Abstract
We synthesized three 20mer caged circular antisense oligodeoxynucleotides (R20, R20B2 and R20B4) with a photocleavable linker and an amide bond linker between two 10mer oligodeoxynucleotides. With these caged circular antisense oligodeoxynucleotides, RNA-binding affinity and its digestion by ribonuclease H were readily photomodulated. RNA cleavage rates were upregulated ∼43-, 25- and 15-fold for R20, R20B2 and R20B4, respectively, upon light activation in vitro. R20B2 and R20B4 with 2- or 4-nt gaps in the target RNA lost their ability to bind the target RNA even though a small amount of RNA digestion was still observed. The loss of binding ability indicated promising gene photoregulation through a non-enzymatic strategy. To test this strategy, three caged circular antisense oligonucleotides (PS1, PS2 and PS3) with 2′-OMe RNA and phosphorothioate modifications were synthesized to target GFP expression. Upon light activation, photomodulation of target hybridization and GFP expression in cells was successfully achieved with PS1, PS2 and PS3. These caged circular antisense oligonucleotides show promising applications of photomodulating gene expression through both ribonuclease H and non-enzyme involved antisense strategies.
INTRODUCTION
Chemical approaches for controlling biomolecular functions have become increasingly important in biological and biomedical research. Photolabile groups (photocages) represent one of most effective approaches to achieve high spatiotemporal resolution with light activation. It has been successfully used to cage many functional oligonucleotides to photomodulate gene expression in vitro and in vivo, such as ribozymes, DNAzymes, complementary RNA or DNA oligonucleotides and their analogs during mRNA transcription, processing, and ribosomal binding and translation (1–26). Currently, there exist several strategies to modulate gene expression by light activation. One is based on using multiple caging groups to kinetically inhibit the access of a target RNA. Functions of DNA plasmids, messenger RNAs, antisense oligodeoxynucleotides (asODNs) and small interfering RNAs (siRNAs) can also be photoregulated with light using above caging strategy (15,27–31). The second strategy is to introduce an inhibitor strand to temporarily block the antisense strand of an oligonucleotide. Removal of the inhibitor strand activates the antisense oligonucleotide and recovers gene silencing activities. Photomodulation of specific gene expression using caged antisense oligonucleotides and their analogs has been achieved in leukemia cells, zebrafish embryos and Caenorhabditis elegans (16–20,32,33). Applying the similar strategy, a photolabile inhibitor strand, caged sense strand, was used to temporarily hybridize antisense strand and block antisense activities for photomodulation of in vitro protein synthesis and green fluorescent protein (GFP) expression in zebrafish (14,34).
For the methods described above, caged nucleic acids with multiple caging moieties sterically inhibited the interaction with their targets. However, incomplete uncaging was inevitable due to limited intensity and duration of light irradiation. A single caging group with the above design could partially solve the problem of light irradiation. However, relative thermostability of the caged oligonucleotide, the duplex of sense/antisense oligonucleotides and the duplex of antisense oligonucleotide/target mRNA were difficult to balance one or the other, especially, for in vivo study. The number and position of paired bases, the length and type of photo linkers and many other factors all had distinct effect on photomodulation of gene regulation in zebrafish (16). Uncaged antisense ODNs were not quickly and fully accessible to mRNA binding, and the optimized candidates in vitro might not be the best one in vivo due to complicated environments in biological systems (18). In addition, cleaved inhibitor strands may cause off-target effects or unexpected side effects. Hence, currently there is still a clear need for the design of new caged nucleic acid tools for gene regulation in vivo.
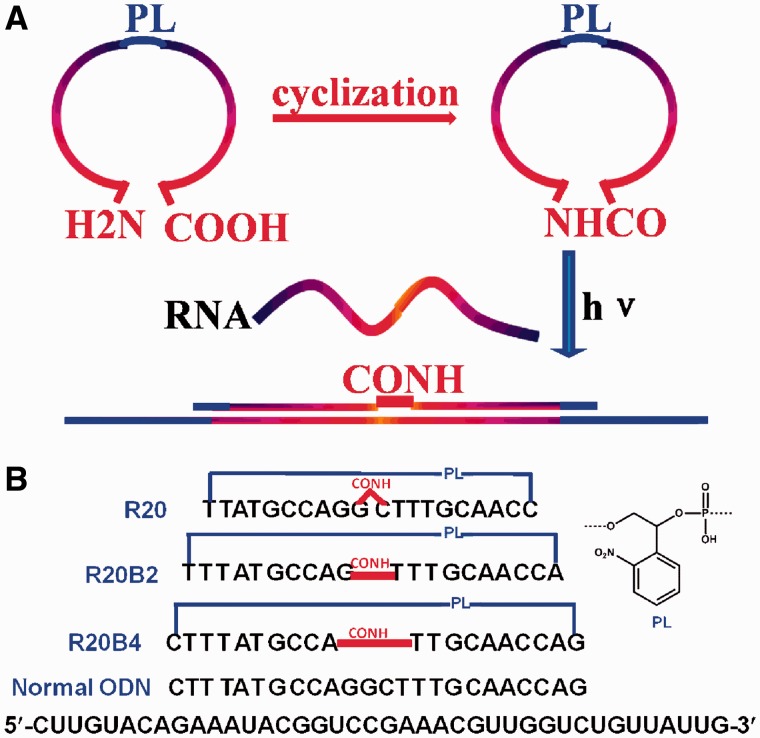
The most recent strategy tried to overcome the problems of cleaved caged moieties or inhibitor strands by cyclizing two ends of an asODN with photocleavable linker (PL). Dmochowski co-workers (35) inserted a PL into the middle of two short ODN sequences which were then ligated into a circular DNAzyme by T4 ligase for photomodulation of RNA digestion. This method is limited only for native DNA/RNA oligonucleotides. Recently, both Chen’s and our laboratory designed circular morpholino oligomers for photoregulating gene expression in zebrafish embryos (36,37). We previously also synthesized a series of different size of caged ODNs with PL attached at 5′-end and then cyclized two ends of ODNs. Our results found the ring size of caged asODNs to be the most important factor in photomodulation of target RNA digestion by ribonuclease H (RNase H) (38). Different from previous work about caged circular asODNs, we designed a new series of caged circular asODNs, as shown in Scheme 1A. These asODNs were composed of two 10mer ODNs, which were linked to each other with PL in the middle of sequence during solid-phase synthesis. The new 3′-end of the linear ODN was modified with an amine group, while the new 5′-end was attached with a carboxylic acid group. After cyclization through amide bond formation, the caged circular asODN can be cleaved in the middle of the previous linear ODN and the sequence recovered was then hybridized with a complementary target RNA with the flexible linker in the middle. We synthesized three such circular caged asODNs (R20, R20B2 and R20B4) according to the above design. For R20, there was no nucleotide gap between the uncaged asODN and the complementary target RNA. While for R20B2 and R20B4, there existed 2- and 4-nt gaps between the two 10mer ODNs, respectively, as shown in Scheme 1B. We expect this design may help minimize the binding of caged circular ODNs with target sequences due to the non-base linkers (PL and amide linker). Upon light activation, the binding ability can be restored even though the flexible linker and nucleotide gap exists in the middle of linear ODNs (39). This will help block protein translation through a non-enzymatic antisense strategy, such as 2′-OMe RNA oligonucleotides and morpholino oligomers. Based on the results of our in vitro assay, we further designed three circular antisense oligonucleotides (PS1, PS2 and PS3) with 2′-OMe RNA and phosphorothioate modifications (Supplementary Figure S1) and successfully photomodulated their binding ability with target sequence and GFP expression in cells.
Scheme 1.
(A) Strategy for photoregulating RNA digestion using caged circular asODNs. (B) Sequences of caged circular asODNs and complementary target RNA. R20B2 and R20B4 have 2 - and 4-nt gaps in the middle when they bind their target RNA.
MATERIALS AND METHODS
General methods
All single-stranded ODNs with a PL were custom synthesized with amine-modified CPG on ABI 394 DNA/RNA synthesizer. Deprotected ODNs were purified with an Agilent 1200 HPLC system using a reverse-phase HPLC column (reverse-phase C18, 4.6 × 250 mm, 9.4 × 150 mm, 5 μm beads). Fluorescence experiments were carried out with a Varian Cary 300 fluorescence spectrometer. Gels of RNA digestion were imaged using an Amersham Biosciences Storm 840 phosphorimager and quantified with ImageQuant software (ImageQuant™ TL v2005, GE Healthcare). All photoirradiation in vitro experiments with ODN samples were carried out with a xenon lamp (450 W) and monochromator (20-nm slit centered at 350 nm, 30 mW/cm2 at the sample). Standard 1× RNase H reaction buffer was defined as 20 mM Tris–HCl, 20 mM KCl, 10 mM MgCl2, 0.1 mM EDTA and 0.1 mM dithiothreitol, pH 8.0. Photoirradiation of HeLa cells was achieved using UV lamp (365 nm, 11 mW/cm2, B-100SP, UVP, LLC) and cell imaging was done on Nikon confocal laser scanning microscope (A1RSi).
Synthesis of photolabile circular asODNs (R20, R20B2 and R20B4)
Controlled pore glass (CPG) with 3′-end amine modification was used for ODN synthesis. ODNs (R20, R20B2 and R20B4) were custom synthesized according to the standard DNA synthesis on ABI 394. [1-(o-Nitrophenyl)-2-dimethoxytrityloxy] ethoxy-N,N-diisopropylamino-2-cyanoethoxyphosphine was synthesized as the PL according to the previous report (40) and incorporated in the middle of 20mer ODN sequences instead of incorporation of PL at the 5′-end of an oligonucleotide sequence in our previous report (41). The 5′-end of ODNs was further coupled with hexylidene amino group [4-methoxyphenyl)diphenylmethyl (MMT) protected] through phosphodiester bonds. MMT was then removed by 4% trifluoacetic acid (TFA) in CH2Cl2 to release 5′-end amine group. Before cleavage from the resin, the amine group reacted with ∼100-fold excess succinic anhydride and diisopropylethylamine (DIPEA) in 0.5 ml dimethylformamide (DMF) at room temperature overnight.
Sequences used in RNase H assay study were as follows:
-
Normal sequence (std): 5′-CCAACGTTTCGGACCGTATT
R20-COOH: 5′-HOOCCH2CH2CONH-(CH2)6-GGACCGTATT-PL-CCAACGTTTC-(CH2)7-NH2-3′
R20B2-COOH: 5′-HOOCCH2CH2CONH-(CH2)6-GACCGTATTT-PL-ACCAACGTTT-(CH2)7-NH2-3′
R20B4-COOH: 5′-HOOCCH2CH2CONH-(CH2)6-ACCGTATTTC-PL-GACCAACGTT-(CH2)7-NH2-3′
RNA target: 5′-CUUGUACAGAAAUACGGUCCGAAACGUUGGUCUGUUAUUG-3′
ODNs were then cleaved and deprotected using concentrated ammonium hydroxide. After the removal of ammonia, these ODNs were further purified with HPLC under reverse-phase conditions: A, 0.05 M triethylammonium acetate (TEAA); B, acetonitrile; B, 0–15% in 30 min, 15–45% in 30 min. Product fractions were collected and characterized by electrospray ionization mass spectrometry (ESI-MS) under negative mode (1% triethylamine (TEA) in H2O/CH3CN). Mass-to-charge ratio (m/z) for R20-COOH: calculated: 6801.0; measured: 6800.0; m/z for R20B2-COOH: calculated: 6800.0, measured: 6799.0; m/z for R20B2-COOH: calculated: 6785.0, measured: 6784.8.
ODNs with amine and acid groups were dried and ethanol precipitated with 3 M NaCl to remove the TEAA residue. The precipitated ODNs (∼20 nmol) were dissolved in 2 ml 0.1 M 2-morpholinoethanesulfonic acid buffer (pH 6.5), 0.3 M NaCl and 10 mM MgCl2 with final concentration of 5 mM 1-hydroxybenzotrizole. Then ∼1 mg 1-ethyl-3 -(3-dimethylaminopropyl) carbodiimide, hydrochloride was added to the solutions. The mixture was vortexed and stood at room temperature for 10–14 h.
The above solutions were then desalted using NAP-10 column, followed by reverse-phase HPLC purification (A, 0.05 M TEAA; B, acetonitrile; B, 0–15% in 30 min, 15–45% in 30 min, running temperature, 40°C). Retention time of product fractions was usually 1–2 min longer than those of starting ODNs. The collected products were subjected to dryness in vacuum and later characterized by ESI-MS. The yields were calculated by dissolving caged circular ODNs in water and measuring the absorbance at 260 nm. The isolated yields for caged circular ODNs were 30–40% under our synthetic conditions, which is similar to our previous results. ESI-MS was carried out under negative mode (1% TEA in H2O/CH3CN). m/z for R20, calculated: 6783.0; measured: 6782.0, 6782.0 + n Na+; m/z for R20B2, calculated: 6782.0, measured: 6781.4, 6781.4 + n Na+; m/z for R20B4, calculated: 6767.0, measured: 6766.4, 6766.4 + n Na+.
Synthesis of photolabile circular antisense phosphorothioated 2′-OMe RNA oligonucleotides (PS1, PS2 and PS3)
In order to photomodulate GFP expression in cells, we designed another five phosphorothioated 2′-OMe RNA oligonucleotides (PS, PS-MS, PS1, PS2 and PS3). PS (matched) and PS-MS (mismatched) were purchased from Sangon Biotech (Shanghai) Co., Ltd as control sequences. According to the same design of R20, R20B2 and R20B4, PS1, PS2 and PS3 were synthesized with two short 10mer 2′-OMe RNA sequences and PL as linkage and there were 0-, 2- and 4-nt gaps for targeting sequence after cyclization and photocleavage (Supplementary Figure S1).
All phosphorothioated 2′-OMe RNA oligonucleotide sequences used in molecular beacon and cell study:
-
PS: GCUCCUCGCCCUUGCUCACC
PS-MS: GCUGCUCCCCCAUGCACACC
PS1-COOH: 5′-HOOCCH2CH2CONH-(CH2)6-CUUGCUCACC-PL-GCUCCUCGCC-(CH2)6-NH2-3′
PS2-COOH: 5′-HOOCCH2CH2CONH-(CH2)6-UUGCUCACCA-PL-AGCUCCUCGC-(CH2)6-NH2-3′
PS3-COOH: 5′-HOOCCH2CH2CONH-(CH2)6-UGCUCACCAU-PL-CAGCUCCUCG-(CH2)6-NH2-3′
The linear phosphorothioated 2′-OMe RNA oligonucleotides (PS1, PS2 and PS3) were prepared according to standard RNA synthesis on ABI 394 using amine-modified CPG and bis(phenylacetyl)disulfide as sulfur substitution agent. Linear PS1-COOH, PS2-COOH and PS3-COOH after cleavage and deprotection were first separated by HPLC under reverse-phase conditions: A, 0.05 M TEAA; B, acetonitrile; B, 10–35% in 35 min, 35–70% in 5 min, 70–70% in 2 min, 70–10% in 3 min. Product fractions were collected and characterized by ESI-MS under negative mode [100 mM NH4Ac in H2O/CH3OH (75:25)]. m/z for PS1-COOH: calculated: 7526.9; measured: 7527.2; m/z for PS2-COOH: calculated: 7576.2, measured: 7574.7; m/z for PS3-COOH: calculated: 7576.2, measured: 7575.5.
Ten nanomoles of phosphorothioated oligonucleotides with amine and acid groups were dried and ethanol precipitated with 3 M NaCl to remove the TEAA residue. The precipitated oligonucleotides were dissolved in 1 ml DMF solutions containing 100 µl DIPEA and 100 mM 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium chloride. The solution was vortexed and stood at room temperature for 24–40 h. The reaction mixtures were desalted by dialysis and separated by 20% denaturing PAGE (acrylamide:bisacrylamide, 19:1) containing 7 M urea (Supplementary Figure S2A). For each separate gel, the side sample lane of the gel was cut and stained with 1× SYBR Gold (Invitrogen), the location of caged circular oligonucleotides was marked and relocated. The gel zones at the same location were cut, crumbled into tiny particles and immersed into a solution containing 0.5 M ammonium acetate, 10 mM magnesium acetate and 1 mM EDTA at 37°C over 12 h. The solutions were further salted out by dialysis and gel chromatography (NAP-10 column). The collected products were dried in vacuum and later characterized by ESI-MS. The isolated yields for purified caged circular phosphorothioated oligonucleotides were 20–30% under our synthetic conditions. ESI-MS was carried out under negative mode [100 mM NH4Ac in H2O/CH3OH (75:25)]. m/z for PS1: calculated: 7508.9; measured: 7509.0; m/z for PS2: calculated: 7558.2, measured: 7554.9; m/z for PS3: calculated: 7558.2, measured: 7556.5.
RNase H assays
40mer RNA target sequence (5′-CUUGUACAGAAAUACGGUCCGAAACGUUGGUCUGUUAUUG-3′) in HPLC pure form was purchased from Shanghai GenePharma Co., Ltd. Recombinant RNase H from Escherichia coli and the reaction buffer were purchased from Epicentre Biotechnologies. Standard procedure for RNase H assay was as follows: caged circular asODNs were dissolved in 1× RNase H reaction buffer at 37°C, then [γ-32P]-labeled RNA oligonucleotide (200-fold excess) was added and incubated at 37°C for 20 min to allow RNA/DNA duplex formation, followed by the addition of two units RNase H and incubation at 37°C. Total reaction volume was 20 µl, and the final concentrations of asODNs and RNA were 0.02 and 4 µM, respectively.
To evaluate RNA degradation with caged asODN by RNase H after light activation, RNase H assays were performed as described above except that caged asODNs initially illuminated by xenon lamp through monochromator (350 nm UV light, ∼30 mW/cm2) were used as antisense templates. Time points were taken at 2, 5, 10, 15 and 30 min by sampling 4 µl of the reaction mixture, adding 6 µl gel loading buffer (50 mM EDTA, 90% formamide with bromphenol blue and xylene cyanol, total volume = 10 μl) and then heating the solutions to 95°C for 3 min to terminate the reaction.
All of the resulting solutions were then subjected to electrophoresis on a 20% polyacrylamide gel containing 7 M urea. The intensities of gel bands were integrated in ImageQuant for each band with automated lane and band finding using a local method background correction in the gel lane. The relative amount of RNA digestion was determined by dividing the intensities of the bands corresponding to cleaved RNA by the total intensity of the cleaved and uncleaved RNA bands. Kinetic data for 40mer RNA cleavage in the presence of R20 and RNase H were obtained by quantifying the percentage of cleaved target RNA at time t and fitting the data using the linear function, [S] = A + kt, for a caged circular asODN (42) and a single exponential decay function, [S] = A(1−e−kt), for an uncaged asODN, where [S] is the fraction of cleaved RNA at time t and k is rate constant of photocleavage.
Gel shift assays
Gel mobility shift assays were performed as described in the literature to determine the binding of RNA to caged circular as ODNs (43). A fixed concentration of [γ-32P]-labeled 40mer RNA (0.02 μM) with increased concentrations of caged circular asODNs was used for the assays in 9 μl standard 1× RNase H reaction buffer. Samples were annealed at 37°C for 30 min and then were put in ice-cold water. Before the samples were loaded into the gel, 1 μl glycerol with bromphenol blue and xylene cyanol was added. All 10 μl solutions were loaded into 12% non-denaturing polyacrylamide gels. The gels were then electrophoresed at 100 V for 2 h at 10°C, using 1× Tris-borate-EDTA (TBE) buffer (pH 8.2). Gels were exposed and then imaged with Storm phosphorimager.
Kinetic binding study of uncaged circular oligonucleotides
A molecular beacon (MB, 5′-FAM-TTAGAATGGTGAGCAAGGGCGAGGAGCTGTCTAA-DABCYL-3′) with a large loop complementary to PS and uncaged PS1, PS2 and PS3 was designed and purchased from Sangon Biotech (Shanghai) Co., Ltd. Fluorescence spectra were determined using Cary Eclipse fluorescence spectrophotometer with the excitation wavelength at 488 nm and emission wavelength at 515 nm. A solution of 100 nM MB in 250 μl 1× PBS buffer stood at 37°C for 3 min, and then 4-fold excess caged or pre-irradiated antisense oligonucleotide samples were added to MB solutions. The increase in fluorescence intensity at 515 nm for each sample solutions was monitored at 37°C.
Second-order rate constants for hybridization, kh (M−1s−1), of MB to the complementary linear PS or the uncaged PS1, PS2 and PS3, were calculated by using the linearization procedures for a second-order reaction with non-stoichiometric proportions of reactants (44).
We used the following equation:
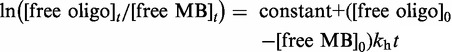 |
(1) |
Thus, for a hybridization reaction, a plot of ln([free oligo]t/[free MB]t) versus t is linear with the slope being equal to ([free oligo]0 − [free MB]0)kh.
Cell study
HeLa cells were grown at 37°C in a humidified atmosphere with 5% CO2 in RPMI Medium 1640 (Gibco), supplemented with 10% fetal calf serum (Zhejiang Tianhang Biological Technology Co., Ltd.), penicillin (100 mg/ml) and streptomycin (100 mg/ml). Cells were trypsinized and resuspended in medium without antibiotics and transferred to Petri dishes at a density of 2.5 × 104 cells per well in a volume of 2000 μl and grown to ∼70% confluency within 24 h. The media were changed to OptiMEM (Gibco). Plasmid DNA coding for the GFP fusion protein (pEGFP-N1, Beijing DingGuo ChangSheng Biotechnology Co., Ltd), control red fluorescent protein (RFP) plasmid (pDsRed2-N1, Biovector Co., Ltd) and caged circular 2′-OMe RNA oligonucleotides were mixed with Lipofectamine 2000 (Invitrogen). For each transfection, 0.3 μg of pEGFP-N1, 0.3 μg pDsRed2-N1 and 8.25 pmol caged circular phosphorothioate oligonucleotides were mixed with 75 μl OptiMEM. In a separate tube, 0.6 μl Lipofectamine 2000 per reaction was added to 75 μl OptiMEM and incubated for 5 min at room temperature. Both solutions were mixed and incubated for additional 20 min at room temperature to allow complex formation. The solutions were added to the cells in Petri dishes, giving an end volume of 150 μl. Cells were incubated at 37°C in the presence of transfection solutions for 6 h. The transfection media were removed and 150 μl PBS buffer (Gibco) was added. The cells in Petri dishes were then irradiated with a hand-held UV lamp (365 nm, 11 mW/cm2) for 10 min. The PBS buffer were replaced with fresh standard grown media without penicillin/streptomycin, the cells were incubated for an additional 42 h. The cotransfected cells were then washed with PBS buffer, followed by nuclei staining with Hoechst 33 258 (Invitrogen) solution for 30 min.
Confocal microscopy
Confocal microscopy was carried out using a Nikon confocal laser scanning microscope (A1RSi). All images were taken using a ×10 objective lens. Hoechst fluorescence was excited at a wavelength of 401 nm and detected at 450 nm, GFP fluorescence was excited at a wavelength of 488 nm and detected at 525 nm, RFP fluorescence was excited at a wavelength of 561 nm and detected at 595 nm, each with a pinhole radius of 19.45 μm. For fluorescence images, identical operation and image processing were done for each channel. Monochrome images generated with charge-couple device (CCD) camera were pseudocolored to the appropriate color. Following confocal microscopy, image analysis was performed using NIS-Elements AR 3.2. The mean fluorescence intensity of equally divided squares for each channel was calculated.
RESULTS AND DISCUSSION
Photomodulation of RNA digestion by RNase H with caged circular asODNs
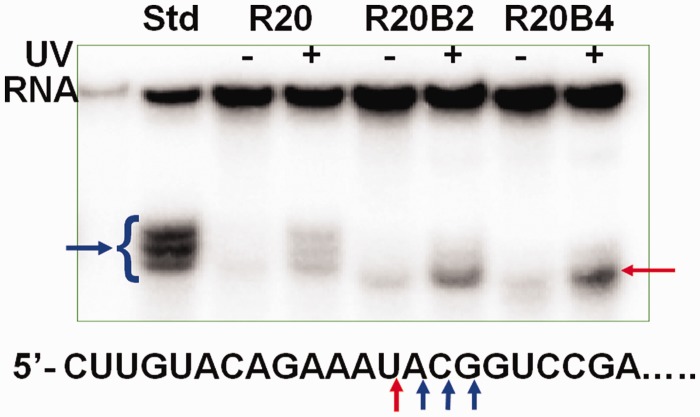
Without its complementary ODN template, RNA cannot be digested by RNase H (45) under our photolysis conditions (350 nm, 30 mW/cm2). The effectiveness of photomodulating target RNA digestion by RNase H was tested with the inclusion of caged circular DNA templates after UV irradiation. 40mer target RNA was mixed with the caged circular asODNs (R20, R20B2 and R20B4, and their uncaged forms) in the presence of RNase H under our RNase H assay conditions. Digestion of the complementary target 40mer RNA was evaluated for all the caged circular asODNs (R20, R20B2 and R20B4) before and after UV irradiation. As shown in Figure 1, a very small amount of target 40mer RNA was digested if no light was applied; however, increased amounts of cleaved RNA bands were clearly observed with 10-min light irradiation under our photolysis conditions. For R20, light triggered 6-fold increase in RNA digestion from 4 to 24% in 30 min. Under the same conditions, RNA digestion increased from 4.5 to 23.2% for R20B2 and 8 to 27% for R20B4, and the photomodulation ratios were 5.1 and 3.3 for R20B2 and R20B4, respectively, which generally has the same photomodulation efficiency as hairpin-like asODNs (18,19). Interestingly, the cleaved patterns of RNA for R20B2 and R20B4 by RNase H were also different from R20, as shown in Figure 1. For R20, the cleaved pattern was similar to that of normal linear 20mer asODN (std). Three mainly cleaved phosphodiester bonds were located inside ApCpGpG which was the fourth to seventh nucleotide of the duplex of R20/target RNA. However, main cutting position was inside ApC instead of CpG for std. While for R20B2 and R20B4, the cleaved positions were both shifted toward 5′-end of target RNA with one nucleotide and mainly cleaved the phosphodiester bond between UpA, which was located between the fourth and fifth nucleotide for duplex R20B2/RNA and the fifth and sixth nucleotide for the duplex R20B4/RNA. As indicated in the literature (46), RNase H has the preference to cleave a certain phosphodiester bond in the target sequence, which leads to lower cutting yields for R20, R20B2 and R20B4 than std (∼50%).
Figure 1.
Denaturing PAGE (20%) analysis of target RNA digestion in 30 min with 4 µM [γ-32P]-labeled 40mer RNA, 0.02 µM caged asODNs in 20 µl RNase H buffer with two units RNase H at 37°C. Arrows show the cleavage positions for different antisense oligonucleotide templates. std represents the normal 20mer asODN complementary to the middle 20 nt of 40mer target RNA. Samples were photocleaved with UV light (350 nm, ∼30 mW/cm2) for 10 min.
Kinetics of target RNA digestion with caged circular asODNs
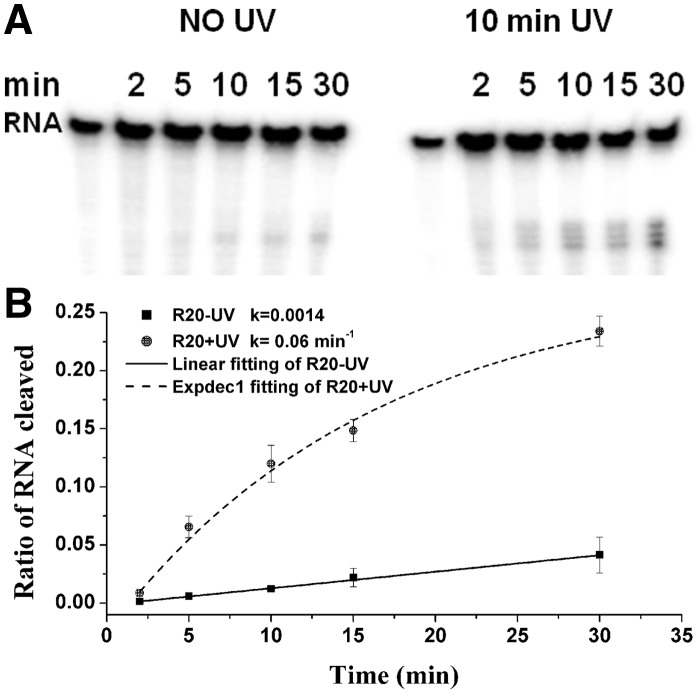
Time dependence of target RNA cleavage with RNase H and caged circular asODNs was also performed under 1× RNase H buffer, before and after light activation. As shown in Figure 2, the percentage of cleaved band with R20 in the PAGE gel increased with time. Kinetic data for 40mer RNA cleavage in the presence of R20 and RNase H were obtained by quantifying the percentage of cleaved target RNA at time t and fitting the data. The cleavage rate constants (k) were 0.0014 and 0.06 min−1 for R20 before and after light activation. The rate increased up to 43-fold. For the other two caged circular asODNs (R20B2 and R20B4, Supplementary Figure S3), the rate constants of RNA cleavage were 0.0017 and 0.0032 in the dark and 0.043 and 0.049 min−1 upon light activation, respectively. Based on the kinetics, light upregulated 25- and 15-fold RNA digestion rates for R20B2 and R20B4. Comparing to above results for caged circular asODNs before light activation, the rate constants increased from 0.0014 for R20, to 0.0017 and 0.0032 for R20B2 and R20B4 upon introducing the gap with different number of nucleotides in the duplex of RNA/caged circular asODN, which were consistent with the results of the percentages of RNA cleavage in 30 min which were 4, 4.5 and 8% for R20, R20B2 and R20B4.
Figure 2.
Denaturing PAGE (20%) analysis of RNA digestion with 4 µM [γ-32P]-labeled 40mer target RNA, 0.02 µM caged circular asODN (R20) and two units RNase H in 20 µl RNase H buffer at 37°C. Uncaged R20 had been irradiated with UV light (350 nm, ∼30 mW/cm2) for 10 min. The charts are cleavage percentage of the 40mer RNA by RNase H at different time points quantified with ImageQuant. Each point is the average of three separate trials.
Binding of RNA with caged circular asODNs
Comparing to the caged circular ODN C20 in our previous work (41), amounts of RNA digestion after UV irradiation were roughly the same; however, the background RNA cleavages before light activation were different. For C20, there was almost no background RNA digestion, but for R20, R20B2 and R20B4, the background signals of RNA digestion were 4, 4.5 and 8%, respectively. This may be due to the long flexible linkage in the middle of antisense sequence, which can easily twist to deny the access to target RNA and/or RNase H. To study the binding of 40mer target RNA with caged circular asODNs, gel mobility shift assays were performed as shown in Figure 3. These results showed that even more than 200-fold excess of caged circular R20 could not fully bind the target 40mer RNA. By quantifying the gels, the ratios of RNA/R20 binding were obtained and then used for the determination of Kd RNA/R20 by interpolation of non-linear regression hyperbola using Origin software. The dissociation constant (Kd RNA/R20) of R20 was 1.6 μM which was higher than that of C20 (Kd RNA/C20 = 1.0 μM) (Supplementary Figure S4), which indicated the binding of R20 to target RNA was less efficient than that of C20 due to the presence of linker in the middle. Interestingly, the results of gel mobility shift assays for R20B2 and R20B4 were completely different from R20. Caged circular R20B2 and R20B4 completely lost their binding affinity for target RNA under our experimental conditions. However, these results are not quite surprising. Due to the gaps in the middle of duplexes of the target RNA with R20B2 and R20B4, π interaction of duplex base stack is interrupted and a bulge will be formed if target RNA binds to caged circular R20B2 and R20B4, which further destabilizes the possible hybrids. Figure 3 also indicates that RNA/caged R20B2 and RNA/caged R20B4 hybrids were not formed under our experimental conditions. However, R20B2 and R20B4 could still be templates for the access of target RNA transiently binding and target RNA digestion at the presence of RNase H. Even though no stable hybrid duplex is formed for RNA with structured asODNs, such as hairpin, aptamer or other complicated structures. RNase H can recruit small amount of structured asODN/RNA hybrids, which promotes the equilibrium shift to RNA cleavage by RNase H (46,47). The results of RNA cleavage and gel mobility shift assays for R20B2 and R20B4 with the complementary target RNA were consistent with a literature report (46,47) even under conditions where hybridization of RNA/R20B2 and RNA/R20B4 was thermodynamically unfavorable. Although caged circular R20B2 and R20B4 work less efficiently for the photomodulation of RNA cleavage by RNase H, their hybridization with target RNA could be efficiently inhibited in the caged state. Light activation can cleave PL linker, and the flexible amide linker in the middle of antisense ODNs has little effect on its binding ability to target RNA (39). Thus, we expect that light activation of asODN-binding ability to target RNA can be used to photoregulate specific genes using non-enzyme-mediated antisense oligonucleotide strategy.
Figure 3.
Gel-shift assay of the 40mer RNA with different concentrations of R20, R20B2 and R20B4 using non-denaturing PAGE gels. The solutions of the 40mer RNA (0.02 μM) with different concentration ratios of R20, R20B2 and R20B4 (from 1:2 to 1:200) in 10 μl RNase H buffer was used for binding assays. The mixtures were first mixed in 9 μl RNase H buffer and then incubated at 37°C for 30 min. After that, the solution was put inside ice water and added ∼1 μl glycerol with bromphenol blue and xylene cyanol. Then the mixtures were loaded into a 12% native gel and the gel was run under 100 V at 10°C.
Photomodulation of the binding of target MB with caged circular antisense oligonucleotides (PS1, PS2 and PS3)
Caged PS1, PS2 and PS3 with 2′-OMe RNA and phosphorothioate modifications were designed according to the same strategy mentioned above. We first evaluated the photolysis of caged circular PS1, PS2 and PS3 using PAGE gel. As shown in Supplementary Figure S2B, caged circular PS1, PS2 and PS3 moved faster than their uncaged ones due to the difference of conformational structure of circular and linear oligonucleotides. To further evaluate photomodulation of their interactions with the target sequence, we designed a MB (5′-FAM-TTAGAATGGTGAGCAAGGGCGAGGAGCTGTCTAA-DABCYL-3′) whose loop sequence was complementary to uncaged PS1, PS2 and PS3 with 0 -, 2 - and 4-nt bulge, respectively. As shown in Figure 4 and Supplementary Figure S5, MB solution itself showed low fluorescence emission before addition of the target oligonucleotide. When PS, a fully complementary phosphorothioated antisense oligonucleotide with no linkage in middle, was added into MB solution, we observed an obvious increase in fluorescence emission, while the addition of a mismatch phosphorothioated oligonucleotide (PS-MS) did not trigger fluorescence enhancement regardless of light irradiation, which indicated the binding was due to sequence-specific interaction and light independence. When caged PS1 was mixed with MB, we did not observe increase in fluorescence emission; however, the addition of light-irradiated PS1 induced 5-fold enhancement in fluorescence intensity which was similar to the addition of PS. Similar results were also found for caged PS2 and PS3, where 3.6 - and 3.8-fold increase in fluorescence intensity with the addition of uncaged PS2 and PS3. These results indicated light activation can uncage these circular antisense oligonucleotides and recover the binding of PS1, PS2 and PS3 to their target sequence.
Figure 4.
Relative fluorescence intensity of MB with the addition of PS, PS-MS, PS1, PS2 and PS2 before and after light irradiation. [MB] = 100 nM, [oligonucleotide] = 400 nM. Oligonucleotide solutions were irradiated for 10 min (365 nm, 11 mW/cm2).
We further evaluated the binding kinetics of MB with PS and uncaged PS1, PS2 and PS3. For caged circular oligonucleotides, light activation would cleave PL and quickly release their linear ones. To study hybridization kinetics of MB with uncaged PS1, PS2 and PS3, fluorescence emission of MB was monitored with the addition of above antisense oligonucleotides. As shown in Supplementary Figure S6, the binding MB with PS reached the maximum fluorescence intensity in 6 min upon the addition of PS, while it took ∼15 min for uncaged circular PS1, PS2 and PS3. By fitting the data using the linearization procedure for a second-order reaction with non-stoichiometric proportions of reactants, the on-rate constants of oligonucleotide hybrids were obtained (Supplementary Figure S6). For PS without any linkage in the middle, it showed the fastest hybridization kinetics with the on-rate constant kh = 2.62 × 104 M−1s−1, while for light-irradiated PS1, PS2 and PS3, the on-rate constants (kh) were 1.27 × 104, 1.28 × 104 and 1.21 × 104 M−1s−1, respectively. The slow on-rate hybridization constants may be due to the flexible amide linkage and 0–4 nt bulge in the duplex of MB and PS1, PS2 and PS3.
Photomodulation of GFP expression in HeLa cells with caged circular antisense phosphorothioated oligonucleotides
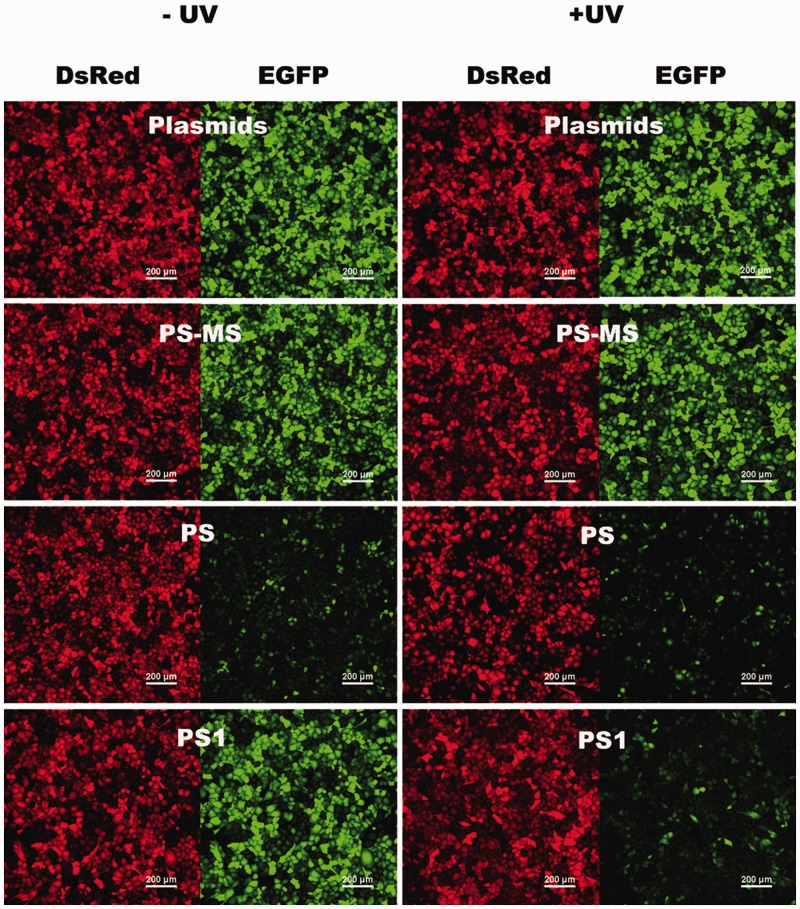
To test the efficacy of these caged circular asODNs (PS1, PS2 and PS3) at regulating GFP knockdown with light activation, HeLa cells were cotransfected with pEGFP-N1 and pDsRed2-N1 expressing plasmids, together with caged circular PS1, PS2 or PS3, respectively. After light irradiation, PS1, PS2 and PS3 were designed to bind GFP mRNA with a bulge of 0, 2 and 4 nt when the duplexes were formed. Another two antisense oligonucleotides (PS and PS-MS) were also tested as positive and negative control experiments. Two groups of cell experiments were always carried out under the same conditions at the same time. Cells in one group of experiments were washed with PBS buffer and irradiated after 6 h transfection. After irradiation, fresh media were applied and cells were allowed to culture for another 42 h. In the other group, cells were treated under the same way except light irradiation. GFP and RFP signals of the same cells were examined using confocal laser scanning microscopy, together with bright field and nuclei staining of the cells. All images were taken using the identical imaging conditions for all groups of experiments. As shown in Figure 5, red fluorescence of RFP, a reporter control, showed strong and uniform expression regardless of light irradiation or the addition of other antisense oligonucleotides in this study, which indicates light and other antisense oligonucleotides do not interrupt RFP gene expression. For the cells cotransfected with only pEGFP-N1 and pDsRed2-N1 plasmids, fluorescence signals from both GFP and RFP were also consistent, and light irradiation had little effect on their gene expression (Supplementary Figure S7). As expected, PS, a positive control antisense oligonucleotide, uniformly reduced gene expression of GFP, but not RFP, while PS-MS, a negative control antisense oligonucleotide, had no effect on both GFP and RFP gene expression. The control experiments confirm that knockdown of GFP is sequence specific and light alone does not cause the obvious decrease of GFP or RFP fluorescence signals. Figure 5 demonstrates the representative results of photomodulation of GFP expression with caged circular PS1. Caged circular PS1 had no effect on either GFP or RFP gene expression, while upon light irradiation, it was converted to linear uncaged PS1, which bound GFP mRNA and greatly lowered fluorescence signal of GFP, but not red fluorescence signal of RFP. Similar results were also observed when caged circular antisense oligonucleotides PS2 and PS3 were applied. The knockdown of GFP expression was achieved through non-enzyme involved antisense strategy which is similar to literature report (48) of morpholino oligomers caged with multiple photolabile moieties. Moreover, bright-field images with Hoechst nuclei staining and light irradiation of HeLa cells confirmed the viability of the cells over the course of the experiments (Supplementary Figure S7 and S8).
Figure 5.
Photomodulation of GFP expression with caged circular phosphorothioated 2′-OMe RNA oligonucleotides (PS, PS-MS and PS1). For each channel, all imaging conditions are identical. Scale bars represent 200 μm.
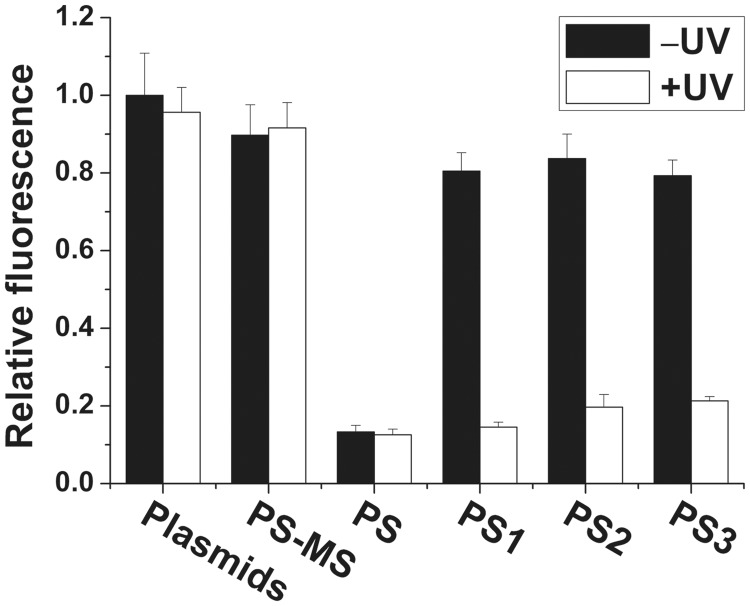
To further quantify photomodulation of GFP expression with caged circular antisense oligonucleotides, the GFP and RFP fluorescence signals were applied for the calculation of photomodulation efficiency. Since RFP gene expression was not affected in the course of our experiments, we applied RFP fluorescence signal as a standard control (Supplementary Figure S9). The ratio of GFP to RFP for each group of experiments was then normalized to that ratio in control cells with pEGFP-N1 and pDsRed2-N1 plasmids only and no light irradiation. Figure 6 shows the results of both groups of experiments in this study. PS strongly knocked down GFP expression with ∼7.5-fold lower fluorescence intensity regardless of light irradiation. Mismatched antisense oligonucleotide, PS-MS, had no effect on GFP expression and cells maintained similar level of GFP signals with GFP plasmids only (a change of GFP expression from 1.0 to 0.9). Similarly, GFP expression in HeLa cells was also maintained at high level of GFP fluorescence signal if caged circular PS1, PS2 or PS3 was cotransfected with GFP plasmid (a change of GFP expression from 1.0 to 0.8, 0.84 and 0.79). However, after 10-min irradiation of these cotransfected cells, GFP signals from these irradiated cells were 5.5 -, 4.2 - and 3.8-fold lower than GFP signals of pre-irradiation for PS1, PS2 and PS3, respectively. This observation shows that photomodulation of GFP gene expression with caged circular antisense oligonucleotides was successfully achieved. Their photomodulation efficiencies of GFP expression are generally a little bit lower than those of siRNA caged with multiple cyclo-dodecyl dimethoxy nitrophenyl ethyl (CD-DMNPE) moieties (49), but higher than those of caged DNAzymes (50). And irradiated PS1 could reach the same low level of GFP expression as PS, which indicated long linkage itself in the middle of PS1 did not have an effect on knockdown of GFP. For PS2 and PS3 cotransfected cells, the GFP signals were not reduced to the level as low as that of PS cotransfected cells. This is probably due to the bulge of extra 2 or 4 nt in the hybrids of GFP mRNA and uncaged PS2 or PS3, which lowers the strong binding ability between mRNA and asODN in cellular environment. To confirm this, we measured the thermodynamic melting temperatures (Tms) of different antisense oliognucleotides (PS1, PS2 and PS3) with a 44mer target sequence (Supplementary Figure S10). Before light irradiation, no clear transition of absorbance was observed. Upon light activation, the Tms for PS1, PS2 and PS3 with the target sequence were 64.2, 61.2 and 60.2°C in comparison to 66.2°C for the duplex of PS and target sequence. These results are also consistent with in vitro MB experimental data.
Figure 6.
Quantified results of light-activated GFP knockdown with different phosphorothioated 2′-OMe RNA oligonucleotides. All antisense oligonucleotides were 55 nM. All samples were cotransfected with pEGFP-N1 and pDsRed2-N1 plasmids. RFP signal was used as an internal control. Each column represents the ratio of GFP to RFP for each condition after normalization to that ratio in the control cells with pEGFP-N1 and pDsRed2-N1 plasmids only and no light irradiation. Each result is the average of five different determinations. Error bar is indicated as standard derivation.
CONCLUSIONS
A new series of caged circular asODNs were synthesized with two 10mer ODNs linked through a PL and an amide bond linker. These caged circular asODNs with PL and 0–4 nt gaps in the middle were not able to stably bind the target RNA. However, upon light irradiation, uncaged asODNs were quickly released and were available for the efficient binding to target RNA. With this successful asODN design, RNA digestion was also efficiently photomodulated in vitro. Further, three caged circular antisense oligonucleotides with 2′-OMe RNA and phosphorothioate modifications were synthesized and were capable of photoregulating GFP expression in cells. These in vitro and cell studies of caged circular oligonucleotides demonstrate promising applications to spatiotemporal photoregulation of gene expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–12.
FUNDING
The National Science Foundation of China (NSFC) [21072015]; National Basic Research Program of China [‘973’ Program, 2012CB720600]; Program for New Century Excellent Talents in University [NCET-10-0203]; China Postdoctoral Science Foundation (CPSF) [2012M510290 to L.W.]. Funding for open access charge: NSFC [21072015]; CPSF [2012M510290].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Vasudevan Ramesh (University of Manchester, UK) for reading the article, Prof. Demin Zhou for providing GFP plasmid for initial experiments and Dr. Bo Xu for assistance with cell and imaging experiments.
REFERENCES
- 1.Adams SR, Tsien RY. Controlling cell chemistry with caged compounds. Annu. Rev. Physiol. 1993;55:755–784. doi: 10.1146/annurev.ph.55.030193.003543. [DOI] [PubMed] [Google Scholar]
- 2.Asanuma H, Ito T, Yoshida T, Liang X, Komiyama M. Photoregulation of the formation and dissociation of a DNA duplex by using the cis-trans isomerization of azobenzene. Angew. Chem. Int. Ed. Engl. 1999;38:2393–2395. doi: 10.1002/(sici)1521-3773(19990816)38:16<2393::aid-anie2393>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 3.Asanuma H, Tamaru D, Yamazawa A, Liu M, Komiyama M. Photoregulation of the transcription reaction of T7 RNA polymerase by tethering an azobenzene to the promoter. ChemBioChem. 2002;8:786–789. doi: 10.1002/1439-7633(20020802)3:8<786::AID-CBIC786>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 4.Chaulk SG, MacMillan AM. Caged RNA: photo-control of a ribozyme reaction. Nucleic Acids Res. 1998;26:3173–3178. doi: 10.1093/nar/26.13.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dmochowski IJ, Tang X. Taking control of gene expression with light-activated oligonucleotides. Biotechniques. 2007;43:161–171. doi: 10.2144/000112519. [DOI] [PubMed] [Google Scholar]
- 6.Dussy A, Meyer C, Quennet E, Bickle TA, Giese B, Marx A. New light-sensitive nucleosides for caged DNA strand breaks. ChemBioChem. 2002;3:54–60. doi: 10.1002/1439-7633(20020104)3:1<54::AID-CBIC54>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 7.Heckel A, Mayer G. Light regulation of aptamer activity: an anti-thrombin aptamer with caged thymidine nucleobases. J. Am. Chem. Soc. 2005;127:822–823. doi: 10.1021/ja043285e. [DOI] [PubMed] [Google Scholar]
- 8.Hoebartner C, Silverman SK. Modulation of RNA tertiary folding by incorporation of caged nucleotides. Angew. Chem. Int. Ed. Engl. 2005;44:7305–7309. doi: 10.1002/anie.200502928. [DOI] [PubMed] [Google Scholar]
- 9.Casey JP, Blidner RA, Monroe WT. Caged siRNAs for spatiotemporal control of gene silencin. Mol. Pharmaceutics. 2009;6:669–685. doi: 10.1021/mp900082q. [DOI] [PubMed] [Google Scholar]
- 10.Keiper S, Vyle JS. Reversible photocontrol of deoxyribozyme-catalyzed RNA cleavage under multiple-turnover conditions. Angew. Chem. Int. Ed. Engl. 2006;45:3306–3309. doi: 10.1002/anie.200600164. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Sen D. Light-regulated catalysis by an RNA-cleaving deoxyribozyme. J. Mol. Biol. 2004;341:887–892. doi: 10.1016/j.jmb.2004.06.060. [DOI] [PubMed] [Google Scholar]
- 12.Matsunaga D, Asanuma H, Komiyama M. Photoregulation of RNA digestion by RNase H with azobenzene-tethered DNA. J. Am. Chem. Soc. 2004;126:11452–11453. doi: 10.1021/ja0471976. [DOI] [PubMed] [Google Scholar]
- 13.Mikat V, Heckel A. Light -dependent RNA interference with nucleobase-caged siR. RNA. 2007;13:2341–2347. doi: 10.1261/rna.753407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richards JL, Tang X, Turetsky A, Dmochowski IJ. RNA bandages for photoregulating in vitro protein translation. Bioorg. Med. Chem. Lett. 2008;18:6255–6258. doi: 10.1016/j.bmcl.2008.09.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah S, Rangarajan S, Friedman SH. Light-activated RNA interference. Angew. Chem. Int. Ed. Engl. 2005;44:1328–1332. doi: 10.1002/anie.200461458. [DOI] [PubMed] [Google Scholar]
- 16.Ouyang X, Shestopalov IA, Sinha S, Zheng G, Pitt CLW, Li W-H, Olson AJ, Chen JK. Versatile synthesis and rational design of caged morpholinos. J. Am. Chem. Soc. 2009;131:13255–13269. doi: 10.1021/ja809933h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shestopalov IA, Sinha S, Chen JK. Light-controlled gene silencing in zebrafish embryos. Nature Chem. Biol. 2007;3:650–651. doi: 10.1038/nchembio.2007.30. [DOI] [PubMed] [Google Scholar]
- 18.Tang X, Swaminathan J, Gewirtz AM, Dmochowski IJ. Regulating gene expression in human leukemia cells using light-activated oligodeoxynucleotides. Nucleic Acids Res. 2008;36:559–569. doi: 10.1093/nar/gkm1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang X, Dmochowski IJ. Controlling RNA digestion by RNase H with a light-activated DNA hairpin. Angew. Chem. Int. Ed. Engl. 2006;45:3523–3526. doi: 10.1002/anie.200600954. [DOI] [PubMed] [Google Scholar]
- 20.Tang X, Maegawa S, Weinberg ES, Dmochowski IJ. Regulating gene expression in zebrafish embryos using light-activated, negatively charged peptide nucleic acids. J. Am Chem. Soc. 2007;129:11000–11001. doi: 10.1021/ja073723s. [DOI] [PubMed] [Google Scholar]
- 21.Tang X, Dmochowski IJ. Synthesis of light-activated antisense oligodeoxynucleotide. Nat. Protoc. 2007;1:3041–3048. doi: 10.1038/nprot.2006.462. [DOI] [PubMed] [Google Scholar]
- 22.Tang X, Dmochowski IJ. Regulating gene expression with light-activated oligonucleotides. Mol. BioSyst. 2007;3:100–110. doi: 10.1039/b614349k. [DOI] [PubMed] [Google Scholar]
- 23.Young DD, Deiters A. Photochemical hammerhead ribozyme activation. Bioorg. Med. Chem. Lett. 2006;16:2658–2661. doi: 10.1016/j.bmcl.2006.02.034. [DOI] [PubMed] [Google Scholar]
- 24.Richards JL, Seward GK, Wang Y-H, Dmochowski IJ. Turning the 10–23 DNAzyme on and off with light. ChemBioChem. 2010;11:320–324. doi: 10.1002/cbic.200900702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi Y, Koh JT. Light-activated transcription and repression by using photocaged SERMs. ChemBioChem. 2004;5:788–796. doi: 10.1002/cbic.200300823. [DOI] [PubMed] [Google Scholar]
- 26.Ruble BK, Richards JL, Cheung-Lau JC, Dmochowski IJ. Mismatch discrimination and efficient photomodulation with split 10–23 DNAzymes. Inorg. Chem. Acta. 2012;380:386–391. doi: 10.1016/j.ica.2011.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ando H, Furuta T, Tsien RY, Okamoto H. Photo-mediated gene activation using caged RNA/DNA in zebrafish embryos. Nat. Genet. 2001;28:317–325. doi: 10.1038/ng583. [DOI] [PubMed] [Google Scholar]
- 28.Ghosn B, Haselton FR, Gee KR, Monroe WT. Control of DNA hybridization with photocleavable adducts. Photochem. Photobiol. 2005;81:953–959. doi: 10.1562/2004-11-15-RA-373. [DOI] [PubMed] [Google Scholar]
- 29.Monroe WT, McQuain MM, Chang MS, Alexander JS, Haselton FR. Targeting expression with light using caged DNA. J. Biol. Chem. 1999;274:20895–20900. doi: 10.1074/jbc.274.30.20895. [DOI] [PubMed] [Google Scholar]
- 30.Shah S, Jain PK, Kala A, Karunakaran D, Friedman SH. Light -activated RNA interference using double-stranded siRNA precursors modified using a remarkable regiospecificity of diazo-based photolabile groups. Nucleic Acids Res. 2009;37:4508–4517. doi: 10.1093/nar/gkp415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Govan JM, Uprety R, Hemphill J, Lively MO, Deiters A. Regulation of transcription through light-activation and light-deactivation of triplex-forming oligonucleotides in mammalian cells. ACS Chem. Biol. 2012;7:1247–1256. doi: 10.1021/cb300161r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang K, Taylor J-S. A caged ligatable DNA strand break. J. Am. Chem. Soc. 1999;121:11579–11580. [Google Scholar]
- 33.Zheng G, Cochella L, Liu J, Hobert O, Li W. Temporal and spatial regulation of microRNA activity with photoactivatable cantimirs. ACS Chem. Biol. 2011;6:1332–1338. doi: 10.1021/cb200290e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomasini AJ, Schuler AD, Zebala JA, Mayer AN. PhotoMorphsTM: a novel light-activated reagent for controlling gene expression in zebrafish. Genesis. 2009;47:736–743. doi: 10.1002/dvg.20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richards JL, Seward GK, Wang Y-H, Dmochowski IJ. Turning the 10-23 DNAzyme on and off with light. ChemBioChem. 2009;11:320–324. doi: 10.1002/cbic.200900702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamazoe S, Shestopalov IA, Provost E, Leach S, Chen JK. Cyclic caged morpholinos: conformationally gated probes of embryonic gene function. Angew. Chem. Int. Ed. Engl. 2012;51:6908–6911. doi: 10.1002/anie.201201690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Wu L, Wang P, Lv C, Yang Z, Tang X. Manipulation of gene expression in zebrafish using caged circular morpholino oligomers. Nucleic Acids Res. 2012;40:11155–11162. doi: 10.1093/nar/gks840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang X, Su M, Yu L, Lv C, Wang J, Li Z. Photomodulating RNA cleavage using photolabile circular antisense oligodeoxynucleotides. Nucleic Acids Res. 2010;38:3848–3855. doi: 10.1093/nar/gkq079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richard JL, Tang X, Turetsky A, Dmochowski IJ. RNA bandages for photoregulating in vitro protein translation. Bioorg. Med. Chem. Lett. 2008;18:6255–6258. doi: 10.1016/j.bmcl.2008.09.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ordoukhanian P, Taylor J-S. Design and synthesis of a versatile photocleavable DNA building block. Application to phototriggered hybridization. J. Am. Chem. Soc. 1995;117:9570–9571. [Google Scholar]
- 41.Tang X, Su M, Yu L, Lv C, Wang J, Li Z. Photomodulating RNA cleavage using photolabile circular antisense oligodeoxynucleotides. Nucleic Acids Res. 2010;38:3848–3855. doi: 10.1093/nar/gkq079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schulz AR. Enzyme Kinetics from Diastase to Multi-Enzyme Systems. New York: Cambridge University Press; 1994. pp. 3–20. [Google Scholar]
- 43.Gangurde R, Modak MJ. Participation of active-site carboxylates of Escherichia coli DNA polymerase I (Klenow fragment) in the formation of a prepolymerase ternary complex. Biochemistry. 2002;41:14552–14559. doi: 10.1021/bi0264216. [DOI] [PubMed] [Google Scholar]
- 44.Kuhn H, Demidov VV, Coull JM, Fiandaca MJ, Gildea BD, Frank-Kamenetskii MD. Hybridization of DNA and PNA molecular beacons to single-stranded and double-stranded DNA targets. J. Am. Chem. Soc. 2002;124:1097–1103. doi: 10.1021/ja0041324. [DOI] [PubMed] [Google Scholar]
- 45.Tadokoro T, Kanaya S. Ribonuclease H: molecular diversities, substrate binding domains, and catalytic mechanism of the prokaryotic enzymes. FEBS J. 2009;276:1482–1493. doi: 10.1111/j.1742-4658.2009.06907.x. [DOI] [PubMed] [Google Scholar]
- 46.Zamaratski E, Pradeepkumar PI, Chattopadhyaya J. A critical survey o the structure-function of the antisense oligo/RNA heteroduplex as substrate for RNase H. J. Biochem. Biophys. Meth. 2001;48:189. doi: 10.1016/s0165-022x(01)00149-x. [DOI] [PubMed] [Google Scholar]
- 47.Li J, Wartell RM. RNase H1 can catalyse RNA/DNA formation and cleavage with stable hairpin or duplex DNA oligomers. Biochemistry. 1998;37:5154–5161. doi: 10.1021/bi9730801. [DOI] [PubMed] [Google Scholar]
- 48.Deiters A, Garner RA, Lusic H, Govan JM, Dush M, Nascone-Yoder NM, Yoder JA. Photocaged morpholino oligomers for the light-regulation of gene function in zebrafish and Xenopus embryos. J. Am. Chem. Soc. 2010;132:15644–15650. doi: 10.1021/ja1053863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jain PK, Shah S, Friedman SH. Patterning of gene expression using new photolabile groups applied to light activated RNAi. J. Am. Chem. Soc. 2011;133:440–446. doi: 10.1021/ja107226e. [DOI] [PubMed] [Google Scholar]
- 50.Young DD, Lively MO, Deiters A. Activation and deactivation of DNAzyme and antisense function with light for the photochemical regulation of gene expression in mammalian cells. J. Am. Chem. Soc. 2010;132:6183–6193. doi: 10.1021/ja100710j. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.