Abstract
The thermodynamic association of RNA polymerase (RNAP) with DNA is sensitive to salt concentration in vitro. Paradoxically, previous studies of changes in osmolarity during steady-state cell growth found no dependence between the association of RNAP to DNA and K+ concentration in Escherichia coli. We reevaluated this issue by following the interaction of RNAP and genomic DNA in time-course experiments during the hyper-osmotic response. Our results show that the interaction is temporally controlled by the same physical chemistry principle in the cell as in vitro. RNAP rapidly dissociates from the genome during the initial response when the cytoplasmic K+ accumulates transiently, and concurrently the nucleoid becomes hyper-condensed. The freed RNAP re-associates with the genome during a subsequent osmoadaptation phase when organic osmoprotectants accumulate as K+ levels decrease. RNAP first surrounds the hyper-condensed nucleoid forming a sphere of RNAP before it progressively moves in to the center of the nucleoid. Our findings reinterpret the dynamic protein–DNA interactions during osmotic stress response. We discuss the implications of the dissociation/association of RNAP for osmotic protection and nucleoid structure.
INTRODUCTION
Transcription by RNA polymerase (RNAP) depends on the interaction of RNAP and DNA. Because the electrostatic component of the binding free energy is of major importance, the thermodynamics of the association of RNAP with DNA (at promoters, non-specific sites and during promoter search along DNA) (1) is extremely sensitive to salt concentration, and RNAP dissociates from the DNA at high salt concentration in vitro (2–5). By analogy with in vitro studies, it would be expected that an increase in cytoplasmic K+ concentration induced by high osmolarity in the growth media should significantly perturb the interaction of RNAP with DNA in the living cells. However, studies with Escherichia coli cells grown in media of different osmolarities found that this correlation is missing: the association of RNAP and DNA, as determined indirectly by gene expression from promoters, is insensitive to the changes in cytoplasmic K+ concentration (6). This paradox has also been found for the interactions of other nucleic acid-binding proteins and DNA (6,7). To account for the discrepancy between the in vitro and in vivo studies, compensation mechanisms including (i) enhanced macromolecular crowding owing to a decrease of free cytoplasmic water at high osmolarity (8,9), (ii) decreased cytoplasmic putrescine (net charge 2+) that acts competitively with K+ (10) and (iii) increased cytoplasmic anion glutamate that stabilizes initiation complexes of promoters (11) have been postulated. A more general macromolecular crowding mechanism that DNA-binding proteins are always bound regardless of changes in environments (12) has also been proposed. However, analysis of the interactions of RNAP and DNA during hyper-osmotic stress response (for simplicity, hereafter called osmotic stress response) has not been determined.
Osmotic response is essential for cell survival in all kingdoms of life (13). In the E. coli model system, osmotic shock such as exposure to high salt induces a stress response, which involves sequential changes in physiology and gene expression (14–20). Plasmolysis occurs immediately after osmotic stress (21) owing to water leaking out of the cell, which results in reducing the effective concentration of cytoplasmic water (22,23) and growth arrest. To prevent dehydration damage and maintain the osmotic or turgor pressure, exogenous K+ from the media is rapidly transported into the cell, resulting in rapid accumulation of cytoplasmic K+ within minutes (8,14). The K+ uptake is mediated by several transporter systems, most of which are constitutively expressed, but some are induced by changes in turgor pressure (24). To compensate the net charge increase caused by the K+ accumulation, glutamate (net charge −1) is either imported from the media or synthesized de novo, and putrescine (net charge +2) is exported from the cell; thus, the net charge of the cytoplasm is preserved (6,10,14,25). The rapid accumulations of cytoplasmic K+ and glutamate occur almost simultaneously at the outset of osmotic shock (14); however, the concentration of K+ is in substantial excess over glutamate (8). Potassium glutamate inhibits rRNA synthesis during the osmotic response (26) and activates osmotic stress response genes (27). Because K+ is not a compatible solute (high concentration inhibits cell growth), it provides only temporary protection during the initial osmotic stress response.
The subsequent osmoadaptation phase involves the accumulation of compatible organic osmotic solutes (osmolytes) (28) or long-term osomoprotectants, which are small molecules including proline, glycine betaine and trehalose. They can be transported from the medium or synthesized de novo (13,14,29). New protein synthesis is required for these transport systems during the response, as chloramphenicol prevents their accumulation (14). Once present, these osmoprotectants promote the efflux of the K+ to a lower steady-state level characteristic of the osmolarity. The cytoplasmic glutamate level diminishes more gradually (14). The length of the time needed to accumulate osomoprotectants and resume cell growth depends on the intensity of the shock and the type of media (14,30).
A direct measurement of the association of RNAP with the genomic DNA during the dynamic process of osmotic stress response has not been performed in E. coli. Previously, using a fluorescent tagged RNAP, we showed that E. coli RNAP is associated with the bacterial chromosome named nucleoid, and its distribution is sensitive to environmental stimuli (31). Here, we examine the effect of osmotic stress on the distribution of RNAP and determine the interaction of RNAP with the nucleoid in the cell. Our results show that the association of RNAP with the genome is temporally controlled and sensitive to the accumulation of cytoplasmic K+ concentration.
MATERIALS AND METHODS
Bacterial strains, bacterial growth and techniques
The strains used in the study are derivatives of MG1655. The CC72 strain containing a chromosomal rpoC-venus gene fusion was constructed with modifications as described (31). Briefly, the kan gene cassette conferring kanamycin resistance from the pKD4 plasmid (32) was PCR amplified using the primers P1F1 (5′ATGCGAATTCGTGTAGGCTGGAGCTGCTTC3′) and P2R1 (5′ATGCCTCGAGCATATGAATATCCTCCTTAG3′). The PCR product was digested with restriction enzymes EcoRI and XhoI and inserted into the cognate restriction sites of the pCS2-Venus plasmid (33), resulting in the pCC61 plasmid that contains the cassette downstream of the venus gene. A PCR product that contained DNA sequences near the end of the rpoC gene, the venus-kan region of pCC61 and the rpoC downstream region, was amplified using the primers rpoC-venus F1 (5′CCAGCCTGGCAGAACTGCTGAACGCAGGTCTGGGCGGTTCTGATAACGAGCTAGAAATAGTGAGCAAGGGCGAGGAGCT3′) and rpoC-venus R1 (5′CCCCCCATAAAAAAACCCGCCGAAGCGGGTTTTTACGTTATTTGCGGATTACATATGAATATCCTCCTTAG3′). Using this PCR fragment, the chromosomal copy of the rpoC gene was replaced by recombineering (34), and the kanamycin-resistant recombinants were selected. The rpoC-venus <> kan marker was then introduced into MG1655 by P1 transduction, resulting in the strain CC49 (rpoC-venus <> kan). Unlike our previous rpoC-gfp construct (31), which renders cells temperature sensitive for growth, the CC49 strain has a wild-type growth phenotype. The kan gene cassette in the CC49 strain was removed by FLP-mediated recombination as described (32), resulting in the strain CC72 (rpoC-venus). These constructions were confirmed by PCR and/or DNA sequencing. The CC241 strain is a derivative of the TK2420 strain [(ΔkdpFAB)5 Δ(trkA-mscL′) trkD1] (35) into which the kan-linked rpoC-venus protein fusion from CC49 was moved in by P1 transduction. The CC287 strain is a derivative of CC72 containing the double mutations ΔproP737 and ΔbetT756::kan. This strain was constructed in multiple steps by two P1 transductions using the corresponding kan-linked mutations from the Keio collection (36) as donors; the kan gene cassette in the first transductant (ΔproP737::kan) was removed by FLP-mediated recombination (32) before the ΔbetT756::kan mutation was moved in by a second P1 transduction.
The bacterial media and techniques are described (37). Unless otherwise noted, cells were grown to early log (or exponential) phase in LB [tryptone 10 g/l, yeast extract 5 g/l, NaCl 5 g/l (∼0.09 M)] at 37°C. The CC241 strain cells, defective in most of the K+ transporter systems and requiring extra KCl in the media for growth, were grown in LB supplemented with 50 mM KCl. To induce hyper-osmotic response, NaCl was rapidly dissolved into a rapidly growing culture at OD600 ∼0.2 to a final concentration of 0.59 M. An osmolarity of about 0.26 Osm/L is calculated from the salt (0.09 M NaCl) in LB, and the value increases to about 1.18 Osm/L with 0.59 M NaCl (8). Samples were taken immediately before the addition of salt (time 0) and at successive time points thereafter. When indicated, the media was supplemented with rifampicin (rif) 100 mg/l. The M63-K (a K+-free medium) is a modified M63 medium in which KH2PO4 has been replaced by NaH2PO4.
Potassium downshift experiment
Cells (OD600 ∼0.2) grown in LB were pelleted and washed twice with warm (37°C) M63-K (K+-free) medium containing 0.2% glucose to eliminate residual K+ from the LB, followed by re-suspension in the same volume of the wash medium (time 0). For the controls, M63 containing 0.2% glucose was used instead of M63-K. The cells were incubated at 37°C, and the osmotic stress response was monitored as described.
Microscopy
The microscopy was done as previously described (38). Briefly, the cells were fixed using formaldehyde (1–3.7% v/v final) owing to the metastable nature of the transcription foci (31) before being mounted on a microscope slide for imaging. An inverted microscope (Zeiss Axio Observer) with a Plan Apochromat 100×/1.4 oil phase objective and a 1.6 Optovar was used for microscopy. Images were captured with a high signal to noise ratio and high resolution EMCCD camera (Hamamatsu). Three images were taken for each sample: phase contrast for cell shape, YFP channel for RNAP-Venus and DAPI channel for DNA. Each experiment was performed at least three times, and in total, >100 cells for each condition were imaged. The cells illustrated are representative of the majority (>90%) of the observed cells unless otherwise noted. Pictures were processed uniformly and false coloured with Adobe Photoshop.
Nucleoid isolation
Immediately after sampling, the cells were fixed for 20 min at room temperature with formaldehyde (1% v/v final). The cells were centrifuged at 5000g for 5 min, resuspended in 50 µl of 10 mM Tris (pH 8.0), 10 mM EDTA, 20% sucrose, 0.2 µl Ready-lyse lysozyme (Epicenter), and incubated for 30 min at 37°C. The cell lysis was completed by the addition of 50 µl of water. After lysis, a 2 µl aliquot of the lysate containing nucleoids was dropped onto a microscopy slide with 2 µl of 1% warm low melting point agarose + 10 µg/ml Hoescht 33342. The slides were imaged immediately with YFP channel for RNAP-Venus and DAPI channel for DNA.
Determination of the nucleoid-associated proteins during the osmotic stress response
Immediately after sampling, the cells were fixed for 20 min at room temperature with formaldehyde (1% v/v final), then centrifuged at 5000g for 5 min and suspended in 200 µl of 10 mM Tris (pH 8.0), 50 mM NaCl, 10 mM EDTA, 20% sucrose, 0.2 µl Ready-lyse lysozyme (Epicenter), with continued incubation for 30 min at 37°C. The cell lysis was completed by the addition of 200 µl of 200 mM Tris (pH 8.0), 600 mM NaCl, 4% Triton X-100, 1 mM PMSF and incubated 10 min at 37°C. Two hundred microlitres of the lysate was removed and kept as the total protein input while the remaining sample was centrifuged for 10 min at 20 000 g at 4°C. After centrifugation, the pellet mainly contains the nucleoid-associated proteins while the supernatant contains the unbound or soluble proteins. The pellet was resuspended in 200 µl of 105 mM Tris (pH 8.0), 325 mM NaCl, 5 mM EDTA, 10% sucrose, 2% Triton X-100, 0.1 µl Ready-lyse lysozyme (Epicenter) and 0.5 mM PMSF. All three fractions, total protein input, pellet and supernatant, were TCA precipitated, and the precipitate was resuspended in 100 µl of 10 mM Tris (pH 8), 1 mM EDTA, 5% SDS and 20% glycerol. They were incubated for 6 h at 65°C to reverse the crosslinking, followed by the addition of 2,2,2 trifluoroethanol (20% v/v final) to completely solubilize the proteins. The same volumes from each of the three fractions were loaded into a 4–12% acrylamide gel for electrophoresis, and the proteins were identified by Western blot using specific antibodies. E. coli RNAP β′ subunit monoclonal antibody (NT73) was purchased from NeoClone (Madison, WI, USA). The polyclonal antibodies for H-NS were a gift from Dr Darren Sledjeski and for IHF were kindly provided by Dr Steven D. Goodman.
RESULTS
Rational and experimental design
Previously the effects of high osmolarity on the interaction of RNAP with DNA were determined during steady-state growth of E. coli cells in minimal media with different osmolarities. Because osmotic response is a dynamic process involving sequential changes, we decided to follow the interaction of RNAP with the genomic DNA in time-course experiments during the osmotic stress response. To achieve a fast response during optimal growth conditions, nutrient-rich LB was used instead of minimal media to minimize requirements for de novo synthesis of the needed organic osmoprotectants. Another advantage of using fast-growing cells in LB at 37°C is to address whether the response to osmotic stress, which inhibits rRNA synthesis (26), affects the transcription foci (31) or factories (39), in which the majority of the RNAP molecules are concentrated in rRNA transcription (40). Such an optimal growth condition becomes possible with the newly constructed rpoC-venus fusion (CC72).
RNAP rapidly dissociates from the genomic DNA during the initial osmotic stress response while cytoplasmic K+ accumulates
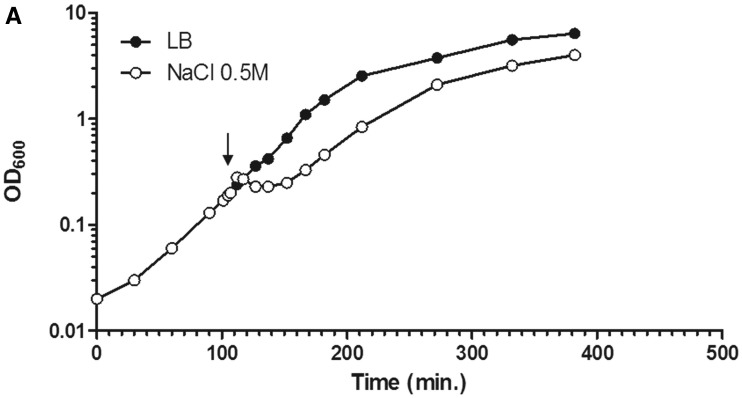
The cells (CC72) were grown in LB at 37°C with a doubling time of 20 min, and the osmotic stress response was induced by the addition of 0.5 M NaCl into the culture at OD600 0.2. Figure 1 shows the growth curve and images of cells before and at different time points after the salt addition. At the earliest time point (5 min) after the salt shock, there is a reproducible slight increase in OD600 compared with the untreated cells, which is quickly followed by a decline in OD600 and growth arrest (Figure 1A). After a lag of ∼30–45 min, cell growth resumes with a slightly reduced growth rate compared with that of untreated cells, similar to previous reports (26,30). The growth curve reflects the sequential changes in bacterial physiology during the stress response. For example, the increase in OD600 5 min after the salt shock signifies a decrease in cytoplasmic volume caused by water loss (14,30). Plasmolysis is apparent at this early time point as indicated by the cytoplasmic membrane collapse away from the cell wall of many cells. This effect is no longer visible 10 min after the shock, suggesting that the efficient K+ influx systems lead to rapid accumulation of cytoplasmic K+ and regaining of the lost water (data not shown).
Figure 1.
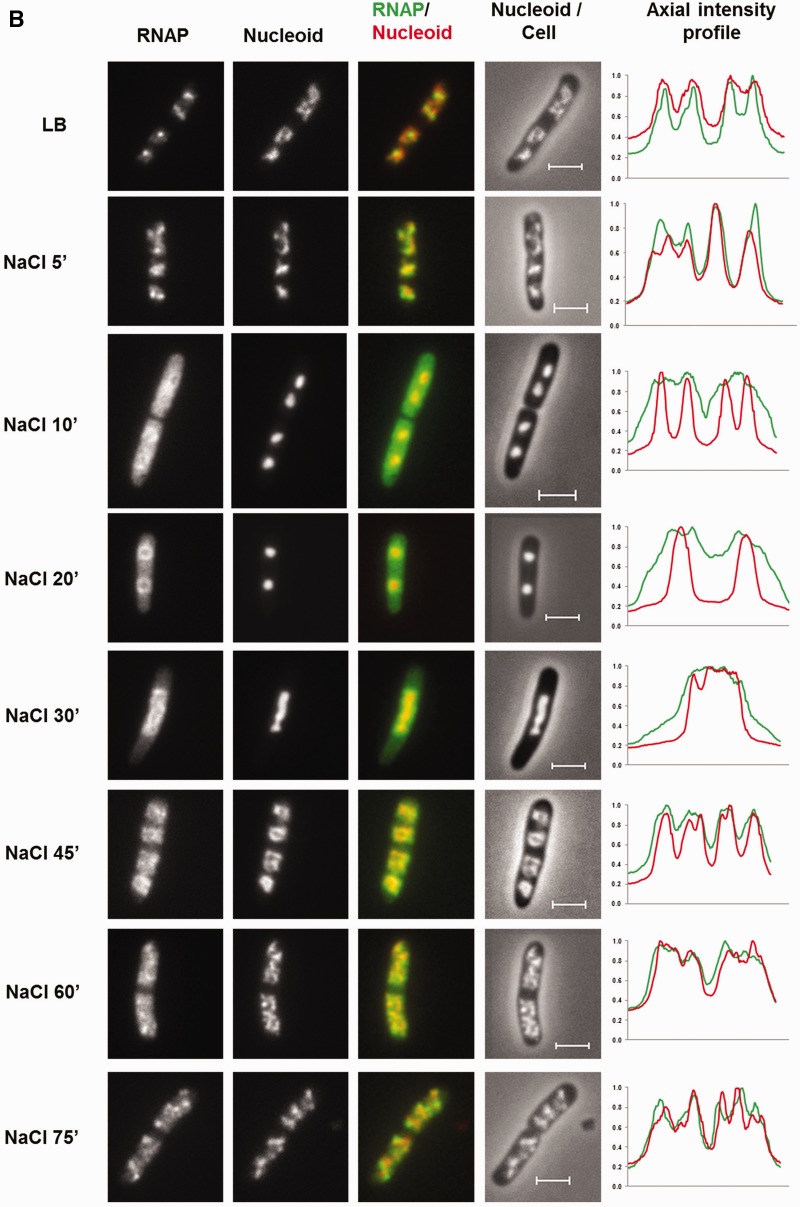
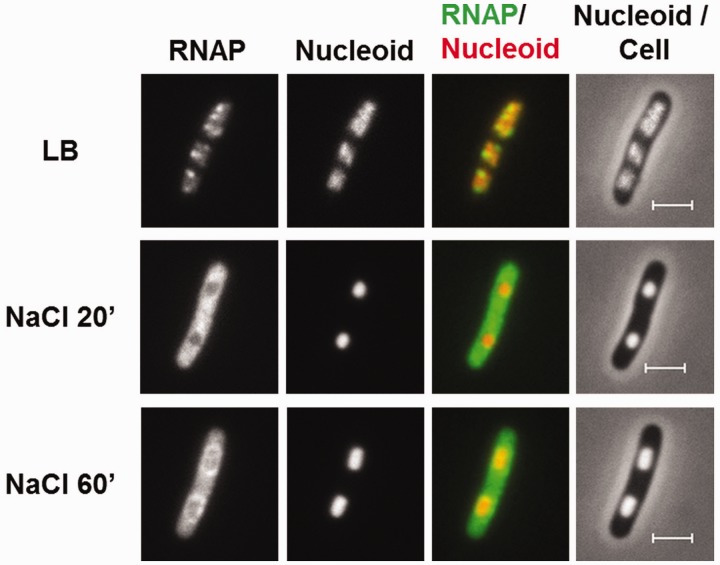
Dynamic interaction of RNA polymerase and DNA during the osmotic stress response. (A) Growth curve of CC72 cells in LB (close circle) and after osmotic shock in mid log phase (open circle). The arrow indicates the induction of the hyperosmotic response.(B) Distribution of RNAP and nucleoid structure during hyper-osmotic response. RNAP-venus protein fusion, nucleoid, overlay of RNAP (green) and nucleoid (red), overlay of nucleoid (white) and cell (black) before (LB) or after osmotic shock (NaCl 5′ to NaCl 75′). Axial intensity profile for RNAP (green) and DNA (red) from the cell shown is also included, in which cumulative relative intensity from the short axis of the cell (y-axis) is plotted against the position along the long axis (pole-to- pole) of the cell (x-axis). The RNAP rapidly dissociates from the nucleoid, and the released RNAP occupies the entire cytoplasmic space shortly after osmotic shock (NaCl 10′). The nucleoid becomes hyper-condensed at the same time. Afterwards, the freed RNAP gradually re-associates with the genome over time by first forming a ring (a 2-D picture) at the surroundings of the hyper-condensed nucleoid (NaCl 10′ to NaCl 30′) before penetrating the interior (NaCl 45′). (C) The nucleoids released from the lysed cells in LB, 20 (NaCl 20′) and 60 min (NaCl 60′) after salt addition maintain the same features as that in the intact cells shown in (B). RNAP, nucleoid and overlay of RNAP (green) and nucleoid (red) are shown. The bar scale represents 2 µm.
The rpoC-venus fusion, which is brighter than rpoC-gfp, coupled with a better imaging system, allows us to observe RNAP and obtain images with greater details and sharpness compared with our previous studies. Both the distribution of RNAP and the interaction of RNAP with the genomic DNA change during the osmotic stress response. Before the salt addition, RNAP forms distinct transcription foci for rRNA synthesis in the nucleoid, which is a signature of the cells growing in optimal conditions, and the overall intensities of RNAP in the remaining vast majority areas of the nucleoid are generally low. The nucleoid is relatively compact in these cells, and it exhibits variations in intensities including apparent voids. Such features have not been described previously (41). At the growth conditions used, each cell contains, on average, four ‘nascent’ nucleoids, two of which are located in each half of the long axis of the cell. Only 5 min after the salt addition, the transcription foci diminish, redistributing RNAP into other areas of the nucleoids, and by 10 min (Figure 1B, NaCl 10′) the transcription foci totally disappear, consistent with the report that rRNA synthesis is inhibited shortly after salt shock (26). After 10 min, significant amounts of RNAP dissociate from the nucleoid; free RNAP molecules relocate and occupy the entire cytoplasmic space. Concurrent with depletion of RNAP from the genome, the nucleoid transforms to a hyper-condensed structure, and the two ‘nascent’ nucleoids in each half of the cell are fused as described (42). At 20 min (Figure 1B, NaCl 20′), RNAP is concentrated at the periphery of the hyper-condensed nucleoid, suggesting that those RNAP molecules are DNA-associated. Our examination of the nucleoids released from the lysed cells after the osmotic stress for 20 min confirms the localization of RNAP at the surface of the nucleoid (compare Figure 1B and C, NaCl 20′). In contrast to the intact cells, the cytoplasmic pool of RNAP molecules is undetected because the unbound RNAP is diluted out during the nucleoid isolation. Because global transcription is active during osmotic stress response (17,19), it is conceivable that many of the expressed genes including those involved in osmotic stress response are located as loose DNA loops surrounding the hyper-condensed nucleoid.
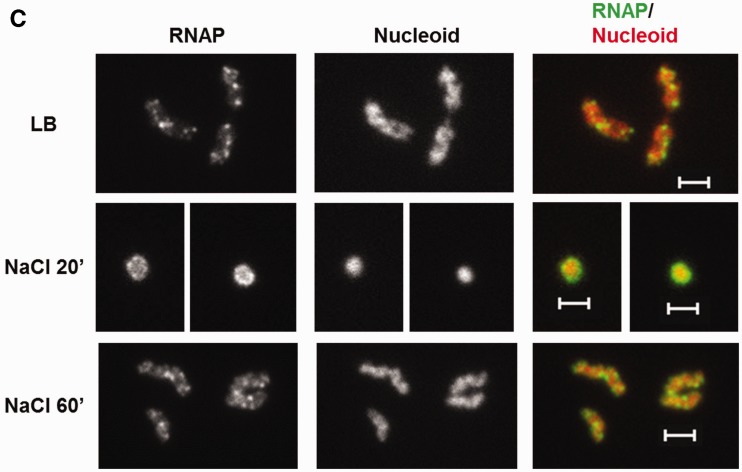
During exponential growth in LB, most of RNAP molecules in the cell are engaged in rRNA synthesis. Because rRNA synthesis is inhibited during initial salt shock (26), it is very likely that RNAP molecules engaged in rRNA synthesis before osmotic stress have contributed to the pool of the cytoplasmic RNAP. Nonetheless, to determine whether the free RNAP is exclusively from the shutdown of the rRNA synthesis during the osmotic stress response, we treated early-log cells with rifampicin (30 min) to inhibit all transcription including rRNA synthesis, and subsequently added salt (0.5 M NaCl, 20 min) to induce the osmotic stress response (Figure 2A). Although rifampicin inhibits transcription initiation by RNAP, it does not block binding of the enzyme to DNA (43,44). Compared with the untreated cells, rifampicin frees up the transcribing RNAP and significantly increases the number of locations occupied by RNAP in the nucleoid, reflecting genome-wide redistribution of RNAP by the antibiotic (45). As previously reported (38), in the rifampicin-treated cells (LB + rif), RNAP associates with the nucleoid, and the nucleoid becomes relatively expanded with significantly reduced cytoplasmic space surrounding the nucleoid compared with untreated cells (LB). Significantly, however, when the rifampicin-treated cells were subjected to osmotic stress (LB + rif + NaCl), significant amounts of RNAP are released from the nucleoid into the cytoplasmic space and the nucleoid contracts significantly as indicated by the increased cytoplasmic space. These results indicate that the released RNAP is not merely from those engaged in rRNA synthesis before the salt addition; rather, dissociation of bound RNAP from the DNA is a general response to the osmotic stress and is independent of transcription.
Figure 2.
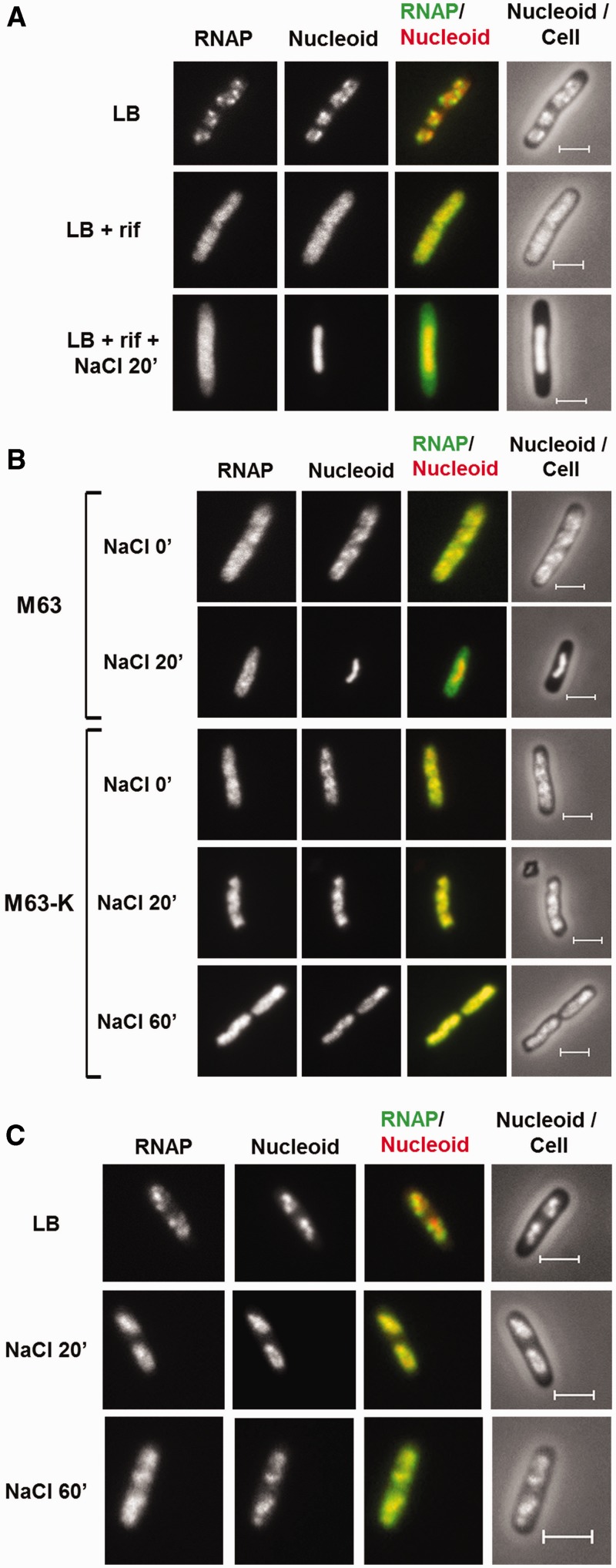
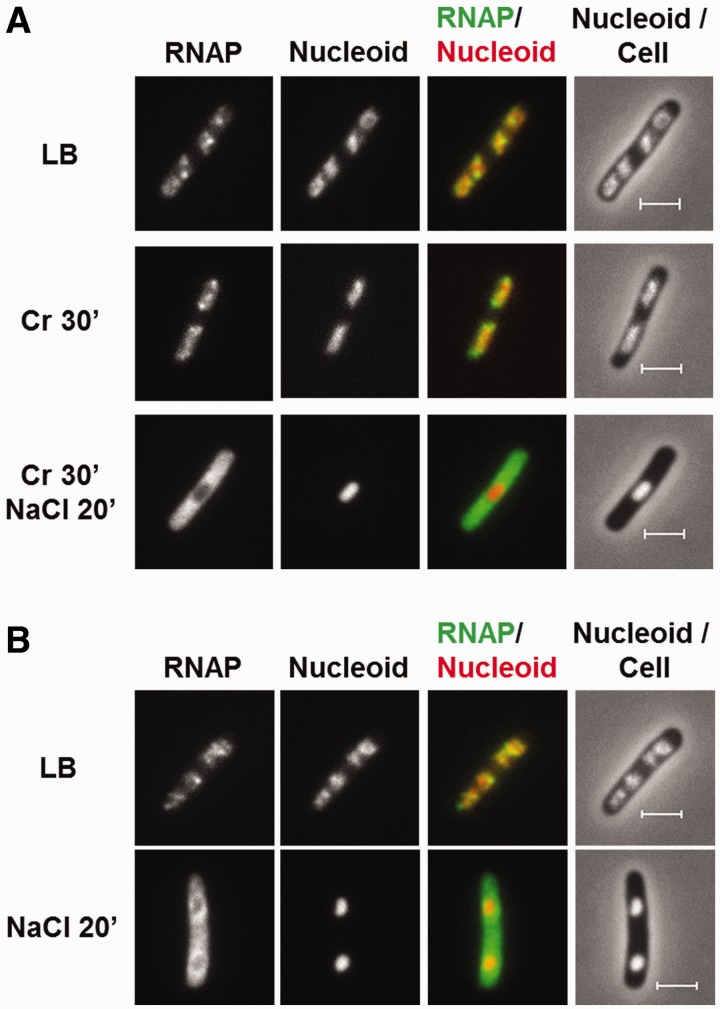
Effects of transcription inhibition and K+ import on the dissociation of RNAP from the nucleoid. (A) RNAP dissociation during osmotic response in rifampicin-treated cells. RNAP-venus protein fusion, nucleoid, overlay of RNAP (green) and nucleoid (red), overlay of nucleoid (white) and cell (black) before the treatment (LB), treated with rifampicin for 30 min only (LB + rif), and sequential treatments of rifampicin (30 min) and salt (20 min) (LB + rif + NaCl 20'). (B) K+ import is required for RNAP dissociation. RNAP-venus protein fusion, nucleoid, overlay of RNAP (green) and nucleoid (red), overlay of nucleoid (white) and cell (black) before (NaCl 0′) or after (NaCl 20′ and NaCl 60′) osmotic shock in the absence (M63-K) or presence (M63) of K+. (C) RNAP is not dissociated from the nucleoid in a K+ import defective strain (ΔkdpFAD ΔtrkAD) during osmotic stress response in LB. RNAP-venus protein fusion, nucleoid, overlay of RNAP (green) and nucleoid (red), overlay of nucleoid (white) and cell (black) before (LB) or after (NaCl 20′ and NaCl 60′) osmotic shock. The bar scale represents 2 µm.
Potassium uptake is essential for the dissociation of RNAP from the nucleoid
To confirm a role for K+ in the release of RNAP, we performed two sets of experiments. First, we induced the osmotic stress response in minimal medium either with (M63) or without K+ (M63-K) (Figure 2B). The nutrient downshift from LB (before osmotic shock) to minimal medium induces the redistribution of RNAP in the chromosome and causes nucleoid expansion (NaCl 0′) (31,46). Similarly to LB, after addition of NaCl to the M63 medium, RNAP dissociates from the nucleoid as indicated by the appearance of the cytoplasmic RNAP, and the nucleoid becomes highly condensed. In contrast to M63, in M63-K medium there is no apparent release of RNAP from the DNA and the nucleoid remains expanded even 60 min after salt addition, although plasmolysis is apparent 20 min after the salt addition (data not shown). These results indicate that K+ is critical for the dissociation of RNAP.
To eliminate complications owing to nutrient downshift into minimal media, we constructed the CC241 strain, a derivative of the TK2420 strain (35) that is deleted for the major K+ uptake systems (ΔkdpFAD ΔtrkAD). We examined these transporter mutant cells during the initial osmotic stress response in LB supplemented with 50 mM KCl, and found that in contrast to wild type, there is no apparent dissociation of RNAP from the DNA and no apparent changes in nucleoid compaction after NaCl addition (compare Figures 1B and 2C, NaCl 20′). Together, these results indicate that RNAP dissociates from the DNA as a consequence on the increase in intracellular K+. The dissociation of RNAP also corresponds to the reported time of peak accumulation of the cytoplasmic K+ after salt addition (14). We conclude that RNAP dissociates from the genomic DNA during the initial osmotic stress response at the time cytoplasmic K+ concentration increases rapidly. This result agrees well with that of the in vitro results of salt effects on the interaction of RNAP with DNA (2–5).
The stepwise re-association of RNAP with the nucleoid during the osmoadaptation phase
The cell starts to release accumulated cytoplasmic K+ by 20–25 min after the salt shock during the osmoadaptation or recovery phase when organic osmoprotectants accumulate (14). At this same time, cytoplasmic RNAP begins to re-associate with the nucleoid in several distinct steps. After 20 min (Figure 1B, NaCl 20′) although RNAP still occupies the cytoplasmic space, RNAP concentrates at the periphery of the fused condensed nucleoid appearing as a distinct ring in a 2-D picture, indicating that RNAP forms a sphere at the periphery of the nucleoid, and by 30 min (Figure 1B, NaCl 30′) the majority of RNAP binds to the surface of the condensed nucleoid, some of which appears to split into four ‘nascent’ nucleoids. Gradually, RNAP penetrates the nucleoid, and concomitantly the nucleoid expands progressively to resemble a global structure observed before salt addition. Transcription foci that are indicative of active rRNA synthesis become evident only after 75 min when steady-state (exponential) growth is re-established, consistent with the time frame reported for recovery of the rRNA synthesis after osmotic stress (26).
The relatively long time needed for the re-association of the freed RNAP with the nucleoid during recovery period may include time for the synthesis of transporters required for accumulation of the osmoprotectants necessary for the K+ efflux. In addition, the diffusion rate for the free RNAP in a macromolecular crowding environment of the cytoplasm is likely to be slower than that of the DNA-associated RNAP (47) and the interior of the hyper-condensed nucleoid may not be easily accessible to RNAP. We speculate that re-association of RNAP with the genome and remodelling of the highly condensed nucleoid are integral parts of the osmoadaptation process.
Proline and glycine betaine are the major osmoprotectants that can be transported from LB by their respective transporters. We determined the effects of the deletion of the genes for the proline/glycine betaine (proP) and choline (betT) (a precursor for the glycine betaine) transporters on the re-association of RNAP during the osmotic stress response in LB (Figure 3). The double mutation (ΔproP and ΔbetT) causes a significant delay in the re-association of RNAP compared with wild type (Figure 1B). After 20 min there is no RNAP associated with the surface of the hyper-condensed nucleoid in the mutant cells; only after 60 min RNAP appears to be concentrated in the periphery of the hyper-condensed nucleoid. These results indicate that under the condition used, the uptake of proline, glycine betaine and choline are important for the re-association of RNAP with the nucleoid. The accumulation of the cytoplasmic proline and glycine betaine from LB promotes the K+ efflux system to reduce the cytoplasmic K+ concentration, which in turn facilitates the re-association of RNAP with the nucleoid.
Figure 3.
The re-association of RNAP with the nucleoid is delayed in the double deletion mutant (ΔproPΔbetT) defective in the accumulation of organic osmoprotectant. RNAP-venus protein fusion, nucleoid, overlay of RNAP (green) and nucleoid (red), overlay of nucleoid (white) and cell (black) before (LB) or after (NaCl 20′ and NaCl 60′) osmotic shock. Note that the appearance of the ring of RNAP surrounding the hyper-condensed nucleoid is delayed compared with wild type (Figure 1B). The scale bar represents 2 µm.
Other nucleoid-associated proteins behave similarly to RNAP during the osmotic stress response
To determine whether the interaction of other nucleoid-associated proteins (NAPs) with DNA is affected during osmotic stress response, we examined the localization of two ‘histone-like’ NAPs (H-NS and IHF) during the initial stress and recovery period. Specially, we looked at time 0 immediately before the salt addition, after 20 min when the dissociation of RNAP is apparent and after 60 min when RNAP has re-associated with the nucleoid. After cells were treated with formaldehyde to cross-link NAPs to DNA and lysed, total cell lysates (T) from each time point were fractionated into soluble (S), which represents cytoplasmic fraction of unbound and weakly bound proteins, and nucleoid bound (N), which represents proteins stably associated with the nucleoid. After de-crosslinking, the proteins were separated by gel electrophoresis and identified by western blot using specific antibodies.
Figure 4 shows that RNAP, as indicated by the β′ subunit staining, localizes exclusively in the nucleoid in cells growing in LB, with no detectable signal from the soluble cytoplasmic fraction. This is expected as RNAP is a strong DNA-binding protein. The transient redistribution of RNAP to the cytoplasm is clearly visible at 20 min after the salt addition. This result confirms that the cytoplasmic RNAP is indeed released from the nucleoid as opposed to being associated with some external DNA loops extruded to the cytoplasm from the nucleoid surface. The freed RNAP then re-associates with the nucleoid 60 min after the salt addition. These data are in complete agreement with the image analyses of Figure 1.
Figure 4.
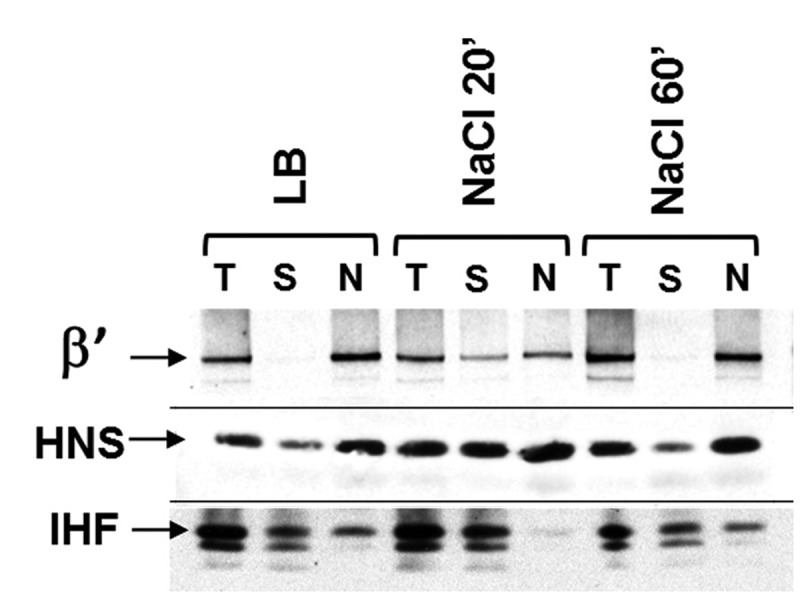
Nucleoid-associated proteins behave similarly as RNAP during the osmotic stress response. Western blot using specific antibodies (β′, H-NS and IHF as indicated) against the total lysate (T), the soluble fraction (S) and the nucleoid fraction (N) from the cells before (LB) or after (NaCl 20′ and NaCl 60′) osmotic shock. After 20 min, increased fractions of RNAP, HNS and IHF are found in the cytoplasm compared with that before salt addition (LB); after 60 min, they become re-associated with the nucleoid. The nature of the other bands in the IHF blot is unknown.
In contrast to RNAP, H-NS and IHF are not exclusively in the nucleoid fraction in LB before salt addition, suggesting that the two small-sized NAPs bind to DNA not as strong as RNAP in the cell, which is consistent with the reported results in vitro (48) and the fact that formaldehyde only crosslinks stably bound proteins to DNA (49). Importantly, however, the soluble fractions of H-NS and IHF increase noticeably at 20 min after the salt addition and return to the condition before the salt addition by 60 min. These results indicate that, similarly to RNAP, the interaction of H-NS and IHF, and possibly other NAPs with the nucleoid, is sensitive to the cytoplasmic K+ concentration during the osmotic stress response.
Hyper-condensed nucleoid during the initial osmotic stress response is not caused by the major factors proposed to affect nucleoid structure
The nucleoid undergoes noticeable dynamic changes during the osmotic stress response. It becomes hyper-condensed when the dissociation of RNAP is mostly apparent 10–20 min after the salt shock (Figure 1B). We attempted to understand the cause(s) for the changes by analysing the contributions of the major factors that have been proposed to affect nucleoid compaction and expansion (50). Although the underlying mechanism responsible for the compaction and expansion of the nucleoid is unknown at this time, we systematically eliminated the possibilities that the observed dynamics of the nucleoid during the osmotic stress response is caused by (i) inhibition of transertion, (ii) NAPs, (iii) DNA supercoiling by gyrase, and (iv) Dps function as detailed below.
Transertion (a process coupling of transcription–translation and translocation of membrane proteins) is proposed to be an expansion force for the nucleoid, which is based on the result that inhibition of transertion by chloramphenicol is correlated with hyper-condensed nucleoid (50). However, inhibition of transertion per se does not lead to nucleoid condensation, as active rRNA synthesis is required for the nucleoid compaction under the conditions in which transertion is inhibited (38). Because rRNA synthesis is inhibited shortly after the salt addition (26), inhibition of transertion is unlikely to be responsible for the hyper-condensed nucleoid. This possibility is further disregarded because RNAP dissociates from the DNA and the nucleoid is hyper-condensed in the salt-shocked cells pre-treated with rifampicin (Figure 2A), which inhibits both transcription and transertion.
Small-sized E. coli ‘histone-like’ proteins are postulated to be involved in nucleoid compaction (41,50,51), which are inferred largely from their biochemical properties in vitro as DNA bridgers and DNA benders causing DNA stiffening (51–56). As shown in Figure 4, at least two of the ‘histone-like’ NAPs (IHF, H-NS) tested also behave like RNAP and are released from the nucleoid during the osmotic stress; thus, it is improbable that the hyper-condensed nucleoid is a consequence of the binding of these NAPs when RNAP is depleted.
Supercoiling by gyrase is postulated to be a nucleoid compaction force in the cell (50), based on the results that supercoiling compacts DNA in vitro (57). Supercoiling by gyrase increases shortly after an osmotic stress response (58). To test whether the increase of supercoiling might be responsible for the hyper-condensed nucleoid during osmotic response, we pre-treated the cells with gyrase inhibitor coumermycin for 30 min before the addition of salt in LB (Figure 5A). By itself coumermycin (Cr 30′) causes no change in nucleoid compaction as reported previously (59). In cells pre-treated with coumermycin and then salt shocked, significant amounts of RNAP are released from the nucleoid into the cytoplasmic space, and the nucleoid becomes hyper-condensed. This result indicates that supercoiling by gyrase is not responsible for the nucleoid compaction during osmotic response.
Figure 5.
Effects of inhibition of DNA supercoiling and dps deletion on the dissociation of RNAP and nucleoid structure during the initial osmotic stress response. (A) The inhibition of supercoiling by gyrase inhibitor coumermycin does not prevent RNAP dissociation nor the nucleoid to condense during the osmotic shock. RNAP-venus protein fusion, nucleoid, overlay of RNAP (green) and nucleoid (red), overlay of nucleoid (white) and cell (black) before (LB), after treatment with coumermycin for 30 min (Cr 30′), and after sequential treatments of coumermycin for 30 min first and then salt (Cr 30′ + NaCl 20′). Note that there is no ring of RNAP surrounding the hyper-condensed nucleoid. (B) The absence of Dps does not prevent RNAP dissociation, nor does it prevent the nucleoid to condense during the osmotic shock. RNAP-venus protein fusion, nucleoid, overlay of RNAP (green) and nucleoid (red), overlay of nucleoid (white) and cell (black) before (LB) or after (NaCl 20′) osmotic shock for the dps deletion mutant cells. The scale bar represents 2 µm.
Because an increase in supercoiling by gyrase is necessary for transcriptional activation of many osmotic stress response genes (17,60), inhibition of supercoiling by gyrase may significantly reduce the expression of the proline and glycine betaine transporter genes. Consistently, there is no ring of RNAP surrounding the nucleoid in the cells treated with coumermycin first and then subjected to osmotic shock (Figure 5A, Cr 30′, NaCl 20′). This phenotype mimics that of the double ΔproP and ΔbetT mutant (Figure 3, NaCl 20′).
Dps is a stress protein that accumulates in late-stationary cells and causes a hyper-condensed nucleoid in starved cells overproducing Dps (61,62). Because the amount of Dps increases during the osmotic stress response (18), we determined whether Dps might be responsible for the hyper-condensed nucleoid under the conditions used. We examined the dps deletion mutant cells during the osmotic stress response and found that, like wild type, RNAP dissociates from the DNA and the nucleoid is hyper-condensed even in the absence of Dps (Figure 5B).
DISCUSSION
Our study emphasizes the importance of following a dynamic process such as the osmotic stress response temporally and demonstrates that the association of RNAP with the nucleoid is sensitive to the cytoplasmic K+ concentration during the osmotic stress response in LB. The cytoplasmic K+ concentration transiently increases through K+ uptake systems and by removing a significant fraction of cytoplasmic water in the initial response phase, which is followed by a decline in K+ owing to accumulation of osmoprotectants including glycine betaine and proline. Consequently, RNAP dissociates shortly after the salt addition followed by the re-association of the freed RNAP with DNA. The accumulation of cytoplasmic glutamate could also contribute to stabilizing RNAP–DNA interaction during the osmoadaptation phase. This work identifies a missing link between in vitro and in vivo studies on the salt-dependent association of RNAP and DNA and confirms that the interaction between RNAP and DNA is governed by the same thermodynamic principle in vivo as it is in vitro. This is also true for the interaction of other NAPs with genomic DNA during the osmotic stress response. It is noteworthy that osmotic stress in yeast also causes an immediate dissociation of many transcription factors (but not of RNA polymerase II) and followed by re-association (63), suggesting a common mechanism in response to salt shock in both prokaryotic and eukaryotic cells. Our findings significantly broaden our understanding of osmotic stress response and provide the framework for the future modeling of the dynamic interaction of RNAP with the genome during the osmotic stress response in E. coli.
Our results show that nucleoid structure undergoes dynamic changes during the osmotic stress response. The nucleoid rapidly becomes hyper-condensed during the initial stress response, followed by a gradual expansion approaching the pre-stress state during the recovery phase. These changes could have potential consequences in global gene regulation and spatial organization of the nucleoid. We speculate that shortly after salt addition, the transient dissociation of RNAP from the genomic DNA with a concurrent nucleoid condensation could be an effective way to silencing genome-wide gene expression while maintaining active expression of osmotic stress response genes, which are presumably oriented at the surface of the hyper-condensed nucleoid. The mechanism underlying the salt-resistant interaction of RNAP and these genes remains to be determined. In addition, changes in global nucleoid compaction will likely affect the cytoplasmic space (volume), which could in turn have potential consequences on macromolecular crowding during the osmotic stress response. A nucleoid with tightly packed DNA is proposed to be a protecting mechanism against DNA damage caused by long-term stress effects (64,65). It remains to be determined whether the nucleoid condensation during the osmotic stress alters response to DNA damage. During the recovery phase, a reversed process is necessary; probably a highly compacted nucleoid would interfere with gene expression and chromosome functions including DNA replication, recombination and segregation.
Our study also raises the question about the potential mechanism underlying the dynamic changes in nucleoid structure during the osmotic stress response. To this end, we examined the contributions of the major factors that have been proposed to be important for nucleoid structure, including transertion, NAPs, DNA supercoiling and Dps, and found that they are not responsible for the effect. One intriguing finding of this study is that there is a correlation between the disassociation of RNAP and the hyper-condensation of the nucleoid during the initial osmotic stress response. Link between the distribution of RNAP, global gene regulation and nucleoid structure has been reported (31,46). Whether the dissociation of RNAP is a cause or a consequence of the nucleoid compaction is unclear; however, we previously showed that the nucleoid compaction does not lead to RNAP dissociation (31,46). All together it is tempting to speculate at this time that dissociation of RNAP may have a direct role in the hyper-compaction of the nucleoid during the initial osmotic stress response. For example, a nucleoid that is depleted of RNAP might be packed into dense aggregates because of a macromolecular crowding environment in the cell (66). And conversely, the re-association of RNAP might be responsible for the de-condensing of the nucleoid during the recovery phase of the response. RNAP binding or transcription from the periphery of the hyper-condensed nucleoid could ‘open up’ the nearby region because RNAP is able to rapidly slide along DNA (67); thus, RNAP could gradually remodel the nucleoid and drive de-condensation. Our study provides the basis for the future study of the role of RNAP in modulating nucleoid structure.
FUNDING
Funding for open access charge: The Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Yan Ning Zhou for many helpful discussions during the course of the study, and thank our colleagues for comments on the manuscript. The authors also thank Darren Sledjeski for the H-NS antibodies, Steven D. Goodman for the IHF antibodies and Lynn Thomason for the TK2420 strain.
REFERENCES
- 1.von Hippel PH, Revzin A, Gross CA, Wang AC. Non-specific DNA binding of genome regulating proteins as a biological control mechanism: part I. The lac operon: equilibrium aspects. Proc. Natl Acad. Sci. USA. 1974;71:4808–4812. doi: 10.1073/pnas.71.12.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaner SL, Melancon P, Lee KS, Burgess RR, Record MT., Jr Ion effects on the aggregation and DNA-binding reactions of Escherichia coli RNA polymerase. Cold Spring Harb. Symp. Quant. Biol. 1983;47(Pt 1):463–472. doi: 10.1101/sqb.1983.047.01.055. [DOI] [PubMed] [Google Scholar]
- 3.Roe JH, Record MT., Jr Regulation of the kinetics of the interaction of Escherichia coli RNA polymerase with the lambda PR promoter by salt concentration. Biochemistry. 1985;24:4721–4726. doi: 10.1021/bi00339a002. [DOI] [PubMed] [Google Scholar]
- 4.deHaseth PL, Lohman TM, Burgess RR, Record MT., Jr Nonspecific interactions of Escherichia coli RNA polymerase with native and denatured DNA: differences in the binding behavior of core and holoenzyme. Biochemistry. 1978;17:1612–1622. doi: 10.1021/bi00602a006. [DOI] [PubMed] [Google Scholar]
- 5.Roe JH, Burgess RR, Record MT., Jr Kinetics and mechanism of the interaction of Escherichia coli RNA polymerase with the lambda PR promoter. J. Mol. Biol. 1984;176:495–522. doi: 10.1016/0022-2836(84)90174-8. [DOI] [PubMed] [Google Scholar]
- 6.Richey B, Cayley DS, Mossing MC, Kolka C, Anderson CF, Farrar TC, Record MT., Jr Variability of the intracellular ionic environment of Escherichia coli. Differences between in vitro and in vivo effects of ion concentrations on protein-DNA interactions and gene expression. J. Biol. Chem. 1987;262:7157–7164. [PubMed] [Google Scholar]
- 7.Winter RB, Berg OG, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids: part 3. The Escherichia coli lac repressor–operator interaction: kinetic measurements and conclusions. Biochemistry. 1981;20:6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- 8.Cayley S, Lewis BA, Guttman HJ, Record MT., Jr Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolarity. Implications for protein-DNA interactions in vivo. J. Mol. Biol. 1991;222:281–300. doi: 10.1016/0022-2836(91)90212-o. [DOI] [PubMed] [Google Scholar]
- 9.Cayley S, Record MT., Jr Large changes in cytoplasmic biopolymer concentration with osmolality indicate that macromolecular crowding may regulate protein-DNA interactions and growth rate in osmotically stressed Escherichia coli K-12. J. Mol. Recognit. 2004;17:488–496. doi: 10.1002/jmr.695. [DOI] [PubMed] [Google Scholar]
- 10.Capp MW, Cayley DS, Zhang W, Guttman HJ, Melcher SE, Saecker RM, Anderson CF, Record MT., Jr Compensating effects of opposing changes in putrescine (2+) and K+ concentrations on lac repressor-lac operator binding: in vitro thermodynamic analysis and in vivo relevance. J. Mol. Biol. 1996;258:25–36. doi: 10.1006/jmbi.1996.0231. [DOI] [PubMed] [Google Scholar]
- 11.Kontur WS, Capp MW, Gries TJ, Saecker RM, Record MT., Jr Probing DNA binding, DNA opening, and assembly of a downstream clamp/jaw in Escherichia coli RNA polymerase-lambdaP(R) promoter complexes using salt and the physiological anion glutamate. Biochemistry. 2010;49:4361–4373. doi: 10.1021/bi100092a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zimmerman SB. Macromolecular crowding effects on macromolecular interactions: some implications for genome structure and function. Biochim. Biophys. Acta. 1993;1216:175–185. doi: 10.1016/0167-4781(93)90142-z. [DOI] [PubMed] [Google Scholar]
- 13.Kempf B, Bremer E. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol. 1998;170:319–330. doi: 10.1007/s002030050649. [DOI] [PubMed] [Google Scholar]
- 14.Dinnbier U, Limpinsel E, Schmid R, Bakker EP. Transient accumulation of potassium glutamate and its replacement by trehalose during adaptation of growing cells of Escherichia coli K-12 to elevated sodium chloride concentrations. Arch Microbiol. 1988;150:348–357. doi: 10.1007/BF00408306. [DOI] [PubMed] [Google Scholar]
- 15.Csonka LN. Physiological and genetic responses of bacteria to osmotic stress. Microbiol. Rev. 1989;53:121–147. doi: 10.1128/mr.53.1.121-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wood JM. Osmosensing by bacteria: signals and membrane-based sensors. Microbiol. Mol. Biol. Rev. 1999;63:230–262. doi: 10.1128/mmbr.63.1.230-262.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheung KJ, Badarinarayana V, Selinger DW, Janse D, Church GM. A microarray-based antibiotic screen identifies a regulatory role for supercoiling in the osmotic stress response of Escherichia coli. Genome Res. 2003;13:206–215. doi: 10.1101/gr.401003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber A, Kogl SA, Jung K. Time-dependent proteome alterations under osmotic stress during aerobic and anaerobic growth in Escherichia coli. J. Bacteriol. 2006;188:7165–7175. doi: 10.1128/JB.00508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber A, Jung K. Profiling early osmostress-dependent gene expression in Escherichia coli using DNA macroarrays. J. Bacteriol. 2002;184:5502–5507. doi: 10.1128/JB.184.19.5502-5507.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood JM. Bacterial osmoregulation: a paradigm for the study of cellular homeostasis. Annu. Rev. Microbiol. 2011;65:215–238. doi: 10.1146/annurev-micro-090110-102815. [DOI] [PubMed] [Google Scholar]
- 21.Ingraham JL, Marr AG. Effect of temperature, pressure, pH, and osmotic stress on growth. In: Neidhardt FC, editor. Escherichia coli and Salmonella. Vol. 2. Washington, DC: ASM Press; 1996. pp. 1570–1578. [Google Scholar]
- 22.Record MT, Jr, Courtenay ES, Cayley DS, Guttman HJ. Responses of E. coli to osmotic stress: large changes in amounts of cytoplasmic solutes and water. Trends Biochem. Sci. 1998;23:143–148. doi: 10.1016/s0968-0004(98)01196-7. [DOI] [PubMed] [Google Scholar]
- 23.Record MT, Jr, Courtenay ES, Cayley S, Guttman HJ. Biophysical compensation mechanisms buffering E. coli protein-nucleic acid interactions against changing environments. Trends Biochem. Sci. 1998;23:190–194. doi: 10.1016/s0968-0004(98)01207-9. [DOI] [PubMed] [Google Scholar]
- 24.Epstein W. The roles and regulation of potassium in bacteria. Prog. Nucleic Acid Res. Mol. Biol. 2003;75:293–320. doi: 10.1016/s0079-6603(03)75008-9. [DOI] [PubMed] [Google Scholar]
- 25.Schiller D, Kruse D, Kneifel H, Kramer R, Burkovski A. Polyamine transport and role of potE in response to osmotic stress in Escherichia coli. J. Bacteriol. 2000;182:6247–6249. doi: 10.1128/jb.182.21.6247-6249.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gralla JD, Vargas DR. Potassium glutamate as a transcriptional inhibitor during bacterial osmoregulation. EMBO J. 2006;25:1515–1521. doi: 10.1038/sj.emboj.7601041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SJ, Gralla JD. Osmo-regulation of bacterial transcription via poised RNA polymerase. Mol. Cell. 2004;14:153–162. doi: 10.1016/s1097-2765(04)00202-3. [DOI] [PubMed] [Google Scholar]
- 28.Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. Living with water stress: evolution of osmolyte systems. Science. 1982;217:1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- 29.Cayley S, Record MT., Jr Roles of cytoplasmic osmolytes, water, and crowding in the response of Escherichia coli to osmotic stress: biophysical basis of osmoprotection by glycine betaine. Biochemistry. 2003;42:12596–12609. doi: 10.1021/bi0347297. [DOI] [PubMed] [Google Scholar]
- 30.Jovanovich SB, Martinell M, Record MT, Jr, Burgess RR. Rapid response to osmotic upshift by osmoregulated genes in Escherichia coli and Salmonella typhimurium. J. Bacteriol. 1988;170:534–539. doi: 10.1128/jb.170.2.534-539.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cabrera JE, Jin DJ. The distribution of RNA polymerase in Escherichia coli is dynamic and sensitive to environmental cues. Mol. Microbiol. 2003;50:1493–1505. doi: 10.1046/j.1365-2958.2003.03805.x. [DOI] [PubMed] [Google Scholar]
- 32.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 34.Sawitzke JA, Thomason LC, Costantino N, Bubunenko M, Datta S, Court DL. Recombineering: in vivo genetic engineering in E. coli, S. enterica, and beyond. Methods Enzymol. 2007;421:171–199. doi: 10.1016/S0076-6879(06)21015-2. [DOI] [PubMed] [Google Scholar]
- 35.Epstein W, Buurman E, McLaggan D, Naprstek J. Multiple mechanisms, roles and controls of K+ transport in Escherichia coli. Biochem. Soc. Trans. 1993;21:1006–1010. doi: 10.1042/bst0211006. [DOI] [PubMed] [Google Scholar]
- 36.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller JH. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 38.Cabrera JE, Cagliero C, Quan S, Squires CL, Jin DJ. Active transcription of rRNA operons condenses the nucleoid in Escherichia coli: examining the effect of transcription on nucleoid structure in the absence of transertion. J. Bacteriol. 2009;191:4180–4185. doi: 10.1128/JB.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cook PR. The organization of replication and transcription. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- 40.Jin DJ, Cagliero C, Zhou YN. Growth rate regulation in Escherichia coli. FEMS Microbiol. Rev. 2012;36:269–287. doi: 10.1111/j.1574-6976.2011.00279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Margolin W. Imaging the Bacterial Nucleoid. In: Dame RT, Dorman CJ, editors. Bacterial Chromatin. London, New York: Springer; 2012. pp. 13–30. [Google Scholar]
- 42.van Helvoort JM, Kool J, Woldringh CL. Chloramphenicol causes fusion of separated nucleoids in Escherichia coli K-12 cells and filaments. J. Bacteriol. 1996;178:4289–4293. doi: 10.1128/jb.178.14.4289-4293.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herring CD, Raffaelle M, Allen TE, Kanin EI, Landick R, Ansari AZ, Palsson BO. Immobilization of Escherichia coli RNA polymerase and location of binding sites by use of chromatin immunoprecipitation and microarrays. J. Bacteriol. 2005;187:6166–6174. doi: 10.1128/JB.187.17.6166-6174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sasse-Dwight S, Gralla JD. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J. Biol. Chem. 1989;264:8074–8081. [PubMed] [Google Scholar]
- 45.Grainger DC, Hurd D, Harrison M, Holdstock J, Busby SJ. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc. Natl Acad. Sci. USA. 2005;102:17693–17698. doi: 10.1073/pnas.0506687102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin DJ, Cabrera JE. Coupling the distribution of RNA polymerase to global gene regulation and the dynamic structure of the bacterial nucleoid in Escherichia coli. J. Struct. Biol. 2006;156:284–291. doi: 10.1016/j.jsb.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 47.Bratton BP, Mooney RA, Weisshaar JC. Spatial distribution and diffusive motion of RNA polymerase in live Escherichia coli. J. Bacteriol. 2011;193:5138–5146. doi: 10.1128/JB.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Azam TA, Ishihama A. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J. Biol. Chem. 1999;274:33105–33113. doi: 10.1074/jbc.274.46.33105. [DOI] [PubMed] [Google Scholar]
- 49.Schmiedeberg L, Skene P, Deaton A, Bird A. A temporal threshold for formaldehyde crosslinking and fixation. PLoS One. 2009;4:e4636. doi: 10.1371/journal.pone.0004636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woldringh CL, Jensen PR, Westerhoff HV. Structure and partitioning of bacterial DNA: determined by a balance of compaction and expansion forces? FEMS Microbiol. Lett. 1995;131:235–242. doi: 10.1111/j.1574-6968.1995.tb07782.x. [DOI] [PubMed] [Google Scholar]
- 51.Macvanin M, Adhya S. Architectural organization in E. coli nucleoid. Biochim. Bbiophys. Acta. 2012;1819:830–835. doi: 10.1016/j.bbagrm.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dame RT. The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol. Microbiol. 2005;56:858–870. doi: 10.1111/j.1365-2958.2005.04598.x. [DOI] [PubMed] [Google Scholar]
- 53.Dillon SC, Dorman CJ. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010;8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 54.Luijsterburg MS, White MF, van Driel R, Dame RT. The major architects of chromatin: architectural proteins in bacteria, archaea and eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2008;43:393–418. doi: 10.1080/10409230802528488. [DOI] [PubMed] [Google Scholar]
- 55.Broyles SS, Pettijohn DE. Interaction of the Escherichia coli HU protein with DNA. Evidence for formation of nucleosome-like structures with altered DNA helical pitch. J. Mol. Biol. 1986;187:47–60. doi: 10.1016/0022-2836(86)90405-5. [DOI] [PubMed] [Google Scholar]
- 56.Liu Y, Chen H, Kenney LJ, Yan J. A divalent switch drives H-NS/DNA-binding conformations between stiffening and bridging modes. Genes Dev. 2010;24:339–344. doi: 10.1101/gad.1883510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bates AD, Maxwell A. DNA Topology. New York, USA: Oxford University Press; 2005. [Google Scholar]
- 58.Hsieh LS, Rouviere-Yaniv J, Drlica K. Bacterial DNA supercoiling and [ATP]/[ADP] ratio: changes associated with salt shock. J. Bacteriol. 1991;173:3914–3917. doi: 10.1128/jb.173.12.3914-3917.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stuger R, Woldringh CL, van der Weijden CC, Vischer NO, Bakker BM, van Spanning RJ, Snoep JL, Westerhoff HV. DNA supercoiling by gyrase is linked to nucleoid compaction. Mol. Biol. Rep. 2002;29:79–82. doi: 10.1023/a:1020318705894. [DOI] [PubMed] [Google Scholar]
- 60.Higgins CF, Dorman CJ, Stirling DA, Waddell L, Booth IR, May G, Bremer E. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell. 1988;52:569–584. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- 61.Almiron M, Link AJ, Furlong D, Kolter R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992;6:2646–2654. doi: 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- 62.Wolf SG, Frenkiel D, Arad T, Finkel SE, Kolter R, Minsky A. DNA protection by stress-induced biocrystallization. Nature. 1999;400:83–85. doi: 10.1038/21918. [DOI] [PubMed] [Google Scholar]
- 63.Proft M, Struhl K. MAP kinase-mediated stress relief that precedes and regulates the timing of transcriptional induction. Cell. 2004;118:351–361. doi: 10.1016/j.cell.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 64.Levin-Zaidman S, Frenkiel-Krispin D, Shimoni E, Sabanay I, Wolf SG, Minsky A. Ordered intracellular RecA-DNA assemblies: a potential site of in vivo RecA-mediated activities. Proc. Natl Acad. Sci. USA. 2000;97:6791–6796. doi: 10.1073/pnas.090532397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Minsky A, Shimoni E, Frenkiel-Krispin D. Stress, order and survival. Nat. Rev. Mol. Cell. Biol. 2002;3:50–60. doi: 10.1038/nrm700. [DOI] [PubMed] [Google Scholar]
- 66.Minton AP. Macromolecular crowding. Curr. Biol. 2006;16:R269–R271. doi: 10.1016/j.cub.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 67.Kabata H, Kurosawa O, Arai I, Washizu M, Margarson SA, Glass RE, Shimamoto N. Visualization of single molecules of RNA polymerase sliding along DNA. Science. 1993;262:1561–1563. doi: 10.1126/science.8248804. [DOI] [PubMed] [Google Scholar]