Abstract
A method of analysis is presented that utilizes matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) to monitor the kinetics and products of RNA cleavage, by use of a program designed to mass-match observed MS peaks with predicted RNA cleavage products. The method is illustrated through application to the study of targeted oxidation of RNA stem loops from HIV-1 Rev Response Element mRNA (RRE RNA) and ribosomal 16S A-site RNA (16S RNA) by metallonucleases. Following incubation of each RNA with catalysts and/or redox co-reactants, reaction mixtures were desalted, and MALDI-TOF MS was used to monitor both time-resolved formation of cleavage products and disappearance of full-length RNA. For each RNA, a unique list was generated that contained the predicted masses of both the full-length, and all of the possible RNA cleavage fragments that resulted from the combination of all possible cleavage sites and each of the six expected overhangs formed at nascent termini adjacent to the cleavage sites. The overhangs corresponded to 2′,3′-cyclic phosphate, 3′-phosphate, 3′-phosphoglycolate, 5′- hydroxyl and 5′- phosphate, which corresponded to differing oxidative, hydrolytic, and/or 2′-OH-mediated-endonucleolytic modes of scission. Each mass spectrum was compared with a corresponding list of predicted masses, and peaks were rapidly assigned by use of a Perl script, with a mass-matching tolerance of 200 ppm. Both time-dependent cleavage mediated by metallonucleases and MALDI-TOF-induced fragmentation were observed, and these were distinguished by time-dependent experiments. The resulting data allowed a semi-quantitative assessment of the rate of formation of each overhang at each nucleotide position. Limitations included artifactual skewing of quantification by mass bias, a limited mass range for quantification, and a lack of detection of secondary cleavage products. Nevertheless, the method presented herein provides a rapid, accurate, highly-detailed and semi-quantitative analysis of RNA cleavage that should be widely applicable.
INTRODUCTION
Ribonuclease mimics that irreversibly cleave or otherwise elicit damage to targeted mRNA transcripts represent a particularly promising therapeutic approach to combat various diseases (1–8). Each mRNA transcript typically precedes many copies of respective protein translation products, and cytoplasmic mRNA targets are generally more accessible to drugs than are nuclear genomic DNA, making mRNA transcripts ideal therapeutic targets. Moreover, there are very few known cellular mechanisms of mRNA repair (9–11). Several families of RNA-targeting model ribonucleases have been developed, and numerous attempts have been made to study their RNA modification chemistry, which typically includes hydrolytic scission, oxidative scission, and other oxidative modifications (1–7,12–15).
Study of these pathways is commonly followed by various analytical separation techniques, antibody-based detection of specific oxidation products, spectroscopic techniques, NMR and other methods (14), but each possesses significant inherent limitations that hinder research. Analytical separation techniques, such as high-performance liquid chromatography (HPLC), capillary electrophoresis (CE) and polyacrylamide gel electrophoresis (PAGE), have been demonstrated to possess up to single-nucleotide resolution in some cases, allowing determination of exact sites of cleavage by ribonucleases (16–20). However, these techniques are critically limited by the number and variable abundance of individual RNA fragments in a single mixture that can be simultaneously monitored. Furthermore, analytical separation techniques typically cannot distinguish between the differing functional groups that may be present at the nascent termini of cleaved RNA fragments, which can provide key evidence regarding the mechanism(s) of strand scission employed by ribonuclease mimics, unless used in conjunction with laborious chemical, enzymatic, radiographic, and/or other techniques (14,21–24). Analytical separation of RNAs under native-like conditions is also hindered by the tendency of RNA to fold into various secondary and tertiary structures.
Traditional methods of nucleic acid analysis typically provide only one or two types of information per experiment, such as approximate size (separation techniques), relative abundance (separation or detection techniques), structural information (NMR and spectroscopic methods), as well as binding and/or the nature of folding (separation and spectroscopic methods). There is certainly a need for a technique that allows simultaneous quantification and exact identification of multiple RNA species in a single complex mixture that possesses high-sensitivity, high-resolution and a wide operating concentration range. One approach that fits these criteria is matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS).
The use of MALDI-TOF MS for the analysis of RNA cleavage chemistry introduces the possibility of simultaneous resolution and exact identification of RNA species within complex mixtures containing many distinct RNA species, as is commonly the case following cleavage of RNA by ribonuclease mimics. Recent studies involving MALDI-TOF MS analysis of RNA have included detection and/or sequencing of RNA constructs (25–28), analysis of small numbers of signature digestion products resulting from controlled digestion by RNases (3,29,30),analysis of specific post-transcriptional modifications (31,32), confirmation of chemical labeling or validation of RNA constructs (26), and elucidation of MALDI-TOF-induced fragmentation pathways introduced by mass spectrometric methods (33–35). The use of MALDI-TOF MS for the analysis of RNA cleavage products resulting from artificial ribonuclease activity has thus far been underused but offers significant advantages as a quantitative tool for kinetic and mechanistic investigations.
Herein we report the use of MALDI-TOF MS to monitor the cleavage chemistry (in the presence of redox co-reactants) of multiple separate RNA species. In particular, targeted oxidative cleavage of HIV Rev Response Element (RRE) RNA stem loop IIB was mediated by Fe–EDTA–Rev, which contains both a redox reactive catalyst and a derivative of the RRE-cognate Rev peptide (2–4,36–41), while oxidative cleavage of the 16S A-site RNA stem loop was mediated by either free Cu2+(aq), Cu-glycylglycylhistidine (Cu–GGH), or other Cu-chelates. Following cleavage of each RNA and detection by MALDI-TOF MS, we observed complex distributions of scission and/or fragmentation products that contained a variety of nascent overhangs consistent with distinct oxidative, hydrolytic, and/or endonucleolytic scission pathways (14,34,35). For each reaction, full-length RNA and cleavage products were identified by use of automated matching of MS peaks with expected masses, and abundances were determined semi-quantitatively using MS peak areas. Additionally, time-dependent experiments allowed semi-quantitative initial rates of formation of each cleavage product to be determined.
By use of MALDI-TOF MS to study catalyst-mediated cleavage of RNA, we have demonstrated the feasibility of large-scale simultaneous identification and quantification of up to several hundred cleavage products per reaction mixture. The technique allowed exact identification of RNA cleavage products of differing lengths and overhangs, providing distinction between cleavage fragments arising from hydrolysis, oxidative scission, or MALDI-TOF-induced fragmentation (14,34,35). The method allows measurement of the time-dependent evolution of all products. The adaptability of the technique is shown by its application to cleavage of three separate RNA constructs. The results of this study demonstrate the utility and universality of MALDI-TOF MS as a tool to study RNA cleavage by ribonucleases mimics, allowing identification and quantification of products, even in the presence of complex product mixtures.
MATERIALS AND METHODS
Chemicals and reagents
The custom RNA constructs Fl-RRE, with sequence 5′-fluorescein-UUGGUCUGGGCGCAGCGCAAGCUGACGGUACAGGCC-3′, Fl-16S, with sequence 5′-fluorescein-GGCGUCACACCUUCGGGUGAAGUCGCC-3′, Fl-IRESSLIV, with sequence 5′-fluorescein-GGACCGUGCACCAUGAGCACGAAUCC-3′, and RRE, with sequence GGUCUGGGCGCAGCGCAAGCUGACGGUACAGGCC-3′, were purchased from Dharmacon. The custom RNA construct Fl-5mer, with sequence 5′-fluorescein(T)-UGUG-3′, was purchased from Integrated DNA Technologies. RNA species were immediately aliquoted into single-use containers and stored at −20°C to minimize freeze–thaw cycles. Catalysts Fe–EDTA–Rev and Cu–GGH were prepared as described previously (2,37). The Fe(II) sulfate heptahydrate and Cu(II) chloride dihydrate salts were purchased from ACROS and J. T. Baker, respectively. Sodium chloride and sodium hydroxide were purchased from Fisher, and HEPES was purchased from Sigma. C18 ZipTips and acetonitrile were obtained from Millipore and Fisher, respectively. Matrix components ammonium citrate and 3-hydroxypicolinic acid were obtained from Aldrich and Sigma-Aldrich, respectively.
RNA cleavage reactions
Reactions of 10 µM catalyst (Fe–EDTA–Rev or Cu–GGH), 1 mM co-reactants (ascorbate and/or H2O2) and 10 µM of either Fl-RRE RNA or Fl-16S RNA were conducted at 37°C in separate tubes, each containing 20 µl total reaction volume. A reaction buffer consisting of 20 mM HEPES, 100 mM NaCl, pH 7.4 was used in all experiments. Prior to reaction, RNA was heated to 90°C for 5 min and allowed to cool to 37°C, and the RNA was immediately added to each pre-incubated tube; for time-dependent reactions, one tube corresponded to one time point, with staggering of start times and quenching of all reactions at the same time. To initiate reactions, catalyst and co-reactants were mixed with pre-incubated RNA, and reactions were performed in a dark incubator. Reactions were quenched by placement on ice and immediately desalted. C18 ZipTips were used for desalting, with initial wetting by 50/50 acetonitrile/water and pre-equilibration with 2M triethylamine acetate, followed by loading of quenched RNA reaction mixtures onto the ZipTips. The RNA that was bound to each ZipTip was washed first three times with 2 M triethylamine acetate and then three times with nanopure water. Elution of RNA from ZipTips was performed using 5 µl ice cold 50/50 acetonitrile/water. Other desalting procedures are addressed in the Supplementary Data. PAGE analysis was performed as described previously (2).
MALDI-TOF MS analysis
Each desalted RNA product mixture, in 5 µl 50/50 acetonitrile/water, was mixed with 2.5 µl of a matrix solution containing 42 mg/ml 3-hydroxypicolinic acid, 6.8 mg/ml ammonium citrate in 30/70 acetonitrile/water. Each target on a Bruker ground steel 96 target microScout plate was spotted using a two-layer approach, where each target was first pre-spotted with 1 µl of the matrix solution and allowed to dry, before spotting with 2.5 µl of the above RNA/matrix mixture and again allowed to dry. A desalted calibration mixture containing three separate RNA species of varying molecular weights (Fl-5mer, Fl-IRESSLIV, and Fl-RRE, with molecular weights of 2057.5, 8861.5 and 12172.5 Da, respectively), bracketing the range of m/z studied, was used to calibrate the instrument prior to each analysis; the calibration mixture was spotted on a separate target on the same plate, and in the same manner, as all other RNA mixtures. All MALDI-TOF MS analysis was performed on a Bruker MicroFlex LRF instrument, equipped with a gridless reflectron, using negative ion and reflectron modes. The pulsed ion extraction time was 1200 ns. At least 1000 shots were summed per spectrum.
Peak assignment and quantification
Using Bruker flexAnalysis software, spectra were smoothed using a Savitzgy-Golay algorithm. Internal calibrations of spectra were performed using a set of regularly-occurring background fragmentation and/or doubly-charged full-length RNA peaks (Supplementary Data), and a peak list was generated for each spectrum that contained m/z values and peak areas. Peak mass matching was performed using our internally developed MassDaddy Perl script, written by Jeff C. Joyner (code and documentation provided in Supplementary Data), that requires user input of a matching tolerance (ppm) and two input files: (i) an expected-mass list file containing all expected RNA cleavage fragments (all positions combined with all overhang types) and corresponding masses (list generated in Microsoft Excel using the masses shown in Tables 1 and 2); (ii) a peak list with all observed masses and respective peak areas (obtained prior to any background subtraction). Assignment of peaks was performed by comparison of each peak list with the theoretical masses for all expected cleavage products, with a matching tolerance of 200 ppm (Supplementary Data). MassDaddy creates one output file for each mass spectrum, with six columns: the theoretical m/z (Da), the observed m/z (Da), the mass matching error (ppm), the nucleotide position of the nascent overhang, the nascent overhang type and the peak area, arranged by overhang type and sequence. Only peaks with m/z >1000 amu were considered in this study, since excessive spectral crowding occurred at lower m/z ranges. Following automated mass-matching of peaks, the peak areas of all mass-matched peaks were summed, and the peak area fraction for each peak was determined; only peaks with a peak area fraction above a threshold of 0.0005 were considered (Supplementary Data). For time-dependent reactions, the change in peak area fraction (semi-quantitative mole fraction) for each species over time was fit to a first order model, and semi-quantitative apparent initial rates (nM/min) were determined.
Table 1.
Mass differences between RNA fragments differing in length by 1 nt and containing the same terminal overhangs
| Nucleotide | Δ mass (amu) |
|---|---|
| G | 345.21 |
| A | 329.21 |
| U | 306.17 |
| C | 305.18 |
Table 2.
Mass differences between observed nascent terminal overhangs, resulting from RNA scission and/or fragmentation, and terminal hydroxyl group overhangsa
| Nascent overhang | Δ mass (amu)a |
|---|---|
| 3′-hydroxylb | 3′-OH + 0.00 |
| 2′,3′-cyclic phosphate | 3′-OH + 61.96 |
| 3′-phosphate | 3′-OH + 79.98 |
| 3′-phosphoglycolate | 3′-OH + 138.02 |
| 5′-hydroxyl | 5′-OH + 0.00 |
| 5′-phosphate | 5′-OH + 79.98 |
aRelative to the terminal 3′- or 5′-hydroxyl group.
bThe 3′-hydroxyl overhang was not typically observed, except for regular occurrence at 3′-termini of full-length RNA.
RESULTS AND DISCUSSION
Reactions studied
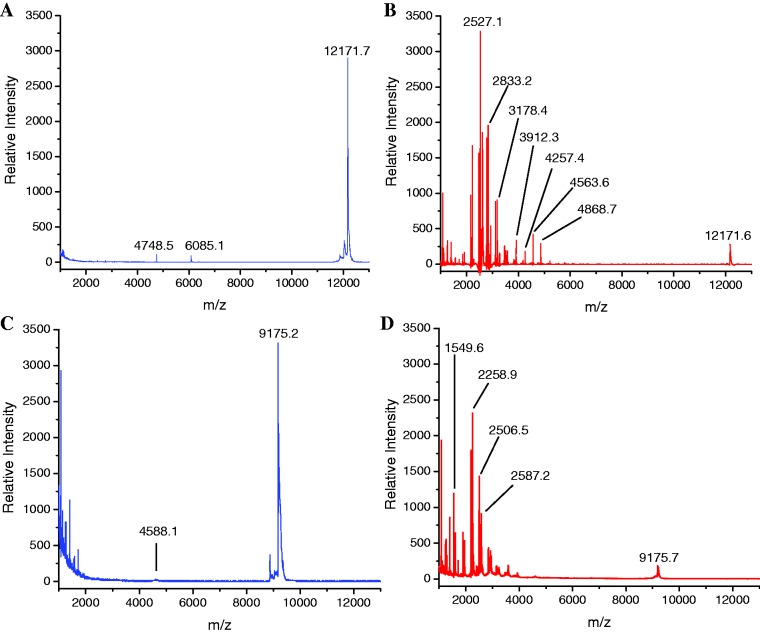
Catalyst-mediated RNA cleavage was performed by incubating each fluorescein-labeled RNA species (Fl-RRE and Fl-16S) with catalyst and redox co-reactants (H2O2 and/or ascorbate). Control incubations were performed that lacked either catalyst, co-reactants, or both. The RRE RNA (Fl-RRE) was incubated with redox co-reactants, with and without Fe–EDTA–Rev. The 16S RNA (Fl-16S) was incubated with redox co-reactants, with and without Cu–GGH. Figure 1 shows mass spectra resulting from these incubations. Time-dependent incubations were performed for several RNA cleavage reactions (in the range of 0–120 min). The 5′-fluorescein end-labels were included on each RNA construct for the purposes of detection in PAGE analysis, thereby allowing comparison of rates obtained by MALDI-TOF MS and PAGE analysis, as discussed in a later section. Cleavage of unlabeled RRE RNA was also studied for comparison (Supplementary Data).
Figure 1.
MALDI-TOF mass spectra for reaction mixtures after 1 h incubation of 10 µM Fl-RRE RNA (12172.5 Da), 1 mM H2O2 and 1 mM ascorbate with either (A) no catalyst or (B) 10 µM Fe–EDTA–Rev, and mass spectra for reaction mixtures after incubation of 10 µM Fl-16S RNA (9176.7 Da), 1 mM H2O2 and 1 mM ascorbate with either (C) no catalyst or (D) 10 µM Cu–GGH.
Analysis of RNA cleavage by MALDI-TOF MS
Following incubation, reaction mixtures were desalted using C18 ZipTips and analyzed by MALDI-TOF MS. Both catalyst-dependent cleavage and MALDI-induced background fragmentation of the RNA were observed. By use of the MassDaddy Perl script (Supplementary Data), the identities of primary cleavage products (resulting from single-cleavage events) were readily determined by automated mass-matching of the observed masses with the masses expected for each primary cleavage product at each position within the corresponding RNA sequence with a mass-matching tolerance of 200 ppm (0.02% of m/z), consistent with other reported mass accuracies for MALDI analysis of small oligonucleotides (28); the root-mean-squared mass-matching error was typically between 100 and 150 ppm (detailed mass-matching and error information shown in Supplementary Data). The apparent abundance of each species was semi-quantitatively determined using the peak area fractions, with a detection threshold peak area fraction of 0.0005 (Supplementary Data). The resulting peak area fractions were arranged by nucleotide position within the RNA and product overhang type (Figure 2), as discussed in a later section. Expected masses for all monitored cleavage products, from both Fl-RRE RNA and Fl-16S RNA, are listed in the Supplementary Data. Automated assignment was required due to the large amount of data collected, ∼216 monitored products per time point per Fl-RRE reaction. While typically 30–40% of the total integrated peak area for a spectrum was contained within assigned peaks, unassigned peak area was largely confined to peaks with low m/z (∼1000 amu), a multitude of peaks with very low individual peak areas, catalyst-related peaks, MALDI-TOF-related background peaks, or low-abundance Na+ and/or NH4+ adducts (not assigned).
Figure 2.
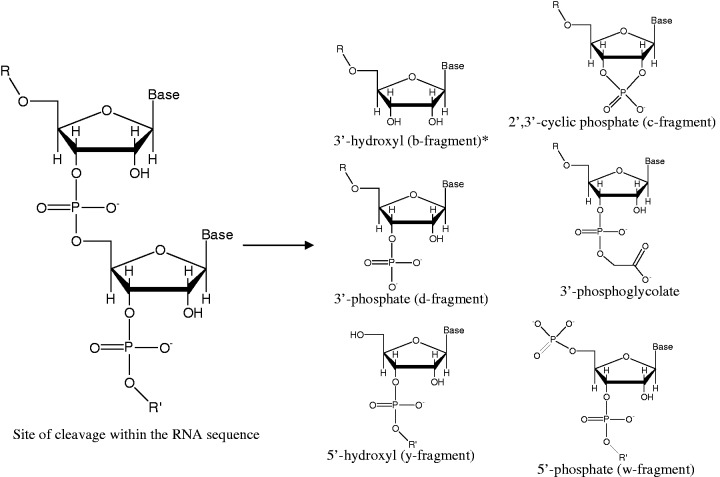
Observed overhangs at nascent termini of fragments resulting from RNA scission and/or fragmentation. R = 5′-fragment. R′ = 3′-fragment. Corresponding mass differences are shown in Table 2. Proposed mechanisms of formation of each overhang are shown in the Supplementary Data. *Monitored, but not a regularly observed cleavage product. An alternative nomenclature commonly used for the corresponding fragment ions in MS is shown where applicable (33–35).
Fragments of differing nucleotide lengths were observed, following RNA strand scission, as evidenced by sequences of peaks with unique mass differences corresponding to single nucleotide units (Table 1), which matched the sequences of the RNA constructs used (Figures 3 and 4). Additionally, each fragment contained one of several distinct overhang types (Figure 2) at the nascent 5′- and 3′-termini, adjacent to the respective site of strand scission, as evidenced by unique mass differences relative to fragments terminated with 5′-OH and 3′-OH groups (Table 2 and Figure 3). At the nascent 3′-termini of 5′-fragments, the observed overhangs were 2′,3′-cyclic phosphate (2′,3′-cPO4), 3′-phosphate (3′-PO4), and 3′-phosphoglycolate (3′-PG) functional groups; at the nascent 5′-termini of 3′-fragments, the observed overhangs were 5′-OH and 5′-PO4 functional groups (Figure 2). The masses of fragments containing nascent 3′-overhangs increased as the cleavage site moved in the direction 5′ to 3′, in increments corresponding to the mass of each nucleotide, while the masses of fragments containing nascent 5′-overhangs conversely increased in mass as the cleavage site moved in the direction 3′ to 5′, as illustrated in Figure 4.
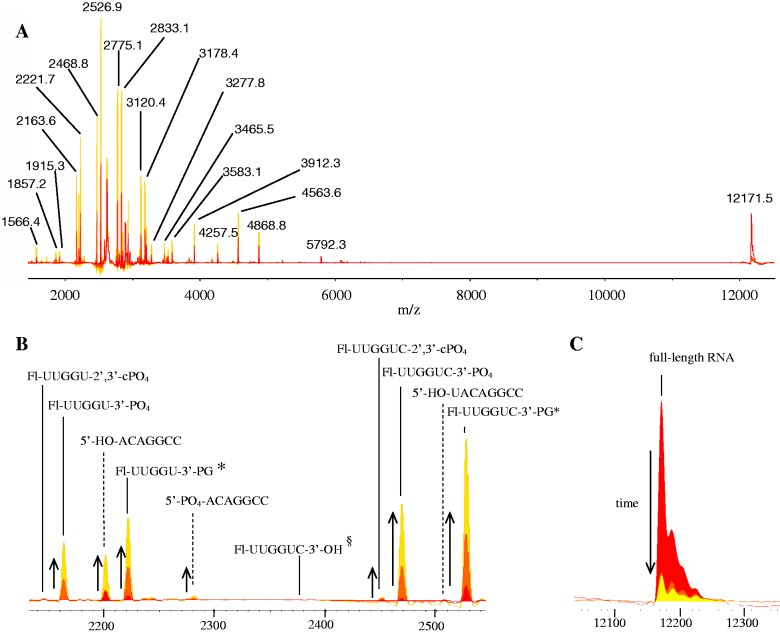
Figure 3.
MALDI-TOF mass spectra overlays and select product assignments for time-dependent cleavage of 10 µM Fl-RRE RNA by 10 µM Fe–EDTA–Rev, 1 mM ascorbate and 1 mM H2O2. (A) Full spectra, with m/z labels, (B) magnified spectra that show the formation of select cleavage products and (C) magnified spectra that show the disappearance of full-length Fl-RRE RNA. The incubation time-points shown here are 0, 2 and 5 min. The time-dependent appearance of cleavage products and disappearance of full-length RNA are indicated by arrows. The sequence of full-length Fl-RRE RNA is: 5′-Fl-UUG GUC UGG GCG CAG CGC AAG CUG ACG GUA CAG GCC-3′. Assigned fragments shown in (B) and (C) contain sequences shown in bold. *PG = phosphoglycolate. §The fragment with a nascent 3′-OH overhang was monitored but not observed.
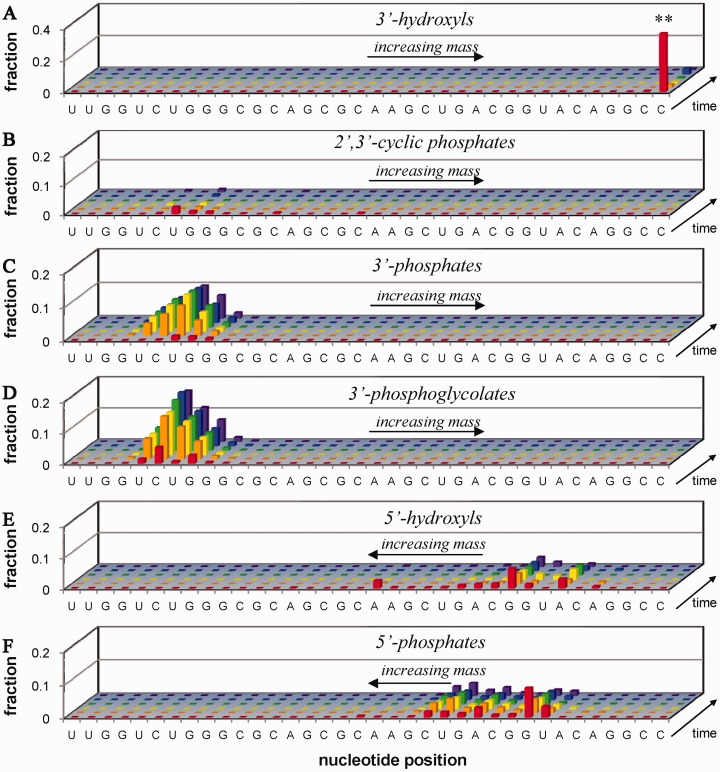
Figure 4.
Time-dependent cleavage of 10 µM Fl-RRE RNA (**) by 10 µM Fe–EDTA–Rev, 1 mM ascorbate and 1 mM H2O2 (see corresponding mass spectra in Figure 3). Cleavage fragments are organized by overhang type (A–F), location of the nascent overhang within the RNA sequence (5′ to 3′, left to right), and time-point within the incubation (0, 2, 5, 10, 60, 120 min, front to back). (A) 3′-hydroxyls (includes unreacted full-length Fl-RRE RNA), (B) 2′,3′-cyclic phosphates, (C) 3′-phosphates, (D) 3′-phosphoglycolates, (E) 5′-hydroxyls and (F) 5′-phosphates. From these data, apparent initial rates of formation were calculated for each product and are shown in the Supplementary Data, presented in the same layout as shown for cleavage of Fl-16S RNA by Cu–GGH/co-reactants in Figure 5C.
Time dependence of catalytic scission
Time-dependent incubations of each RNA with co-reactants, with and without each catalyst, were performed prior to MALDI-TOF MS analysis (Figures 3 and 4), and apparent initial rates of formation of each cleavage product and disappearance of the full-length RNA were determined (Supplementary Data). Product fragments with increasing abundance over the course of incubation were attributed to catalyst-mediated cleavage. Products that were observed at time zero and also had near-zero rates of formation during time-dependent incubations were attributed to either background fragmentation, most likely resulting from the MALDI-TOF MS procedure (discussed below), or cleavage that was too rapid to monitor by the techniques used.
Time-dependent experiments (Figures 3 and 4) possess the advantage of increased confidence in both the assignment and quantification of products, as opposed to single time-point experiments. Although the use of single-time-point experiments is less arduous and allows a more high-throughput approach, initial rates (calculated from the change of peak area fractions over time) provide a more accurate quantification of cleavage data that are less prone to skewing by mass bias and experimental variability. Time-dependent experiments provided a remarkable level of detail regarding the many sites of cleavage, the overhang types, and the approximate rates of formation of each cleavage product.
Background fragmentation
Background fragmentation of RNA species, which was distinct from the catalytic scission by metal complexes, was observed and resulted in the appearance of low levels of the endonucleolytic scission RNA fragments containing nascent 2′,3′-cyclic(PO4) or 5′-OH overhangs, which are well-known RNA fragments (c-fragments and y-fragments, respectively) that commonly result from gas-phase post-source decay (PSD) and/or collision-induced dissociation (33,35,42). These RNA fragments were observed by MALDI-TOF MS even in the absence of catalyst. No significant dependence of fragmentation on base composition was observed (Supplementary Tables SM5 and SM6). In some cases, the presence of catalysts appeared to enhance the formation of these endonucleolytic fragmentation products, during either the desalting, matrix crystallization, and/or MALDI-TOF MS processes. It should be noted that, in limited cases, the c- and y-fragments were also observed to slowly increase in abundance during time-dependent incubations, suggesting mixed causes for formation of these fragments (y-fragments are also proposed to result from hydrolysis). MALDI-PSD analysis has been used previously for RNA sequencing, by analysis of the mass differences between c- and y-fragments (27,32). However, following oxidative cleavage by ribonuclease mimics, we observed that RNA cleavage products containing nascent 3′-phosphate or 3′-phosphoglycolate overhangs, which appeared to arise primarily from catalyst-mediated cleavage, were present at much higher apparent abundance than the c- and y-fragments (potentially useful for RNA sequencing applications).
Limitations
We have observed several limitations to monitoring RNA cleavage by MALDI-TOF MS. The relationship between mass spectral peak areas and relative abundances of RNA fragments was considered only semi-quantitative, for several reasons. Detection was inherently biased toward RNA species with smaller mass (Supplementary Data), which appeared to result from a greater extent of MALDI-induced fragmentation for larger RNA fragments; this effect was partially accounted for during quantification, since the peak areas for artificial fragments were included in the denominators of all calculated peak area fractions (variation of peak areas over time were used to calculate rates of cleavage).
The desalting process was a critical part of each experiment, since removal of Na+ and other adduct-forming species was necessary for detection and quantification of RNA species. However, a tradeoff was observed that involved an incremental loss of smaller and/or less hydrophobic RNA fragments, such as those lacking a 5′-fluorescein end label, that increased with the number of washes with triethylamine acetate (Supplementary Data). Such sample loss was most likely due to a decreased retention of less hydrophobic RNA fragments on the ZipTip. For comparison, ethanol precipitation was used as a desalting method. Although ethanol precipitation appeared to allow identification of some larger RNA fragments that were not recovered following the ZipTip protocol, many of the smaller RNA fragments that were observed following the ZipTip protocol were completely lost during ethanol precipitation (Supplementary Data). Skewing of the apparent abundances by artifacts resulting from desalting and/or mass bias effects was not corrected for herein by any post-analysis calibration techniques, although the impact of these effects is limited as discussed later and in the Supplementary Data (Supplementary Figures SM4, SM5 and SM8).
The following limitation was more inherent to the nature of the experiment. For any given single cleavage event, a pair of primary fragments was expected. However, the apparent abundances of the two fragments were typically very different—the higher abundance fragment was typically that with the lower mass (Supplementary Data). While the aforementioned bias of detection toward fragments with smaller masses could explain this observation, the following explanation is more satisfactory. Since cleavage was typically observed to occur at multiple locations within each RNA, it is likely that the larger primary fragments were further cleaved (secondary cleavage events), at other known sites of cleavage, resulting in the observation of lower abundances of the larger primary cleavage fragments. This is consistent with the fact that larger RNA fragments are inherently more susceptible to secondary cleavage events and/or MALDI-induced fragmentation than are smaller RNA fragments. However, detection of secondary cleavage products was not performed, due to the dramatically increased number and low individual abundance of possible secondary cleavage products, as well as the increased complexity of peak assignment that results if secondary products are considered. While our spectral resolution (∼1000) was sufficient to resolve the ∼216 possible primary cleavage products, a higher-resolution (>50 000; common instruments are compared in Supplementary Figure SM40) mass spectrometer would be required to resolve the ∼2162 possible secondary cleavage products, which may explain a portion of the unassigned peak area seen in each mass spectrum.
An inherent limitation in the matching of MS peaks to expected-mass lists is that the expected-mass lists may not contain all possible products that are actually formed during RNA cleavage reactions, which could result in failure to identify a unique product not contained in the predicted-mass list. Appendages to expected-mass lists are easily made, so that these products may be detected, but the method is ultimately limited by the completeness of the user-defined list of expected products.
Validation of product assignments and semi-quantification
The identities of the detected primary cleavage products were readily assigned, based on both mass-matching within the defined tolerance of 200 ppm and abundance above the defined peak area fraction threshold of 0.0005 (Supplementary Data). Further validation of assignments was enabled by: (i) the ability to sequence the RNA construct in retrospect by use of the single-nucleotide mass differences between fragments (Table 1) with the same terminal overhangs, at different positions within the RNA sequence (Figures 3 and 4), (ii) the ability to verify the mass differences between different overhang products (Table 2), for fragments ending at a given nucleotide position within the RNA sequence (Figure 3) and (iii) monitoring the variation of peak area fractions for each product over time, which provided additional verification of products.
Comparison of MALDI-TOF MS versus PAGE analysis
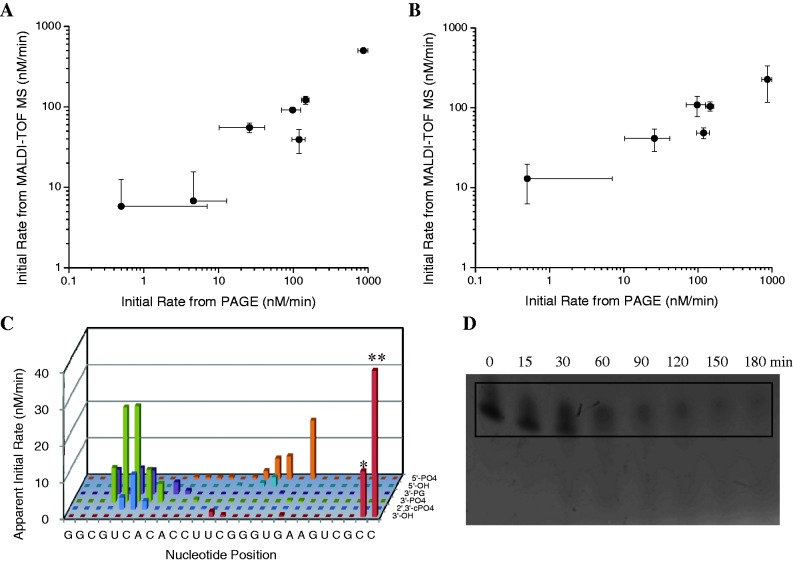
The ability to semi-quantitatively approximate the abundances, and also the rates of formation, of cleavage products and disappearance of full-length RNA was validated by comparison of initial rates determined by MALDI-TOF MS with those determined by 5′-fluorescein-monitored PAGE analysis of time-dependent RNA cleavage (Figure 5) for a set of reactions involving cleavage of the Fl-16S RNA by Cu-containing ribonuclease mimics, in the presence of redox co-reactants (mass spectra and PAGE data shown in the Supplementary Data). For PAGE analysis, disappearance of the full-length Fl-16S RNA was monitored over time, and the initial rate of cleavage was determined. For MALDI-TOF MS, two methods were used: either the initial rate was determined by summation of the initial rates of formation of all 5′ cleavage products (‘method 1’ shown in Figure 5A, where only positive rates were summed), or the initial rate of disappearance of full-length Fl-16S RNA was used (‘method 2’ shown in Figure 5B). The initial rates obtained by MALDI-TOF MS were generally in agreement with initial rates obtained by PAGE analysis (Figure 5). However, quantitative agreement is likely to dissipate for much larger RNA species, as a result of mass bias in MALDI-TOF MS; greater deviations were observed for rates of cleavage (MALDI versus PAGE) of the larger Fl-RRE RNA, although the MALDI-TOF MS-monitored rates were still semi-quantitative (Supplementary Data). Additionally, rates obtained for cleavage of Fl-RRE RNA by MALDI-TOF MS were similar to rates obtained by fluorimetry (Table SM7), using 2-aminopurine-labeled RNA (2).
Figure 5.
Comparison of MALDI-TOF MS versus 5′-fluorescein-monitored PAGE analysis for time-dependent cleavage of Fl-16S RNA by various ribonuclease mimics: quantification of initial rates (A and B) and information content (C and D). For MALDI-TOF MS, initial rates of cleavage were obtained for a series of reactions by use of either (A) method 1 or (B) method 2 (y-axes of A and B). For PAGE analysis, initial rates were obtained for the same reactions as the rate of disappearance of full-length RNA (x-axes of A and B), as shown in (D). For time-dependent cleavage of Fl-16S RNA by Cu–GGH/co-reactants, the MALDI-TOF MS technique (C) was able to resolve, identify, and provide apparent initial rates of formation of ∼100 individual cleavage products; apparent rates of disappearance of the full-length 27-mer Fl-16S RNA (**) and the 26-mer impurity (*) were 126 nM/min (abbreviated in figure) and 12 nM/min, respectively. For the same reaction, PAGE analysis (D) was unable to quantify individual species other than the full-length RNA (boxed). The rates shown in (C) were obtained from MALDI-TOF MS data similar to that shown for cleavage of Fl-RRE RNA by Fe–EDTA–Rev in Figure 4.
A distinct advantage of the MALDI-TOF MS technique, relative to PAGE analysis, is the far greater information content obtained by the MALDI-TOF MS method. To contrast the information that can be obtained by MALDI-TOF MS versus 5′-fluorescein-monitored PAGE analysis, time-dependent cleavage of Fl-16S RNA by the ribonuclease mimic Cu–GGH/co-reactants was monitored at each time point by both methods (in parallel), as shown by the comparison in Figure 5C and D. The MALDI-TOF MS technique provided a superior analysis, since it was able to resolve, identify, and approximate the individual rates of formation of ∼100 cleavage products, while the PAGE analysis could neither resolve nor identify any individual cleavage products (other than full-length RNA). Furthermore, even high-resolution PAGE analysis is both ill-suited for discrimination between nucleotides of the same length, but with differing overhangs (14,21,22), and prone to skewing of electrophoresis migration rates, which may be more prevalent for RNA than for DNA, due to the increased propensity of RNA to adopt secondary/tertiary structures (possibly even under denaturing conditions). The MALDI-TOF MS method circumvents these problems.
CONCLUSIONS
We have demonstrated a robust method to monitor catalyst-mediated scission of RNA by use of MALDI-TOF MS. The technique, which is adaptable to multiple RNA systems, allowed simultaneous identification and semi-quantification of the rate of formation of each RNA cleavage product within a complex mixture of RNA fragments. Additionally, the technique was able to easily distinguish (by mass) RNA fragments of the same nucleotide length, but with differing overhangs at nascent termini adjacent to cleavage sites (in contrast to PAGE analysis). Time-dependent incubations of RNA with ribonuclease mimics/co-reactants, combined with MALDI-TOF MS detection, allowed an approximate rate of formation to be established for each observed RNA cleavage product, allowing a detailed analysis of the reactivity between each ribonuclease mimic and RNA. Several limitations were observed and these included skewing of apparent abundances by mass bias and/or desalting processes. The ability of MALDI-TOF MS to resolve, provide exact identification and provide approximate quantification for hundreds of RNA products within complex mixtures provides a vast advantage over widely employed analytical separation techniques, such as gel electrophoresis or liquid chromatography. Notably, the various partial ribose overhangs formed upon RNA cleavage, monitored at each cleavage site, allow an assessment of the mechanisms of RNA cleavage, a major focus in studies of ribozymes, ribonucleases, ribonuclease mimics, chemical footprinting and RNA damage. The method presented herein provides a powerful analytical tool for use in these studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–7 and Supplementary Figures 1–40.
FUNDING
The National Institutes of Health [HL093446 and AA016712]; National Institutes of Health Chemistry/Biology Interface training [T32 GM08512, to J.C.J.]; Bruker Microflex instrument used for all MALDI-TOF analysis was provided by a grant from the Ohio BioProducts Innovation Center. Funding for open access charge: Departmental fund.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
J.C.J. thanks Dr Michael A. Freitas for sharing his expertise in the analysis of MS data. Seth Bradford provided useful advice during development of the MALDI-TOF MS methodology.
REFERENCES
- 1.Tung C-H, Wei Z, Leibowitz MJ, Stein S. Design of peptide-acridine mimics of ribonuclease activity. Proc. Natl Acad. Sci. USA. 1992;89:7114–7118. doi: 10.1073/pnas.89.15.7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joyner JC, Cowan JA. Targeted cleavage of HIV RRE RNA by rev coupled transition metal chelates. J. Am. Chem. Soc. 2011;133:9912–9922. doi: 10.1021/ja203057z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin Y, Cowan JA. Targeted cleavage of HIV rev response element RNA by metallopeptide complexes. J. Am. Chem. Soc. 2006;128:410–411. doi: 10.1021/ja055272m. [DOI] [PubMed] [Google Scholar]
- 4.Jin Y, Cowan JA. Cellular activity of Rev response element RNA targeting metallopeptides. J. Biol. Inorg. Chem. 2007;12:637–644. doi: 10.1007/s00775-007-0221-2. [DOI] [PubMed] [Google Scholar]
- 5.Prakash TP, Ganesh KN. Ribonuclease mimic: Zn2+ promoted cleavage of C8-histamino-r(UpA) proceeds through 2′,3′-cUMP as intermediate. Chem. Comm. 1994:1357–1358. [Google Scholar]
- 6.Mironova NL, Pyshnyi DV, Shtadler DV, Fedorova AA, Vlassov VV, Zenkova MA. RNase T1 mimicking artificial ribonuclease. Nucleic Acids Res. 2007;35:2356–2367. doi: 10.1093/nar/gkm143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breslow R, Anslyn E, Huang D-L. Ribonuclease mimics. Tetrahedron. 1991;47:2365–2376. [Google Scholar]
- 8.Bradford S, Cowan JA. Catalytic MetalloDrugs targeting HCV IRES RNA. Chem. Comm. 2012;48:3118–3120. doi: 10.1039/c2cc17377h. [DOI] [PubMed] [Google Scholar]
- 9.Bellacosa A, Moss EG. RNA repair: damage control. Curr. Biol. 2003;13:R482–R484. doi: 10.1016/s0960-9822(03)00408-1. [DOI] [PubMed] [Google Scholar]
- 10.Chan CM, Zhou C, Huang RH. Reconstituting bacterial RNA repair and modification in vitro. Science. 2009;326:247. doi: 10.1126/science.1179480. [DOI] [PubMed] [Google Scholar]
- 11.Aas PA, Otterlei M, Falnes PØ, Vågbø CB, Skorpen F, Akbari M, Sundheim O, Bjørås M, Slupphaug G, Seeberg E, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2002;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 12.Cowan JA. Catalytic metallodrugs. Pure Appl. Chem. 2008;80:1799–1810. [Google Scholar]
- 13.Hocharoen L, Cowan JA. Metallotherapeutics: novel strategies in drug design. Chem. Eur. J. 2009;15:8670–8676. doi: 10.1002/chem.200900821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pogozelski W, Tullius T. Oxidative strand scission of nucleic acids: routes initiated by hydrogen abstraction from the sugar moiety. Chem. Rev. 1998;98:1089–1107. doi: 10.1021/cr960437i. [DOI] [PubMed] [Google Scholar]
- 15.Bradford S, Kawarasaki Y, Cowan JA. Copper·Lys-Gly-His-Lys mediated cleavage of tRNAPhe: studies of reaction mechanism and cleavage specificity. J. Inorg. Biochem. 2009;103:871–875. doi: 10.1016/j.jinorgbio.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Thayer JR, McCormick RM, Avdalovic N. In: Methods In Enzymology, Part 2: High Resolution Separation and Analysis of Biological Macromolecules. Karger B, Hancock W, editors. Vol. 271. New York: Academic Press; 1996. pp. 147–174. [Google Scholar]
- 17.O'Neill K, Shao X, Zhao Z, Malik A, Lee ML. Capillary electrophoresis of nucleotides on ucon-coated fused silica columns. Anal. Biochem. 1994;222:185–189. doi: 10.1006/abio.1994.1471. [DOI] [PubMed] [Google Scholar]
- 18.Geldart SE, Brown PR. Analysis of nucleotides by capillary electrophoresis. J. Chromatography A. 1998;828:317–336. doi: 10.1016/s0021-9673(98)00633-5. [DOI] [PubMed] [Google Scholar]
- 19.Swerdlow H, Gesteland R. Capillary gel electrophoresis for rapid, high resolution DNA sequencing. Nucleic Acids Res. 1990;18:1415–1419. doi: 10.1093/nar/18.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murakami H, Borde V, Nicolas A, Keeney S. Gel electrophoresis assays for analyzing DNA double-strand breaks in Saccharomyces cerevisiae at various spatial resolutions. Meth. Mol. Biol. 2009;557:117–142. doi: 10.1007/978-1-59745-527-5_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henner WD, Grunberg SM, Haseltine WA. Sites and structure of gamma radiation-induced DNA strand breaks. J. Biol. Chem. 1982;257:11750–11754. [PubMed] [Google Scholar]
- 22.Henner WD, Rodriguez LO, Hecht SM, Haseltine WA. gamma Ray induced deoxyribonucleic acid strand breaks. 3' Glycolate termini. J. Biol. Chem. 1983;258:711–713. [PubMed] [Google Scholar]
- 23.Chaconas G, van de Sande JH, Church RB. End group labelling of RNA and double stranded DNA by phosphate exchange catalyzed by bacteriophage T4 induced polynucleotide kinase. Biochem. Biophys. Res. Commun. 1975;66:962–969. doi: 10.1016/0006-291x(75)90734-2. [DOI] [PubMed] [Google Scholar]
- 24.Angeloff A, Dubey I, Pratviel G, Bernadou J, Meunier B. Characterization of a 5'-aldehyde terminus resulting from the oxidative attack at C5' of a 2-deoxyribose on DNA. Chem. Res. Toxicol. 2001;14:1413–1420. doi: 10.1021/tx0100800. [DOI] [PubMed] [Google Scholar]
- 25.Spottke B, Gross J, Galla H-J, Hillenkamp F. Reverse Sanger sequencing of RNA by MALDI-TOF mass spectrometry after solid phase purification. Nucleic Acids Res. 2004;32:e97. doi: 10.1093/nar/gnh089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Limbach PA. Indirect mass spectrometric methods for characterizing and sequencing oligonucleotides. Mass Spectrom. Rev. 1996;15:297–336. doi: 10.1002/(SICI)1098-2787(1996)15:5<297::AID-MAS2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 27.Kühn-Hölsken E, Lenz C, Sander B, Lührmann R, Urlaub H. Complete MALDI-ToF MS analysis of cross-linked peptide-RNA oligonucleotides derived from nonlabeled UV-irradiated ribonucleoprotein particles. RNA. 2005;11:1915–1930. doi: 10.1261/rna.2176605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Limbach PA, Crain PF, McCloskey JA. Characterization of oligonucleotides and nucleic acids by mass spectrometry. Curr. Opin. Biotech. 1995;6:96–102. doi: 10.1016/0958-1669(95)80015-8. [DOI] [PubMed] [Google Scholar]
- 29.Hossain M, Limbach PA. Mass spectrometry-based detection of transfer RNAs by their signature endonuclease digestion products. RNA. 2007;13:295–303. doi: 10.1261/rna.272507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polo LM, Limbach PA. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for the analysis of RNase H cleavage products. J. Mass Spectrom. 1998;33:1226–1231. doi: 10.1002/(SICI)1096-9888(199812)33:12<1226::AID-JMS739>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 31.Meng Z, Limbach PA. Mass spectrometry of RNA: linking the genome to the proteome. Brief Funct. Genomic Proteomic. 2006;5:87–95. doi: 10.1093/bfgp/ell012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirpekar F, Douthwaite S, Roepstorff P. Mapping posttranscriptional modifications in 5S ribosomal RNA by MALDI mass spectrometry. RNA. 2000;6:296–306. doi: 10.1017/s1355838200992148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andersen TE, Kirpekar F, Haselmann KF. RNA fragmentation in MALDI mass spectrometry studied by H/D-exchange: mechanisms of general applicability to nucleic acids. J. Am. Soc. Mass Spectr. 2006;17:1353–1368. doi: 10.1016/j.jasms.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 34.Huang T-Y, Kharlamova A, Liu J, McLuckey SA. Ion trap collision-induced dissociation of multiply deprotonated RNA: c/y-ions versus (a-B)/w-ions. Am. Soc. Mass Spectrom. 2008;19:1832–1840. doi: 10.1016/j.jasms.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 35.Wu J, McLuckey SA. Gas-phase fragmentation of oligonucleotide ions. Int. J. Mass Spectrom. 2004;237:197–241. [Google Scholar]
- 36.Joyner JC, Keuper KD, Cowan JA. DNA nuclease activity of Rev-coupled transition metal chelates. Dalton Trans. 2012;41:6567–6578. doi: 10.1039/c2dt00026a. [DOI] [PubMed] [Google Scholar]
- 37.Joyner JC, Reichfield J, Cowan JA. Factors influencing the DNA nuclease activity of iron, cobalt, nickel, and copper chelates. J. Am. Chem. Soc. 2011;133:15613–15626. doi: 10.1021/ja2052599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Battiste J, Mao H, Rao N, Tan R, Muhandiram D, Kay L, Frankel A, Williamson J. Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex. Science. 1996;273:1547. doi: 10.1126/science.273.5281.1547. [DOI] [PubMed] [Google Scholar]
- 39.Daly TJ, Cook KS, Gray GS, Maione TE, Rusche JR. Specific binding of HIV-1 recombinant Rev protein to the Rev-responsive element in vitro. Nature. 1989;342:816–819. doi: 10.1038/342816a0. [DOI] [PubMed] [Google Scholar]
- 40.Lacourciere KA, Stivers JT, Marino JP. Mechanism of neomycin and Rev peptide binding to the Rev responsive element of HIV-1 as determined by fluorescence and NMR spectroscopy. Biochemistry. 2000;39:5630–5641. doi: 10.1021/bi992932p. [DOI] [PubMed] [Google Scholar]
- 41.Luedtke NW, Tor Y. Fluorescence-based methods for evaluating the RNA affinity and specificity of HIV-1 Rev-RRE inhibitors. Biopolymers. 2003;70:103–119. doi: 10.1002/bip.10428. [DOI] [PubMed] [Google Scholar]
- 42.Krivos KL, Addepalli B, Limbach PA. Removal of 3′-phosphate group by bacterial alkaline phosphatase improves oligonucleotide sequence coverage of RNase digestion products analyzed by collision-induced dissociation mass spectrometry. Rapid Commun. Mass Spectrom. 2011;25:3609–3616. doi: 10.1002/rcm.5266. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.