Abstract
Ty1, a long terminal repeat retrotransposon of Saccharomyces, is structurally and functionally related to retroviruses. However, a differentiating aspect between these retroelements is the diversity of the replication strategies used by long terminal repeat retrotransposons. To understand the structural organization of cis-acting elements present on Ty1 genomic RNA from the GAG region that control reverse transcription, we applied chemoenzymatic probing to RNA/tRNA complexes assembled in vitro and to the RNA in virus-like particles. By comparing different RNA states, our analyses provide a comprehensive structure of the primer-binding site, a novel pseudoknot adjacent to the primer-binding sites, three regions containing palindromic sequences that may be involved in RNA dimerization or packaging and candidate protein interaction sites. In addition, we determined the impact of a novel form of transposon control based on Ty1 antisense transcripts that associate with virus-like particles. Our results support the idea that antisense RNAs inhibit retrotransposition by targeting Ty1 protein function rather than annealing with the RNA genome.
INTRODUCTION
The long terminal repeat (LTR)-retrotransposon Ty1 is a mobile genetic element that replicates through an RNA intermediate and shares several important structural and functional characteristics with retroviruses [see reviews (1–3)]. These include packaging of a dimeric RNA genome tethered by a non-covalent linkage and sequestration of a host-coded tRNA (initiator methionyl-tRNA: tRNA-Meti) that primes minus-strand DNA synthesis within a virus-like particle (VLP) (4–6). VLPs comprise a Gag-like structural protein and the POL-encoded enzymes protease, reverse transcriptase (RT) and integrase (IN), required for particle maturation, replication and integration of the resulting duplex DNA into the yeast genome, respectively. However, Ty1 elements differ from retroviruses in several important respects. Ty1 VLPs lack an envelope protein and are not infectious. Ty1 Gag shows no obvious sequence homology with its retroviral Gag counterparts, although it displays RNA chaperone activities reminiscent of the nucleocapsid protein (NC) (7). Also, Ty1 Gag is not processed to form the characteristic retroviral Gag proteins matrix, capsid and NC.
Retroelement genomic transcripts contain internal structures fundamental to gene expression and propagation. Prominent among these are motifs required for programmed ribosomal frameshifting, initiation of reverse transcription and packaging of dimeric RNA into particles. The diversity of priming strategies among LTR-retrotransposons includes tRNA-independent self-priming in Tf1 of Schizosaccharomyces pombe, priming from an internal site of tRNA in copia of Drosophila melanogaster or bipartile primer binding site (PBS) sequences separated by ∼4800 nt in Ty3 [reviewed in (8)]. Retroviruses typically have a PBS of 18 contiguous nt complementary to the 3′ acceptor stem of their cognate tRNA primer that extends into the TΨC arm (9). Additional interactions between the tRNA primer and the PBS region of Rous sarcoma and human immunodeficiency virus (HIV) also contribute to efficient minus-strand synthesis (10–12). The PBS region of Ty1 contains only 10 contiguous nt complementary to the 3′-acceptor stem of initiator tRNA-Meti (13,14). However, extensive regions of complementarity with additional segments of the tRNA adjacent to the PBS may help position the tRNA for efficient priming. A long-range interaction between a region (Complementary sequence: CYC5) adjacent to the extended PBS with sequences near the 3′-end of genomic RNA (CYC3) is also required for reverse transcription and transposition (15).
Dimerization and packaging of retroviral RNA has been intensively studied using genetic, biochemical and structural approaches [reviewed in (16)]. HIV-1 RNA seems to be selectively packaged as a dimer, and genome dimerization and packaging are mechanistically coupled (17). Elements required for these processes are located in the highly structured 5′-untranslated region (UTR). The dimer initiation site (DIS) is a conserved hairpin with a palindromic (PAL) sequence at its apex allowing two RNA molecules to interact through a ‘kissing-loop’. The organization and complexity of the packaging region (Ψ-site) differs among retroviruses (16). For HIV-1, efficient packaging requires 5′-UTR and possibly extends into GAG coding sequence (18,19).
Little is known about how Ty1 or other LTR-retrotransposons package their genomic RNA. The Ty1 5′-UTR comprises only 53 nt, and mini-Ty1 elements of ∼0.5 kb support retrotransposition when co-expressed with a helper element (20,21). Mutations isolated in these truncated elements block the use of RNA for retrotransposition. However, other than the PBS region and short sequences at the 5′-end of the RNA that anneal with downstream partners (15,20), the regulatory sites remain undefined. Structural information on Ty1 RNA is available only for the PBS region of ∼140 nt in vitro (13), and the proposed structure is inconsistent with mutational data (15).
Multiple Ty1 antisense (AS) RNAs are transcribed from a region containing sequences required for copy number control (CNC) and cis-acting signals required for Ty1 mobility (22,23). AS transcript I (∼900 nt) and II (624 nt) have been characterized. These transcripts associate with VLPs and dramatically reduce the level of mature IN and, to a lesser extent, the level of additional Ty1 proteins. The Ty1AS RNAs act stoichiometrically to confer CNC, as they inhibit transposition in a dose-dependent manner, and Saccharomyces lacks the machinery for RNA interference (24). However, whether AS transcripts inhibit retrotransposition by annealing with genomic RNA, which would affect RNA-interactions required for replication, or antagonize VLP protein function is unclear.
Here, we explore the structure of Ty1 RNA in different biological states using selective 2′-hydroxyl acylation analysed by primer extension or SHAPE (25–27) [also see a recent review on the advantages and limitations of SHAPE (28)]. Our results suggest a coherent structure of the PBS region that incorporates mutational and additional RNA interactions and identifies candidate motifs that may be important for dimerization and packaging. Finally, we do not find evidence for annealing of AS RNAs with genomic RNA in VLPs, suggesting the novel possibility that the AS RNAs compromise Ty1 protein function.
MATERIALS AND METHODS
VLP isolation and characterization
VLPs isolated from ∼50 l of YEM524 (CNC−) or YEM525 (CNC+) were pooled for SHAPE analysis. Cells were grown in 4–6 l batches and fractionated as previously described (23,29). To monitor consistency of each VLP preparation, the level of Gag, IN and RT activity was determined by immunoblotting and an exogenous RT assay with added primer/template (30). CNC− and CNC+ VLPs were also analysed for reverse transcription intermediates that result from endogenous synthesis in vitro (4,6). Endogenous tRNA labelling was performed similar to the endogenous synthesis in vitro, but in the presence of a subset of nucleoside triphosphates (NTPs).
RNA analyses
RNA was extracted from VLPs as described previously (23). Estimating the amount of Ty1 AS and genomic RNA present in VLPs by northern hybridization is described in the Suplementary Materials and Methods section. Ty1 RNA packaging during VLP assembly was monitored by benzonase sensitivity (31,32). RNA protection assays were performed in triplicate from independent cultures. RNA transcripts were synthesized with the SP6-MEGAscript (Life Technologies, Carlsbad, CA, USA) following the manufacturer’s protocol. BamHI-cleaved pJef953 (kindly provided by Jef Boeke) was used as template. The template for the tRNA-Meti mimic was obtained by polymerase chain reaction using overlapping oligonucleotides. RNA was recovered by LiCl precipitation.
N-methyl isatoic anhydride modification of VLPs
Concentrated samples of purified CNC− or CNC+ particles (500 µl) in VLP storage buffer (20 mM of HEPES pH7.6, 140 mM of KCl, 0.5 mM of ethylenediaminetetraaceticacid (EDTA) and 10% of glycerol) were treated with NMIA (N-methyl isatoic anhydride; 50 µl, 100 mM in DMSO) or DMSO alone (50 µl) for 45 min at 37°C.
Extraction of Ty1 genomic RNA from NMIA-modified particles
VLP samples (550 µl) were incubated with 67.5 µg of proteinase K (Novagen-Merck KGaA, Darmstadt, DE) in extraction buffer [50 mM of Tris–HCl (pH 7), 10 mM of EDTA, 100 mM of NaCl and 1% sodium dodecyl sulphate] for 30 min at 37°C. Nucleic acids were purified by three extractions with phenol/CHCl3, followed by precipitation with LiCl and washing with 70% ethanol. Samples were dissolved in DNase I solution (Life Technologies, Carlsbad, CA, USA) and were treated with 1 µl of DNase I at 37°C for 30 min. RNA was phenol/CHCl3 extracted (3×), CHCl3 extracted (3×), precipitated with LiCl and rinsed with 70% ethanol. RNA was resuspended in 25 µl of 5 mM of Tris–HCl pH7.0, 0.5 mM of EDTA and was stored at −80°C until use.
NMIA treatment of Ty1 RNA from VLPs
VLPs were treated with proteinase K and DNase I as described previously. Deproteinized genomic RNAs were resuspended in folding buffer (50 mM of Tris pH 8, 250 mM of KCl and 5 mM of MgCl2) and equilibrated at 37°C for 10 min. RNA was divided equally between two tubes, and DMSO or NMIA (final concentration 1 mM) were added. The modification reaction was allowed to proceed for 45 min at 37°C and was terminated by ethanol precipitation. Modification of in vitro transcript, detection of 2′-O-adducts and data processing are described in the Supplementary Materials and Methods and Table S1.
RESULTS AND DISCUSSION
Characterization of the model system
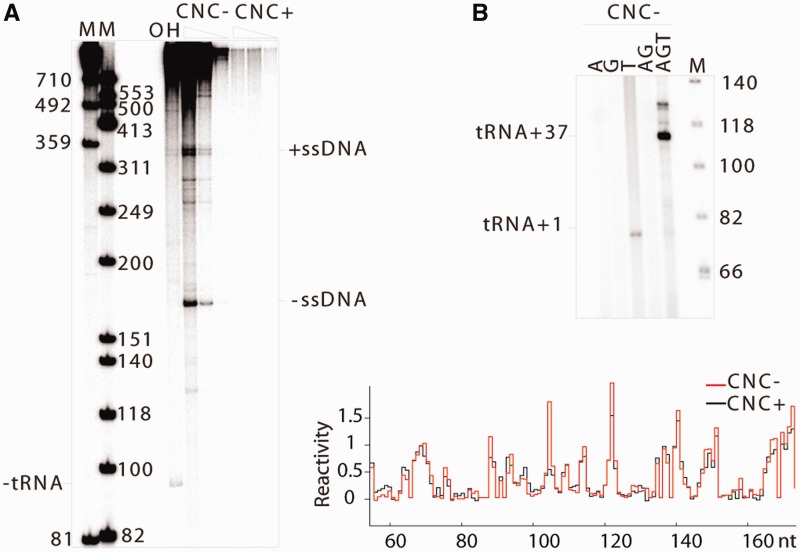
VLPs were isolated from closely related Saccharomyces strains containing low (CNC−) or high levels of Ty1AS RNAs (CNC+) and a GAL1-promoted Ty1 element integrated into the genome (23). To study the structures of genomic RNAs in either context, it was necessary to establish that these RNA populations were structurally homogenous, as the presence of replication intermediates that alter RNA structure would complicate the analysis. Endogenous reverse transcription reactions were conducted with CNC− and CNC+ VLPs in the presence of the full complement of dNTPs (30). No minus-strand strong stop (−ssDNA) or plus-strand strong stop (+ssDNA) intermediates and low amounts of longer reverse transcription products were detected in CNC+ VLPs (Figure 1A). The absence of full-length cDNA was demonstrated previously (23), where the inability to reverse transcribe Ty1 RNA in CNC+ VLPs was attributed to a low level of mature IN, whose association with RT is required for reverse transcription in vivo (33,34). We also confirmed that although CNC+ VLPs lack IN, they contain active RT when supplemented with an exogenous primer/template (23). Productive endogenous reverse transcription was observed in CNC− VLPs, with −ssDNA, +ssDNA and longer reverse transcription products detectable in significant amounts (Figure 1A). Treatment of reaction products with NaOH revealed the presence of a shorter −ssDNA product, demonstrating that tRNA-Meti is hybridized with the PBS, and Ty1 genomic RNA in CNC− VLPs is poised to initiate reverse transcription.
Figure 1.
Analysis of endogenous reaction products from Ty1 VLPs. (A) Endogenous reverse transcription in CNC− and CNC+ VLPs (VLP amount decreasing from left to right). M: mass markers, and OH: NaOH treatment after reverse transcription (CNC− VLPs, intermediate amount). (B) Products of endogenous reverse transcription in CNC− VLPs in the presence of limiting dNTPs. A, G, T corresponds to the type(s) of dNTP ([α-32P]-labelled and unlabelled) present in the reaction mixture. (C) Plots of NMIA reactivity of Ty1 ribonucleotides in and around the PBS obtained from CNC− (red) and CNC+ (black) VLPs.
To evaluate whether tRNA-Meti in CNC− VLPs is extended, we conducted endogenous reactions in the presence of a subset of dNTPs (Figure 1B). No products were detected in reactions in which only dATP, dGTP or both deoxynucleotides were included (Figure 1B; lanes A, G and AG), whereas a ∼77-nt product was generated in reactions containing dTTP (Figure 1B; lanes T and AGT). These results support the expectations, based on the Ty1 sequence, that thymidine is the first nucleotide added to PBS-bound tRNA-Meti, and reverse transcription does not initiate from other primer(s) aberrantly hybridized to alternative sites on the Ty1 genome. Similarly, a ∼113-nt product consistent with tRNA extended by 37 nt was generated in the presence of dATP, dGTP and dTTP, but lacking dCTP (Figure 1B; lane AGT). Again, this result is consistent with DNA synthesis primed by tRNA hybridized to the PBS, where dCTP is first incorporated 38 nt from its 3′ terminus. Together, these results demonstrate that in the majority of VLPs, tRNA extension products are not randomly distributed and unextended tRNA is present.
SHAPE profiles of the PBS region within CNC− and CNC+ VLPs were very similar (Figure 1C), despite the strong replication defect of CNC+ VLPs. These results indicate that most RNAs from CNC− and CNC+ VLPs share similar structures in their PBS region, and they are not actively being reversed transcribed. In addition, although a strength of SHAPE is discrimination between flexible regions resulting from loops, bulges and junctions in RNA and inflexible double-stranded or protein bound regions [see review (27)], we found no evidence for extensive annealing between the AS transcripts and the PBS region. As expected, Ty1 sense/antisense dsRNA formed in vitro was non-reactive towards NMIA (Supplementary Figure S1).
Model of PBS/tRNA complex in Ty1 VLP
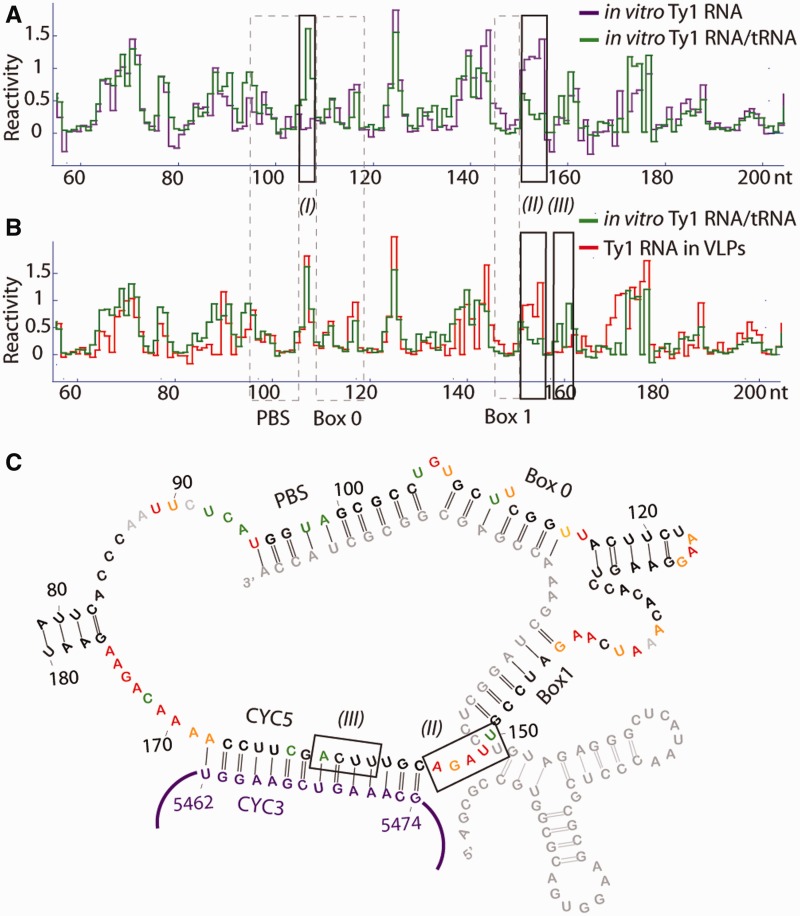
Two structural models for the Ty1 PBS region have been reported, based on enzymatic probing, computational and mutational analyses (13,15). These models differ in one critical respect, where the same stretch of nucleotides in Ty1 RNA is proposed to interact either with tRNA-Meti (Box 2.1) (13) or with the 3′-terminus (CYC3) (15), effectively cyclizing the Ty1 genome. To generate a comprehensive model of the PBS region, we analysed the PBS/tRNA structure in vitro by heat-annealing an RNA that mimics tRNA-Meti to full-length RNA synthesized through transcription in vitro. Formation of the Ty1 RNA/tRNA complex was confirmed by differences in NMIA modification patterns between Ty1 RNA hybridized to the tRNA-Meti mimic and Ty1 RNA alone (Figure 2A). The most pronounced differences are in regions (I) and (II). Specifically, nucleotides 105–107, which were unreactive in the absence of tRNA (Supplementary Figure S2A), gain reactivity in the complex, whereas nucleotides 151–155 became unreactive. Box 0 and 2.1 interactions were observed in our in vitro complex (Supplementary Figure S2B), consistent with a previous model showing an extended interaction between Ty1 RNA and tRNA outside the 10 nt PBS (13,35).
Figure 2.
Structural analysis of the Ty1 PBS region. (A and B) NMIA reactivity plots of ribonucleotides in the PBS region of in vitro transcribed, full-length Ty1 RNA hybridized to tRNA (green), in comparison with that obtained from (A) the same RNA folded in the absence of tRNA (violet), or (B) CNC− VLPs in virio (red). Nucleotide positions corresponding to PBS, Box 0 and Box 1 (13,35) are marked (grey, dashed line). Regions displaying significant changes between each state are noted (I), (II), (III). (C) Model of Ty1 PBS region structure within VLPs. The CYC3 region (15) of the Ty1 genomic RNA is noted (violet).
In contrast, the NMIA modification pattern of the same complex from CNC− VLPs was strikingly different (Figure 2B). Although the region encompassing the PBS and Box 0 showed reactivities similar to the in vitro complex, confirming tRNA hybridization, significant differences were observed downstream of Box 1 [Figure 2B, (II) and (III)]. Nucleotides 153–155, which were hybridized to tRNA in vitro, were reactive in virio, whereas nucleotides 158–161, which were reactive in vitro, became constrained in virio. In virio reactivity patterns suggest a model where tRNA-Meti not only hybridizes with the Ty1 PBS but also occludes additional nucleotides downstream of this region (Figure 2C; bottom panel). Also, the Box 2.1 interaction proposed by Friant et al. (13,35) was not confirmed, and the CYC3/CYC5 motif is present.
Structure of the CNC region
To characterize the functional and structural organization of Ty1 RNA and address whether AS transcripts anneal with the CNC region, which contains nucleotides 136–1702 of Ty1 DNA (23), genomic RNA was modified with NMIA within CNC− and CNC+ VLPs (in virio) and in the protein-free state (ex virio) after gentle extraction from purified CNC− VLPs. In parallel, we determined the structure of the in vitro transcript (Supplementary Figure S3).
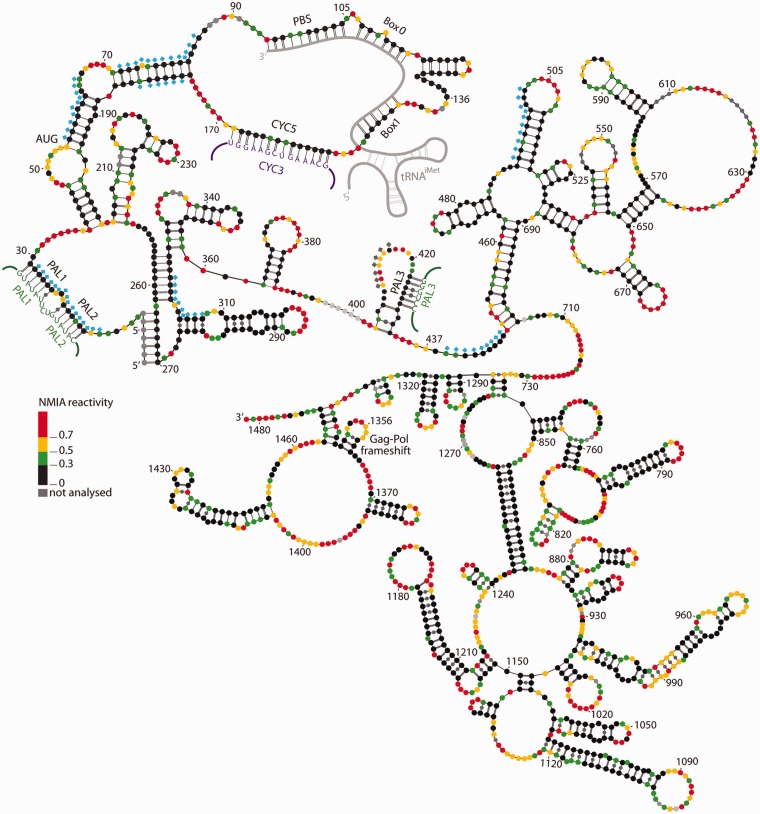
The structure of a 440-nt segment of Ty1 RNA, based on an in silico structure prediction and mutational analysis, has been reported (20), whereas our analysis was conducted on full-length Ty1 RNA (∼5.6 kb). The structure we obtained for the first 440 nt in the context of the full-length genome folded in vitro was similar (20). However, significant differences arose when we determined the in virio conformation (Figure 3). In addition to extensive changes in the PBS region, we detected a novel pseudoknot structure where nucleotides 1–7 and 256–262 pair with nucleotides 264–270 and 319–325, respectively. Although this pseudoknot was previously undetected, such a structure is supported by extensive mutational analyses that affect reverse transcription and transposition (Huang et al., submitted for publication) (20). When these mutants are combined with compensatory changes, retrotransposition is restored.
Figure 3.
RNA secondary structure of the nucleotides 1–1482 of Ty1 genomic RNA (CNC region) in VLPs. This model is based on SHAPE constraints derived from the protein-free state (ex virio) (see ‘Materials and Methods’ section). Colour-coding corresponds to in virio reactivities. Positions (diamonds) within the first 500 nt of Ty1 genomic RNA where reactivity differs in the protein-bound and -free states are marked (blue: increased, grey: decreased). The following regions are annotated: PAL sequences, including reciprocal interstrand interactions (green), and cyclization mediated by CYC5 and CYC3 (violet).
This pseudoknot structure was absent from in vitro transcribed full-length Ty1 RNA. Unreactive nucleotides 256–262, which in virio constitute part of the pseudoknot, were highly reactive in vitro (Supplementary Figure S4). Although Mg2+ has been demonstrated as important for tertiary folding (36), the Ty1 pseudoknot within full-length Ty1 RNA in vitro failed to fold in the presence of Mg2+, despite raising the divalent cation concentration to 25 mM.
Hierarchical folding is specific for some RNAs (37), but formation of tertiary structure can also change the fold achieved during transcription (38). We investigated the possibility that the conformational change following tRNA hybridization to the PBS facilitated pseudoknot formation within the full-length Ty1 transcript. Although we failed to observe reactivity changes in the pseudoknot region in vitro in the presence of the tRNA mimic, the in vitro assembled complex differs from that present in virio (Figure 2). In particular, the CYC3/CYC5 interaction is absent, raising the possibility that this interaction is required to maintain a specific 3D architecture governing proper pseudoknot folding within the full-length transcript. Protein-mediated RNA chaperone activity may also be involved in this process.
We noted differences between the in vitro and in virio states in three regions containing PAL sequences, namely nucleotides 15–20 (PAL1), 24–29 (PAL2) and 423–428 (PAL3) (Figure 3). The NMIA reactivity pattern in vitro is consistent with these intramolecular structures. A hairpin structure with a 5-nt apical loop reflects intramolecular PAL1/PAL2 annealing, whereas PAL3 is involved in forming a hairpin stem with nucleotides 408–413 (Supplementary Figure S3). The intramolecular character of these structures is supported by native polyacrylamide gel electrophoresis analyses showing the Ty1 RNA transcript is monomeric under our NMIA probing conditions. However, changes in NMIA reactivity observed in native VLPs support the model where the PALs are involved in intermolecular interactions. Only U residues in the -U-C-U- sequence (nucleotides 21–23) separating PAL1 and PAL2 are reactive in virio (Figure 3). PAL3 remains unreactive in virio, whereas reactivity of its counterpart increases significantly. As NMIA sensitivity of neighbouring nucleotides remains unaffected, arguing against a global alteration in secondary structure, the observed changes may reflect intermolecular interaction. Self-complementary sequences are involved in dimerization of retroviral genomes [reviewed in (16)], and although Ty1 RNA was shown to be dimeric in VLPs (5), its DIS remains undefined. Interestingly, PAL1 and PAL2 are comprised of tandem repeats. For murine leukaemia virus, tandem -U-C-U-G- motifs are critical for packaging (39–41). Therefore, the PAL motifs are attractive candidates for mediating Ty1 RNA dimerization and packaging into VLPs.
As the region mediating Ty1 translational frameshifting is conserved in the in vitro and in virio states, we examined it for stable structures that influence translational recoding in viral systems [reviewed in (42)]. Ty1 +1 frameshifting requires the GAG heptamer -C-U-U-A-G-G-C- (43,44). Our structural analysis (Figure 3) showed this sequence is not embedded within a particularly stable structure. Therefore, our model supports the extensive mutational analyses of the heptameric sequence, suggesting that Ty1 frameshifting does not require a specific RNA structure.
Finally, the overall structure of the in virio RNA from CNC− and CNC+ VLPs was similar throughout the CNC region (Supplementary Data set S1), again indicating a lack of extensive annealing between the AS transcripts and genomic RNA.
Protein interaction sites on the Ty1 RNA genome
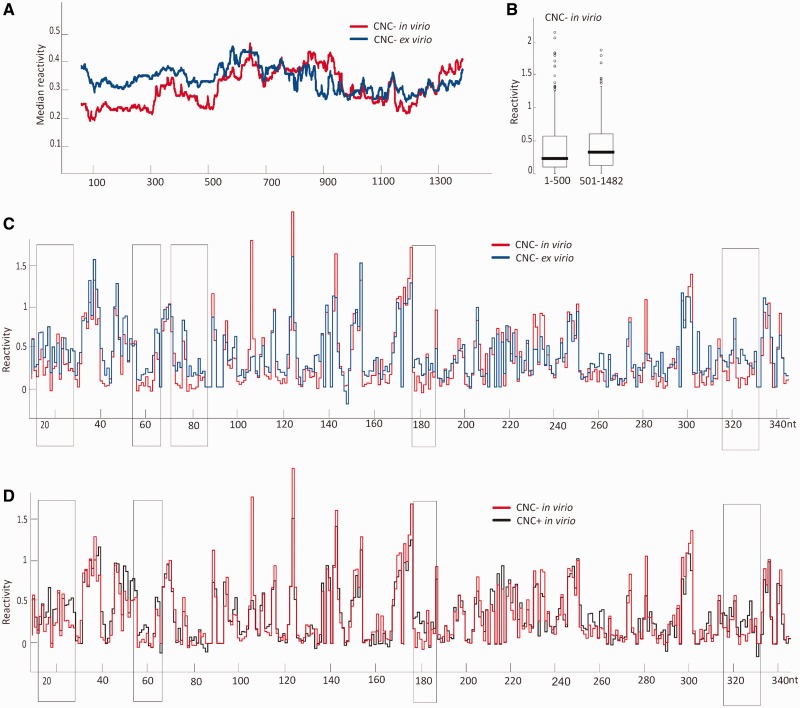
By comparing the protection of Ty1 genomic RNA from chemical modification in virio and ex virio, we explored RNA sequences and/or structural motifs that might interact with Gag or possibly other proteins. We demonstrated that in vitro transcribed full-length Ty1 RNA displayed different structural characteristics than in virio RNA. Therefore, Ty1 RNA was extracted from VLPs and modified in the protein-free state. Median reactivity, reflecting levels of flexibility, was lower with in virio (0.29) than with ex virio (0.32) RNA. This smaller-than-anticipated difference resulted in part from the fact that local nucleotide flexibility decreases and increases in distinct RNA regions after removal of proteins (Supplementary Figure S5). Alterations detected in these different biological states can reflect local structural changes and the effects of direct binding of proteins to the RNA. We observed prominent changes in reactivity (P < 3.844e-06) within ∼500 nt adjacent to the RNA 5′-end (Figure 4A), whereas differences in the remainder of the CNC region were not significant (P = 0.933). This result indicates that disrupting RNA–protein interactions impacts mainly the 5′-end of Ty1 RNA. Median reactivities for nucleotides 1–500 and 501–1482 in virio (Figure 4B) were 0.22 and 0.32, respectively, whereas the corresponding values ex virio were 0.32. These results raise the possibility that Ty1 Gag is required to stabilize the 5′-end of the genome. Furthermore, the 5′-end of Ty1 RNA may contain the major recognition signals for dimerization, packaging and initiating replication, extending observations that first 380 nt are required in cis for transposition (20,21).
Figure 4.
Effects of protein binding on Ty1 RNA structure. (A) Median reactivities for the CNC region of Ty1 RNA in virio (red) and ex virio (blue) calculated for a rolling window of 155 nt. (B) Analysis of reactivity distributions for +1–500 nt (left) and +501–1482 (right). Boxes outline the middle 50% of each data set with medians (bar) and outliers (circles) shown. (C and D) Reactivity plots of in virio CNC− Ty1 RNA (red) in comparison with (C) ex virio CNC− RNA (blue) and (D) in virio CNC+ (black). Regions showing increased reactivity in ex virio CNC− RNA or in virio CNC+ RNA are boxed.
We examined the 5′-end in more detail, as the functions of this region are likely carried out by the interplay of RNA structure and protein recognition. The overall reactivity profile was comparable for RNA in VLPs and in the protein-free state (Figure 4C), arguing against the notion that removal of proteins results in a global structural change. Several regions displayed small but reproducible reactivity differences spanning several consecutive nucleotides. In particular, nucleotides within specific sites (Figure 4C; boxed regions) showed highly significant (P < 8.218e-06) increases in susceptibility to acylation ex virio. Those motifs were located in double stranded regions, with two sites on opposite strands of the helical region adjacent to the PBS. A strong effect was observed in the pseudoknot and in PAL1 and PAL2. Comparing in vitro and in virio SHAPE profiles pointed to the 6-nt PAL sequences as possibly mediating intermolecular interactions. PAL instability ex virio also suggests these motifs participate in intermolecular interactions. Whether the alterations in Ty1 RNA SHAPE reactivity ex virio result from previous chaperone activity remains to be determined. However, it is well known that retroviral NC can help form stable RNA structures that persist when the NC is removed (15,45).
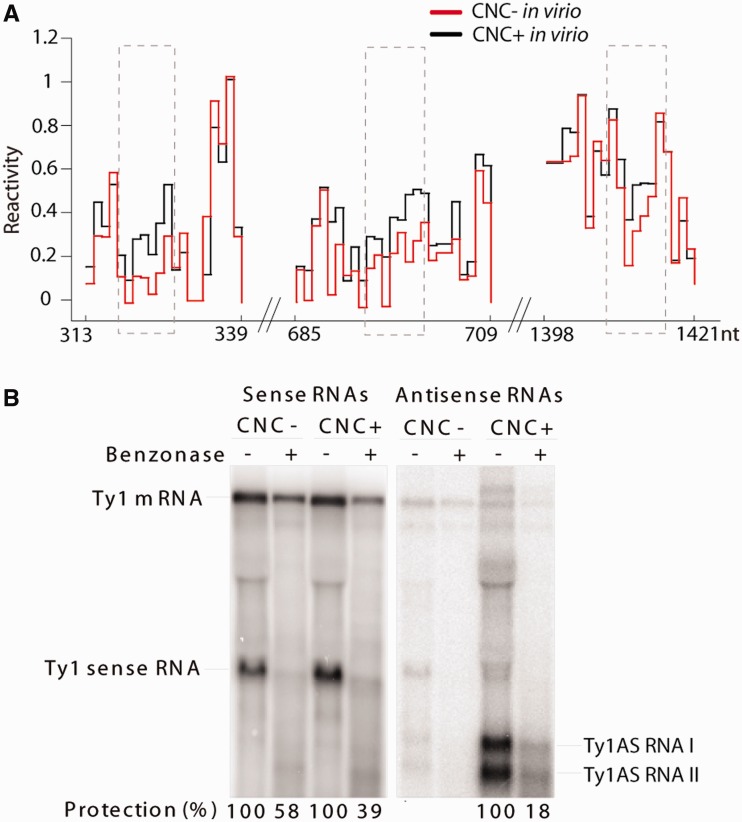
Regions destabilized in CNC− RNA ex virio are destabilized in CNC+ RNA in virio, despite its presence within VLPs (Figure 4D; boxed). The exception is a region preceding the PBS, which could reflect an interaction with factors required for reverse transcription. As the most abundant VLP ligand is Gag, we hypothesize that the destabilization reflects interactions between Gag and Ty1 RNA. However, possible alterations in Gag–RNA binding did not change the diameter of CNC+ VLPs, as determined by electron microscopy (Supplementary Figure S6). Examination of NMIA reactivity profiles from CNC− and CNC+ VLPs revealed that the sequence -A-U-G-A-U-G-A- at all three positions in the CNC region showed higher reactivity in CNC+ VLPs (Figure 5A). NMIA sensitivity in those regions was also altered in CNC− RNA ex virio, strengthening the idea that RNA/protein interactions are critical for Ty1AS RNA-mediated interference.
Figure 5.
Impact of Ty1AS RNA. (A) Reactivity plots of in virio CNC− Ty1 RNA (red) in comparison with in virio CNC+ (black) for the regions encompassing the A-U-G-A-U-G-A heptameric sequence (nucleotides 324–330, 697–703, 1409–1415; dashed rectangles). (B) Ty1 genomic RNA (denoted mRNA) and AS RNA protection. Whole-cell extracts from strains YEM524 (CNC−) and YEM525 (CNC+) were spiked with in vitro transcribed Ty1 sense RNA from the CNC region (∼1.5 kb). Equal aliquots were incubated with (+) or without (−) the nuclease benzonase. RNA extracted from each sample was subjected to northern analysis using 32P-labelled RNA probes specific for genomic RNA and the added sense RNA or AS RNA I and II. Protection is expressed as a ratio of treated to untreated RNA.
Impact of Ty1AS RNAs
Ty1AS transcripts may target genomic RNA, as these non-coding RNAs specifically associate with VLPs (23). If Ty1AS RNAs anneal with Ty1 genomic RNA, the reactivity profile of Ty1 RNA would be drastically altered in CNC+ VLPs. The PBS region is a plausible target for Ty1AS RNAs because of defects in reverse transcription, and the PBS is covered by the AS transcripts. However, SHAPE profiles of the PBS region of CNC− and CNC+ VLPs were similar (Figure 1C), indicating that Ty1AS RNAs do not extensively anneal with the Ty1 PBS in CNC+ VLPs. Furthermore, the in virio structure throughout the CNC region is similar for both types of VLPs (Figure 3 and Supplementary Data set S1).
If AS transcripts anneal with localized regions of genomic RNA or interact stoichiometrically with the RNA, the AS RNAs should be present in an equivalent molar amount as genomic RNA and should be specifically packaged into the VLPs. The molar amounts of genomic RNA and AS RNAs I and II in samples of CNC+ VLPs were estimated by quantitative northern analysis, using known amounts of in vitro synthesized sense and antisense RNAs (Supplementary Figure S7A and B). If we assume that all Ty1 RNA in VLPs is dimeric (5), and account for a putative RNA degradation product present in the preparations from VLPs, the CNC+ sample contains ∼3.5 fmol of dimeric Ty1 RNA (Supplementary Figure S7C). Conversely, the CNC+ VLP sample contained ∼1.1 and 1.6 fmol of AS RNA I and II, respectively. As these transcripts are apparently required to inhibit transposition and confer CNC (23), it is unlikely that both transcripts are present in every VLP.
To monitor packaging of Ty1 RNA and AS transcripts, cellular extracts containing CNC− or CNC+ VLPs were treated with benzonase (Figure 5B), a non-specific endonuclease that degrades RNA and DNA (32,46). Total VLP RNA was subjected to northern blot analyses using strand-specific probes to detect Ty1 RNA or the AS RNAs. Hybridization signals were normalized to mock-treated samples treated in parallel. Benzonase activity was monitored by degradation of a Ty1 sense RNA synthesized in vitro and added to the CNC− extract. The level of protected AS transcripts (18%) decreased at least 2-fold relative to protection of Ty1 genomic RNA (39%) in the CNC+ strain. In addition, the level of protected mRNA decreased ∼1.5-fold in the CNC+ strain (39%) when compared with the CNC− strain (58%). These results indicate that the AS RNAs become more susceptible to degradation than the genomic RNA in the CNC+ strain, and raise the possibility that although the AS RNAs specifically associate with the VLPs (23), these transcripts may not interact with Gag or other Ty1 proteins in the same way as the genomic RNA. Our results also reinforce the idea that AS RNAs target another Ty1 product, such as Gag.
CONCLUSIONS
We provide the first structure for ∼1.5 kb of Ty1 genomic RNA under several different biological states. The in virio structure of a ∼900 nt portion of HIV-1 genomic RNA (47) and the structure of full-length ex virio RNA (26) has been reported. Although several important observations arose from those studies, comparing two evolutionarily distant relatives that undergo similar RNA transactions should also help understand replication strategies of both retroelement classes. In particular, retroviral genome cyclization (48) may stimulate strand-transfer and is required for HIV-1 replication (49–51). However, the structure reported for HIV-1 genomic RNA lacks cyclization motifs (26). Perhaps the Ty1 cyclization domains can be used to help identify their HIV-1 counterpart, once more is known about the Ty1 packaging and dimerization signals, which may reside in the newly discovered PAL or pseudoknot motifs. Furthermore, we investigated a unique antisense RNA strategy for CNC (23). In light of the observation that Saccharomyces cerevisiae and close congeners have lost the conserved RNAi pathways to silence endogenous transposons (24), this process is even more intriguing. Our data support a novel mechanism where AS RNAs do not anneal with genomic RNA in VLPs but interfere with protein functions before reverse transcription.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–7, Supplementary Materials and Methods, Supplementary Data set 1 and Supplementary References [52–57].
FUNDING
Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research, University of Georgia Research Foundation and NIH [GM095622 to D.J.G.]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. The contents of this publication do not necessarily reflect the views of the Department of Health and Human Services, and mention of trade names, commercial products or organizations does not imply endorsement by the US government.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Jef Boeke for helpful discussions, sharing unpublished data and plasmid pJef953, Ferri Soheilian for the assistance with TEM and Jason Rausch for fruitful discussions.
REFERENCES
- 1.Voytas DF, Boeke JD. Ty1 and Ty5 of Saccharomyces cerevisiae. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 614–630. [Google Scholar]
- 2.Lesage P, Todeschini A. Happy together: the life and times of Ty retrotransposons and their hosts. Cytogenet. Genome Res. 2005;110:70–90. doi: 10.1159/000084940. [DOI] [PubMed] [Google Scholar]
- 3.Maxwell P, Curcio M. Host factors that control long terminal repeat retrotransposons in Saccharomyces cerevisiae: implications for regulation of mammalian retroviruses. Eukaryot. Cell. 2007;6:1069–1080. doi: 10.1128/EC.00092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapman K, Bystrom A, Boeke J. Initiator methionine tRNA is essential for Ty1 transposition in yeast. Proc. Natl Acad. Sci. USA. 1992;89:3236–3240. doi: 10.1073/pnas.89.8.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng Y, Moore S, Garfinkel D, Rein A. The genomic RNA in Ty1 virus-like particles is dimeric. J. Virol. 2000;74:10819–10821. doi: 10.1128/jvi.74.22.10819-10821.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilhelm M, Wilhelm F-X, Keith G, Agoutin B, Heyman T. Yeast Ty1 retrotransposon: the minus-strand primer binding site and a cis-acting domain of the Ty1 RNA are both important for packaging of primer tRNA inside virus-like particles. Nucleic Acids Res. 1994;22:4560–4565. doi: 10.1093/nar/22.22.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cristofari G, Ficheux D, Darlix JL. The GAG-like protein of the yeast Ty1 retrotransposon contains a nucleic acid chaperone domain analogous to retroviral nucleocapsid proteins. J. Biol. Chem. 2000;275:19210–19217. doi: 10.1074/jbc.M001371200. [DOI] [PubMed] [Google Scholar]
- 8.Le Grice SFJ. In the beginning: initiation of minus strand DNA synthesis in retroviruses and LTR-containing retrotransposons. Biochemistry. 2003;42:14349–14355. doi: 10.1021/bi030201q. [DOI] [PubMed] [Google Scholar]
- 9.Telesnitsky A, Goff SP. Revers transcriptase and generation of retroviral DNA. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. New York: Cold Spring Harbor Laboratory Press; 1997. pp. 121–160. [PubMed] [Google Scholar]
- 10.Beerens N, Berkhout B. The tRNA primer activation signal in the human immunodeficiency virus type 1 genome is important for initiation and processive elongation of reverse transcription. J. Virol. 2002;76:2329–2339. doi: 10.1128/jvi.76.5.2329-2339.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldschmidt V, Rigourd M, Ehresmann C, Le Grice SF, Ehresmann B, Marquet R. Direct and indirect contributions of RNA secondary structure elements to the initiation of HIV-1 reverse transcription. J. Biol. Chem. 2002;277:43233–43242. doi: 10.1074/jbc.M205295200. [DOI] [PubMed] [Google Scholar]
- 12.Morris S, Leis J. Changes in Rous sarcoma virus RNA secondary structure near the primer binding site upon tRNATrp primer annealing. J. Virol. 1999;73:6307–6318. doi: 10.1128/jvi.73.8.6307-6318.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friant S, Heyman T, Wilhelm ML, Wilhelm FX. Extended interactions between the primer tRNAi(Met) and genomic RNA of the yeast Ty1 retrotransposon. Nucleic Acids Res. 1996;24:441–449. doi: 10.1093/nar/24.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keeney J, Chapman K, Lauermann V, Voytas D, Astrom S, von Pawel-Rammingen U, Bystrom A, Boeke J. Multiple molecular determinants for retrotransposition in a primer tRNA. Mol. Cell Biol. 1995;15:217–226. doi: 10.1128/mcb.15.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cristofari G, Bampi C, Wilhelm M, Wilhelm FX, Darlix JL. A 5′-3′ long-range interaction in Ty1 RNA controls its reverse transcription and retrotransposition. EMBO J. 2002;21:4368–4379. doi: 10.1093/emboj/cdf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu K, Heng X, Summers MF. Structural determinants and mechanism of HIV-1 genome packaging. J. Mol. Biol. 2011;410:609–633. doi: 10.1016/j.jmb.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Russell RS, Liang C, Wainberg MA. Is HIV-1 RNA dimerization a prerequisite for packaging? Yes, no, probably? Retrovirology. 2004;1:23. doi: 10.1186/1742-4690-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buchschacher GL, Jr, Panganiban AT. Human immunodeficiency virus vectors for inducible expression of foreign genes. J. Virol. 1992;66:2731–2739. doi: 10.1128/jvi.66.5.2731-2739.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luban J, Goff SP. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J. Virol. 1994;68:3784–3793. doi: 10.1128/jvi.68.6.3784-3793.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolton E, Coombes C, Eby Y, Cardell M, Boeke J. Identification and characterization of critical cis-acting sequences within the yeast Ty1 retrotransposon. RNA. 2005;11:308–322. doi: 10.1261/rna.7860605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu H, Boeke JD. Localization of sequences required in cis for yeast Ty1 element transposition near the long terminal repeats: analysis of mini-Ty1 elements. Mol. Cell Biol. 1990;10:2695–2702. doi: 10.1128/mcb.10.6.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morillon A. A cryptic unstable transcript mediates transcriptional silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev. 2008;22:615–626. doi: 10.1101/gad.458008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuda E, Garfinkel DJ. Posttranslational interference of Ty1 retrotransposition by antisense RNAs. Proc. Natl Acad. Sci. USA. 2009;106:15657–15662. doi: 10.1073/pnas.0908305106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, Bartel DP. RNAi in budding yeast. Science. 2009;326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 26.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weeks KM, Mauger DM. Exploring RNA structural codes with SHAPE chemistry. Acc. Chem. Res. 2011;44:1280–1291. doi: 10.1021/ar200051h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sztuba-Solinska J, Le Grice SF. Probing retroviral and retrotransposon genome structures: the “SHAPE” of things to come. Mol. Biol. Int. 2012;2012:530754. doi: 10.1155/2012/530754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eichinger DJ, Boeke JD. The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: cell-free Ty1 transposition. Cell. 1988;54:955–966. doi: 10.1016/0092-8674(88)90110-9. [DOI] [PubMed] [Google Scholar]
- 30.Garfinkel DJ, Boeke JD, Fink GR. Ty element transposition: reverse transcriptase and virus-like particles. Cell. 1985;42:507–517. doi: 10.1016/0092-8674(85)90108-4. [DOI] [PubMed] [Google Scholar]
- 31.Dutko JA, Kenny AE, Gamache ER, Curcio MJ. 5′ to 3′ mRNA decay factors colocalize with Ty1 gag and human APOBEC3G and promote Ty1 retrotransposition. J. Virol. 2010;84:5052–5066. doi: 10.1128/JVI.02477-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin JH, Levin HL. Reverse transcription of a self-primed retrotransposon requires an RNA structure similar to the U5-IR stem-loop of retroviruses. Mol. Cell. Biol. 1998;18:6859–6869. doi: 10.1128/mcb.18.11.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilhelm M, Wilhelm FX. Role of integrase in reverse transcription of the Saccharomyces cerevisiae retrotransposon Ty1. Eukaryot. Cell. 2005;4:1057–1065. doi: 10.1128/EC.4.6.1057-1065.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilhelm M, Wilhelm FX. Cooperation between reverse transcriptase and integrase during reverse transcription and formation of the preintegrative complex of Ty1. Eukaryot. Cell. 2006;5:1760–1769. doi: 10.1128/EC.00159-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friant S, Heyman T, Bystrom AS, Wilhelm M, Wilhelm FX. Interactions between Ty1 retrotransposon RNA and the T and D regions of the tRNA(iMet) primer are required for initiation of reverse transcription in vivo. Mol. Cell Biol. 1998;18:799–806. doi: 10.1128/mcb.18.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soto AM, Misra V, Draper DE. Tertiary structure of an RNA pseudoknot is stabilized by “diffuse” Mg2+ ions. Biochemistry. 2007;46:2973–2983. doi: 10.1021/bi0616753. [DOI] [PubMed] [Google Scholar]
- 37.Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- 38.Wu M, Tinoco I., Jr RNA folding causes secondary structure rearrangement. Proc. Natl Acad. Sci. USA. 1998;95:11555–11560. doi: 10.1073/pnas.95.20.11555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gherghe C, Lombo T, Leonard CW, Datta SA, Bess JW, Jr, Gorelick RJ, Rein A, Weeks KM. Definition of a high-affinity Gag recognition structure mediating packaging of a retroviral RNA genome. Proc. Natl Acad. Sci. USA. 2010;107:19248–19253. doi: 10.1073/pnas.1006897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mely Y, Cornille F, Fournie-Zaluski MC, Darlix JL, Roques BP, Gerard D. Investigation of zinc-binding affinities of Moloney murine leukemia virus nucleocapsid protein and its related zinc finger and modified peptides. Biopolymers. 1991;31:899–906. doi: 10.1002/bip.360310709. [DOI] [PubMed] [Google Scholar]
- 41.Mougel M, Barklis E. A role for two hairpin structures as a core RNA encapsidation signal in murine leukemia virus virions. J. Virol. 1997;71:8061–8065. doi: 10.1128/jvi.71.10.8061-8065.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giedroc DP, Cornish PV. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res. 2009;139:193–208. doi: 10.1016/j.virusres.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belcourt MF, Farabaugh PJ. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990;62:339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clare JJ, Belcourt M, Farabaugh PJ. Efficient translational frameshifting occurs within a conserved sequence of the overlap between the two genes of a yeast Ty1 transposon. Proc. Natl Acad. Sci. USA. 1988;85:6816–6820. doi: 10.1073/pnas.85.18.6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat. Rev. Mol. Cell Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- 46.Larsen LSZ, Zhang M, Beliakova-Bethell N, Bilanchone V, Lamsa A, Nagashima K, Najdi R, Kosaka K, Kovacevic V, Cheng J, et al. Ty3 capsid mutations reveal early and late functions of the amino-terminal domain. J. Virol. 2007;81:6957–6972. doi: 10.1128/JVI.02207-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Darlix JL. Control of Rous sarcoma virus RNA translation and packaging by the 5′ and 3′ untranslated sequences. J. Mol. Biol. 1986;189:421–434. doi: 10.1016/0022-2836(86)90314-1. [DOI] [PubMed] [Google Scholar]
- 49.Berkhout B, Das AT, van Wamel JL. The native structure of the human immunodeficiency virus type 1 RNA genome is required for the first strand transfer of reverse transcription. Virology. 1998;249:211–218. doi: 10.1006/viro.1998.9321. [DOI] [PubMed] [Google Scholar]
- 50.Beerens N, Kjems J. Circularization of the HIV-1 genome facilitates strand transfer during reverse transcription. RNA. 2010;16:1226–1235. doi: 10.1261/rna.2039610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balakrishnan M, Roques BP, Fay PJ, Bambara RA. Template dimerization promotes an acceptor invasion-induced transfer mechanism during human immunodeficiency virus type 1 minus-strand synthesis. J. Virol. 2003;77:4710–4721. doi: 10.1128/JVI.77.8.4710-4721.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. ShapeFinder: a software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA. 2008;14:1979–1990. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad. Sci. USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilkinson KA, Vasa SM, Deigan KE, Mortimer SA, Giddings MC, Weeks KM. Influence of nucleotide identity on ribose 2′-hydroxyl reactivity in RNA. RNA. 2009;15:1314–1321. doi: 10.1261/rna.1536209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Butcher SE, Pyle AM. The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc. Chem. Res. 2011;44:1302–1311. doi: 10.1021/ar200098t. [DOI] [PubMed] [Google Scholar]
- 56.Laing C, Jung S, Iqbal A, Schlick T. Tertiary motifs revealed in analyses of higher-order RNA junctions. J. Mol. Biol. 2009;393:67–82. doi: 10.1016/j.jmb.2009.07.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laing C, Schlick T. Analysis of four-way junctions in RNA structures. J. Mol. Biol. 2009;390:547–559. doi: 10.1016/j.jmb.2009.04.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.