Abstract
A remarkable number of guanine-rich sequences with potential to adopt non-canonical secondary structures called G-quadruplexes (or G4 DNA) are found within gene promoters. Despite growing interest, regulatory role of quadruplex DNA motifs in intrinsic cellular function remains poorly understood. Herein, we asked whether occurrence of potential G4 (PG4) DNA in promoters is associated with specific function(s) in bacteria. Using a normalized promoter-PG4-content (PG4P) index we analysed >60 000 promoters in 19 well-annotated species for (a) function class(es) and (b) gene(s) with enriched PG4P. Unexpectedly, PG4-associated functional classes were organism specific, suggesting that PG4 motifs may impart specific function to organisms. As a case study, we analysed radioresistance. Interestingly, unsupervised clustering using PG4P of 21 genes, crucial for radioresistance, grouped three radioresistant microorganisms including Deinococcus radiodurans. Based on these predictions we tested and found that in presence of nanomolar amounts of the intracellular quadruplex-binding ligand N-methyl mesoporphyrin (NMM), radioresistance of D. radiodurans was attenuated by ∼60%. In addition, important components of the RecF recombinational repair pathway recA, recF, recO, recR and recQ genes were found to harbour promoter-PG4 motifs and were also down-regulated in presence of NMM. Together these results provide first evidence that radioresistance may involve G4 DNA-mediated regulation and support the rationale that promoter-PG4s influence selective functions.
INTRODUCTION
Guanine-rich sequences are known to adopt non-canonical secondary structure forms known as guanine quadruplex or G4 DNA motifs. These are four-stranded, Hoogsten base-paired self-assembly of DNA strands in parallel/antiparallel orientation stabilized by charge coordination with monovalent cations (Figure 1) (1–4). Intramolecular quadruplex motifs result from folding of a single-nucleotide chain with the guanine tetrads linked by loops of varying sizes and can adopt multiple conformations (5). On the other hand, combinations of nucleotide chains that contribute towards tetrad formation give intermolecular motifs. Sequence with potential to form quadruplex motifs are present in various regions of the genome including telomeres (3,6) and promoters (7–12). In telomeres, quadruplex motifs have been implicated in mechanisms that reduce activity of the ribonucleoprotein telomerase (13) and also in telomere capping in S accharomyces cerevisiae (14). In promoters work from our and other groups demonstrate quadruplex motifs as potential regulatory elements that influence gene expression (15–21). Furthermore, recent findings predict role of quadruplex motifs in chromatin packaging (12,22), recombination (23), CpG methylation (24) and genomic translocations in cancer tissues (25). Following the finding that potential G4 (PG4) motifs are enriched in promoters of E scherichia coli and several other bacteria (7,11), several reports showed that promoters of many other species including human are not only replete with quadruplex-forming sequences (7–9,11,12) but that such motifs are also conserved across human, chimpanzee, mouse and rat promoters (10).
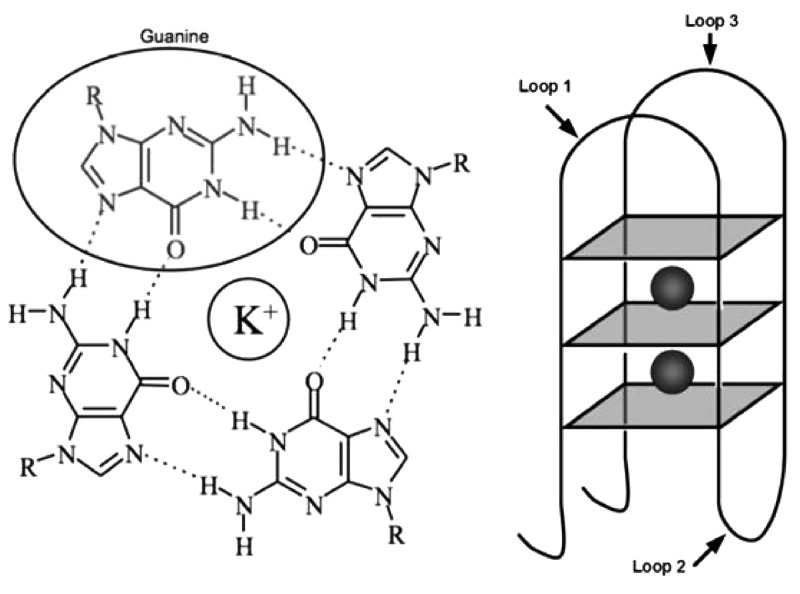
Figure 1.
Structure of the guanine quadruplex motif. Hydrogen-bonded self-assembly of guanine bases stabilized by monovalent cations form tetrads (left) that make the core of the four-stranded structure. Intramolecular quadruplex motifs are made of guanine tetrads linked by three loops of variable nucleotide length comprised of any of the four bases A, T, G or C (right).
c-MYC was the first in vitro case where a G-quadruplex upstream of the P1 promoter was shown to affect transcription (15). This finding was further substantiated by other observations showing gene expression was influenced by G-quadruplexes within the core promoter of human c-KIT (26,27) and k-RAS (16) oncogenes. In addition, promoter-quadruplex motifs were reported for many genes, including VEGF, PDGF, HIF1α, BCL-2, RB, RET (28,29), HRAS (30) and human telomerase hTERT (31,32). Furthermore, recently we found a non-canonical quadruplex motif, formed by two guanine repeats instead of three, to be functionally active in case of human thymidine kinase 1 (33).
Further studies using chromatin immunoprecipitation (ChIP) experiments demonstrated that the non-metastatic factor NM23-H2 associates with the c-MYC promoter through a G-quadruplex motif providing relatively direct evidence in support of G-quadruplex-mediated transcription (17). In addition, interaction of recombinant hnRNP A1/Up1 with the KRAS promoter G-quadruplex (34); Myc-associated zinc finger protein (MAZ)/poly(ADP-ribose) polymerase 1 (PARP-1) binding to the G-quadruplex element in the murine KRAS promoter (35); and binding of nucleolin/hnRNP proteins to the G-quadruplex-forming sequences of the VEGF promoter (36) built more support for quadruplex-mediated transcription. Similarly, quadruplex motifs in the promoters of human sarcomeric mitochondrial creatine kinase, muscle creatine kinase and integrin α-7 of mouse were also shown to associate with the dimeric form of MyoD in vitro (37,38). In line with these reports, transcriptome profiling in presence of intracellular G-quadruplex-binding ligands suggested a wide spread regulatory role of quadruplex motifs in transcription (18).
These studies gave credence to the possibility that quadruplex motifs, like many other regulatory elements, hold functional significance. However, unlike most established regulatory elements, association of quadruplex motifs with intrinsic cellular function(s) is poorly understood. But, given the complexity of eukaryote gene regulation, it is possible that regulatory role, if any, of quadruplex motifs can be better understood from the analysis of relatively less complex bacterial transcription. With this in mind, we sought to study possible links between gene function and the presence in gene promoters of motifs that potentially fold into quadruplex structures. This was done by analysing the relationship between functional classes of genes and quadruplex occurrence in their promoters, both at a genome-wide level and in individual genes (Figure 2a). Interestingly, this showed that promoter-quadruplex motifs occur in a fashion that is likely to impart specific functional attributes in species. We tested this prediction experimentally in Deinococcus radiodurans and Deinococcus geothermalis which withstand high levels of radiation. Findings suggest that quadruplex motifs present in promoters of key genes may play a critical role in response to radiation.
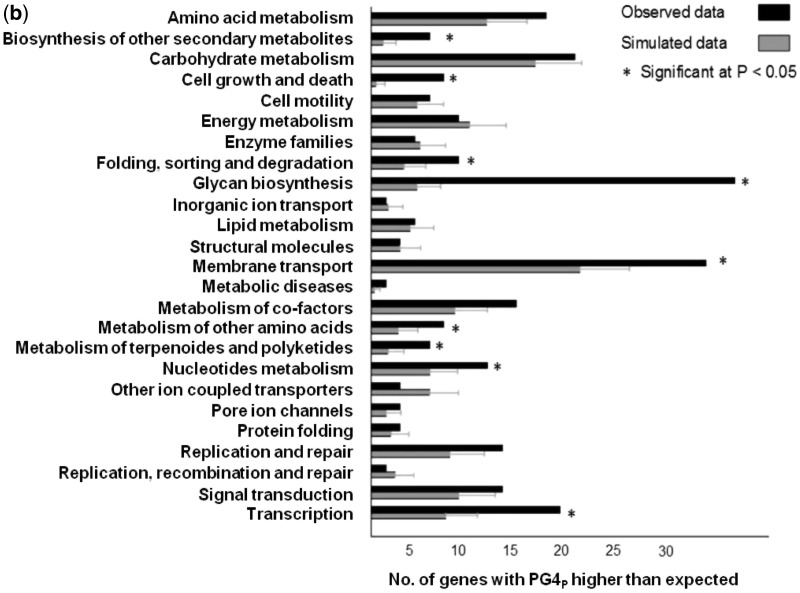
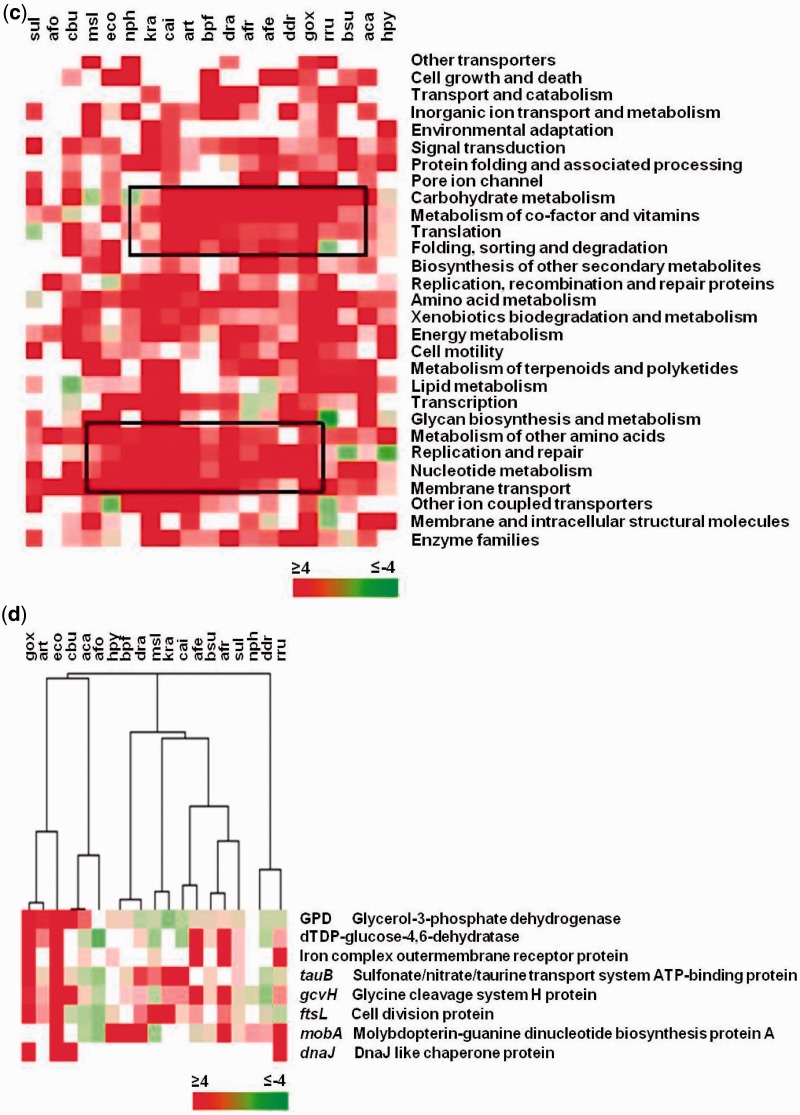
Figure 2.
Promoter PG4 motifs define distinct functional classes in organisms. (a) Scheme for quadruplex-occurrence analysis in function classes (left panel) and individual genes (right panel). The index PG4P was used for normalized content of potential quadruplex-forming sequence within individual promoters. Enrichment of promoters with significant PG4P in a functional class was computed using the mean of randomly expected number drawn from 1000 simulations as described in Materials and Methods section. (b) Functional classes with higher than expected number of promoters having significant presence of PG4 motifs in E. coli; classes at least two standard deviation above expected are denoted with asterisk. (c) Heat map representation of cluster showing enrichment (z-score) of genes with significant PG4P in functional classes across different organisms; blank squares indicate no gene with higher than expected PG4P was found. (d) Heat map of clustering showing promoters of individual genes that have significant PG4P across organisms. Escherichia coli was chosen as the reference organism and orthologues of the E. coli gene were used to construct gene-groups across 19 other organisms. Gene-groups where significant PG4P (z > 2.0) was observed in at least three organisms (in addition to E. coli) are shown; Aca, Acidobacterium capsulatum; Afe, Acidithiobacillus ferrooxidans (ATCC 53993); Afo, Acidimicrobium ferrooxidans; Afr, Acidithiobacillus ferrooxidans (ATCC 23270); Art, Arthrobacter sp.; Bpf, Bacillus pseudofirmus; Bsu, Bacillus subtilis; Cai, Catenulispora acidiphila; Cbu, Catenulispora burnetii; Ddr, Deinococcus deserti; Dra, Deinococcus radiodurans; Eco, Escherichia coli; Gox, Gluconobacter oxydans; Hpy, Helicobacter pylori; Kra, Kineococcus radiotolerans; Msl, Methylocella silvestris; Nph, Natronomonas pharaonis; Rru, Rhodospirillum rubrum; Sul, Sulfurihydrogenibium sp.
MATERIALS AND METHODS
Quadruplex detection
An algorithm written in Java was developed to identify sequence patterns with quadruplex-forming potential which was designed to find quadruplex and loop length combinations and count as well as perform sequence randomizations that were required for computing statistical significance of the results (see below). The algorithm is based on previous developed strategies (7) from our group and uses a tree structure. Briefly it assumes that:
-
-
the stem size is constant in a single quadruplex and between 2 and 5;
-
-
stems are only made of G (C on the complementary strand);
-
-
the loop sizes are between 1 and 7; and
-
-
one quadruplex is made of four stems and three loops.
For a given sequence, the algorithm checks every sliding window with size equal to
which is the maximum size of a quadruplex. After the end of a given sequence is reached, all detected quadruplexes were returned with specific loop and stem size combinations. When quadruplexes with more than four stems were found, the first four stems were considered as a single quadruplex, the extra stem was considered for the subsequent sliding window.
Promoter-wise PG4 content—PG4P
In order to compare PG4 motif content across genes and organisms we devised a method of attributing each promoter with a normalized value of PG4 motif content (PG4P) based on PG4 motif density in a particular promoter which was controlled for both GC% and sequence content of the promoter. To compute significance of the presence of PG4 motifs we shuffled each promoter 100 times (while sequence content and GC% of individual promoters were maintained) and 100 simulated PG4P values were used to find the simulated mean m and standard deviation σ. Assuming that the distribution was normal in random condition, PG4P of a particular promoter was considered statistically significant when it was at least two standard deviations above the simulated average PG4P for that promoter
Z-score was computed for PG4P and functional class enrichment (see below) using the classical formula:
where obs is the observed value of the variable (PG4P or occurrence of genes for functional class analysis); μ is the expected means and σ is the standard deviation of the variable were obtained from the simulations of PG4P or functional classes enrichment.
Functional classes enrichment
For functional class annotations, 19 well-annotated bacterial genomes were obtained from KEGG (39). In order to avoid very general classifications, the second layer in KEGG annotation hierarchy was used for all analysis. For each gene the KEGG orthologue annotation was extracted and manually curated to avoid redundancy. In case of multiple annotations all possible functions were considered. To compute whether a particular function class was enriched for genes having significant PG4P first the number of such genes in a particular class was found, which comprised the actual or observed set. Identical number of genes was randomly pulled from the same genome and 1000 such sets were prepared (randomly expected sets)—for each of the 1000 sets, number of genes with significant PG4P were calculated; average number of genes across 1000 sets was used as the randomly expected number. A function class was considered to be enriched in genes with significant PG4P when the actual or observed number was at least two standard deviations above the randomly expected average (that is z-score > 2).
Orthologous groups analysis
All E. coli genes with PG4P higher than expected by at least two standard deviations from simulation (see above) were selected. Orthologues of these genes across the remaining 18 genomes of our set were identified using KEGG orthologous cluster information. Fifty groups of orthologues were obtained in this way. For all these genes, z-score of PG4P by comparison with simulation was computed. When an organism had more than one orthologue corresponding to the E. coli gene, only the gene with highest z-score was considered. In the same way orthologues of genes involved in radioresistance in D. radiodurans [taken from (40–42)] were selected using KEGG orthologous cluster information.
For cluster analysis we used ‘Cluster’ developed by Eisen et al . (43).
Growth conditions/treatment with quadruplex-binding ligands and gamma irradiation
To directly probe for the role of PG4P in D. radiodurans IAM 12271 (MTCC 4465) and D. geothermalis (DSM 11300), porphyrin derivatives, N-methyl mesoporphyrin (NMM) or its un-methylated analogue mesoporphyrin IX dihydrochloride (MIX), and 5,10,15,20-tetrakis-(N-methyl-4-pyridyl)porphyrin (TMPyP4) or its positional isomer 5,10,15,20-tetra-(N-methyl-2-pyridyl)porphyrin (TMPyP2) were used. D. radiodurans and D. geothermalis were grown at 32°C in TGY (0.5% tryptone, 0.3% yeast extract, 0.1% glucose) broth containing respective concentrations of NMM or MIX (25 nM and 50 nM) and TMPyP4 or TMPyP2 (1.5 µM and 3 µM). Only cultures in exponential growth (OD600 nm = 0.2–0.5) were evaluated for their ability to survive ionizing gamma radiation. Exponential phase bacterial cultures in TGY medium with or without NMM (or MIX) and TMPyP4 (or TMPyP2) were exposed to gamma irradiation (see Supplementary Figure S1 for details of method used).
Radiation resistance was evaluated as described earlier (44). Briefly, mid log-phase culture (OD600 nm = 0.3) was divided into 40 ml aliquots, placed in 50 ml falcon tubes and were exposed to 5 kGy or 10 kGy of 60Co γ-rays, at a dose rate of 2.57 kGy/h using 60Co gamma chamber (Gamma Cell 5000, BRIT, Mumbai, India) installed at Nuclear Research Laboratory, IARI, Delhi. All the irradiation experiments were done at 20°C. Another aliquot, kept outside the radiation source at 20°C, served as control. All the irradiated and unirradiated control samples were transferred into fresh TGY broth and incubated on a rotary shaker at 200 rev.min− 1 at 32°C. For gene expression analysis RNA was isolated at 3 h following irradiation based on earlier observations that found up-regulation/down-regulation of most genes after gamma irradiation was at 3 h post-irradiation recovery (45). Bacterial growth was observed by measuring turbidity at 600 nm of liquid cultures (TGY broth) and the viability of irradiated cells were evaluated after 24–90 h of post-irradiation recovery at 32°C as described earlier (46). All the assays were performed in triplicates.
E scherichia coli was grown at 37°C in Luria-Bertani (LB) broth containing 2 µM or 4 µM of NMM (or MIX) and TMPyP4 (or TMPyP2), which did not alter the growth of E. coli, and exposed to 1 kGy or 2 kGy of 60Co γ-rays. The viability of E. coli in response to gamma irradiation was evaluated using the procedure described above, after 6–20 h of post-irradiation recovery at 37°C because of its lesser doubling time, as compared to D. radiodurans.
Gene expression analysis following irradiation
All samples were harvested in exponential phase by centrifugation at 10 000 rev.min− 1 for 10 min for RNA extraction. Total RNA was extracted from irradiated (at 3 h post-irradiation recovery) and unirradiated cultures using RNeasy RNA isolation kit (Qiagen) following manufacturer’s protocol. Total RNA derived from each sample condition was treated with DNase I (Fermentas) and RNA quality and quantity were evaluated by determining UV absorbance at 260 nm and 280 nm. Two micrograms of each DNase I treated and purified RNA sample were reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) as described in manufacturer’s protocol. PCR primers were designed (Supplementary Table S1) to amplify each open reading frame based on fully sequenced D. radiodurans R1 and E. coli K-12 MG1655 genomes. Expression of recA, recF, recO, recR and recQ along with 16S rRNA transcript (endogenous control), expression of which was unaffected by ionizing radiation (47), was determined before and after irradiation. To analyse the relative intensity of the PCR bands, gel image of the PCR products was scanned and then analysed using AlphaEaseFC 4.0 software (Alpha Innotech, USA). All experiments were performed in triplicates.
RESULTS
To investigate the connection between promoter-PG4 motifs and function of genes, each promoter was assigned a normalized value of promoter-PG4 motif content, PG4P, which was based on motif density (see Materials and Methods section). Significance of PG4P was computed relative to 100 simulated PG4P values obtained for respective promoters. For analysing function-PG4P relationships we undertook two complementary approaches: (i) function class-specific, where PG4P enrichment within orthologous function groups built from the KEGG database was analysed (Figure 2a, left panel) and (ii) gene-specific, where genes with significant PG4P in a reference organism were used to query across other organisms (Figure 2a, right panel). It was necessary that we studied only well-annotated organisms so that associations with function could be analysed with relative confidence, therefore 19 well-annotated organisms were considered.
Genes with PG4 motifs in promoters influence specific functions including carbohydrate metabolism
For each annotated function class in KEGG, we first determined the number of genes with significant PG4P in a given organism. Next a rigorous method was used to ascertain whether the function class was enriched for PG4P-genes by mere chance. Each class was randomly populated 1000 times to estimate the proportion of PG4P-genes expected by chance; enrichment was considered significant when actual occurrence of PG4P in any gene was at least two standard deviations above the random expectation (z-score > 2; see Materials and Methods section). All 19 organisms were analysed in this way to check for enriched function classes; a representative example for E. coli is shown in Figure 2b.
Next, we checked for correlation between enriched function classes, determined using PG4P, and organisms: 29 function classes, found to be enriched in at least one organism, were clustered based on z-scores for enrichment of genes with significant PG4P (Figure 2c, see Materials and Methods section for z-score analysis). Furthermore, to avoid any bias from sparsely populated function classes, we excluded ones that had <5 genes in any of the 19 species. Two major clusters were evident. The first cluster comprising ‘carbohydrate metabolism’, ‘metabolism of co-factor and vitamins’, ‘translation’ and ‘folding, sorting and degradation’ was enriched in 11 species. The second group of classes—‘metabolism of other amino acids’, ‘replication and repair’, ‘metabolism of nucleotides’ and ‘membrane transport’ were enriched in 13 species. Furthermore, we noted four functional classes to be largely predominant across most of the 19 organisms —‘carbohydrate metabolism’, ‘amino acid metabolism’, ‘membrane transport’ and ‘energy metabolism’. As ‘carbohydrate metabolism’ involves glucogenesis, this function is closely linked to ‘energy metabolism’, therefore, it was interesting to find that both the classes were represented in majority of the organisms analysed. Taken together, it was apparent that metabolism of several essential entities like, carbohydrate, vitamins and amino acids appear to be important in the context of PG4 motif-controlled regulation.
Genes with promoter-PG4 motifs are organism specific
As mentioned above, in the second approach for function analysis we focused on individual genes that had significant PG4P (Figure 2a, right panel). E scherichia coli was used as the reference organism and genes that had significant PG4P (z > 2.0) in E. coli were selected and corresponding orthologues in the 18 other organisms identified using KEGG classifications; 150 gene-groups were obtained in this way. In cases where an organism had more than one gene corresponding to the E. coli gene, the one with the highest z-score for PG4P was considered. In order to see if there were any gene-groups that had significant PG4 content across organisms we clustered all the 150 groups across 19 organisms (Supplementary Figure S3a). Interestingly, most gene-groups were found to have only few organisms with z-score above two suggesting that it was unlikely that there was any particular gene(s) that had significant PG4P that could be considered across several species. To ascertain that this was not due to the choice of E. coli as the reference organism, we performed similar analyses using either C atenulispora acidiphila (cai), K ineococcus radiotolerans (kra) or G luconobacter oxydans (gox) as reference organisms; again, in each case we found very few gene-groups that were significant across organisms (Supplementary Figure S3b–d).
Next, we focused on gene-groups made with E. coli as reference that had at least three organisms with z-score of 2 or more, in addition to E. coli. Eight out of 150 orthologue groups that remained after this selection were clustered (Figure 2d), which revealed that for all genes except 1, G . oxydans (gox) and E. coli (eco) had high PG4P. Similarly, Sulfurihydrogenibium sp. and Bacillus subtilis had significantly high PG4P for at least three genes (glucose hydratase, an iron outer membrane receptor and a protein required for glycine cleavage). Overall, we noted that many genes with high PG4P did not share this feature with other organisms suggesting that PG4P enrichment of genes was likely to be specific to a particular organism.
Genes imparting resistance to radiation have enriched promoter-PG4 motifs in radioresistant bacteria
The observations described above suggest possible role of promoter-PG4 motifs in imparting specific functions to organism(s). Therefore, we reasoned, if one selects genes specific for a function and uses PG4P to discriminate, it should be possible to segregate organisms. On the other hand, if PG4P is not important one should not see any difference in the segregation pattern.
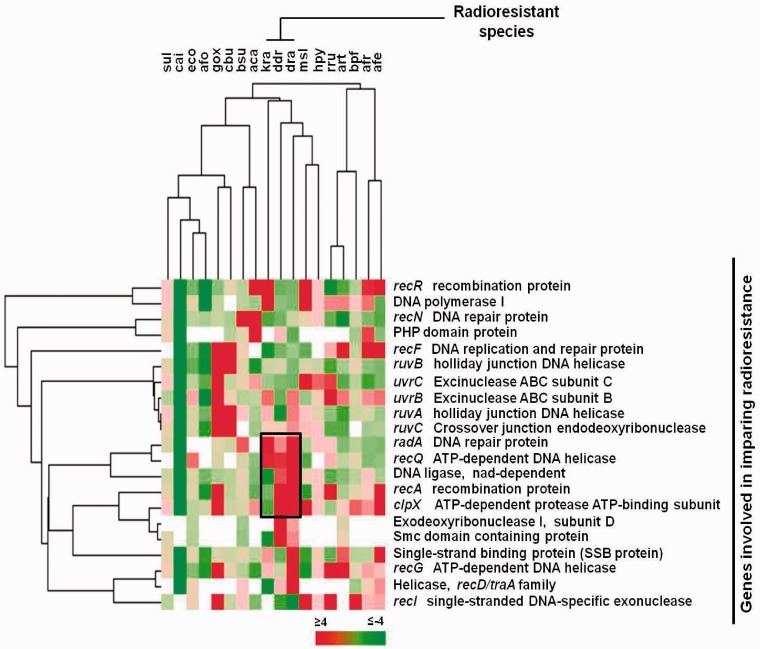
As a test case we selected radiation resistance because it has been extensively studied and genes for replication, repair and recombination delineated in radioresistant as well as other species (40–42). Furthermore, we reasoned that response to radiation as a functional readout would be relatively distinct. Twenty-one genes directly involved in radiation resistance were considered and orthologues of the selected genes in the remaining 18 bacteria from our set were determined using KEGG, giving 21 gene-groups. Next, we used PG4P-z score for clustering, as done earlier, using both organisms and gene-groups (Figure 3). In doing so, we noted with interest that three radioresistant bacteria (dra, D. radiodurans; ddr, Deinococcus desertii and kra, K . radiotolerans) clustered together. Interestingly, using this analysis we found both recA and recQ, demonstrated to be important in imparting radioresistance, as part of a cluster which also had other important DNA repair genes like radA and the ATP-dependent protease clpX (48). On the other hand, and perhaps more importantly, it was evident that most organisms that were not radioresistant did not have enriched PG4P in these genes. Taken together, it was noteworthy that unbiased clustering with respect to genes involved in radiation resistance could segregate radioresistance bacteria vis-à-vis function.
Figure 3.
PG4P-based analysis independently clusters radiation resistance species. Heat map representation of cluster diagram showing promoters of individual genes involved in imparting radiation resistance. Cluster was drawn for 18 organisms based on PG4P of genes, with E. coli as the reference organism.
Radiation resistance is compromised in presence of quadruplex-binding ligands
Based on above findings, we reasoned if quadruplex motifs are important for radiation resistance a ligand that could interact with quadruplex motifs inside cells would adversely affect radiation sensitivity. This was tested using the ligand NMM which specifically binds G-quadruplex motifs intracellularly and has no detectable binding to other nucleic acid structures, including ssDNA, dsDNA, triplex DNA, Z-DNA, duplex RNA and DNA–RNA hybrids (49,50). MIX, an unmethylated analogue of NMM, which does not bind to G-quadruplex motifs was used as a negative control (14,51). In addition, we used a second intracellular G-quadruplex-binding ligand TMPyP4 and its positional isomer TMPyP2, which does not bind G-quadruplex motifs (18,52).
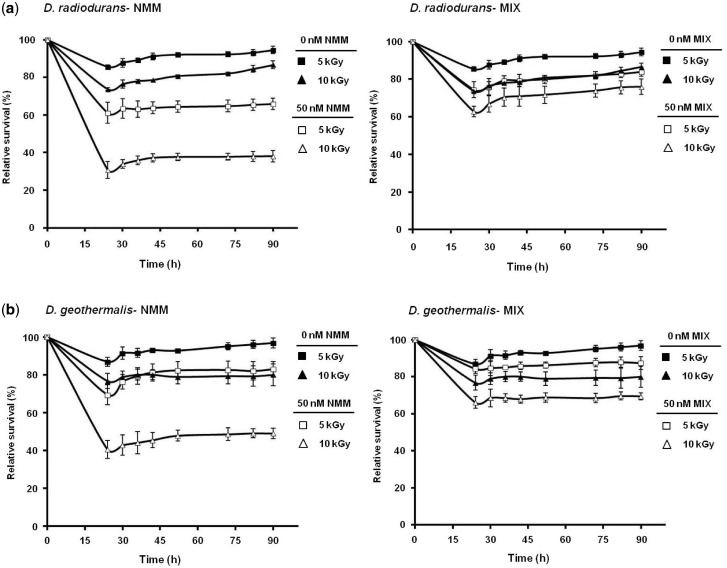
We selected D. radiodurans and D. geothermalis, the best studied species among the members of Deinococcus for further studies (40–42,53). Experiments were performed at two doses of gamma irradiation, 5 or 10 kGy in presence or absence of NMM (or MIX) and TMPyP4 (or TMPyP2), as described under Materials and Methods section. We found that relative survival of D. radiodurans and D. geothermalis decreased in a dose-dependent fashion in presence of NMM: while ∼65–80% of the bacteria survived at 5 kGy, at 10 kGy survival dropped to around 40–45% in presence of 50 nM NMM (Figure 4a and b, left panel). Relative survival of D. radiodurans and D. geothermalis also decreased in presence of TMPyP4; ∼45–55% of the bacteria survived at 5 kGy whereas at 10 kGy survival dropped to about 20–35% in presence of 3 µM TMPyP4 (Supplementary Figure S4a and b, left panels). On the other hand, relative survival (%) of D. radiodurans and D. geothermalis was minimally affected in presence of MIX (Figure 4a and b, right panel) or TMPyP2 (Supplementary Figure S4a and b, right panel) in response to 5–10 kGy irradiation. As a control organism for radioresistance we used E. coli, which is far more sensitive to ionizing radiation than D. radiodurans and D. geothermalis (54); survival of E. coli was relatively unaffected in presence of ligands NMM/MIX and TMPyP4/TMPyP2 in response to irradiation (Supplementary Figures S5 and S6).
Figure 4.
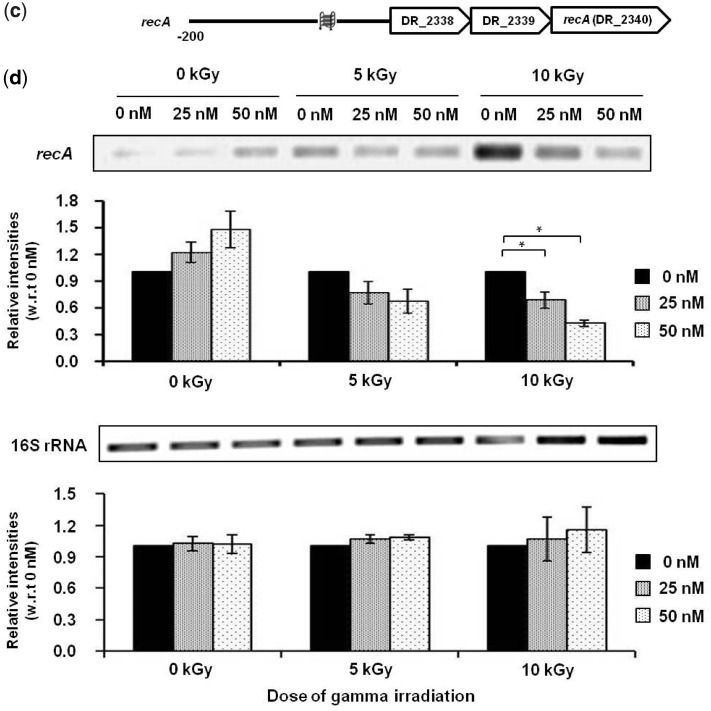
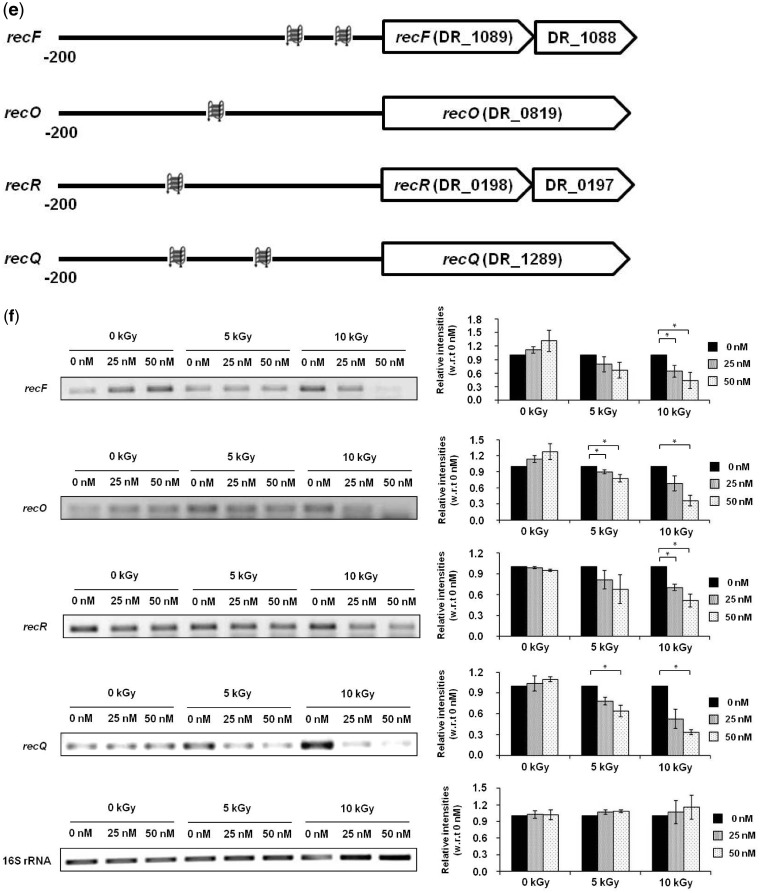
Quadruplex-binding ligands and radiation resistance. NMM attenuates growth of (a) D. radiodurans and (b) D. geothermalis, following exposure to gamma irradiation; D. radiodurans and D. geothermalis levels at 0 kGy were used to estimate relative survival following 24–90 h of post-irradiation recovery at 32°C in presence of 0 nM or 50 nM NMM (left panel) and MIX (right panel), respectively. Values are mean ± standard error of three independent experiments. (c) Scheme showing PG4 motif positions within putative regulatory region of the recA operon. (d) Expression levels of recA and 16S rRNA genes in presence/absence of NMM in response to gamma irradiation (upper panel); quantification of relative intensities of the recA and 16S rRNA RT-PCR bands with respect to 0 nM NMM at respective radiation doses (lower panel). (e) Scheme showing PG4 motif in putative promoters of respective opreons with recF, recO, recR and recQ genes. (f) Expression levels of recF, recO, recR, recQ and 16S rRNA genes in presence/absence of NMM in response to gamma irradiation (left panel); relative quantification of the recF, recO, recR, recQ and 16S rRNA RT-PCR results with respect to 0 nM NMM at respective radiation doses (right panel). All PCR assays were performed in triplicates and values are presented as mean ± standard error; * denotes P < 0.05.
Promoters of key genes that confer radiation resistance harbour quadruplex motifs and are repressed in presence of quadruplex-binding ligand
Next, we sought to find out if compromised radiation resistance of D. radiodurans and D. geothermalis in presence of NMM was due to altered expression of key genes. In earlier studies we found that D. radiodurans in absence of recA, an important component of the DNA double-strand repair pathway, exhibits extreme sensitivity to ionizing radiation (55–57). Furthermore, as expected, we noted that recA expression was up-regulated on irradiation of D. radiodurans at 5 kGy or 10 kGy in a manner that was dependent on the dosage of irradiation, supporting the role of recA in radiation resistance (Supplementary Figure S7). Based on this and our cluster analysis, which indicated significant PG4P for recA (Figure 3), we first focused on recA. A closer analysis revealed presence of one/two distinct PG4 motifs within 200 bases of the first gene start site in the recA operon in both D. radiodurans and D. geothermalis, respectively (Figure 4c and Supplementary Figure S8a). To directly test whether presence of NMM affects expression of recA, D. radiodurans was treated with either 25 or 50 nM of NMM. In presence of NMM, down-regulation of recA was clearly observed whereas expression of the 16S rRNA (endogenous control) gene remained unchanged (Figure 4d). Furthermore, we noted that inhibition of recA expression was dependent on the concentration of NMM; higher NMM levels resulted in relatively increased inhibition at both 5 kGy and 10 kGy irradiation. To further test whether the effect of NMM was due to G-quadruplex DNA binding, we treated E. coli-K12 MG1655, where recA is devoid of PG4P motifs (Supplementary Figure S8b), with NMM. In contrast to D. radiodurans, we did not find any decrease in expression of recA in presence of 2 or 4 µM NMM following irradiation (Supplementary Figure S9).
In case of D. radiodurans apart from recA, recF, recO and recR genes are also essential for radioresistance as demonstrated by greatly impaired growth in mutant strains devoid of these genes (42,58,59). Additionally, recQ mutants were also found to be radiation sensitive (60) though this has been contradicted by a recent study (58). Taking cue from our computational predictions, we independently analysed the promoters of recF, recO, recR and recQ operons and found multiple PG4 motifs within 200 bp upstream of the first gene of operons (Figure 4e and Supplementary Figure S8a). Based on this we tested expression of all four genes in presence/absence of NMM following irradiation, though other than recQ, PG4P of the recF, recO and recR promoters were not above the statistical threshold considered in our computational analysis. Interestingly, NMM significantly repressed expression of all the four genes, recF, recO, recR and recQ, and not the endogenous control 16S rRNA gene, such that higher NMM levels resulted in relatively increased inhibition at both 5 kGy and 10 kGy irradiation (Figure 4f).
DISCUSSION
Herein independent lines of analyses revealed characteristics of promoter-G-quadruplex motifs that suggest role in specific functions. Not only did we find putative involvement in regulation of gene-classes related to selected functions, we also noted that the type of function was not analogous across organisms. On asking whether PG4 motif content of individual promoters (the PG4P index) is significantly associated with function, we first found that functional groups could be segregated based on PG4P. Secondly, we noted with interest that this was specific for related group of species. In a complementary approach, using individual genes instead of functional classes, we again found that genes clustered in a fashion that was specific to organisms.
In order to experimentally test these predictions, we performed a case study using D. radiodurans and D. geothermalis, which have remarkable DNA repair systems that can withstand lethal doses of ionizing radiation (40,42,53). There are mainly two RecA-dependent recombinational DNA repair pathway in bacterial populations; the RecBCD and RecFOR pathways, which normally operates independently. The RecFOR pathway comprising recA, recF, recO, recR, recQ, recJ, recN, ruvA, ruvB and ruvC, is used mainly during recombinational repair in D. radiodurans, as it lacks the RecBCD system like many other bacteria (41,46,59,61). Furthermore, D. radiodurans also does not encode homologs of the SbcB nuclease, which is an inhibitor of the RecF pathway (62). In support of our predictions we found that radiation resistance of D. radiodurans and D. geothermalis was attenuated by >50% in presence of 50 nM NMM, a ligand that is known to specifically bind G-quadruplex motifs inside cells. We also found that recA, recF, recO, recR and recQ genes have promoter-PG4 motifs and are repressed in a dose-dependent fashion in presence of NMM in D. radiodurans. Together, these findings support involvement of G-quadruplex-mediated regulatory mechanisms in radioresistance.
We observed relative increase in recA, recF and recO on NMM treatment in absence of irradiation. Though up-regulation of genes with promoter-quadruplex motifs has been reported in presence of NMM (22), this contrasts our observations related to NMM-mediated suppression of genes that confer radioresistance to D. radiodurans following irradiation. On the other hand, in case of E. coli, which does not have any G-quadruplex motif in the recA proximal promoter, also we found enhanced expression of recA on NMM treatment in absence of irradiation (Supplementary Figure S9), though no change was detected following irradiation. Considered together with the specific intracellular G-quadruplex binding reported for NMM earlier (22,50), it is possible that the NMM-induced specific effects are observed on irradiation while the pre-irradiation changes are more general effects of the ligand treatment. It is also possible that the role of quadruplex motifs become more effective following irradiation, however further experiments will be required to test this speculation.
In an earlier study, we reported the genomic distribution of PG4 motifs in bacterial genomes. This provided first evidence for possible regulatory role of quadruplex motifs in any organism based on remarkable prevalence of PG4 motifs in bacterial promoters (7), which we subsequently found was a hallmark across more than 140 bacterial species (11). Furthermore, our genome-wide analyses also suggested that promoters with PG4 motifs in E. coli could be induced by supercoiling that leads to destabilization of the duplex DNA. In the current work, we have extended this initial finding to ask whether typical gene(s) pertaining to a particular functional class(es) have higher propensity for promoter PG4 content. This was done by focusing on relative estimation of PG4 motif content of promoters, both within and across several organisms, using the normalized index PG4P, which also allowed us to test the significance of PG4 motif occurrence in promoters across functional classes.
Despite arguments against the stability/formation of G-quadruplex motifs of stem size 2, we opted to consider them. This was primarily because in previous studies we have experimentally tested and found that such motifs randomly selected from E. coli readily adopt G-quadruplex motifs in solution (7). Further support stems from work demonstrating that a two-tetrad non-canonical quadruplex motif regulates expression of human thymidine kinase 1 (33) and recent work from Mergny group showing that G-quadruplex motifs with stem size of 2 remain stable in vitro (63). Moreover, one of the most studied quadruplex sequence, the thrombin aptamer constitutes of a stem size of 2 bases (64).
It is possible that radiation resistance involves PG4 motifs as a fortuitous connection due to high PG4P content. By a similar analogy radioresistant species would be expected to cluster with E. coli when genes with high PG4P were considered. This was not the case. Moreover, promoter-level analysis showed that PG4 motifs may have functional roles that were specific for at least a group of similar organisms. This also argues against a random association of PG4 motifs with radiation resistance genes. Furthermore, though unexpected, in addition to radioresistant species, we observed segregation of acidophiles when clustered for genes involved in radiation resistance. This further suggests that it is unlikely radioresistant species were found merely due to PG4-content. Finally when experimentally tested we found that indeed occurrence of G-quadruplex motif and its interaction with a specific G4-binding ligand impacts how D. radiodurans withstands radiation damage.
In summary, the notion of selective advantage prompted this study of promoter-wise PG4 motif content with the understanding that a finer analysis would provide distinct indication about possible PG4 motif-associated functions. This was first substantiated by cluster analysis of functional classes, where the distribution of promoters having significant PG4P appeared to be non-random and associated to specific function(s). Secondly, a closer analysis of the cluster (Figure 2d) suggested an interesting pattern: groups of genes with high PG4P appeared to be specific to related organisms. Keeping this in mind, we hypothesized that PG4 motif-function relationships may be interesting when considered with respect to functions that are specific to related organisms. Unbiased clustering of genes involved in radiation resistance segregated radioresistance organisms lending credence to this understanding. Taken together, based on these results, it is tempting to speculate that emergence of PG4 motifs as regulatory units not only influences function but also imparts directed advantage to counter environmental pressures. Further work addressing this possibility in different organisms and in relation to functional advantages acquired by the organism will be required to better understand this aspect of G-quadruplex function.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–9.
FUNDING
Council of Scientific & Industrial Research (postdoctoral fellowship for foreign scholars (to N.B.); financial assistant [OLP 6101 to H.K.G.]; Department of Science and Technology (Swarnajayanti Fellowship for research grant (to S.C.); University Grants Commission, India (Junior Research Fellowship to R.P.). Funding for open access charge: Swarnajayanti Fellowship by Department of Science and Technology.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Authors acknowledge Munia Ganguli for careful reading and editing of the manuscript and thank members of the Chowdhury group for helpful discussions. The authors are also grateful to University Grants Commission, India.
REFERENCES
- 1.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc. Natl Acad. Sci. USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 3.Sundquist WI, Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 4.Balagurumoorthy P, Brahmachari SK. Structure and stability of human telomeric sequence. J. Biol. Chem. 1994;269:21858–21869. [PubMed] [Google Scholar]
- 5.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williamson JR. G-quartet structures in telomeric DNA. Annu. Rev. Biophys. Biomol. Struct. 1994;23:703–730. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- 7.Rawal P, Kummarasetti VB, Ravindran J, Kumar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du Z, Zhao Y, Li N. Genome-wide analysis reveals regulatory role of G4 DNA in gene transcription. Genome Res. 2008;18:233–241. doi: 10.1101/gr.6905408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verma A, Halder K, Halder R, Yadav VK, Rawal P, Thakur RK, Mohd F, Sharma A, Chowdhury S. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 2008;51:5641–5649. doi: 10.1021/jm800448a. [DOI] [PubMed] [Google Scholar]
- 11.Yadav VK, Abraham JK, Mani P, Kulshrestha R, Chowdhury S. QuadBase: genome-wide database of G4 DNA–occurrence and conservation in human, chimpanzee, mouse and rat promoters and 146 microbes. Nucleic Acids Res. 2008;36:D381–D385. doi: 10.1093/nar/gkm781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halder K, Halder R, Chowdhury S. Genome-wide analysis predicts DNA structural motifs as nucleosome exclusion signals. Mol. Biosyst. 2009;5:1703–1712. doi: 10.1039/b905132e. [DOI] [PubMed] [Google Scholar]
- 13.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 14.Smith JS, Chen Q, Yatsunyk LA, Nicoludis JM, Garcia MS, Kranaster R, Balasubramanian S, Monchaud D, Teulade-Fichou MP, Abramowitz L, et al. Rudimentary G-quadruplex-based telomere capping in Saccharomyces cerevisiae . Nat. Struct. Mol. Biol. 2011;18:478–485. doi: 10.1038/nsmb.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thakur RK, Kumar P, Halder K, Verma A, Kar A, Parent JL, Basundra R, Kumar A, Chowdhury S. Metastases suppressor NM23-H2 interaction with G-quadruplex DNA within c-MYC promoter nuclease hypersensitive element induces c-MYC expression. Nucleic Acids Res. 2009;37:172–183. doi: 10.1093/nar/gkn919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verma A, Yadav VK, Basundra R, Kumar A, Chowdhury S. Evidence of genome-wide G4 DNA-mediated gene expression in human cancer cells. Nucleic Acids Res. 2009;37:4194–4204. doi: 10.1093/nar/gkn1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar P, Yadav VK, Baral A, Kumar P, Saha D, Chowdhury S. Zinc-finger transcription factors are associated with guanine quadruplex motifs in human, chimpanzee, mouse and rat promoters genome-wide. Nucleic Acids Res. 2011;39:8005–8016. doi: 10.1093/nar/gkr536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baral A, Kumar P, Halder R, Mani P, Yadav VK, Singh A, Das SK, Chowdhury S. Quadruplex-single nucleotide polymorphisms (Quad-SNP) influence gene expression difference among individuals. Nucleic Acids Res. 2012;40:3800–3811. doi: 10.1093/nar/gkr1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hershman SG, Chen Q, Lee JY, Kozak ML, Yue P, Wang LS, Johnson FB. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae . Nucleic Acids Res. 2008;36:144–156. doi: 10.1093/nar/gkm986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mani P, Yadav VK, Das SK, Chowdhury S. Genome-wide analyses of recombination prone regions predict role of DNA structural motif in recombination. PLoS One. 2009;4:e4399. doi: 10.1371/journal.pone.0004399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halder R, Halder K, Sharma P, Garg G, Sengupta S, Chowdhury S. Guanine quadruplex DNA structure restricts methylation of CpG dinucleotides genome-wide. Mol. Biosyst. 2010;6:2439–2447. doi: 10.1039/c0mb00009d. [DOI] [PubMed] [Google Scholar]
- 25.De S, Michor F. DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat. Struct. Mol. Biol. 2011;18:950–955. doi: 10.1038/nsmb.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernando H, Reszka AP, Huppert J, Ladame S, Rankin S, Venkitaraman AR, Neidle S, Balasubramanian S. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006;45:7854–7860. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat . Rev. Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Membrino A, Cogoi S, Pedersen EB, Xodo LE. G4-DNA formation in the HRAS promoter and rational design of decoy oligonucleotides for cancer therapy. PLoS One. 2011;6:e24421. doi: 10.1371/journal.pone.0024421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palumbo SL, Ebbinghaus SW, Hurley LH. Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands. J. Am. Chem. Soc. 2009;131:10878–10891. doi: 10.1021/ja902281d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim KW, Lacroix L, Yue DJ, Lim JK, Lim JM, Phan AT. Coexistence of two distinct G-quadruplex conformations in the hTERT promoter. J. Am. Chem. Soc. 2010;132:12331–12342. doi: 10.1021/ja101252n. [DOI] [PubMed] [Google Scholar]
- 33.Basundra R, Kumar A, Amrane S, Verma A, Phan AT, Chowdhury S. A novel G-quadruplex motif modulates promoter activity of human thymidine kinase 1 . FEBS J. 2010;277:4254–4264. doi: 10.1111/j.1742-4658.2010.07814.x. [DOI] [PubMed] [Google Scholar]
- 34.Paramasivam M, Membrino A, Cogoi S, Fukuda H, Nakagama H, Xodo LE. Protein hnRNP A1 and its derivative Up1 unfold quadruplex DNA in the human KRAS promoter: implications for transcription. Nucleic Acids Res. 2009;37:2841–2853. doi: 10.1093/nar/gkp138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cogoi S, Paramasivam M, Membrino A, Yokoyama KK, Xodo LE. The KRAS promoter responds to Myc-associated zinc finger and poly(ADP-ribose) polymerase 1 proteins, which recognize a critical quadruplex-forming GA-element. J. Biol. Chem. 2010;285:22003–22016. doi: 10.1074/jbc.M110.101923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uribe DJ, Guo K, Shin YJ, Sun D. Heterogeneous nuclear ribonucleoprotein K and nucleolin as transcriptional activators of the vascular endothelial growth factor promoter through interaction with secondary DNA structures. Biochemistry. 2011;50:3796–3806. doi: 10.1021/bi101633b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yafe A, Shklover J, Weisman-Shomer P, Bengal E, Fry M. Differential binding of quadruplex structures of muscle-specific genes regulatory sequences by MyoD, MRF4 and myogenin. Nucleic Acids Res. 2008;36:3916–3925. doi: 10.1093/nar/gkn340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shklover J, Weisman-Shomer P, Yafe A, Fry M. Quadruplex structures of muscle gene promoter sequences enhance in vivo MyoD-dependent gene expression. Nucleic Acids Res. 2010;38:2369–2377. doi: 10.1093/nar/gkp1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanehisa M, Goto S, Hattori M, oki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blasius M, Sommer S, Hubscher U. Deinococcus radiodurans: what belongs to the survival kit? Crit . Rev. Biochem. Mol. Biol. 2008;43:221–238. doi: 10.1080/10409230802122274. [DOI] [PubMed] [Google Scholar]
- 41.Makarova KS, Aravind L, Wolf YI, Tatusov RL, Minton KW, Koonin EV, Daly MJ. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 2001;65:44–79. doi: 10.1128/MMBR.65.1.44-79.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slade D, Radman M. Oxidative stress resistance in Deinococcus radiodurans . Microbiol. Mol. Biol. Rev. 2011;75:133–191. doi: 10.1128/MMBR.00015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta AK, Pathak R, Singh B, Gautam HK, Kumar R, Kumar R, Arora R, Gautam H. Proteomic analysis of global changes in protein expression during exposure of gamma radiation in Bacillus sp. HKG 112 isolated from saline soil. J. Microbiol. Biotechnol. 2011;21:574–581. [PubMed] [Google Scholar]
- 45.Liu Y, Zhou J, Omelchenko MV, Beliaev AS, Venkateswaran A, Stair J, Wu L, Thompson DK, Xu D, Rogozin IB, et al. Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc. Natl Acad. Sci. USA. 2003;100:4191–4196. doi: 10.1073/pnas.0630387100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu G, Wang L, Chen H, Lu H, Ying N, Tian B, Hua Y. RecO is essential for DNA damage repair in Deinococcus radiodurans . J. Bacteriol. 2008;190:2624–2628. doi: 10.1128/JB.01851-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan M, Chen M, Zhang W, Lu W, Wang J, Yang M, Zhao P, Tang R, Li X, Hao Y, et al. Genome sequence and transcriptome analysis of the radioresistant bacterium Deinococcus gobiensis: insights into the extreme environmental adaptations. PLoS One. 2012;7:e34458. doi: 10.1371/journal.pone.0034458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choi W, Harshey RM. DNA repair by the cryptic endonuclease activity of Mu transposase. Proc. Natl Acad. Sci. USA. 2010;107:10014–10019. doi: 10.1073/pnas.0912615107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren J, Chaires JB. Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry. 1999;38:16067–16075. doi: 10.1021/bi992070s. [DOI] [PubMed] [Google Scholar]
- 50.Cahoon LA, Seifert HS. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae . Science. 2009;325:764–767. doi: 10.1126/science.1175653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nicoludis JM, Barrett SP, Mergny JL, Yatsunyk LA. Interaction of human telomeric DNA with N-methyl mesoporphyrin IX. Nucleic Acids Res. 2012;40:5432–5447. doi: 10.1093/nar/gks152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han H, Langley DR, Rangan A, Hurley LH. Selective interactions of cationic porphyrins with G-quadruplex structures. J. Am. Chem. Soc. 2001;123:8902–8913. doi: 10.1021/ja002179j. [DOI] [PubMed] [Google Scholar]
- 53.Makarova KS, Omelchenko MV, Gaidamakova EK, Matrosova VY, Vasilenko A, Zhai M, Lapidus A, Copeland A, Kim E, Land M, et al. Deinococcus geothermalis: the pool of extreme radiation resistance genes shrinks. PLoS One. 2007;2:e955. doi: 10.1371/journal.pone.0000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krisko A, Radman M. Protein damage and death by radiation in Escherichia coli and Deinococcus radiodurans . Proc. Natl Acad. Sci. USA. 2010;107:14373–14377. doi: 10.1073/pnas.1009312107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daly MJ, Ouyang L, Fuchs P, Minton KW. In vivo damage and recA-dependent repair of plasmid and chromosomal DNA in the radiation-resistant bacterium Deinococcus radiodurans . J. Bacteriol. 1994;176:3508–3517. doi: 10.1128/jb.176.12.3508-3517.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Misra HS, Khairnar NP, Kota S, Shrivastava S, Joshi VP, Apte SK. An exonuclease I-sensitive DNA repair pathway in Deinococcus radiodurans: a major determinant of radiation resistance. Mol. Microbiol. 2006;59:1308–1316. doi: 10.1111/j.1365-2958.2005.05005.x. [DOI] [PubMed] [Google Scholar]
- 57.Khairnar NP, Misra HS. DNA polymerase X from Deinococcus radiodurans implicated in bacterial tolerance to DNA damage is characterized as a short patch base excision repair polymerase. Microbiology. 2009;155:3005–3014. doi: 10.1099/mic.0.029223-0. [DOI] [PubMed] [Google Scholar]
- 58.Bentchikou E, Servant P, Coste G, Sommer S. A major role of the RecFOR pathway in DNA double-strand-break repair through ESDSA in Deinococcus radiodurans . PLoS Genet. 2010;6:e1000774. doi: 10.1371/journal.pgen.1000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Webb BL, Cox MM, Inman RB. Recombinational DNA repair: the RecF and RecR proteins limit the extension of RecA filaments beyond single-strand DNA gaps. Cell. 1997;91:347–356. doi: 10.1016/s0092-8674(00)80418-3. [DOI] [PubMed] [Google Scholar]
- 60.Huang L, Hua X, Lu H, Gao G, Tian B, Shen B, Hua Y. Three tandem HRDC domains have synergistic effect on the RecQ functions in Deinococcus radiodurans . DNA Repair. 2007;6:167–176. doi: 10.1016/j.dnarep.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 61.Rocha EP, Cornet E, Michel B. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet. 2005;1:e15. doi: 10.1371/journal.pgen.0010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli . Microbiol. Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guedin A, Gros J, Alberti P, Mergny JL. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010;38:7858–7868. doi: 10.1093/nar/gkq639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kankia BI, Marky LA. Folding of the thrombin aptamer into a G-quadruplex with Sr(2+): stability, heat, and hydration. J. Am. Chem. Soc. 2001;123:10799–10804. doi: 10.1021/ja010008o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.