Abstract
Holliday junction (HJ) resolvases are structure-specific endonucleases that cleave four-way DNA junctions (HJs) generated during DNA recombination and repair. Bacterial RuvC, a prototypical HJ resolvase, functions as homodimer and nicks DNA strands precisely across the junction point. To gain insights into the mechanisms underlying symmetrical strand cleavages by RuvC, we performed crystallographic and biochemical analyses of RuvC from Thermus thermophilus (T.th. RuvC). The crystal structure of T.th. RuvC shows an overall protein fold similar to that of Escherichia coli RuvC, but T.th. RuvC has a more tightly associated dimer interface possibly reflecting its thermostability. The binding mode of a HJ-DNA substrate can be inferred from the shape/charge complementarity between the T.th. RuvC dimer and HJ-DNA, as well as positions of sulfate ions bound on the protein surface. Unexpectedly, the structure of T.th. RuvC homodimer refined at 1.28 Å resolution shows distinct asymmetry near the dimer interface, in the region harboring catalytically important aromatic residues. The observation suggests that the T.th. RuvC homodimer interconverts between two asymmetric conformations, with alternating subunits switched on for DNA strand cleavage. This model provides a structural basis for the ‘nick-counter-nick’ mechanism in HJ resolution, a mode of HJ processing shared by prokaryotic and eukaryotic HJ resolvases.
INTRODUCTION
Homologous recombination plays critical roles in generating genetic diversity and repairing DNA lesions including double-strand breaks (1). A key intermediate formed during the homologous recombination process is a four-way junction DNA structure known as the Holliday junction (HJ) (2). In a Holliday junction, two homologous duplex DNA molecules are linked by crossovers as a result of strand exchange. The HJs are resolved by structure-specific endonucleases called HJ resolvases that cleave the two crossover strands across the junction point (3). The enzymatic resolution of HJ ensures that the canonical linear double-stranded DNA structure is restored on completion of the recombination reaction, which is critical for the DNA molecules to segregate at cell division. HJs formed by replication fork reversal are also resolved by HJ resolvases, as part of a mechanism to rescue stalled forks during DNA replication (4). Thus, HJ resolvases are important in maintaining genome integrity and are found across all kingdoms of life from bacteriophages to humans (5–7).
In most bacteria, HJs are processed by the RuvA, RuvB and RuvC proteins originally identified through mutations that confer genetic defects in ultraviolet-induced DNA damage repair (8,9). The RuvA–RuvB complex facilitates an adenosine triphosphate–dependent branch migration of HJ, modulating the size of the hetroduplex region during homologous recombination (10,11). According to a proposed model, two homologous duplex arms in a four-way junction exchange their pairing partners by an unwinding-rewinding process when passing through the RuvA octamer, whereas RuvB complex functions as a pump to pull DNA duplex arms (12). The branch migration allows relocation of the junction point to any cleavable sequences. RuvC, the HJ resolvase, then resolves the HJ structure into duplex products via a pair of symmetrical incisions across the junction point (13,14). The dual incisions turn the HJs into two nicked duplex products that can be directly repaired by DNA ligases (15).
Although the Escherichia coli RuvC does not bind to HJ-DNA in a sequence-specific fashion (16), it exhibits sequence selectivity in DNA cleavage and preferentially nicks at 3′ side of Thy bases (17–19). It has been suggested that the rate-limiting step during HJ resolution is the first strand cleavage; once RuvC nicks one strand, it quickly cuts the opposing strand to complete the resolution reaction (20,21). Thus, HJ resolution by RuvC entails two sequential strand cleavages. An elegant biochemical experiment by Birkenbihl and Kemper demonstrated that the bacteriophage T4 endonuclease VII also resolves HJ-DNA by sequential nicks within a stable enzyme–DNA complex (22). Similarly, eukaryotic HJ resolvases cut pre-nicked HJs much more readily than intact HJ substrate, suggesting that the nick-counter-nick mechanism may be a common strategy employed by HJ resolvases (23,24). However, the molecular bases for the sequence selectivity and the asymmetric sequential DNA cleavages by RuvC are not well understood.
The crystal structure of the RuvC resolvase from E. coli at 2.5 Å resolution revealed that RuvC forms a symmetric homodimer of 19 kDa subunit with an elongated overall shape (25). The catalytic center of each subunit is composed of four acidic residues, Asp7, Glu66, Asp138 and Asp141, clustered at the bottom of a basic cleft (26). These carboxylate residues coordinate divalent metal cations to catalyze hydrolysis of the phosphodiester bond in DNA backbones, which is a common feature shared by many nucleases (27). The two catalytic centers in the RuvC dimer are positioned on the same side of a dimer surface, defining the ‘front’ side of the enzyme responsible for the DNA binding. The region between the two active centers on the surface of the RuvC dimer includes Phe69, a residue critical in the HJ-DNA binding and resolution (28). During junction cleavage by RuvC, the aromatic ring of phenylalanine 69 is thought to stack on a DNA nucleobase and stabilize the open conformation of HJ to facilitate its resolution (29).
Even though the E. coli RuvC has long served as a model system of HJ resolvase, mechanisms of HJ recognition and symmetrical strand cleavages by RuvC are not fully understood. Despite the importance of RuvC in DNA recombination and repair across gram-negative bacteria, there is essentially no biochemical study reported for RuvC from bacteria other than E. coli, and the crystal structure of the E. coli RuvC (25) remains to be the only structure of RuvC from any organism. We reasoned that additional structural information would help better understand how RuvC specifically recognizes a HJ-DNA and resolves it by two symmetrical but sequential strand cleavages, and therefore performed crystallographic studies of RuvC from Thermus thermophilus (T.th. RuvC). A high-resolution structure of T.th. RuvC also reveals unique structural features that may contribute to the increased thermostability of this enzyme.
MATERIALS AND METHODS
Purification of the Thermus thermophilus RuvC protein
A codon-optimized synthetic gene for the full-length T.th. RuvC protein (Uniprot ID: Q5SJC4) was inserted into the pET11a vector to generate the expression plasmid used in this study. The expression plasmids for the D146N, F73A, F74A and Y75A point mutants were generated by the standard site-directed mutagenesis procedure. The proteins were overexpressed in the E. coli strain BL21 (DE3). Transformed bacterial cells were grown in 4L of LB medium supplemented with 0.4 g of ampicillin to an OD600 of ∼0.5, at which point isopropyl-β-d-thio-galactoside was added to a final concentration of 1 mM to induce protein expression. The cells were further incubated at 20°C for 18 h and then collected by centrifugation. The collected cells were re-suspended in 40 ml of buffer A (20 mM HEPES-NaOH pH 7.5, 0.25 M NaCl), disrupted by sonication, and spun at 59 000g for 1 h. The supernatant was then heated at 70°C for 30 min and centrifuged again at 59 000g for 40 min. The supernatant was filtered through a surfactant-free cellulose acetate membrane with a 0.2 µm pore-size. The filtered supernatant was then applied onto a Hi-trap Heparin-Sepharose column pre-equilibrated with buffer A. Elution by a linear NaCl concentration gradient from 0.25–1.0 M yielded a single peak corresponding to the purified T.th. RuvC protein.
HJ-DNA resolution assay
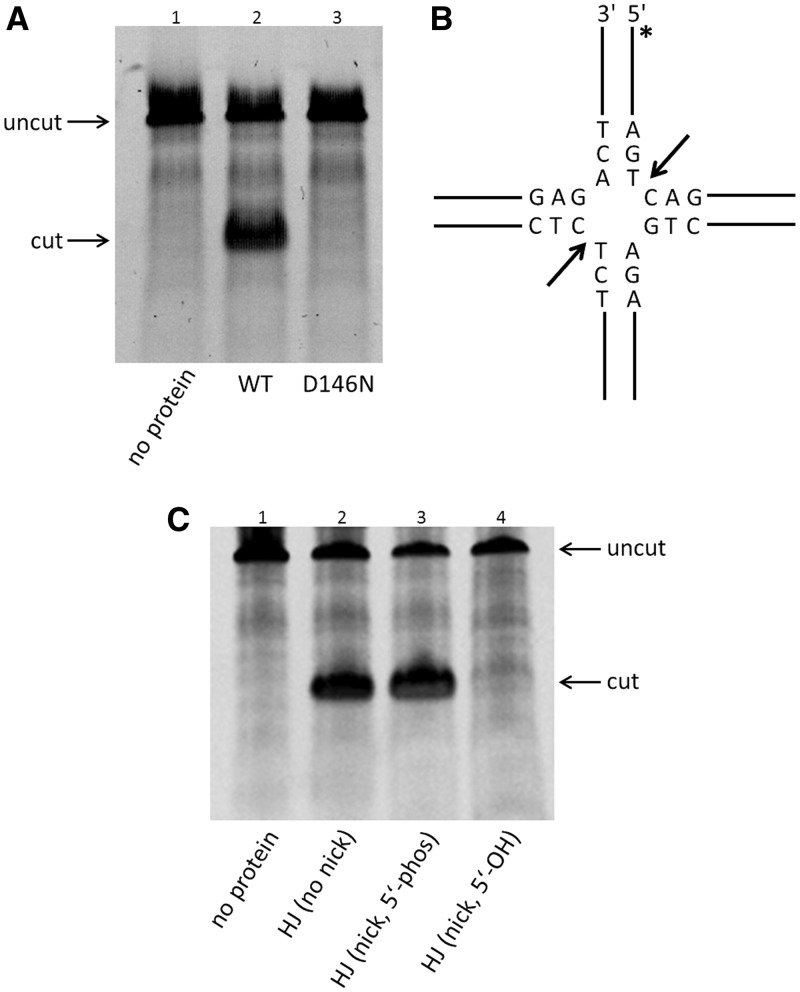
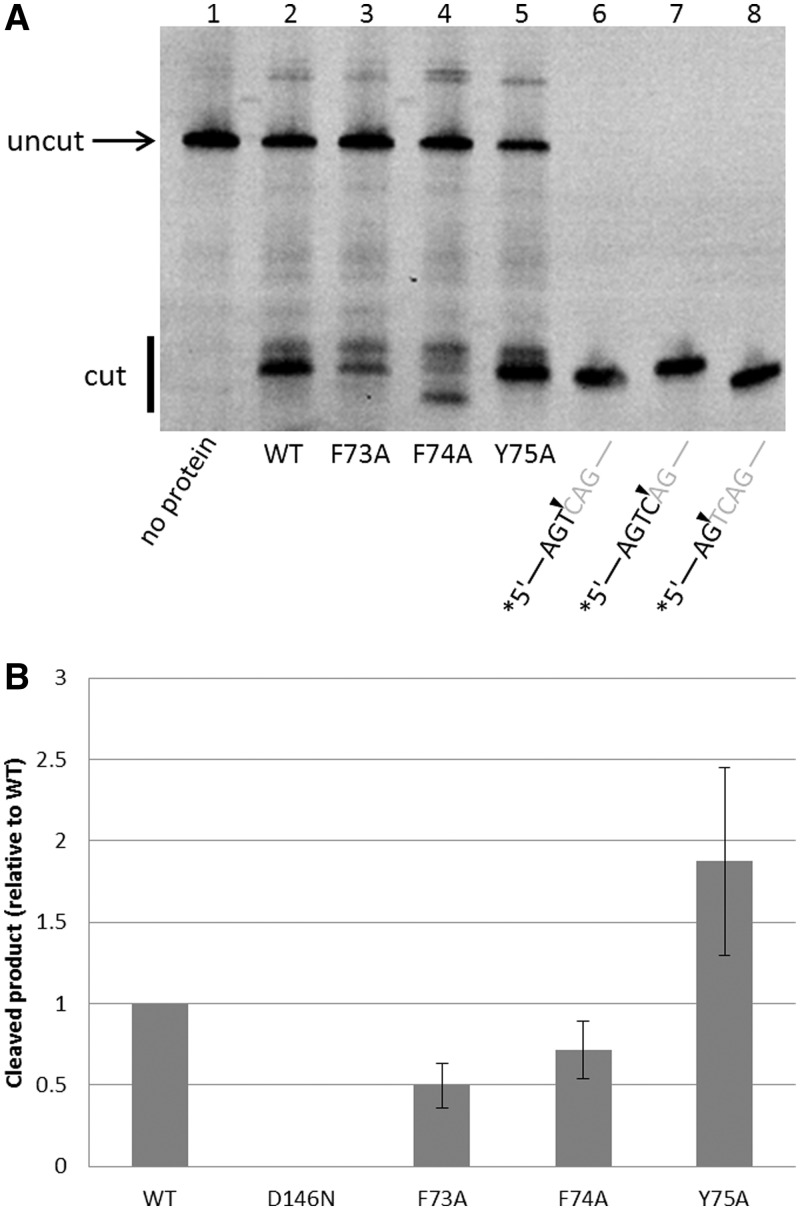
The reaction mixture (50 µl) containing a synthetic HJ-DNA (100 nM) in which one of the four strands is 6-carboxyfluorescein (6-FAM) labeled at the 5′-end, T.th. RuvC protein (500 nM), 20 mM HEPES-NaOH (pH 7.5), 50 mM NaCl, 5 mM MnCl2, 1 mM tris(2-carboxyethyl)phosphine, 5% (v/v) Glycerol and 15% (v/v) dimethyl sulfoxide was incubated for 60 min at an optimized reaction temperature (58°C). The reactions were stopped by the addition of 1 µl 10% sodium dodecyl sulphate and 50 µl denaturing polyacrylamide gel electrophoresis gel-loading buffer (95% formamide and 0.25% bromophenol blue), denatured by heating and analyzed on 15% polyacrylamide TBE-Urea gels (Invitrogen). The gels were scanned by a FUJIFILM Fluorescent Image Analyzer. The sequences of the oligonucleotides used to form the HJ substrate were as follows: FAM_RUV_X64_1 (5′-ATTCTACCAGTGCCTTGCTA GCCACAGCCA GTCAGCCGAT TGCGGGACAT CTTTGCCCAC CTGC-3′), RUV_X64_2 (5′-GCAGGTGGGC AAAGATGTCC CGCAATCGGC TGAGACCGAGCACGATCTGTTGTAATCGTCAAGC-3′), RUV_X64_3 (5′-GCTTGACGAT TACAACAGAT CGTGCTCGGT CTCTCGGCAG ATGCCATGGA GCTGTCTAGA GGAT-3′), RUV_X64_4 (5′-ATCCTCTAGA CAGCTCCATG GCATCTGCCG AGACTGGCTG TGGCTAGCAA GGCACTGGTA GAAT-3′). The annealed HJ substrate is a ‘bimobile’ junction (18) with limited central mobility (Figure 1B), and E. coli RuvC had been reported to cleave between the T and C nucleotides highlighted by underlines. To generate the nicked HJ-DNA substrate, RUV_X64_3 was replaced by two 32-base oligonucleotides, either with or without the 5′-phosphate group at the junction. The positions of the cut site by T.th. RuvC were determined by comparing the mobilities of the cleaved products with those of fluorescently labeled oligonucleotides with various lengths (Figure 6A, lanes 6–8).
Figure 1.
T.th. RuvC functions as a HJ resolvase in vitro. (A) Denaturing polyacrylamide gel analysis of HJ-DNA resolution by the wild-type and the D146N mutant T.th. RuvC proteins. The top band corresponds to the fluorescently labeled uncut 64-base oligonucleotide (uncut), whereas the bottom band corresponds to the cleaved product (cut). The mutation of a catalytic residue Asp146 abolishes the DNA cleavage activity. (B) Structure of the ‘bi-mobile’ HJ substrate and the sequences near the junction point. The dominant cutting sites by the wild-type T.th. RuvC are indicated by arrows. The asterisk (*) indicates the position of the 6-FAM label. (C) Cleavage of unnicked or pre-nicked HJ substrates (5′-end at the nick having either phosphate or hydroxyl group) by the wild-type T.th. RuvC, analyzed on a denaturing gel. The result shows that resolution of the pre-nicked HJ requires presence of the 5′-phosphate group at the nick. All reactions shown in Figure 1 were carried out at a protein concentration of 500 nM and 100 nM of HJ-DNA substrate.
Figure 6.
HJ-DNA resolution by the T.th. RuvC mutants. (A) Resolution of the unnicked HJ-DNA substrate by the wild-type and mutant T.th. RuvC proteins, analyzed on a denaturing gel. Mutations of the aromatic residues in the asymmetric loop region differently affect the HJ resolvase activity. Lanes 6–8 have fluorescently labeled oligonucleotides corresponding to cleaved products, as size markers. The F74A mutant (lane 4) shows a distinct pattern with unique sequence selectivity in DNA cleavage. The enzyme and HJ-DNA substrate concentration was 500 and 100 nM, respectively. (B) Quantification of the total products generated, normalized against the wild-type. The error bars show standard deviations between three experiments.
Crystallization
T.th. RuvC crystals were grown by the hanging drop vapor diffusion method at 20°C. The wild-type RuvC crystals (form I) were obtained by mixing the protein at ∼2 mg/ml in 20 mM HEPES (pH 7.5), 0.25 M NaCl and 2.5 mM 2-mercaptoethanol with an equal volume of well solution consisting of 35% polyethylene glycol (PEG) 3350, 0.2 M Li2SO4 and 0.1 M Tris–HCl (pH 8.5). Crystals typically appeared after ∼48 h and continued to grow for a week. The crystals were cryoprotected by gradually introducing glycrol into the drops to a final concentration of 20%, and then flash cooled in liquid nitrogen. X-ray diffraction data were collected at the beamlines 24ID-C and 24ID-E (Advanced Photon Source, Argonne, IL). The crystals typically were clusters of smaller crystals. Thus, datasets were collected by shooting edges of the crystals with an X-ray beam collimated to a 10 µm diameter (Supplementary Figure S1). The best crystal diffracted to a Bragg spacing of ∼1 Å. The crystal belonged to the space group P212121, with the unit cell parameters of 36.8 Å, 51.2 Å and 134.9 Å. The asymmetric unit (ASU) contained two T.th. RuvC molecules. Side chains for the following residues were modeled as multiple conformers. A chain: P40, K45, F73 (only the major conformation is shown in the Figure 4B, 5B and 5D for clarity), K111, L127, E137, I150, B chain: K45, V67, R76, L127, M128, I150, M160.
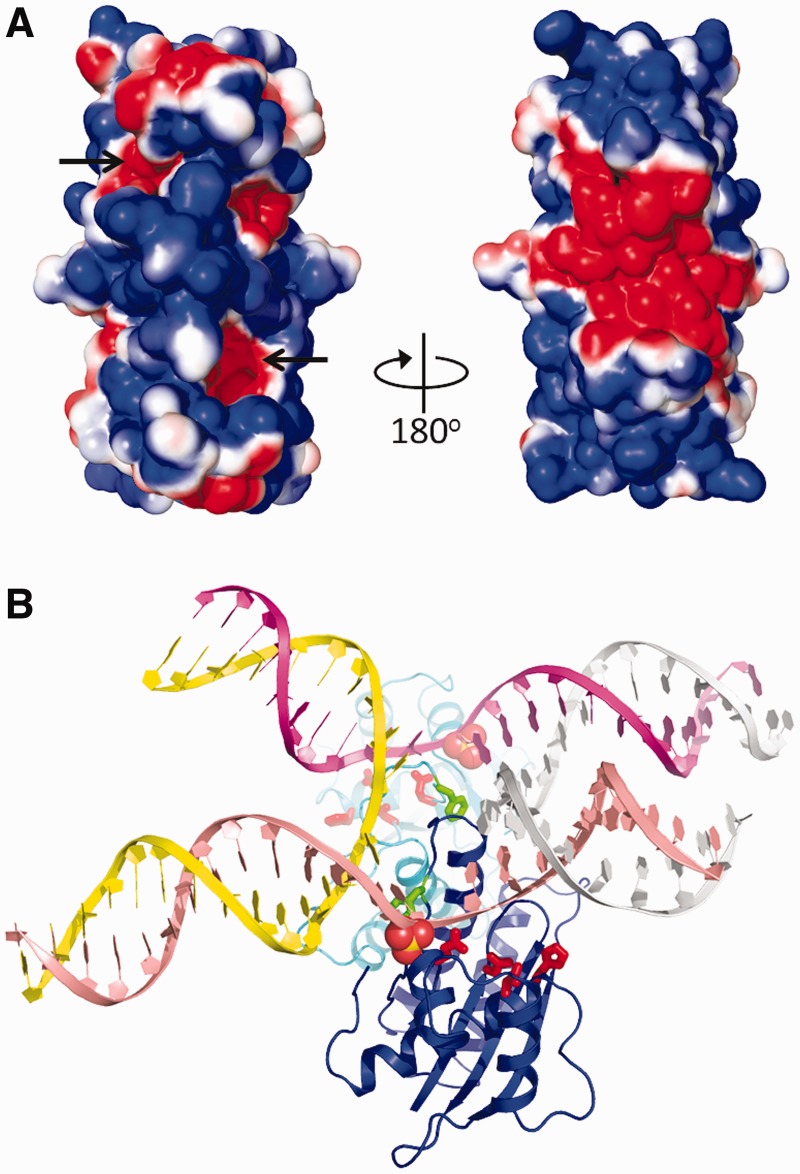
Figure 4.
Surface potential of the T.th. RuvC dimer and a possible mode of HJ-DNA binding. (A) Solvent accessible surface of the T.th. RuvC dimer colored according to the electrostatic surface potential (red: −1 kT/e to blue: +1 kT/e), shown in two orientations rotated by 180°. The location of the active sites is indicated by arrows. (B) A hypothetical model of T.th. RuvC dimer bound to a HJ-DNA. The spheres represent sulfate ions bound on the protein surface (in the crystal form II), whereas the active site residues are shown in red sticks. The Phe73 side chains are shown by green sticks. A close-up view around the junction point is shown in Supplementary Figure S7.
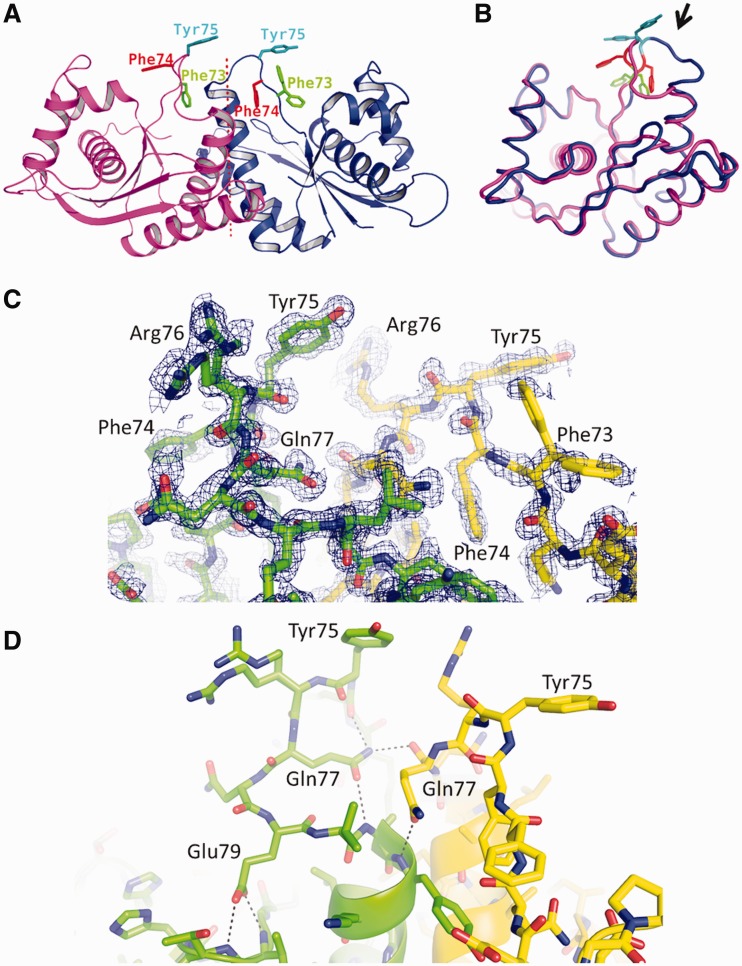
Figure 5.
The asymmetric loop. (A) Ribbon diagram of the T.th. RuvC dimer, with the aromatic residues in the asymmetric loop region Phe73, Phe74 and Tyr75 shown in sticks. (B) Superposition of the two T.th. RuvC molecules from the asymmetric dimer, with Phe73, Phe74 and Tyr75 colored as in (A). The loop region with the greatest conformational difference between the two molecules is highlighted by an arrow. Deviations of the Cα positions in this superposition are shown in Supplementary Figure S6. (C) Electron density for the asymmetric loops. Simulated annealing composite omit 2Fo-Fc map within 1.7 Å from the protein atoms is shown, contoured at 1.0σ. Arg76 from one of the molecules and Phe73 from the other molecule show multiple conformations. (D) Hydrogen-bonding network involving Gln77 and Glu79 stabilizes the asymmetric conformations of the loops. The hydrogen bonds are indicated by the dotted lines.
For the D146N T.th. RuvC crystals (form II), the well solution consisted of 32% PEG400, 0.4 M Li2SO4 and 0.1 M sodium acetate (pH 4.2). Although crystallization in this condition required the presence of DNA (a HJ with alternating arm lengths of 10 and 14 bp), there was no DNA in the crystal. The crystals belonged to the space group I212121, with the unit cell parameters of 35.2 Å, 60.0 Å and 135.7 Å. The asymmetric unit contained a RuvC monomer. Both of the T.th. RuvC crystal forms had relatively low solvent content, 30.9% and 39.1% for form I and II, respectively.
Structure determination and refinement
X-ray diffraction frames were processed using the HKL2000 suite (30). A molecular replacement calculation was performed with PHASER (31) on the higher resolution wild-type T.th. RuvC dataset (crystal form I) using the E. coli RuvC (25) as the search model, yielding a clear solution. Iteration of phase-restrainted refinement using REFMAC5 (32) and manual model building in COOT (33) eventually generated a model consisting of residues 1∼166 for two T.th. RuvC molecules and 414 water molecules. The final model has been refined at 1.28 Å resolution to a free R-factor of 19.1% using PHENIX (34). No non-crystallographic symmetry restraint was used throughout the refinement. The structure of the D146N mutant (form II) was determined employing the refined T.th. RuvC model from the form I as the molecular replacement search model, and was refined to a final free R-factor of 26.1% at 2.08 Å resolution. A summary of X-ray diffraction data and model refinement statistics is available in Table 1.
Table 1.
Data collection and refinement statistics
| Form I (wild-type) | Form II (D146N) | |
|---|---|---|
| PDB_ID | 4EP4 | 4EP5 |
| Data collection | ||
| Space group | P212121 | I212121 |
| Unit cell dimensions (Å) | 36.78, 51.15, 134.91 | 35.18, 60.02, 135.73 |
| Number of molecules in ASU | 2 | 1 |
| Resolution range (Å) | 50.00–1.28 (1.30–1.28) | 50.00–2.08 (2.12–2.08) |
| Total number of reflections | 356 967 | 55 406 |
| Number of unique reflections | 65 468 | 8463 |
| Rsym (%) | 4.8 (42.3) | 6.0 (50.6) |
| Completeness (%) | 98.4 (96.7) | 93.3 (69.8) |
| I/σ(I) | 28.0 (2.1) | 13.0 (2.3) |
| Redundancy | 5.5 (2.7) | 6.5 (5.0) |
| Structural refinement | ||
| Resolution range | 35.48–1.28 | 34.16–2.08 |
| Work set reflections | 63 393 | 7600 |
| Test set reflections | 2000 | 844 |
| Rwork (%) | 15.30 | 20.78 |
| Rfree (%) | 19.10 | 26.07 |
| Root-mean-square deviation | ||
| Bond length (Å) | 0.005 | 0.006 |
| Bond angle (°) | 1.008 | 0.974 |
| Ramachandran plot | ||
| Preferred/allowed (%) | 100 | 100 |
| Disallowed | 0 | 0 |
RESULTS AND DISCUSSION
T.th. RuvC is a HJ resolvase
We recombinantly expressed and purified the full-length T.th. RuvC protein and demonstrated that it functions as a HJ-DNA resolvase in vitro. T.th. RuvC cleaved DNA strands in a synthetic HJ-DNA substrate designed to have limited mobility of the branch point (Figure 1A, 1B and 6A, Supplementary Figure S2). The strand cleavage on this particular HJ-DNA substrate occurred predominantly on the 3′ side of a Thy base, as had been demonstrated for E. coli RuvC (17–19). Also, similar to E. coli RuvC, T.th. RuvC exhibited the resolvase activity on a pre-nicked HJ-DNA substrate in which one of the four strands is nicked at the branch point, provided that the 5′-end of DNA at the nick is phosphorylated (Figure 1C) (21). We detected the activities of T.th. RuvC only at 52°C or 58°C out of five temperature points tested (37°C, 52°C, 58°C, 65°C, 72°C), and the protein was found to be more active at 58°C in our assay condition. No endonuclease activity was detected at 37°C, confirming that the observed activities were not due to a contaminating E. coli enzyme. The D146N mutant of T.th. RuvC that has a mutation in one of the catalytic carboxylate residues corresponding to Asp141 of E. coli RuvC showed no HJ resolvase activity (Figure 1A, lane 3), consistent with its critical role in DNA cleavage.
Structure of T.th. RuvC
We have determined crystal structures of T.th. RuvC HJ resolvase in two different crystal forms at 1.28 Å and 2.08 Å resolution. The first crystal form that gave the higher resolution diffraction (form I; Supplementary Figure S1) was obtained with the wild-type T.th. RuvC and PEG3350 as the precipitant, and contained one RuvC homodimer in the asymmetric unit. The two protein molecules in the RuvC homodimer are related by a pseudo 2-fold axis but their conformations do not follow a perfect 2-fold symmetry. The second crystal form, obtained with the D146N mutant and PEG400 as the precipitant (form II), had one RuvC molecule in the asymmetric unit and thus the RuvC homodimer obeys a perfect 2-fold symmetry. Molecular packing arrangements in the two crystal forms are otherwise similar to each other.
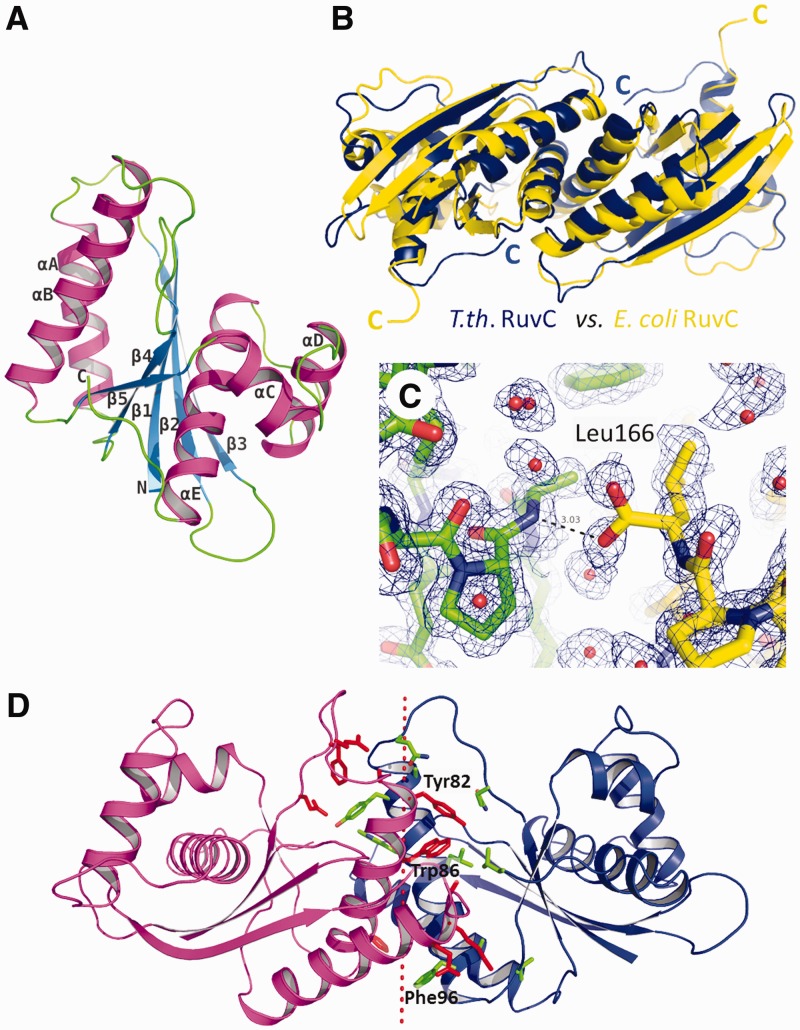
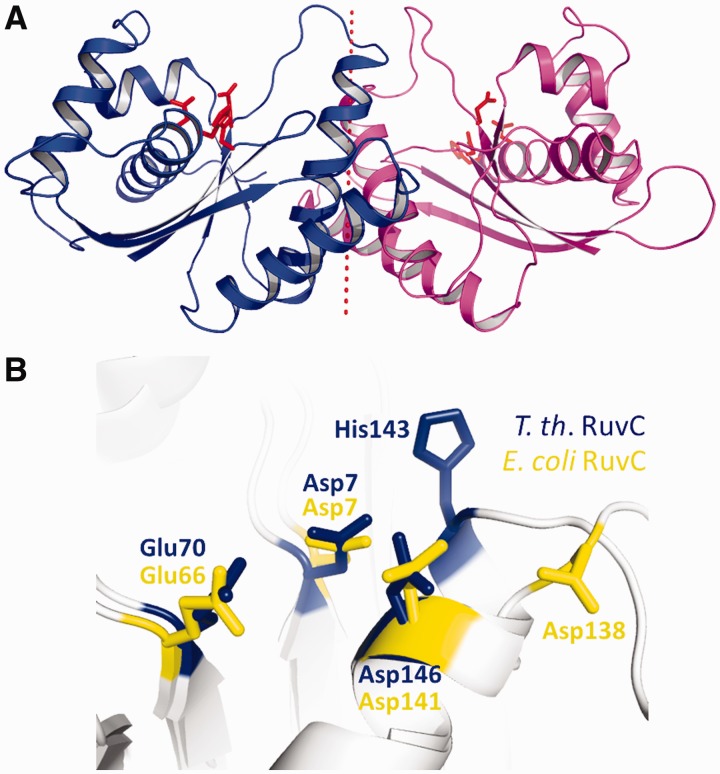
T.th. RuvC has a protein fold consisting of a single layer of β-sheet sandwiched between α-helices, characteristic of enzymes from the retrovirus integrase superfamily (Figure 2A and Supplementary Figure S3) (35). The overall structure of the T.th. RuvC homodimer is similar to that of E. coli RuvC (25), as expected from the reasonably high amino acid sequence identity (∼35%) between the two proteins (Figure 2B). The RuvC homodimers from E. coli and T.th. can be superimposed with a root-mean-square deviation for the backbone Cα atoms of 1.6 Å (over 130 atoms per each monomer). The structural deviations are attributed mostly to the loop regions between the secondary structure elements and the N- and C-termini of the protein. For instance, in contrast to the flexible C-terminal end of E. coli RuvC, the C-terminal ends of T.th. RuvC are fully ordered with the last residue Leu166 involved in dimerization interactions. The terminal carboxyl group of Leu166 forms a hydrogen bond with the backbone amide group of Ala44 from the other molecule, whereas its side chain is part of the hydrophobic dimer interface (Figure 2C). Although many of the secondary structure elements superimpose well between the E. coli and T.th. RuvC dimers, notable differences were found in the positions of the long α-helices in the core of the protein dimer (α-helices A and B; Figure 2A), such that T.th. RuvC has a more tightly associated dimer interface (Figure 2B). This is reflected in the difference in total buried accessible surface areas at the dimer interface, 1896 Å2 for E. coli RuvC versus 2432 Å2 for T.th. RuvC. In addition, the dimer interface of T.th. RuvC is richer in aromatic residues, with Tyr82, Trp86 and Phe96 involved in hydrophobic interactions (Figure 2D). These structural differences may contribute to the improved thermostability of T.th. RuvC.
Figure 2.
Structure of T.th. RuvC. (A) T.th. RuvC monomer shown in a ribbon diagram, with the secondary structure elements labeled. (B) T.th. RuvC dimer in blue superimposed on the E. coli RuvC dimer in yellow. The α-helices are packed more tightly at the dimer interface in the T.th. RuvC dimer, with the C-termini of the proteins tucked in. (C) The C-terminal residue Leu166 of T.th. RuvC is well ordered, with the carboxyl group involved in hydrogen bonding across the dimer interface. The two different molecules within the T.th. RuvC dimer are colored differently (yellow and green) to highlight the intermolecular interaction. The simulated annealing composite omit 2Fo-Fc map is contoured at 1.0σ. (D) Residues involved in the dimer interface are shown, with the aromatic residues Tyr82, Trp86 and Phe96 highlighted. The dotted line represents the pseudo 2-fold axis relating the two molecules in the T.th. RuvC dimer.
Active site and the DNA-binding mode
The catalytic center of T.th. RuvC is composed of Asp7, Glu70, His143 and Asp146 clustered at the bottom of a cleft that traverses the protein surface (Figure 3A). The third metal-chelating residue His143 corresponds to Asp138 of E. coli RuvC (Figure 3B and Supplementary Figure S4). Having a histidine residue at this position is a feature shared by many RuvC orthologs according to a sequence alignment (Supplementary Figure S5). Based on analogy to other enzymes in the retroviral integrase family such as RNaseH1 (27,36) the tetrad of active site residues are expected to chelate two metal ions to catalyze hydrolysis of a DNA backbone phosphodiester bond. The active centers of T.th. RuvC are surrounded by broad positively charged surfaces, as expected for an enzyme known to bind DNA in a non-sequence–specific fashion (Figure 4A) (16).
Figure 3.
The enzyme active site of T.th. RuvC. (A) Ribbon diagram of the T.th. RuvC dimer, with the catalytic residues Asp7, Glu70, His143 and Asp146 shown in red sticks to indicate the location of the active sites. (B) Superposition of the catalytic residues from E. coli RuvC (yellow) and T.th. RuvC (blue).
Based on the overall shape, surface electrostatic potential, and the location of the enzyme active site on the T.th. RuvC dimer, the mode of its HJ-DNA binding could be predicted. We therefore built a hypothetical model of how the T.th. RuvC dimer may bind a HJ-DNA by docking the ‘H-shaped’ HJ-DNA bound to the T4 endonuclease VII (5) onto the T.th. RuvC dimer (Figure 4B). The H-shaped HJ-DNA was oriented according to the biochemical studies demonstrating that RuvC cleaves the continuous (non-crossover) strands in an unfolded HJ-DNA (37). In the structure of T.th. RuvC crystallized in a higher concentration of Li2SO4 (crystal form II), a total of four sulfate ions per RuvC monomer were found to be bound on the protein surface. One of these four sulfate ions is located at the edge of the basic cleft that harbors the catalytic residues, possibly mimicking the backbone phosphate group of a bound DNA. Thus the paths of DNA strands in the RuvC–HJ-DNA model were adjusted accordingly. The open and planer conformation of the bound HJ-DNA is consistent with that in the RuvC–DNA complex models proposed in earlier studies (25,29), and the positioning of HJ-DNA relative to the enzyme active sites is consistent with the observation that RuvC preferentially nicks the phosphodiester bond one nucleotide 3′ to the crossover point (38).
Asymmetry in the T.th. RuvC dimer
Unexpectedly, the structure of the T.th. RuvC homodimer refined at 1.28 Å resolution shows distinct asymmetry in residues 71-81 that span the dimer interface (Figure 5A and B, Supplementary Figure S6). This region, comprising the N-terminus of α-helix B and the preceding loop, forms a lobe located between the two active centers on the surface of the RuvC dimer. The lobe fits into the central opening of the HJ-DNA in our model of the T.th. RuvC–HJ-DNA complex (Figure 4B). In one of the molecules within the RuvC dimer, the α-helix B starts from Ala81and the preceding loop hangs over the active site cleft. In the other molecule, the helix starts from Glu79 and the preceding residues are positioned farther from the active site. The asymmetric conformations of these residues are stabilized by intermolecular as well as intramolecular hydrogen-bonding network involving Gln77 and Glu79 (Figure 5C and D). The corresponding region in the strictly 2-fold symmetrical RuvC homodimer (form II) appears to be disordered, as the 2.08 Å resolution 2Fo-Fc map does not show interpretable electron density. Taken together, these observations suggest that the residues 71-81 of T.th. RuvC interconvert between two alternate conformations in a concerted fashion within the protein dimer to break the 2-fold symmetry. This ‘flip-flop’ motion has an important implication in the mechanism of HJ resolution as discussed below. Of note, the loop encompassing residues 68-72 of E. coli RuvC was reported to show different conformations between the four molecules in an asymmetric unit, suggesting conformational flexibility (25). The residues 68-72 of E. coli RuvC correspond to residues 72-76 of T.th. RuvC (Supplementary Figure S5), being consistent with the observed asymmetry in the T.th. RuvC structure.
Possible mechanism for HJ resolution
The presence of sequence selectivity in DNA strand cleavage by E. coli RuvC implies that RuvC makes base contacts near the cutting site. Phe69 of E. coli RuvC was proposed to play a critical role in base stacking near the junction point, based on the observation that F69A mutation causes a severe defect in HJ resolution (29). The corresponding residue Phe73 of T.th. RuvC is located in the loop region that undergoes the conformation switch, projecting into the active-site cleft in one of the molecules within the dimer while pointing away from the active site in the other molecule and is partially buried. In our model of the T.th. RuvC dimer bound to a HJ-DNA, Phe73 from one of the molecules is positioned in close proximity to DNA near the crossover point poised for making base-stacking interactions (Figure 4B and Supplementary Figure S7). Curiously, Phe73 of T.th. RuvC is followed by Phe74 and Tyr75, where these aromatic residues all take different conformations between the two molecules in the dimer (Figure 5). It seemed likely that some or all of these aromatic residues play roles in positioning DNA during the strand cleavage analogously to Phe69 of E. coli RuvC, and if so, the observed asymmetric conformation implies that only one of the molecules within the T.th. RuvC dimer can have a productive active site for DNA cleavage at any given time. We therefore tested the in vitro HJ-DNA cleavage activities of T.th. RuvC with single point mutations F73A, F74A and Y75A. The three mutant proteins showed an interesting spectrum of activities (Figure 6). The first mutant F73A showed a moderate defect in HJ-DNA processing, although it was not completely inactive like the corresponding F69A mutant of E. coli RuvC. The second mutant F74A also generated a slightly reduced amount of cleaved DNA products than the wild-type. In addition, this mutant showed altered sequence selectivity. In contrast to the wild-type protein that cuts preferentially at the 3′ side of the Thy base, F74A did not cut at the 3′ side of the Thy base and instead cut the DNA strand only at neighboring positions. The third mutant Y75A appeared to be hyperactive, and cleaved DNA more efficiently than the wild-type. The same trend was observed when these three mutants were tested for cleavage on the pre-nicked HJ substrate, except that F74A cuts the pre-nicked HJ-DNA at the same site as the wild-type (Supplementary Figure S8). Taken together, the results are consistent with the idea that these aromatic residues are involved in DNA interactions near the scissile site, modulating DNA cleavage by T.th. RuvC.
It was shown previously that the two enzyme active sites within the RuvC dimer can operate independently of each other (39) and the resolution of a HJ-DNA by RuvC is likely achieved by two sequential symmetrical strand cleavages across the junction point (20). On the other hand, the strict requirement for the 5′-phosphate group after the first strand cleavage in resolving the nicked HJ-DNA intermediate, as revealed by the earlier work for E. coli RuvC (21) and this study for T.th. RuvC, argues that the two active sites are functionally somehow coupled, rather than being completely independent of each other. Our structural work raises an intriguing possibility that the two active sites in the T.th. RuvC dimer may in fact catalyze DNA strand cleavages neither in a concerted fashion nor independently of each other. Rather, the two molecules switch roles via coupled conformational changes (flip-flop motion) during the sequential strand cleavages. The local structural asymmetry and flexibility in an otherwise 2-fold symmetrical HJ resolvase dimer could be a common theme for broader classes of HJ resolvases, including those from eukaryotic organisms that employ the nick-counter-nick mechanism in HJ resolution (23,24).
ACCESSION NUMBERS
The atomic coordinates and structure factors have been deposited in the Protein Data Bank with the accession codes 4EP4 (form I) and 4EP5 (form II).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–8.
FUNDING
National Institutes of Health [GM095558 and AI087098 to H.A.]; Undergraduate Research Opportunity Program of the University of Minnesota (to L.C.). Funding for open access charge: NIH.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff of Sector-24 (NE-CAT) of the Advanced Photon Source (APS) for help in x-ray data collection, and Kayo Kurahashi for technical assistance. Computer resources were provided by the Basic Sciences Computing Laboratory of the University of Minnesota Supercomputing Institute. This work is based on research conducted at the APS on the NE-CAT beamlines, which are supported by award RR15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357.
REFERENCES
- 1.West SC. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 2.Holliday R. A mechanism for gene conversion in fungi. Genet. Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- 3.Lilley DM, White MF. The junction-resolving enzymes. Nat. Rev. Mol. Cell Biol. 2001;2:433–443. doi: 10.1038/35073057. [DOI] [PubMed] [Google Scholar]
- 4.Michel B, Boubakri H, Baharoglu Z, LeMasson M, Lestini R. Recombination proteins and rescue of arrested replication forks. DNA Repair. 2007;6:967–980. doi: 10.1016/j.dnarep.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 5.Biertümpfel C, Yang W, Suck D. Crystal structure of T4 endonuclease VII resolving a Holliday junction. Nature. 2007;449:616–620. doi: 10.1038/nature06152. [DOI] [PubMed] [Google Scholar]
- 6.Hadden JM, Déclais AC, Carr SB, Lilley DM, Phillips SE. The structural basis of Holliday junction resolution by T7 endonuclease I. Nature. 2007;449:621–624. doi: 10.1038/nature06158. [DOI] [PubMed] [Google Scholar]
- 7.Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- 8.Shinagawa H, Iwasaki H. Processing the Holliday junction in homologous recombination. Trends Biol. Sci. 1996;21:107–111. [PubMed] [Google Scholar]
- 9.Sharples GJ, Ingleston SM, Lloyd RG. Holliday junction processing in bacteria: insights from the evolutionary conservation of RuvABC, RecG, and RusA. J. Bacteriol. 1999;181:5543–5550. doi: 10.1128/jb.181.18.5543-5550.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwasaki H, Takahagi M, Nakata A, Shinagawa H. Escherichia coli RuvA and RuvB proteins specifically interact with Holliday junctions and promote branch migration. Genes Dev. 1992;6:2214–2200. doi: 10.1101/gad.6.11.2214. [DOI] [PubMed] [Google Scholar]
- 11.Tsaneva IR, MuÈller B, West SC. ATP-dependent branch migration of Holliday junctions promoted by the RuvA and RuvB proteins of E. coli. Cell. 1992;69:1171–1180. doi: 10.1016/0092-8674(92)90638-s. [DOI] [PubMed] [Google Scholar]
- 12.Parsons CA, Stasiak A, Bennett RJ, West SC. Structure of a multisubunit complex that promotes DNA branch migration. Nature. 1995;374:375–378. doi: 10.1038/374375a0. [DOI] [PubMed] [Google Scholar]
- 13.Iwasaki H, Takahagi M, Shiba T, Nakata A, Shinagawa H. Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure. EMBO J. 1991;10:4381–4389. doi: 10.1002/j.1460-2075.1991.tb05016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunderdale HJ, Benson FE, Parsons CA, Sharples GJ, Lloyd RG, West SC. Formation and resolution of recombination intermediates by E. coli RecA and RuvC proteins. Nature. 1991;354:506–510. doi: 10.1038/354506a0. [DOI] [PubMed] [Google Scholar]
- 15.Bennett RJ, Dunderdale HJ, West SC. Resolution of Holliday junctions by RuvC resolvase: cleavage specificity and DNA distortion. Cell. 1993;74:1021–1031. doi: 10.1016/0092-8674(93)90724-5. [DOI] [PubMed] [Google Scholar]
- 16.Takahagi M, Iwasaki H, Shinagawa H. Structural requirements of substrate DNA for binding to and cleavage by RuvC, a Holliday junction resolvase. J. Biol. Chem. 1994;269:15132–15139. [PubMed] [Google Scholar]
- 17.Shah R, Bennett RJ, West SC. Genetic recombination in E. coli: RuvC protein cleaves Holliday junctions at resolution hotspots in vitro. Cell. 1994;79:853–864. doi: 10.1016/0092-8674(94)90074-4. [DOI] [PubMed] [Google Scholar]
- 18.Shida T, Iwasaki H, Saito A, Kyogoku Y, Shinagawa H. Analysis of substrate specificity of the RuvC Holliday junction resolvase with synthetic Holliday junction. J. Biol. Chem. 1996;42:26105–26109. doi: 10.1074/jbc.271.42.26105. [DOI] [PubMed] [Google Scholar]
- 19.Eggleston AK, West SC. Cleavage of Holliday junctions by the Escherichia coli RuvABC complex. J. Biol Chem. 2000;275:26467–26476. doi: 10.1074/jbc.M001496200. [DOI] [PubMed] [Google Scholar]
- 20.Fogg JM, Lilley DM. Ensuring productive resolution by the junction-resolving enzyme RuvC: large enhancement of the second-strand cleavage rate. Biochemistry. 2000;39:16125–16134. doi: 10.1021/bi001886m. [DOI] [PubMed] [Google Scholar]
- 21.Osman F, Gaskell L, Whitby MC. Efficient second strand cleavage during Holliday junction resolution by RuvC requires both increased junction flexibility and an exposed 5′ phosphate. PLoS One. 2009;4:e5347. doi: 10.1371/journal.pone.0005347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birkenbihl RP, Kemper B. High affinity of endonuclease VII for the Holliday structure containing one nick ensures productive resolution. J. Mol. Biol. 2002;321:21–28. doi: 10.1016/s0022-2836(02)00594-6. [DOI] [PubMed] [Google Scholar]
- 23.Gaillard PH, Noguchi E, Shanahan P, Russell P. The endogenous Mus81-Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol. Cell. 2003;12:747–759. doi: 10.1016/s1097-2765(03)00342-3. [DOI] [PubMed] [Google Scholar]
- 24.Rass U, Compton SA, Matos J, Singleton MR, Ip SC, Blanco MG, Griffith JD, West SC. Mechanism of Holliday junction resolution by the human GEN1 protein. Genes Dev. 2010;24:1559–1569. doi: 10.1101/gad.585310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ariyoshi M, Vassylyev DG, Iwasaki H, Nakamura H, Shinagawa H, Morikawa K. Atomic structure of the RuvC resolvase: a holliday junction-specific endonuclease from E. coli. Cell. 1994;78:1063–1072. doi: 10.1016/0092-8674(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 26.Saito A, Iwasaki H, Ariyoshi M, Morikawa K, Shinagawa H. Identification of four acidic amino acids that constitute the catalytic center of the RuvC Holliday junction resolvase. Proc. Natl Acad. Sci. USA. 1995;92:7470–7474. doi: 10.1073/pnas.92.16.7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang W. Nucleases: diversity of structure, function and mechanism. Q. Rev. Biophys. 2011;44:1–93. doi: 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ichiyanagi K, Iwasaki H, Hishida T, Shinagawa H. Mutational analysis on structure-function relationship of a Holliday junction specific endonuclease RuvC. Genes Cells. 1998;3:575–586. doi: 10.1046/j.1365-2443.1998.00213.x. [DOI] [PubMed] [Google Scholar]
- 29.Yoshikawa M, Iwasaki H, Shinagawa H. Evidence that Phenylalanine 69 in Escherichia coli RuvC resolvase forms a stacking interaction during binding and destabilization of a Holliday Junction DNA substrate. J. Biol. Chem. 2001;276:10432–10436. doi: 10.1074/jbc.M010138200. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tool for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nowotny M. Retroviral integrase superfamily: the structural perspective. EMBO Rep. 2009;2:144–151. doi: 10.1038/embor.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nowotny M, Yang W. Stepwise analyses of metal ions in RNase H catalysis: from substrate destabilization to product release. EMBO J. 2006;25:1924–1933. doi: 10.1038/sj.emboj.7601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bennett RJ, West SC. RuvC protein resolves Holliday junctions via cleavage of the continuous (noncrossover) strands. Proc. Natl Acad. Sci. USA. 1995;92:5635–5639. doi: 10.1073/pnas.92.12.5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sha R, Iwasaki H, Liu F, Shinagawa H, Seeman NC. Cleavage of symmetric immobile DNA junctions by Escherichia coli RuvC. Biochemistry. 2000;39:11982–11988. doi: 10.1021/bi001037z. [DOI] [PubMed] [Google Scholar]
- 39.Shah R, Cosstick R, West SC. The RuvC protein dimer resolves Holliday junctions by a dual incision mechanism that involves base-specific contacts. EMBO J. 1997;16:1464–1472. doi: 10.1093/emboj/16.6.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.