Abstract
TRIM-NHL proteins are conserved regulators of development and differentiation but their molecular function has remained largely elusive. Here, we report an as yet unrecognized activity for the mammalian TRIM-NHL protein TRIM71 as a repressor of mRNAs. We show that TRIM71 is associated with mRNAs and that it promotes translational repression and mRNA decay. We have identified Rbl1 and Rbl2, two transcription factors whose down-regulation is important for stem cell function, as TRIM71 targets in mouse embryonic stem cells. Furthermore, one of the defining features of TRIM-NHL proteins, the NHL domain, is necessary and sufficient to target TRIM71 to RNA, while the RING domain that confers ubiquitin ligase activity is dispensable for repression. Our results reveal strong similarities between TRIM71 and Drosophila BRAT, the best-studied TRIM-NHL protein and a well-documented translational repressor, suggesting that BRAT and TRIM71 are part of a family of mRNA repressors regulating proliferation and differentiation.
INTRODUCTION
The family of TRIM-NHL ubiquitin ligases is characterized by an N-terminal tripartite motif [TRIM; consisting of a RING domain, B-box and coiled-coil (CC) regions] and a C-terminal NHL domain (1). TRIM is also found associated with other C-terminal domains and defines a superfamily of TRIM proteins (2), many of which are functional ubiquitin ligases. TRIM-NHL proteins are conserved among metazoa and are key regulators of development and differentiation. Mammals express four such proteins: TRIM2, TRIM3, TRIM32 and TRIM71. However, apart from Drosophila BRAT, which acts as a translational repressor (3–5), the molecular functions of TRIM-NHL proteins are not well-defined.
Several TRIM-NHL proteins have been identified recently as modulators of microRNA (miRNA)-mediated repression in mammals, flies and worms (6–9). miRNAs, in association with Argonautes (AGO) and other proteins of miRNA-protein particles (miRNPs), repress translation of mRNAs and accelerate their decay by base-pairing with mRNA 3′-untranslated regions (3′-UTRs). Interaction of several TRIM-NHL proteins with AGOs and other miRNP components has been demonstrated but the functional consequences vary: while mammalian TRIM32 and Caenorhabditis elegans NHL-2 are enhancers of miRNA-mediated repression (6,9), mammalian TRIM71 and Drosophila Mei-P26 act as negative pathway regulators (7,8).
TRIM-NHL proteins themselves are targets of miRNA repression and C. elegans LIN-41 was the first known target of let-7 miRNA (10). In C. elegans, LIN-41 and let-7 both regulate developmental timing events, LIN-41 preventing premature differentiation of epidermal skin cells and let-7 promoting their differentiation (10). The regulatory relationship of let-7 with LIN-41 orthologs, TRIM71 in vertebrates and Dappled in flies, is conserved (11–15). However, while the molecular function of let-7 as a key regulator driving differentiation in development and disease is well-defined (16), little is known about the function of LIN-41/TRIM71.
TRIM71 is highly expressed in undifferentiated cells, such as embryonic stem (ES) cells, but becomes rapidly down-regulated upon differentiation, when let-7 levels rise (8). Like other proteins that sustain stemness and pluripotency (e.g. MYC, LIN-28 or SALL4), expression of TRIM71 is not only negatively regulated by let-7 but also indirectly up-regulated by ES-cell-specific miRNAs (17). This suggests a role for TRIM71 in the maintenance of stem cell identity or inhibition of premature differentiation. Although gene targeting in mice revealed that TRIM71 is essential for embryonic viability (18), its molecular function remains unknown.
Here we report that the association of mammalian TRIM71 with mRNAs results in translational repression and mRNA degradation. The NHL domain and the central part of the protein mediate RNA interaction and translational repression, respectively. We have identified targets in human HEK293 and mouse ES (mES) cells that indicate a role for TRIM71 in ES cell maintenance. TRIM71 shares many targets with miRNAs and full repression of these targets requires expression of both TRIM71 and miRNAs.
MATERIALS AND METHODS
Cell culture, transfection, RNA interference and generation of stable cell lines
Human HEK293 and HEK293 Flp-In (Invitrogen) cells were grown in DMEM (Gibco-BRL) supplemented with 2 mM L-glutamine and 10% heat inactivated FCS. mES cells (E14TG2a.4) were cultured as described previously (19). For time course experiments cells were treated with 5 µg/ml Actinomycin D (Sigma), 10 µg/ml Cycloheximide (Sigma) or 10 µM MG132 (Calbiochem). Transfections were performed using nanofectin (PAA Laboratories), nanofectin siRNA (PAA Laboratories) or attractene (Qiagen) transfection reagents according to manufacturer’s instructions. siRNAs were ordered from Microsynth and sequences are listed in Supplementary Table S3. To generate HEK293 Flp-In stable cell lines, parental cells were transfected with 5.4 µg pOG44, 0.6 µg of the respective pCDN5/FRT plasmid and 19.2 µl nanofectin per 10 cm dish. Cells were split 48 h post-transfection and selection started 72–96 h post-transfection. Selection of HEK293 Flp-In stable cell lines was carried out using 100 µg/ml hygromycin B (Invitrogen) and for the maintenance of HEK293 Flp-In stable cell lines 50 µg/ml hygromycin B was added to the culture medium. Knock-down of Trim71 in mES cells was performed in 6-well plates using a mixture of 3 different siRNAs specific to Trim71 or allstar negative control siRNA (Qiagen) (at 25 nM final) and 4 µl dharmafect1 (Dharmacon) according to manufacturer’s instructions. Medium was changed 4 h and 24 h post-transfection and cells were harvested 72 h post-transfection.
Plasmids
Tethering reporter plasmid pRL-5BoxB and the control plasmid pGL-FL, as well as plasmids encoding NHA- or HA-AGO2 and NHA-LacZ have been described (20,21). Constructs expressing NHA-or HA-TRIM-NHL proteins or mutants thereof were generated by PCR amplification of respective ORFs or fragments thereof and subsequent cloning into pCIneo vectors that contain NHA or HA-tags (20) using SalI and NotI sites, except for full-length TRIM71 which is described below. For the generation of NHA- or HA-TRIM71-NHL32 and TRIM32-NHL71 respective fragments were PCR amplified using PCR primer that introduced SalI/XbaI and XbaI/NotI sites. Reporter plasmids pMIR-HMGA2, containing the 3′UTR of HMGA2 with either wt or mutated (mut) let-7 sites, and pMIR-KRAS, containing the 3′UTR of KRAS, have been described (22). All other 3′UTR reporters were generated from these vectors by exchange of the respective 3′UTRs using SacI/NaeI sites. Respective 3′UTRs were PCR amplified from cDNA of HEK293 or mES cells using primers listed in Supplementary Table S3.
pEGFPC1-TRIM71 was generated by releasing the EcoRI-EcoRI and EcoRI-KpnI fragments from Image clone AA 169781 (geneservice), and cloning into the pEGFPC1 vector (Clontech). pCDNA5/FRT-FHA-TRIM71 was generated in three steps: first, a HindIII/KpnI fragment from pEGFPC1-TRIM71 was cloned into pCDNA5/FRT (Invitogen); second, annealed oligonucleotides coding for the FLAG-HA-tag and containing a 3′ end proximal SalI site was inserted via the HindIII site thereby destroying the 5′ HindIII site but leaving the 3′ HindIII site intact; third, the HindIII-HindIII fragment remaining from the HindIII/KpnI cleavage of pEGFPC1-TRIM71 was inserted. All other full-length TRIM71 constructs were generated by releasing TRIM71 from this construct using SalI/NotI sites. pIRESneoFHA-AGO2 and pIRESneoFHA-TRIM32 have been described (23). To obtain pGFPC1- and pHIS-MYC-TRIM32, the ORF of TRIM32 was PCR amplified and cloned into the respective vectors via EcoRI/BamHI or SalI/NotI sites, respectively. To construct pCDNA5/FRT-FHA-TRIM32, the ORF of TRIM32 including FLAG-HA-tag were released from pIRESneo-FHA-TRIM32 and inserted into the pCDNA5/FRT vector using KpnI/BamHI sites. For generation of pCDNA5/FRT-FHA-TRIM71 deletion mutants TRIM32 was released from pCDNA5/FRT-FHA-TRIM32 using SalI/NotI sites, leaving the FLAG-HA-tag, and fragments of TRIM71 were PCR amplified and inserted into the vector. Point mutations were introduced by site-directed mutagenesis (24). In case of TRIM71, point mutations were first introduced in TRIM71-NHLonly constructs; SmaI/NotI fragments were then released and introduced into full-length TRIM71. All constructs were verified by sequencing.
Luciferase assays
Transfections were performed in 12 - or 24-well formats and done in triplicate. For tethering reporter experiments transfection mixtures contained 500 ng pGL-FL, 10 ng RL-5BoxB, 20 ng NHA- or HA-fusion-protein expressing plasmids and 1.6 µl nanofectin per 3 wells of a 12-well plate. For 3′UTR reporter experiments transfection mixtures contained 500 ng pMIR-3′UTR-reporter constructs, 20 ng protein expressing plasmids and 1.6 µl nanofectin per 3 wells of a 12-well plate. Transfection mixtures for reporter gene experiments with simultaneous knock-down of AGO or Pumilio (PUM) proteins contained 2 µg pMIR-3′UTR reporter, 50 ng protein coding plasmids, siRNAs directed against AGO1, AGO2, PUM1 or PUM2 or allstar negative control siRNA (at 100 nM final) and 8 µl nanofectin siRNA reagent per 3 wells of a 12-well plate. Transfection mixtures for miR-302 co-transfection experiments contained 1.2 µg pMIR-E2F7 reporter, 20 ng protein expressing plasmids, miR-302 b mimic (Dharmacon) or allstar negative control siRNA (at 10 nM final) and 4.5 µl attractene per 3 wells of a 24-well plate. Transfections of reporter gene assays in mES cells were done with attractene. Transfection mixtures contained 2 µg pGL-FL, 400 ng RL-5BoxB, 200 ng NHA- or HA-expressing plasmids and 9 µl of attractene per 3 wells of a 12-well plate or 1.2 µg pMIR 3′UTR-reporter, siRNAs directed against Trim71 or allstar negative control siRNA (100 nM final) and 4.5 µl attractene per 3 wells of a 24-well plate. Cells were lysed 48 h or, in case of knock-down experiments, 72 h post-transfection using passive lysis buffer (Promega) and firefly (FL) and renilla luciferase (RL) activities were measured with the Dual-Luciferase Reporter Assay System (Promega). For all luciferase assays, values represent means of two to five independent experiments, each performed in triplicate, and error bars show standard error of the mean (s.e.m.).
Immunoprecipitation and western blotting
Transient-transfected HEK293 cells or HEK293 Flp-In cells constitutively expressing FLAG-HA-tagged proteins were washed twice with ice-cold PBS, lysed in whole cell extraction buffer (WCE) [50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5% NP-40, 10% Glycerol, 0.2 mM EDTA, 2 mM EGTA, 50 mM NaF, 10 mM Glycerophosphat, 1 mM NaVanadate, 2 mM DTT and Complete protease inhibitor mix (Roche)] for 30 min on ice and cleared by centrifugation at 13 000g for 15 min at 4°C. The cleared lysate was incubated with FLAG M2 beads (Roche) for 2 h at 4°C on a rotating wheel. When indicated, 250 µg/ml RNase A (Roche) was added to the reaction. Beads were washed 5× with WCE or, if samples were used for ms analyses, 2× with WCE and 3× with TBS (20 mM Tris-HCl, pH 7.4, 150 mM NaCl). Bound proteins were eluted with 150 ng/µl FLAG peptide (Roche) in either TBS or WCE for 30 min at 4°C.
For western blot analyses, lysates or immunoprecipitates (IPs) were boiled in Laemmli buffer, separated by SDS-PAGE and electro-transferred to PDVF membranes. The following primary antibodies were used: anti-FLAG M2, 1:5000 (Sigma); anti-GFP, 1:5000 (Roche, 1814460); anti-HA (9F10), 1:2000 (Roche); anti-AGO2 (M01), 1:1000 (Abnova); anti-AGO2 (11A9), 1:1000 (Ascenion); anti-DDX3X, 1:500 (Milipore, 09-80); anti-HSP-90 (D19), 1:2000 (Santa Cruz, sc-1057); anti-HSP-70, 1:5000 (Santa Cruz, sc-32239); anti-MOV10, 1:1000 (Novus, NB100-77314); anti-MYC (9E10), 1:200 (Santa Cruz, sc-40); anti-PABP1, 1:5000 (Cell Signaling Technology); anti-PUM1, 1:5000 (Bethyl, A300-201A); anti-PUM2, 1:2000 (Bethyl, A300-202A); anti-α-tubulin, 1:10 000 (Sigma T5168); anti-TRIM71, 1:500 (kind gift of G. Wulczyn) (8); anti-Ubiquitin (P4D1), 1:1000 (Santa Cruz, sc-8014), anti-XRN1, 1:5000 (Bethyl, A300-443A) and anti-XRN2, 1:5000 (Bethyl, A301-102A).
RNA-immunoprecipitation
RNA-immunoprecipitation (RIP) methodology was adapted from previously described reports (25,26). Briefly, HEK293 Flp-In cells stably expressing FLAG-HA-tagged proteins or the parental cells were washed twice with ice-cold PBS, scraped of the plates in PBS and pelleted by centrifugation at 3000g for 5 min at 4°C. Cells were lysed in lysis buffer (10 mM HEPES pH 7, 100 mM KCl, 2 mM EDTA, 0.5% NP-40, 2 mM DTT, 50 U/ml RNaseOUT (Invitrogen) and Complete protease inhibitor cocktail) for 15 min on ice and the lysate was cleared by centrifugation at 13 000g for 15 min at 4°C. Protein concentration was determined using Bradford reagent. Lysates were incubated with FLAG M2 beads for 2 h at 4°C on a rotating wheel. Beads were washed 5× with NT2-RIP buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl, 0.05% NP-40 and 2 mM DTT) and bound proteins were eluted with 150 ng/µl FLAG peptide in NT2-RIP buffer containing 50 U/ml RNaseOUT and complete protease inhibitor cocktail for 30 min at 4°C. RNA was extracted using Trizol (Invitrogen) and used for RT-qPCR or microarray analysis. In later case RNA was further treated with RNase-free DNaseI and cleaned up using RNeasy columns (Qiagen).
Results
TRIM71 is associated with mRNPs
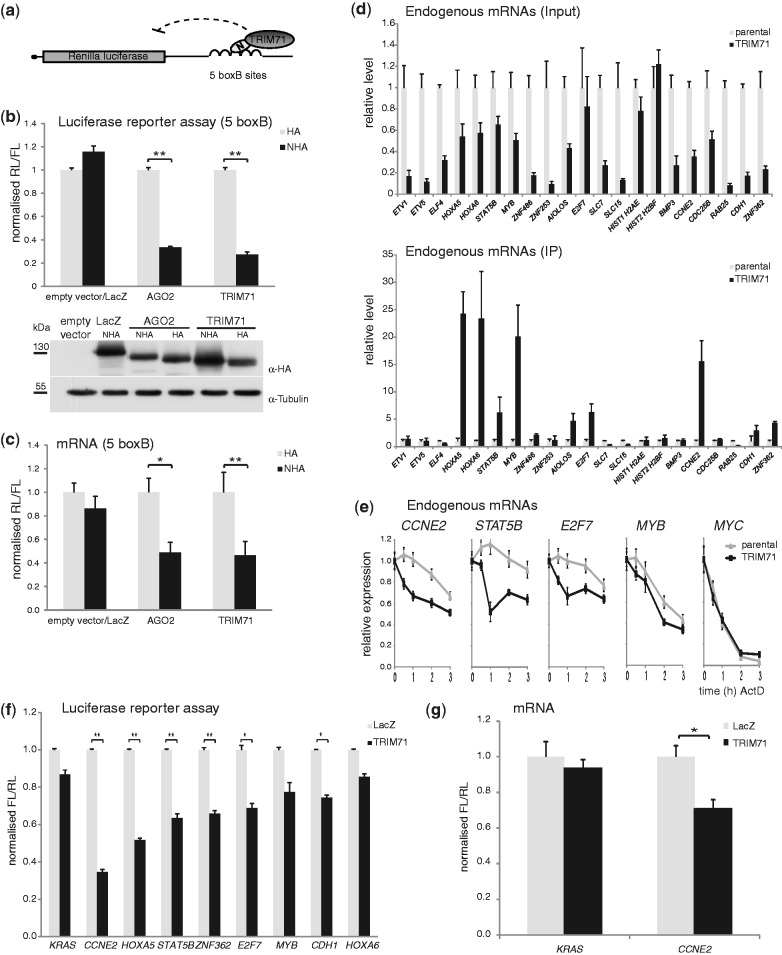
To investigate TRIM71 function, we constructed HEK293 cell lines stably expressing FLAG-HA-tagged-TRIM71. Immunofluorescence analyses of these cells revealed granular and perinuclear cytoplasmic staining of TRIM71 and its partial co-localization with AGO2 and DDX6 (also known as RCK/p54) (Supplementary Figure S1), resembling its previously reported localization (8). To identify TRIM71-associated proteins, we immunopurified FLAG-HA-tagged-TRIM71 and characterized co-purified proteins by mass spectrometry (MS). TRIM71 associated with many proteins involved in aspects of mRNA function (Supplementary Table S1). The association of some proteins was confirmed by western blotting (Figure 1). In addition to proteins identified by MS, PUM1 and PUM2 were included in the analysis as their Drosophila homolog PUM is an interacting partner of Drosophila BRAT (5). All analysed proteins, except HSP70 and HSP90, associated with TRIM71 in an RNA-dependent fashion; their association decreased upon treatment with RNase A.
Figure 1.
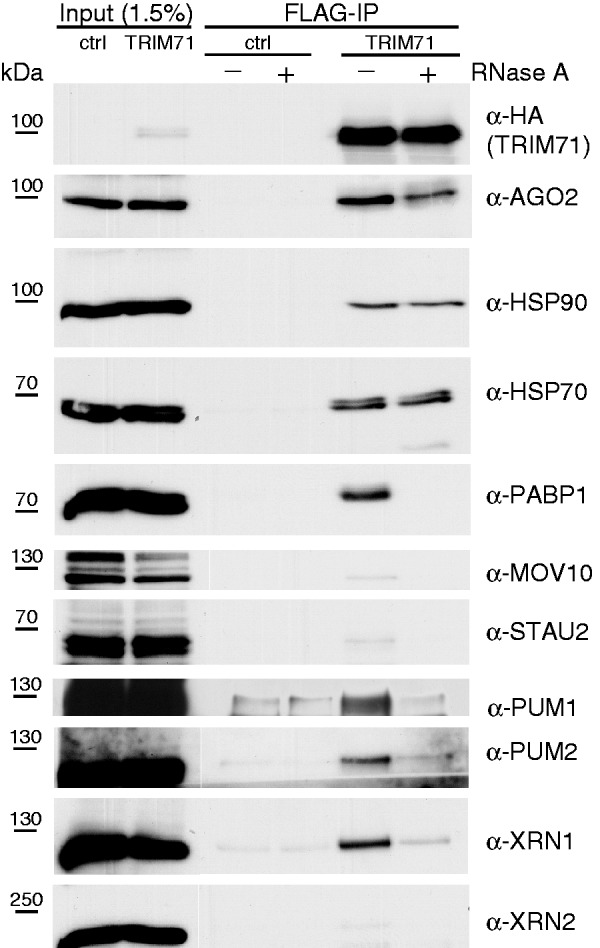
RNA-dependent association of TRIM71 with mRNA regulatory proteins. Immunoprecipitation experiment with anti-FLAG M2 affinity gel (FLAG-IP) from HEK293 cells constitutively expressing FLAG-HA-tagged TRIM71 (TRIM71) or the parental cell line (ctrl). Where indicated (+) RNase A was added to the reaction. Western blots were probed with antibodies against the indicated endogenous proteins or HA.
TRIM71 is a repressor of mRNA function
The RNA-dependent association of TRIM71 with many mRNA-binding proteins and the established function of BRAT as a translational repressor prompted us to examine possible TRIM71 activity in regulating protein synthesis. We fused HA-TRIM71 to the λ phage N-peptide (resulting in the protein NHA-TRIM71), which specifically recognizes boxB hairpins and thus can target the fusion-protein to a RL reporter mRNA containing boxB sites in its 3′UTR (RL-5boxB) (Figure 2a). As shown in Figure 2b, expression of NHA-TRIM71 repressed RL activity, but not HA-TRIM71 or NHA-LacZ control proteins when expressed at similar levels. The repression was comparable to that of NHA-AGO2. Measurements of mRNA levels by RT-qPCR showed that the decrease in RL level was accompanied by a partial decrease in RL-5boxB mRNA (Figure 2c). Thus, inhibition of protein synthesis by tethered TRIM71 may results from translational repression and mRNA degradation.
Figure 2.
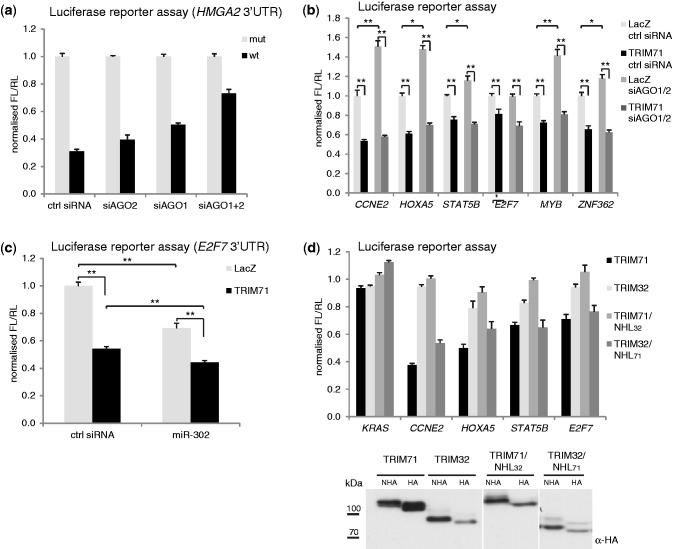
TRIM71 is a repressor of mRNA function. (a) Schematic representation of the RL-5boxB reporter used in this study. The RL-5boxB reporter contains RL coding sequence and a 3′UTR with five boxB sites. (b) (top) Repression of RL-5boxB by NHA-AGO2 or NHA-TRIM71. HEK293 cells were co-transfected with plasmids expressing RL-5boxB, the indicated NHA- or HA-fusion-proteins and a FL control plasmid. RL was normalized to FL and values of normalized RL produced in the presence of the proteins without λN-peptide (HA) were set to 1. Tethering of ß-galactosidase (LacZ) served as an additional control. (bottom) Protein expression was confirmed by western blotting. (c) RT-qPCR to estimate the RL and FL mRNA levels from the experiment described in panel (b). (d) RIP experiment. Levels of indicated endogenous mRNAs in HEK293 cells stably expressing FLAG-HA-TRIM71 (TRIM71) were compared to those of the parental control cell line (parental) before (Input) and after immunoprecipitation with FLAG-M2 affinity gel (IP). RT-qPCR values were normalized for GAPDH mRNA. Values obtained from parental cells were set to 1. Western blots showing expression of FLAG-HA-TRIM71 and IP efficiency are shown in Supplementary Figure S2a. (e) TRIM71 accelerates mRNA decay. HEK293 cells stably expressing FLAG-HA-TRIM71 (TRIM71) or parental control cells (Parental) were treated with 5 µg/ml Actinomycin D (ActD) to stop transcription. Levels of indicated endogenous mRNAs were analysed at indicated time points after addition of ActD by RT-qPCR. RT-qPCR values were normalized to GAPDH and values obtained in the absence of ActD were set to 1. (f) Repression of indicated FL-3′UTR reporter constructs by TRIM71. HEK293 cells were co-transfected with plasmids expressing the indicated FL reporters together with RL control and plasmids expressing either TRIM71 or LacZ as control. FL was normalized to RL and values of normalized FL produced in the presence of LacZ were set to 1. (g) Determination of the effect of TRIM71 on RNA level of the FL-3′UTR reporters by RT-qPCR. HEK293 cells were co-transfected with plasmids expressing the indicated FL reporters together with RL control and plasmids expressing either TRIM71 or LacZ as control. Panels (b), (c), (f) and (g) *P-value < 0.05; **P-value < 0.01.
Identification of TRIM71 targets in HEK293 cells
As TRIM71-mediated repression of protein synthesis was accompanied by a decrease in mRNA level, we sought to identify potential TRIM71 targets using mRNA arrays. HEK293 cells do not express endogenous TRIM71; hence we compared the mRNA profile of HEK293 cells stably expressing FLAG-HA-TRIM71 to that of parental cells. TRIM71 expression led to down-regulation of 524 and up-regulation of 297 transcripts by at least 1.5-fold (P < 0.01) (Supplementary Table S2).
To identify mRNAs down-regulated due to direct targeting by TRIM71, as opposed to mRNAs misregulated due to secondary effects, we performed RIP, expecting direct TRIM71 targets to associate with the protein. Specifically, we immunopurified FLAG-HA-TRIM71 from HEK293 cells stably expressing the protein, isolated bound RNA, and quantified transcripts selected from the list of mRNAs down-regulated by TRIM71 (Supplementary Table S2) by RT-qPCR (for western analysis of protein input and IPs, see Supplementary Figure S2a). Of the 21 transcripts analysed, 18 were down-regulated in cells expressing TRIM71, compared with parental HEK293 cells (Figure 2d, top), confirming the array results. Moreover, nine transcripts showed enrichment in the FLAG-HA-TRIM71 precipitate compared with control IP (Figure 2d, bottom). These transcripts likely represent bonafide TRIM71 targets as they were associated with TRIM71 and their mRNA levels decreased upon TRIM71 expression.
To determine if the observed decrease in mRNA levels mediated by TRIM71 is a result of accelerated mRNA degradation, we compared decay rates of several identified TRIM71 target mRNAs in control HEK293 cell line and the cell line stably expressing TRIM71. Transcription was stopped with actinomycin D and cytoplasmic mRNA levels were measured at different time points up to 3 h after addition of the drug. As shown in Figure 2e, decay rates of endogenous CCNE2, STAT5B, E2F7 or MYB were markedly accelerated in the presence of TRIM71, while the decay of MYC, which was not identified as a TRIM71 target, was unaffected.
Many mRNA regulatory proteins bind to and regulate mRNA targets via their 3′UTR. To test whether this applies to TRIM71-mediated regulation, we constructed FL reporters containing 3′UTRs of the identified bonafide TRIM71 targets. The FL-3′UTR reporter plasmids (also expressing an RL control reporter) were co-transfected with plasmids encoding either TRIM71 or LacZ. The 3′UTR of KRAS, which was not identified as a TRIM71 target in the array analyses, served as an additional control. Expression of TRIM71 led to a more than 3-fold repression of the CCNE2 3′UTR reporter and to a 1.6 - to 2-fold repression of HOXA5, STAT5B, ZNF362 and E2F7 reporters (Figure 2f), indicating that TRIM71 acts via the 3′UTR of these target mRNAs. Repression of reporters bearing 3′UTRs of MYB, CDH1 and HOXA6 was less pronounced. Measurement of the RNA level of the CCNE2 3′UTR reporter revealed that the decrease in the reporter FL activity induced by TRIM71 was accompanied by a partial decrease in the reporter mRNA level (Figure 2g), while the level of the KRAS 3′UTR control reporter was unaffected. Hence, the effect of TRIM71 on the function of 3′UTR reporters appears to result from a combination of translational repression and mRNA degradation, similarly as in the case of 5boxB reporters (Figure 2c).
Collectively, these data indicate that TRIM71 associates with 3′UTRs of many mRNAs and that this association represses protein synthesis, accompanied by the decrease in mRNA abundance due to accelerated mRNA decay.
Identification of TRIM71 targets in mES cells
TRIM71 is highly expressed in stem cells, including ES cells (8,12,18). To examine whether TRIM71 acts as a repressor of mRNAs in a more physiological context, we first tested the effect of tethering TRIM71 to the RL-5boxB reporter in mES cells. Tethering of TRIM71 led to repression of the reporter similar to that found in HEK293 cells (Figure 3a), indicating that the protein also represses mRNA function in mES cells. The effect of TRIM71 on RL-5boxB mRNA levels was however less pronounced in mES cells than in HEK293 cells (Figure 3b).
Figure 3.
TRIM71 acts as a repressor of mRNAs in mES cells. (a) Repression of RL-5boxB by NHA-TRIM71 in mES cells. The experiment was performed as described in Figure 2b. (b) RT-qPCR to estimate RL and FL mRNA levels from the experiment described in (a). (c) Knock-down of TRIM71 in mES cells. mES cells were transfected with a combination of three siRNAs against Trim71 (TRIM71 KD) or a non-targeting control siRNA (ctrl). 72 h post-transfection cells were lysed and western blots were probed with indicated antibodies. (d) Luciferase reporter assay with FL reporters containing 3′UTRs of candidate TRIM71 targets. Luciferase reporter assays were performed in HEK293 cells as described in Figure 2f. (e) De-repression of TRIM71 target reporters upon TRIM71 knock-down in mES cells. mES cells were co-transfected with indicated FL-3′UTR reporters and siRNAs against Trim71 (TRIM71 KD) or non-targeting control siRNA (ctrl). FL was normalized to RL and values of normalized FL from cells treated with the ctrl siRNA were set to 1. *P-value < 0.05; **P-value < 0.01.
To identify TRIM71 targets in mES cells, we knocked-down TRIM71 using a combination of three different siRNAs. The siRNAs strongly decreased TRIM71 protein level (Figure 3c). The effect of TRIM71 knock-down on global mRNA levels was relatively mild, consistent with the mild effect of TRIM71 on RL-5boxB mRNA levels observed in the tethering reporter assay (Figure 3b), potentially indicating an involvement of cell-type-specific factors in mediating the TRIM71 response. We detected an up-regulation of 77 and down-regulation of 41 genes by at least 1.5-fold (P < 0.05) (Supplementary Table S2). Of the TRIM71 target genes identified in HEK293 cells, only CCNE2 and E2F7 were slightly up-regulated upon TRIM71 knock-down in mES cells.
To identify TRIM71 targets relevant for the proposed function of TRIM71 in ES cell maintenance, we constructed FL reporters bearing 3′UTRs of several candidate target mRNAs. Candidate mRNAs were selected on the following criteria: up-regulation upon TRIM71 knock-down, up-regulation upon ES cell differentiation when TRIM71 levels decline (J. Krol and W.F. unpublished results), or previous implication in maintenance or repression of stem cell identity. Expression of TRIM71 in HEK293 cells led to strong repression of the Rbl1, Rbl2 and Ccne2 reporters and intermediate repression of the Ccnd2 reporter (Figure 3d). Importantly, the Rbl1, Rbl2, Ccnd2 and Ccne2 reporters also showed significant up-regulation upon knock-down of TRIM71 in mES cells (Figure 3e), indicating their repression by endogenous TRIM71.
The TRIM71 RING domain is dispensable for repression
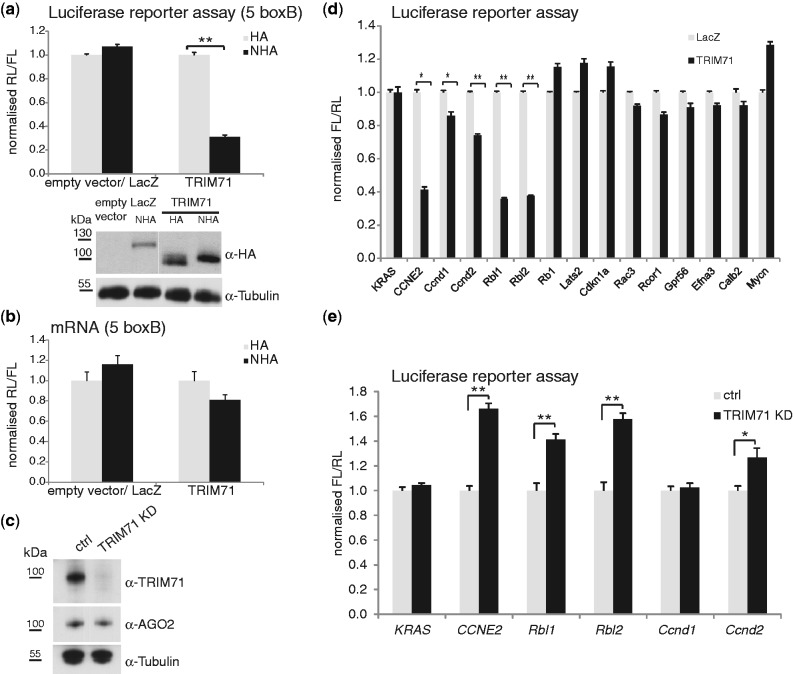
Having established that TRIM71 represses mRNA function in both HEK293 and mES cells, we asked which domains of the protein mediate this effect. To facilitate the analysis, HEK293 cells were used as they lack endogenous TRIM71. In TRIM71, the TRIM motif consists of a RING, two B-boxes and a CC region, followed by filamin (Fil) homology and NHL domains (Figure 4a). We tested mutants lacking the RING domain and B-box1 (ΔRING/B1), the entire TRIM motif (thus lacking the RING domain, the two B-boxes and the CC region; ΔTRIM), or the NHL domain (ΔNHL) in tethering and 3′UTR-reporter assays. Deletion of TRIM strongly attenuated repression whereas deletion of either RING/B1 or NHL had no effect when mutants were tethered to the mRNA (Figure 4b). Thus, RING/B1 and NHL domains do not contribute to repression. The ΔRING/B1 mutant repressed 3′UTR reporters of CCNE2, HOXA5, STAT5B or E2F7 genes as efficiently as the full-length protein but, in marked contrast, mutants lacking either TRIM or NHL failed to repress the 3′UTR reporters (Figure 4c).
Figure 4.
The NHL domain targets TRIM71 to RNA. (a) Schematic representation of human TRIM71 and its deletion mutants. The RING domain (R) is shown in red, the two B-boxes (B1 and B2) in orange, the CC domain in yellow, the Fil domain in blue and the NHL domain, composed of 6 NHL repeats, in green. The RING domain, the B-boxes and the CC domain constitute the TRIM motif. Note that ΔRING/B1 mutant has also a part of B-box2 deleted. (b) (top) Repression of RL-5boxB by NHA-TRIM71 and deletion mutants thereof in HEK293 cells. Experiments were performed as described in Figure 2b. (bottom) Expression of TRIM71 and its deletion mutants was estimated by western blotting with antibodies against HA. (c) Repression of the indicated FL-3′UTR reporters by TRIM71 and deletion mutants thereof in HEK293 cells. The experiments were performed as described in Figure 2f. (d) RIP experiment. Levels of indicated endogenous mRNAs in HEK293 cells stably expressing FLAG-HA-TRIM71 (TRIM71), FLAG-HA-ΔTRIM (ΔTRIM), FLAG-HA-ΔNHL (ΔNHL) were compared to those of the parental control cell line not expressing any FLAG-HA-tagged protein (parental) before (Input) and after immunoprecipitation with FLAG-M2 affinity gel (IP). RT-qPCR values were normalized for GAPDH mRNA. Values obtained from parental cells were set to 1. Western blots showing protein expression and IP efficiency are shown in Supplementary Figure S2b. Note that, in contrast to other mRNAs, the HOXA5 mRNA level was decreased in cells expressing FLAG-HA-ΔTRIM, for reasons not yet identified. (e) Immunoprecipitations with anti-FLAG M2 affinity gel (FLAG-IP) from HEK293 cells transiently transfected with FLAG-HA-tagged TRIM71 (TRIM71), FLAG-HA-tagged TRIM71 deletion mutants (ΔNHL, ΔRING/B1, ΔTRIM) or empty control vector (ctrl). Western blots were probed with antibodies against the indicated endogenous proteins or HA.
These results indicate that the RING/B1 domain, and thus ubiquitin ligase activity (8), is dispensable for TRIM71-mediated repression. While the TRIM domain was essential in both assays, the NHL domain was required for repression of the 3′UTR reporters but not when the protein was tethered to mRNA, suggesting that the NHL domain targets TRIM71 to RNA.
The NHL domain is needed to target TRIM71 to RNA
To test the hypothesis that NHL targets TRIM71 to RNA, we generated cell lines stably expressing FLAG-HA-ΔTRIM and -ΔNHL mutants and performed RIP (Figure 4d and Supplementary Figure S2b). In RT-qPCR analyses on input material (Figure 4d, top) both mutants failed to destabilize TRIM71 targets, confirming the results of activity assays obtained with 3′UTR reporters (Figure 4c). However, analysis of RNA samples immuno-selected by ΔTRIM and ΔNHL proteins revealed different reasons for their failure to repress targets. The ΔTRIM mutant associated with RNA whereas the ΔNHL mutant did not (Figure 4d, bottom).
Next, we examined the ability of deletion mutants to pull down identified TRIM71-associated proteins (Figure 1). Deletion of RING/B1 or the entire TRIM domain did not affect association with any protein tested (Figure 4e). However, deletion of NHL abolished association with all proteins whose interaction was sensitive to RNase A (Figure 1). Moreover, these proteins were pulled down by the isolated NHL domain (NHLonly) (Supplementary Figure S3a). This is consistent with the notion that NHL is necessary and sufficient for association of TRIM71 with RNA and that it mediates the RNA-dependent association of TRIM71 with other mRNA-binding proteins.
In conclusion, our data demonstrate that the NHL domain is necessary and sufficient to target TRIM71 to RNA.
A sub-region of TRIM mediates repression together with the Fil domain
Having shown that deletion of the entire TRIM region but not of either RING/B-box1 or NHL abolishes TRIM71-mediated repression in the tethering assay, we identified the minimal region needed for repression (Supplementary Figure S4b). Surprisingly, the tethered TRIM domain (TRIMonly) repressed protein synthesis only weakly. The TRIMonly mutant differs from the fully repressive ΔNHL mutant by the absence of the Fil domain. Therefore, we tested a further mutant encompassing part of B-box2, the CC and Fil region (B2*-CC-Fil) (Supplementary Figure S4b). This mutant induced considerable repression and we concluded that the minimal domain needed to execute repression consists of the CC and Fil domains.
Single-point mutations within the NHL domain abolish TRIM71-mediated repression
Association of Drosophila BRAT with RNA occurs via its NHL domain, mediated by the RNA-binding protein (RBP) PUM (5). The crystal structure of the BRAT NHL domain revealed a six-bladed ß-propeller with each blade consisting of one NHL repeat (27). The domain has an electropositive ‘top’ and an electronegative ‘bottom’ surface. Residues at the top are crucial for the interaction with PUM and their mutation can result in the brat mutant phenotype (27,28), underscoring their importance. Alignment of BRAT and TRIM71 showed conservation of residues important for the BRAT and PUM interaction (Supplementary Figure S5a).
Individual mutation of each of the four conserved residues completely abrogated repression of 3′UTR reporters by TRIM71, demonstrating the importance of its NHL domain and functional similarities between TRIM71 and BRAT (Figure 5a). However, when the point mutants were tested in the tethering assay, where RNA binding is dispensable, two mutants showed repression similar to that of wild-type (wt) TRIM71 but, surprisingly, repression was partially relieved for the other two (Figure 5b). The observation that deletion of the entire NHL domain does not affect repression while two single-point mutations (GD and HL) within it partially abrogate repression suggests that mutation of the NHL domain may cause a conformational change that affects intra- or intermolecular interactions of the protein. All single-point mutants were expressed at levels similar to that of the wt protein (Figure 5b, bottom). However, the two mutants showing only partial repression in the tethering assay also displayed a stronger interaction with HSP70 and HSP90 (Supplementary Figure S5b), possibly indicative of misfolding. Notably, one point mutant (YA) but not others, still pulled down most of the RNA-dependent interacting partners, suggesting that the properties of the four point mutants are not identical although they are all affected in the ability to repress target genes.
Figure 5.
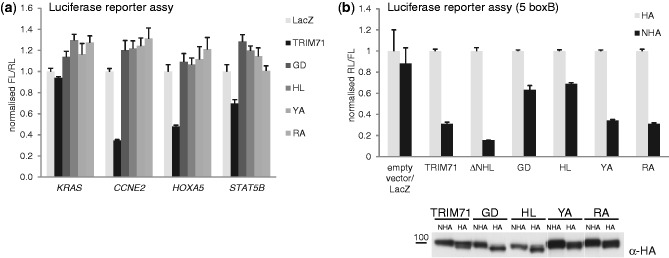
Individual mutations of conserved residues within the NHL domain abrogate TRIM71 function. (a) Repression of indicated FL-3′UTR reporter constructs by TRIM71 and indicated TRIM71 point mutants in HEK293 cells. HEK293 cells were co-transfected with plasmids expressing the indicated FL-3′UTR reporter constructs together with RL control and plasmids expressing either TRIM71, indicated TRIM71 point mutants or LacZ as a control. FL was normalized to RL and values of normalized FL produced in the presence of LacZ were set to 1. (b) (top) Repression of RL-5boxB reporter by TRIM71 and indicated TRIM71 mutants in HEK293 cells. HEK293 cells were co-transfected with plasmids expressing RL-5boxB, the indicated NHA- or HA-fusion-proteins, and a FL control plasmid. RL was normalized to FL and values of normalized RL produced in the presence of the proteins without N-peptide (HA) were set to 1. (bottom) Protein expression was confirmed by western blotting with antibodies against HA. For identity of point mutants, see Supplementary Figure S5a.
We conclude that NHL domain residues implicated in the association between BRAT and PUM and conserved between TRIM71 and BRAT are also important for TRIM71 mediation of repression.
Knock-down of PUM proteins does not affect TRIM71-mediated repression
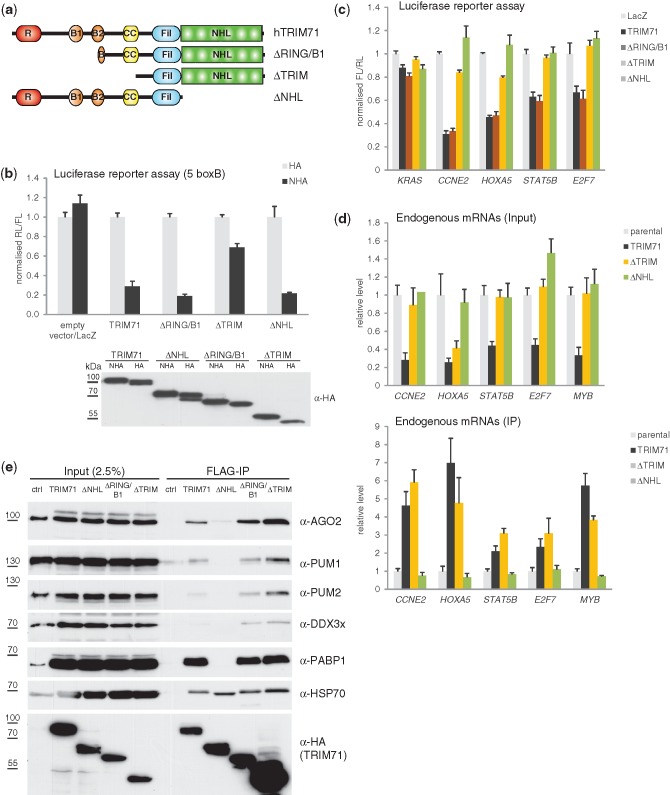
Similarity of effects of the NHL domain point mutations on activity of TRIM71 and BRAT, and the detected NHL-dependent association of TRIM71 with PUM1 and PUM2 (Figure 4e), prompted us to investigate whether PUM proteins are involved in targeting TRIM71 to RNA. Mammals express two close homologs of Drosophila PUM, PUM1 and PUM2, which both bind to an 8-nt consensus motif UGUA(A/U/C)AUA and share an overlapping set of mRNA targets (26,29). We first analysed the occurrence of the PUM consensus motif in transcripts affected by TRIM71 expression [either overexpression in HEK293 cells (Supplementary Table S2) (Figure 6a) or knock-down in mES cells (Supplementary Table S2) (data not shown)]. The motif was not found to be enriched among predicted TRIM71 targets (Figure 6a).
Figure 6.
Repression by TRIM71 is independent of mammalian PUM1 and PUM2. (a) The PUM consensus motif is not enriched in 3′UTRs of TRIM71 targets. TRIM71 versus parental mRNA expression changes for mRNAs (Supplementary Table S2) with no (black), low scoring (blue) and high scoring (red) PUM sites in the respective 3′UTRs, depicted as cumulative densities. To control for motif enrichment potentially caused by 3′UTR length differences in regulated versus non-regulated genes we included a low scoring PUM site category. These are mRNAs that contain highly degenerate PUM sites unlikely to bind PUM1/2. (b) Knock-down of PUM1 and PUM2 does not affect TRIM71-mediated repression. HEK293 cells were co-transfected with indicated FL-3′UTR reporters, plasmids expressing TRIM71 or LacZ and a combination of siRNAs against PUM1 and PUM2, or a non-targeting control siRNA as indicated. FL was normalized to RL and values of normalized FL produced from samples expressing LacZ and the non-targeting control siRNA were set to 1. *P-value < 0.05; **P-value < 0.01.
Although the bioinformatic analysis argues against TRIM71 being recruited to RNA via the PUM motif, it does not exclude involvement of PUM1/2 in targeting TRIM71 to RNA, possibly to sites other than the consensus (26). To address this, we analysed individual TRIM71 targets. CCNE2, which shows the strongest repression by TRIM71 among genes tested, is associated with PUM1 and PUM2 in HeLa (26) and with PUM2 in HEK293 cells (29), and contains two PUM motifs. Indeed, knock-down of either PUM protein alone (Supplementary Figure S6) or in combination (Figure 5b) relieved repression of the CCNE2 reporter. Thus, CCNE2 is repressed by PUM1/2 in HEK293 cells. However, knock-down of PUM proteins had no effect on TRIM71-mediated repression (Figure 6b and Supplementary Figure S6).
We analysed two further TRIM71 targets, E2F7 and STAT5B. Although E2F7 has no PUM motif, it is associated with PUM2 in HEK293 cells (29) and, consistently, showed appreciable relief of repression when PUM proteins were knocked-down (Figure 6b and Supplementary Figure S6). STAT5B is considered not to be a PUM target (26,29) and, as expected, showed no repression relief upon PUM1/2 knock-down. Importantly, for E2F7 and STAT5B, knock-down of PUM1/2 did not affect TRIM71-mediated repression of the reporters (Figure 6b and Supplementary Figure S6).
Thus, PUM proteins are dispensable for TRIM71-mediated repression and most likely do not mediate TRIM71 association with RNA.
TRIM71 and the miRNA pathway repress common targets
TRIM-NHL proteins have been shown to directly or indirectly interact with AGO proteins, either positively or negatively affecting miRNA-mediated repression in various organisms (6–9). The molecular details of TRIM-NHL enhancement of miRNA-mediated repression and the factors determining positive or negative regulation are not well-understood.
We detected a robust interaction between TRIM71 and AGO2 that was mediated by the NHL domain (Figure 4e and Supplementary Figure S3). Since this interaction was weakened but not completely abolished by RNase A treatment (with 10–40% remaining; n = 4), we examined the possibility that association of TRIM71 with RNA is mediated by AGO proteins. Mammals express four different AGOs (AGO1–4) that participate in miRNA-mediated repression and TRIM71 interacts with all tested AGOs (8). To investigate the potential involvement of AGO proteins in TRIM71-mediated repression, we knocked-down AGO1 and AGO2, the most abundant AGO proteins in HEK293 cells (30), monitoring knock-down efficiency using the established let-7 miRNA reporter HMGA2 (22). As expected, simultaneous AGO1/2 knock-down partially relieved HMGA2 repression (Figure 7a). Likewise, knock-down of AGO1/2 caused significant relief of the CCNE2, HOXA5, STAT5B, MYB and ZNF362 3′UTR reporters (Figure 7b), indicating that these mRNAs are under the control of miRNAs in HEK293 cells. Indeed CCNE2, HOXA5 and MYB were identified previously as miRNA targets by AGO/TNRC6 immunoprecipitation in HEK293 cells (31). In marked contrast, AGO1/2 knock-down did not relieve TRIM71-mediated repression of the CCNE2, HOXA5, STAT5B, E2F7, MYB and ZNF362 reporters (Figure 7b). Thus, repression by TRIM71 appears to be independent of AGO proteins.
Figure 7.
TRIM71 and miRNAs pathway repress common targets. (a) Knock-down of AGO1 and AGO2 partially relieves miRNA-mediated repression. To monitor AGO knock-down efficiency, HEK293 cells were co-transfected with FL reporter constructs containing the 3′UTR of the let-7 target HMGA2 either (wt) or with its let-7 binding sites mutated (mut), a plasmid expressing pri-let-7 and siRNAs against AGO1 and AGO2 (either individually or in combination), or a non-targeting control siRNA as indicated. Values of normalized FL produced from reporters containing the mutated 3′UTR were set to 1. (b) TRIM71-mediated repression is independent of AGO proteins. HEK293 cell were co-transfected with indicated FL-3′UTR reporter constructs, plasmids expressing either TRIM71 or LacZ, and either a combination of siRNAs against AGO1 and AGO2 (siAGO1/2) or a non-targeting control siRNA (ctrl siRNA) as indicated. FL was normalized to RL and values of normalized FL produced in the presence of LacZ and the non-targeting control siRNA were set to 1. (c) Co-repression of the E2F7 FL-3′UTR reporter by TRIM71 and miR-302. The reporter was co-transfected to HEK293 cells with plasmids expressing either TRIM71 or LacZ and either miR-302 b mimic (miR-302) or a non-targeting siRNA as a control. Values of normalized FL produced in the presence of LacZ and the control siRNA were set to 1. (d) NHL domain-swap experiment. (top) HEK293 cells were co-transfected with indicated FL-3′UTR constructs and plasmids expressing TRIM71, TRIM32, indicated TRIM71-TRIM32 chimeras, or with plasmids expressing LacZ (not shown). FL was normalized to RL and values of normalized FL produced in the presence of LacZ were set to 1. (bottom) Protein expression was estimated by western blotting with antibodies against HA. *P-value < 0.05; **P-value < 0.01.
To further investigate the potential co-regulation of TRIM71 targets by miRNAs, we turned to the E2F7 3′UTR reporter, which was identified by us as a TRIM71 target. E2F7 showed no repression relief upon AGO1/2 knock-down in HEK293 cells (Figure 7c), suggesting that it is not repressed by miRNAs in these cells. However, E2F7 was identified as a target of miR-302 (17), an ES-cell-specific miRNA. Indeed, expression of miR-302 mimic in HEK293 cells led to repression of the E2F7 reporter as compared to an unrelated siRNA used as a control mimic (Figure 7c). Additional expression of TRIM71 resulted in stronger repression than that seen in the presence of miR-302 alone. Thus, both TRIM71 and miR-302 repress the E2F7 mRNA, probably independently.
Taken together, the data suggest that some TRIM71-targeted mRNAs are also under the control of the miRNA pathway but that inhibition by the two classes of repressors is independent. This conclusion is consistent with our observation that, despite the interaction between AGO2 and TRIM71 being relatively robust, AGO2 is not found enriched in TRIM71 IPs (Figure 1), indicating that only a minor fraction of AGO2 is associated with TRIM71.
TRIM71 and TRIM32 act via a similar molecular mechanism
Our results suggested that full repression of some mRNAs requires activity of both TRIM71 and miRNAs, a situation that could be perceived as enhancement of miRNA-mediated repression by TRIM71. A similar effect (an enhancement of miRNA-mediated repression with no effect on miRNA levels) has been described for the mammalian TRIM71 paralog TRIM32 and the C. elegans TRIM-NHL protein NHL-2 (6,9). We hypothesized that the molecular mechanisms of action of TRIM32 and TRIM71 are similar. Indeed tethering of TRIM32 (but not TRIM2 or TRIM3) also led to reporter repression, albeit to a lesser extent than for TRIM71 (Supplementary Figure S7b). However, expression of TRIM32 did not or only weakly repressed the TRIM71 target genes CCNE2, HOXA5, STAT5B or E2F7 (Figure 7d), indicating that the target repertoires of TRIM71 and TRIM32 differ. Given that the NHL domain was found to be necessary and sufficient to target TRIM71 to RNA, we performed a domain-swap experiment, replacing the NHL domain of TRIM71 with that of TRIM32 and vice versa. A chimera of TRIM32 with the NHL domain of TRIM71 (TRIM32/NHL71) repressed the TRIM71 targets, whereas TRIM71/NHL32 did not (Figure 7d), arguing that both TRIM32 and TRIM71 have the potential to repress mRNAs and that target specificity is provided by the NHL domain.
In contrast to TRIM32, which enhances miRNA repression (9), TRIM71 was described previously as a negative regulator of miRNA-mediated repression, facilitating ubiquitination of AGO2 and targeting it for proteasomal degradation (8). Although we also observed ubiquitination of AGO2, facilitated by either TRIM71 or TRIM32 (Supplementary Figure S8a and b), neither TRIM71 nor TRIM32 affected AGO2 stability (Supplementary Results, Figure 3c, Supplementary Figure S8c and d). This argues against a general negative effect of TRIM71 or TRIM32 on AGO-mediated repression.
In conclusion, our data support a model of TRIM71 as a repressor of mRNA function that shares targets with miRNAs, but with the two classes of repressors functioning independently.
DISCUSSION
TRIM71 as a repressor of mRNAs
Our results identify the mammalian TRIM-NHL protein TRIM71 as a repressor of mRNA function. TRIM71 associates with mRNAs, resulting in translational repression and mRNA degradation. We have identified several TRIM71 targets and shown that TRIM71 acts via their 3′UTR. This novel role of TRIM71 as a repressor of mRNA function is reminiscent of the documented role of the Drosophila TRIM-NHL protein BRAT, which inhibits translation of several mRNAs in the early embryo (5), the female germline (32), and the nervous system (4). Strikingly, the similarities between TRIM71 and BRAT extend to the fact that mutations of individual conserved residues within the NHL domain, known to abrogate BRAT function (27), also abolish the ability of TRIM71 to repress its targets.
The role of TRIM71 as a repressor of mRNAs is consistent with the proposed role of LIN-41, the C. elegans TRIM71 ortholog, to negatively and post-transcriptionally regulate expression of the Zn-finger transcription factor lin-29 (10). Very recently, the Drosophila TRIM-NHL protein Mei-P26 was shown to repress mRNAs in ovarian germline stem cells (33). Given the structural similarities of TRIM-NHL proteins (2), it is conceivable that other family members also repress mRNAs. Our results indicate that at least mammalian TRIM32 has the potential to do so when tethered to RNA. The activity of TRIM-NHL proteins in repressing mRNAs is further supported by the observation that several TRIM-NHL proteins, including mammalian TRIM32 and TRIM71, Drosophila BRAT, Mei-P26 and Dappled, and C. elegans NHL-2, interact directly or indirectly with AGO proteins and other miRNA pathway components (6–9,33).
Physiological role of TRIM71
LIN-41 prevents premature differentiation of epidermal stem cells by negatively regulating the differentiation-promoting transcription factor lin-29 in C. elegans (10). The high expression of mammalian TRIM71 in undifferentiated cells (8), its negative regulation by let-7 (8,15,18), and its indirect up-regulation by ES-cell-specific miRNAs (17) suggest that the activity of TRIM71/LIN-41 to maintain an undifferentiated state is conserved; and indeed recently, mouse TRIM71 was shown to promote self-renewal and proliferation of embryonic neuronal progenitor cells and to prevent their premature differentiation (34). Consistent with this role, we have identified Rbl1 and Rbl2 as TRIM71 targets in mES cells. RBL1 and RBL2 are transcription factors that negatively regulate the expression of cell cycle-dependent genes and their down-regulation by the action of ES-cell-specific miRNAs was shown previously to be important for stem cell maintenance and differentiation (19,35,36).
We also detected moderate repression of Ccnd2 by TRIM71 in both HEK293 and mES cells. Although an involvement of CCND2 down-regulation in mES cell regulation has not been reported, CCND2 was shown previously to be targeted by an ES-cell-specific miRNA in human ES cells (37). Furthermore, Ccnd2 mRNA levels are strongly up-regulated upon neuronal differentiation of mES cells (J. Krol and W.F., unpublished results). Thus, keeping CCND2 levels low might contribute to stem cell maintenance or inhibition of premature differentiation. It remains to be determined whether the TRIM71 targets identified in HEK293 cells, particularly CCNE2, are physiologically relevant. Interestingly, ∼15% of transcripts down-regulated by TRIM71 in HEK293 cells are Zn-finger transcription factors (compared with 2% of up-regulated genes; Supplementary Table S2). These include Znf362, the closest mammalian homolog of Lin-29, which represents the only proposed target of LIN-41. Znf362 mRNA was also enriched in TRIM71 IPs and its 3′UTR was repressed 1.5-fold by TRIM71, suggesting that some TRIM71/LIN-41 targets are evolutionary conserved.
Notably, although TRIM71 and BRAT (and possibly other TRIM-NHL proteins) appear very similar at the molecular level, their target repertoires and thus their physiological activities differ. TRIM71 and LIN-41 are highly expressed in undifferentiated cells and repress targets like Rbl1/2 and possibly lin-29, respectively, thereby maintaining the undifferentiated state. The opposite is true for BRAT, which is absent in undifferentiated cells and up-regulated upon differentiation (32,38,39). Of the other TRIM-NHL proteins, TRIM32 and NHL-2 have also been shown to drive differentiation (6,9,40), while Mei-P26 was reported to either prevent (33) or promote differentiation (7), depending on the cellular context. Thus, knowing what determines the specificity of TRIM-NHL proteins for mRNA targets will be important for understanding their physiological roles.
The NHL domain
We have identified the NHL domain, the defining feature of TRIM-NHL proteins, as necessary and sufficient to target TRIM71 to RNA. Domain-swap experiments between TRIM71 and TRIM32 indicated that the NHL domain provides specificity to target repression. Interestingly, five out of seven alleles identified in the initial screen recognizing lin-41 as a heterochronic gene have single amino acid substitutions within the NHL domain, emphasizing its importance for protein activity (10). Also in the case of BRAT, most mutations abrogating its activity have been mapped to NHL (5,27,28).
Interestingly, mutation of the NHL residues conserved between TRIM71 and BRAT that are important for BRAT– PUM interaction and BRAT activity also abolished the ability of TRIM71 to repress its targets. Moreover, NHL of TRIM71 was found to interact in an RNA-dependent manner with the two mammalian PUM homologs PUM1 and PUM2. Despite these apparent similarities, two lines of evidence make it unlikely that mammalian PUM1/2 mediate association of TRIM71 with RNA: the PUM consensus motif is not enriched in the 3′-UTRs of identified TRIM71 targets, and simultaneous knock-down of both PUM1/2 does not abolish TRIM71-mediated repression. Therefore, we favor the possibility that the TRIM71 and PUM1/2 interaction is mediated by RNA and that TRIM71 and PUM1/2 share many mRNA targets. In support of this hypothesis, some TRIM71 targets we identified (CCNE2, HOXA5, E2F7, MYB and CCND2) have been found associated with PUM2 in HEK293 cells (29). Consistently, repression of several tested TRIM71 targets was partially relieved upon PUM1/2 knock-down in the absence of TRIM71 in HEK293 cells.
It is conceivable that TRIM71 interacts directly with RNA. The NHL domain forms a ß-propeller structure that resembles structures of WD-40 domains (27). Lau et al. found that the WD-40 domain of Gemin5 contacts RNA directly (41) and more recently the WD-40 motif has also been identified as putative new RNA-binding platform by Castello et al. (42). Hence, it is possible that the NHL domain mediates association of TRIM-NHL proteins with mRNAs by directly contacting RNA. Interestingly, all identified mutations that abrogate BRAT (27), TRIM71 (our work) and Lin-41 (10) activity lie at the electropositive ‘top’ surface of the NHL ß-propeller structure. We attempted to identify the RNA motif that is responsible for TRIM71 association with mRNA. However, neither the dissection of the 3′UTR of the TRIM71 target CCNE2 nor a bioinformatic motif enrichment analysis of TRIM71-associated mRNAs revealed a candidate sequence motif (data not shown), possibly indicating that association of TRIM71 involves some structural RNA features.
TRIM71 and miRNA-mediated repression
Individual mRNAs are regulated by the interplay of a multitude of different RBPs and miRNAs that control mRNA function. Moreover, single RBPs or miRNAs can target hundreds of mRNAs, forming complex regulatory networks that fine-tune gene expression (43). Not surprisingly, we found that many TRIM71 targets are also subject to miRNA repression: their expression increased upon knock-down of AGO1/2 in HEK293 cells. Furthermore, many of them have been shown previously to be associated with AGO proteins (29,31), indicating that TRIM71 and the miRNA pathway share common targets. Consistent with this, the TRIM71 targets we identified in mES cells are either predicted (Rbl1) or confirmed (Rbl2 and CCND2) targets of ES-cell-specific miRNAs (19,36,37). As exemplified by E2F7, full repression of these targets requires expression of both miRNA and TRIM71.
Our model that full repression of some miRNA targets requires expression of TRIM71 resembles observations made for mouse TRIM32 and C. elegans NHL-2, which were shown to enhance miRNA-mediated repression by an as yet unknown mechanism without affecting miRNA levels (6,9). While these reports suggested a role for TRIM-NHL proteins as co-factors or modulators of miRNPs (6), our results indicate that TRIM71 (and possibly other TRIM-NHL proteins) repress mRNAs independent of the miRNA pathway. Consistent with TRIM-NHL proteins being independent repressors sharing some targets with miRNAs is a recent report identifying Mei-P26 as a repressor of brat and orb mRNAs (33). While repression of orb required both AGO1 and Mei-P26, repression of brat by Mei-P26 was miRNA-independent. A common mechanism for TRIM-NHL proteins is also inferred from observations that the NHL domain, which is necessary for TRIM71 and BRAT association with RNA, is likewise required for TRIM32 enhancement of miRNA-mediated repression, but the RING/E3-ligase domain, which is dispensable for TRIM71 function (and absent in BRAT), is also dispensable for the enhancement of miRNA-mediated repression by TRIM32 (9).
In contrast to the examples of TRIM-NHL proteins augmenting the effects of miRNAs, there are also reports of Mei-P26 and TRIM71 as negative miRNA-pathway regulators (7,8). Notably, TRIM71 was shown to ubiquitinate AGO2, targeting it for proteasomal degradation and thereby antagonizing miRNA function (8). This effect is not consistent with the repressive activity of TRIM-NHL proteins identified by us and others (6,9,32,33) and accordingly a more recent report found no evidence for an effect of TRIM71 on AGO2 stability or ubiquitination (34). Although we found that both TRIM71 and TRIM32 can facilitate AGO2 ubiquitination, we also did not detect negative effects of TRIM71 or TRIM32 expression on AGO2 stability, either endogenous or when overexpressed. We mapped several ubiquitination sites in AGO2 but have been unable so far to identify their significance (unpublished results). Ubiquitination of endogenous AGO proteins by a global approach has also been reported recently (44). AGO protein ubiquitination is certainly not required for the activity of TRIM71 as a repressor of mRNAs, given that its RING domain, and thus ubiquitin ligase activity, is dispensable for TRIM71-mediated repression. This property is shared with BRAT, an incomplete TRIM protein lacking the RING domain.
When our article was ready for submission, Chang et al. (45) reported that Cdkn1a mRNA is co-repressed by TRIM71 and miR-302 in mES cells. Similar to our results, they also found that TRIM71 does not cause destabilization of AGO2.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–8, Supplementary Materials and Methods, Supplementary Results and Supplementary References [46–50].
FUNDING
I.L. was the recipient of a DFG fellowship. Work done in the laboratory of W.F. was supported by the European Community FP6 Program ‘Sirocco’. The FMI is supported by the Novartis Research Foundation. Funding for open access charge: Friedrich-Miescher-Institute.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank G. Wulczyn (Charité Berlin) for antibodies; T.C. Roloff and S. Thiry for array analysis; L. Gelman for help with microscopy experiments; H. Grosshans and P. King for critical reading of the manuscript; L. Jaskiewicz, J. Krol, M. Chekulaeva and J. Bethunè for experimental and intellectual support; members of the W.F. group for discussions. I.L. designed and performed experiments. D.G. analysed array data and performed PUM-enrichment analyses. R.S. performed and analysed MS experiments. W.F. supervised the project, and I.L and W.F. analysed data and wrote the article.
REFERENCES
- 1.Slack FJ, Ruvkun G. A novel repeat domain that is often associated with RING finger and B-box motifs. Trends Biochem. Sci. 1998;23:474–475. doi: 10.1016/s0968-0004(98)01299-7. [DOI] [PubMed] [Google Scholar]
- 2.Sardiello M, Cairo S, Fontanella B, Ballabio A, Meroni G. Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol. Biol. 2008;8:225. doi: 10.1186/1471-2148-8-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho PF, Gamberi C, Cho-Park YA, Cho-Park IB, Lasko P, Sonenberg N. Cap-dependent translational inhibition establishes two opposing morphogen gradients in Drosophila embryos. Curr. Biol. 2006;16:2035–2041. doi: 10.1016/j.cub.2006.08.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muraro NI, Weston AJ, Gerber AP, Luschnig S, Moffat KG, Baines RA. Pumilio binds para mRNA and requires Nanos and Brat to regulate sodium current in Drosophila motoneurons. J. Neurosci. 2008;28:2099–2109. doi: 10.1523/JNEUROSCI.5092-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sonoda J, Wharton RP. Drosophila Brain Tumor is a translational repressor. Genes Dev. 2001;15:762–773. doi: 10.1101/gad.870801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V. nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell. 2009;136:926–938. doi: 10.1016/j.cell.2009.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neumuller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, Cohen SM, Knoblich JA. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature. 2008;454:241–245. doi: 10.1038/nature07014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rybak A, Fuchs H, Hadian K, Smirnova L, Wulczyn EA, Michel G, Nitsch R, Krappmann D, Wulczyn FG. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat. Cell Biol. 2009;11:1411–1420. doi: 10.1038/ncb1987. [DOI] [PubMed] [Google Scholar]
- 9.Schwamborn JC, Berezikov E, Knoblich JA. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell. 2009;136:913–925. doi: 10.1016/j.cell.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 11.Kanamoto T, Terada K, Yoshikawa H, Furukawa T. Cloning and regulation of the vertebrate homologue of lin-41 that functions as a heterochronic gene in Caenorhabditis elegans. Dev. Dyn. 2006;235:1142–1149. doi: 10.1002/dvdy.20712. [DOI] [PubMed] [Google Scholar]
- 12.Lancman JJ, Caruccio NC, Harfe BD, Pasquinelli AE, Schageman JJ, Pertsemlidis A, Fallon JF. Analysis of the regulation of lin-41 during chick and mouse limb development. Dev. Dyn. 2005;234:948–960. doi: 10.1002/dvdy.20591. [DOI] [PubMed] [Google Scholar]
- 13.Lin YC, Hsieh LC, Kuo MW, Yu J, Kuo HH, Lo WL, Lin RJ, Yu AL, Li WH. Human TRIM71 and its nematode homologue are targets of let-7 microRNA and its zebrafish orthologue is essential for development. Mol. Biol. Evol. 2007;24:2525–2534. doi: 10.1093/molbev/msm195. [DOI] [PubMed] [Google Scholar]
- 14.O'Farrell F, Esfahani SS, Engstrom Y, Kylsten P. Regulation of the Drosophila lin-41 homologue dappled by let-7 reveals conservation of a regulatory mechanism within the LIN-41 subclade. Dev. Dyn. 2008;237:196–208. doi: 10.1002/dvdy.21396. [DOI] [PubMed] [Google Scholar]
- 15.Schulman BR, Esquela-Kerscher A, Slack FJ. Reciprocal expression of lin-41 and the microRNAs let-7 and mir-125 during mouse embryogenesis. Dev. Dyn. 2005;234:1046–1054. doi: 10.1002/dvdy.20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maller Schulman BR, Liang X, Stahlhut C, DelConte C, Stefani G, Slack FJ. The let-7 microRNA target gene, Mlin41/Trim71 is required for mouse embryonic survival and neural tube closure. Cell Cycle. 2008;7:3935–3942. doi: 10.4161/cc.7.24.7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowicz W. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2008;15:259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 20.Pillai RS, Artus CG, Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA. 2004;10:1518–1525. doi: 10.1261/rna.7131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 22.Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beitzinger M, Meister G. Experimental identification of microRNA targets by immunoprecipitation of Argonaute protein complexes. Methods Mol. Biol. 2011;732:153–167. doi: 10.1007/978-1-61779-083-6_12. [DOI] [PubMed] [Google Scholar]
- 26.Galgano A, Forrer M, Jaskiewicz L, Kanitz A, Zavolan M, Gerber AP. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS One. 2008;3:e3164. doi: 10.1371/journal.pone.0003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edwards TA, Wilkinson BD, Wharton RP, Aggarwal AK. Model of the brain tumor-Pumilio translation repressor complex. Genes Dev. 2003;17:2508–2513. doi: 10.1101/gad.1119403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arama E, Dickman D, Kimchie Z, Shearn A, Lev Z. Mutations in the beta-propeller domain of the Drosophila brain tumor (brat) protein induce neoplasm in the larval brain. Oncogene. 2000;19:3706–3716. doi: 10.1038/sj.onc.1203706. [DOI] [PubMed] [Google Scholar]
- 29.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petri S, Dueck A, Lehmann G, Putz N, Rudel S, Kremmer E, Meister G. Increased siRNA duplex stability correlates with reduced off-target and elevated on-target effects. RNA. 2011;17:737–749. doi: 10.1261/rna.2348111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris RE, Pargett M, Sutcliffe C, Umulis D, Ashe HL. Brat promotes stem cell differentiation via control of a bistable switch that restricts BMP signaling. Dev. Cell. 2011;20:72–83. doi: 10.1016/j.devcel.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Maines JZ, Tastan OY, McKearin DM, Buszczak M. Mei-P26 regulates the maintenance of ovarian germline stem cells by promoting BMP signaling. Development. 2012;139:1547–1556. doi: 10.1242/dev.077412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, Lai F, Niswander L. The ubiquitin ligase mLin41 temporally promotes neural progenitor cell maintenance through FGF signaling. Genes Dev. 2012;26:803–815. doi: 10.1101/gad.187641.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benetti R, Gonzalo S, Jaco I, Munoz P, Gonzalez S, Schoeftner S, Murchison E, Andl T, Chen T, Klatt P, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat. Struct. Mol. Biol. 2008;15:998. doi: 10.1038/nsmb0908-998b. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee NS, Kim JS, Cho WJ, Lee MR, Steiner R, Gompers A, Ling D, Zhang J, Strom P, Behlke M, et al. miR-302b maintains “stemness” of human embryonal carcinoma cells by post-transcriptional regulation of Cyclin D2 expression. Biochem. Biophys. Res. Commun. 2008;377:434–440. doi: 10.1016/j.bbrc.2008.09.159. [DOI] [PubMed] [Google Scholar]
- 38.Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 39.Lee CY, Wilkinson BD, Siegrist SE, Wharton RP, Doe CQ. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev. Cell. 2006;10:441–449. doi: 10.1016/j.devcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 40.Nicklas S, Otto A, Wu X, Miller P, Stelzer S, Wen Y, Kuang S, Wrogemann K, Patel K, Ding H, et al. TRIM32 regulates skeletal muscle stem cell differentiation and is necessary for normal adult muscle regeneration. PLoS One. 2012;7:e30445. doi: 10.1371/journal.pone.0030445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lau CK, Bachorik JL, Dreyfuss G. Gemin5-snRNA interaction reveals an RNA binding function for WD repeat domains. Nat. Struct. Mol. Biol. 2009;16:486–491. doi: 10.1038/nsmb.1584. [DOI] [PubMed] [Google Scholar]
- 42.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 43.van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat. Rev. Cancer. 2011;11:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 44.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang HM, Martinez NJ, Thornton JE, Hagan JP, Nguyen KD, Gregory RI. Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nat. Commun. 2012;3:923. doi: 10.1038/ncomms1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnston M, Geoffroy MC, Sobala A, Hay R, Hutvagner G. HSP90 protein stabilizes unloaded argonaute complexes and microscopic P-bodies in human cells. Mol. Biol. Cell. 2010;21:1462–1469. doi: 10.1091/mbc.E09-10-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Locke M, Tinsley CL, Benson MA, Blake DJ. TRIM32 is an E3 ubiquitin ligase for dysbindin. Hum. Mol. Genet. 2009;18:2344–2358. doi: 10.1093/hmg/ddp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smyth GK. Limma: Linear Models for Microarray Data. New York: Springer; 2005. [Google Scholar]
- 50.Bethune J, Artus-Revel CG, Filipowicz W. Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep. 2012;13:716–723. doi: 10.1038/embor.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.