK, Schopoff S, Jantsch MF. Regulation of glutamate receptor B pre-mRNA splicing by RNA editing. Nucleic Acids Res. 2007;35:3723–3732. doi: 10.1093/nar/gkm314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Penn AC, Greger IH. Sculpting AMPA receptor formation and function by alternative RNA processing. RNA Biol. 2009;6:517–521. doi: 10.4161/rna.6.5.9552. [DOI] [PubMed] [Google Scholar]
- 19.Grooms SY, Noh KM, Regis R, Bassell GJ, Bryan MK, Carroll RC, Zukin RS. Activity bidirectionally regulates AMPA receptor mRNA abundance in dendrites of hippocampal neurons. J. Neurosci. 2006;26:8339–8351. doi: 10.1523/JNEUROSCI.0472-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doyle M, Kiebler MA. Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J. 2011;30:3540–3552. doi: 10.1038/emboj.2011.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat. Rev. Neurosci. 2007;8:776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- 22.Orlandi C, La Via L, Bonini D, Mora C, Russo I, Barbon A, Barlati S. AMPA receptor regulation at the mRNA and protein level in rat primary cortical cultures. PLoS One. 2011;6:e25350. doi: 10.1371/journal.pone.0025350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muddashetty RS, Kelic S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J. Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsson C, Grundberg I, Soderberg O, Nilsson M. In situ detection and genotyping of individual mRNA molecules. Nat. Methods. 2010;7:395–397. doi: 10.1038/nmeth.1448. [DOI] [PubMed] [Google Scholar]
- 25.Bagni C, Mannucci L, Dotti CG, Amaldi F. Chemical stimulation of synaptosomes modulates alpha -Ca2+/calmodulin-dependent protein kinase II mRNA association to polysomes. J. Neurosci. 2000;20:RC76. doi: 10.1523/JNEUROSCI.20-10-j0004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunkley PR, Jarvie PE, Robinson PJ. A rapid Percoll gradient procedure for preparation of synaptosomes. Nat. Protoc. 2008;3:1718–1728. doi: 10.1038/nprot.2008.171. [DOI] [PubMed] [Google Scholar]
- 27.Kiebler MA, Lopez-Garcia JC, Leopold PL. Purification and characterization of rat hippocampal CA3-dendritic spines associated with mossy fiber terminals. FEBS Lett. 1999;445:80–86. doi: 10.1016/s0014-5793(99)00077-0. [DOI] [PubMed] [Google Scholar]
- 28.Barbon A, Vallini I, La Via L, Marchina E, Barlati S. Glutamate receptor RNA editing: a molecular analysis of GluR2, GluR5 and GluR6 in human brain tissues and in NT2 cells following in vitro neural differentiation. Brain Res. 2003;117:168–178. doi: 10.1016/s0169-328x(03)00317-6. [DOI] [PubMed] [Google Scholar]
- 29.Barbon A, Fumagalli F, La Via L, Caracciolo L, Racagni G, Riva MA, Barlati S. Chronic phencyclidine administration reduces the expression and editing of specific glutamate receptors in rat prefrontal cortex. Exp. Neurol. 2007;208:54–62. doi: 10.1016/j.expneurol.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 30.Barbon A, Fumagalli F, Caracciolo L, Madaschi L, Lesma E, Mora C, Carelli S, Slotkin TA, Racagni G, Di Giulio AM, et al. Acute spinal cord injury persistently reduces R/G RNA editing of AMPA receptors. J. Neurochem. 2010;114:397–407. doi: 10.1111/j.1471-4159.2010.06767.x. [DOI] [PubMed] [Google Scholar]
- 31.Barbon A, Caracciolo L, Orlandi C, Musazzi L, Mallei A, La Via L, Bonini D, Mora C, Tardito D, Gennarelli M, et al. Chronic antidepressant treatments induce a time-dependent up-regulation of AMPA receptor subunit protein levels. Neurochem. Int. 2011;59:896–905. doi: 10.1016/j.neuint.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 32.Blichenberg A, Schwanke B, Rehbein M, Garner CC, Richter D, Kindler S. Identification of a cis-acting dendritic targeting element in MAP2 mRNAs. J. Neurosci. 1999;19:8818–8829. doi: 10.1523/JNEUROSCI.19-20-08818.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wheeler DG, Cooper E. Depolarization strongly induces human cytomegalovirus major immediate-early promoter/enhancer activity in neurons. J. Biol. Chem. 2001;276:31978–31985. doi: 10.1074/jbc.M103667200. [DOI] [PubMed] [Google Scholar]
- 34.Huang YS, Kan MC, Lin CL, Richter JD. CPEB3 and CPEB4 in neurons: analysis of RNA-binding specificity and translational control of AMPA receptor GluR2 mRNA. EMBO J. 2006;25:4865–4876. doi: 10.1038/sj.emboj.7601322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pavlopoulos E, Trifilieff P, Chevaleyre V, Fioriti L, Zairis S, Pagano A, Malleret G, Kandel ER. Neuralized1 activates CPEB3: a function for nonproteolytic ubiquitin in synaptic plasticity and memory storage. Cell. 2011;147:1369–1383. doi: 10.1016/j.cell.2011.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trotter J, Karram K, Nishiyama A. NG2 cells: properties, progeny and origin. Brain Res. Rev. 2010;63:72–82. doi: 10.1016/j.brainresrev.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bergles DE, Jabs R, Steinhauser C. Neuron-glia synapses in the brain. Brain Res. Rev. 2010;63:130–137. doi: 10.1016/j.brainresrev.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stegmuller J, Werner H, Nave KA, Trotter J. The proteoglycan NG2 is complexed with alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors by the PDZ glutamate receptor interaction protein (GRIP) in glial progenitor cells. Implications for glial-neuronal signaling. J. Biol. Chem. 2003;278:3590–3598. doi: 10.1074/jbc.M210010200. [DOI] [PubMed] [Google Scholar]
- 39.Poon MM, Choi SH, Jamieson CA, Geschwind DH, Martin KC. Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J. Neurosci. 2006;26:13390–13399. doi: 10.1523/JNEUROSCI.3432-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyashiro K, Dichter M, Eberwine J. On the nature and differential distribution of mRNAs in hippocampal neurites: implications for neuronal functioning. Proc. Natl Acad. Sci. USA. 1994;91:10800–10804. doi: 10.1073/pnas.91.23.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ju W, Morishita W, Tsui J, Gaietta G, Deerinck TJ, Adams SR, Garner CC, Tsien RY, Ellisman MH, Malenka RC. Activity-dependent regulation of dendritic synthesis and trafficking of AMPA receptors. Nat. Neurosci. 2004;7:244–253. doi: 10.1038/nn1189. [DOI] [PubMed] [Google Scholar]
- 43.Aoto J, Nam CI, Poon MM, Ting P, Chen L. Synaptic signaling by all-trans retinoic acid in homeostatic synaptic plasticity. Neuron. 2008;60:308–320. doi: 10.1016/j.neuron.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maghsoodi B, Poon MM, Nam CI, Aoto J, Ting P, Chen L. Retinoic acid regulates RARalpha-mediated control of translation in dendritic RNA granules during homeostatic synaptic plasticity. Proc. Natl Acad. Sci. USA. 2008;105:16015–16020. doi: 10.1073/pnas.0804801105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith WB, Starck SR, Roberts RW, Schuman EM. Dopaminergic stimulation of local protein synthesis enhances surface expression of GluR1 and synaptic transmission in hippocampal neurons. Neuron. 2005;45:765–779. doi: 10.1016/j.neuron.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 46.Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, Schuman EM. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74:453–466. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soden ME, Chen L. Fragile X protein FMRP is required for homeostatic plasticity and regulation of synaptic strength by retinoic acid. J. Neurosci. 2010;30:16910–16921. doi: 10.1523/JNEUROSCI.3660-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schutt J, Falley K, Richter D, Kreienkamp HJ, Kindler S. Fragile X mental retardation protein regulates the levels of scaffold proteins and glutamate receptors in postsynaptic densities. J. Biol. Chem. 2009;284:25479–25487. doi: 10.1074/jbc.M109.042663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mori Y, Imaizumi K, Katayama T, Yoneda T, Tohyama M. Two cis-acting elements in the 3′ untranslated region of alpha-CaMKII regulate its dendritic targeting. Nat. Neurosci. 2000;3:1079–1084. doi: 10.1038/80591. [DOI] [PubMed] [Google Scholar]
- 50.Blichenberg A, Rehbein M, Muller R, Garner CC, Richter D, Kindler S. Identification of a cis-acting dendritic targeting element in the mRNA encoding the alpha subunit of Ca2+/calmodulin-dependent protein kinase II. Eur. J. Neurosci. 2001;13:1881–1888. doi: 10.1046/j.0953-816x.2001.01565.x. [DOI] [PubMed] [Google Scholar]
- 51.Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21:741–751. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- 52.Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3'UTR ends. Trends Cell Biol. 2009;19:465–474. doi: 10.1016/j.tcb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 53.Prakash N, Fehr S, Mohr E, Richter D. Dendritic localization of rat vasopressin mRNA: ultrastructural analysis and mapping of targeting elements. Eur. J. Neurosci. 1997;9:523–532. doi: 10.1111/j.1460-9568.1997.tb01629.x. [DOI] [PubMed] [Google Scholar]
- 54.Chiaruttini C, Vicario A, Li Z, Baj G, Braiuca P, Wu Y, Lee FS, Gardossi L, Baraban JM, Tongiorgi E. Dendritic trafficking of BDNF mRNA is mediated by translin and blocked by the G196A (Val66Met) mutation. Proc. Natl Acad. Sci. USA. 2009;106:16481–16486. doi: 10.1073/pnas.0902833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baj G, Leone E, Chao MV, Tongiorgi E. Spatial segregation of BDNF transcripts enables BDNF to differentially shape distinct dendritic compartments. Proc. Natl Acad. Sci. USA. 2011;108:16813–16818. doi: 10.1073/pnas.1014168108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhogal B, Jepson JE, Savva YA, Pepper AS, Reenan RA, Jongens TA. Modulation of dADAR-dependent RNA editing by the Drosophila fragile X mental retardation protein. Nat. Neurosci. 2011;14:1517–1524. doi: 10.1038/nn.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bassell GJ. Fragile balance: RNA editing tunes the synapse. Nat. Neurosci. 2011;14:1492–1494. doi: 10.1038/nn.2982. [DOI] [PubMed] [Google Scholar]
- 58.Mueller O, Hahnenberger K, Dittmann M, Yee H, Dubrow R, Nagle R, Ilsley D. A microfluidic system for high-speed reproducible DNA sizing and quantitation. Electrophoresis. 2000;21:128–134. doi: 10.1002/(SICI)1522-2683(20000101)21:1<128::AID-ELPS128>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 59.Barbon A, Popoli M, La Via L, Moraschi S, Vallini I, Tardito D, Tiraboschi E, Musazzi L, Giambelli R, Gennarelli M, et al. Regulation of editing and expression of glutamate alpha-amino-propionic-acid (AMPA)/kainate receptors by antidepressant drugs. Biol. Psychiatry. 2006;59:713–720. doi: 10.1016/j.biopsych.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 60.Barbon A, Orlandi C, La Via L, Caracciolo L, Tardito D, Musazzi L, Mallei A, Gennarelli M, Racagni G, Popoli M, et al. Antidepressant treatments change 5-HT2C receptor mRNA expression in rat prefrontal/frontal cortex and hippocampus. Neuropsychobiology. 2011;63:160–168. doi: 10.1159/000321593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.