Abstract
High-throughput bisulfite sequencing is widely used to measure cytosine methylation at single-base resolution in eukaryotes. It permits systems-level analysis of genomic methylation patterns associated with gene expression and chromatin structure. However, methods for large-scale identification of methylation patterns from bisulfite sequencing are lacking. We developed a comprehensive tool, CpG_MPs, for identification and analysis of the methylation patterns of genomic regions from bisulfite sequencing data. CpG_MPs first normalizes bisulfite sequencing reads into methylation level of CpGs. Then it identifies unmethylated and methylated regions using the methylation status of neighboring CpGs by hotspot extension algorithm without knowledge of pre-defined regions. Furthermore, the conservatively and differentially methylated regions across paired or multiple samples (cells or tissues) are identified by combining a combinatorial algorithm with Shannon entropy. CpG_MPs identified large amounts of genomic regions with different methylation patterns across five human bisulfite sequencing data during cellular differentiation. Different sequence features and significantly cell-specific methylation patterns were observed. These potentially functional regions form candidate regions for functional analysis of DNA methylation during cellular differentiation. CpG_MPs is the first user-friendly tool for identifying methylation patterns of genomic regions from bisulfite sequencing data, permitting further investigation of the biological functions of genome-scale methylation patterns.
INTRODUCTION
In mammalian genomes, DNA methylation primarily occurs symmetrically at cytosine residues followed by guanine (CpG) on both DNA strands, and ∼70–80% of CpG dinucleotides are methylated (1,2). DNA methylation patterns of genomic regions are associated with gene transcription, development, aging and tumorigenesis (3). Tissue-specific methylation patterns of CpG islands are strongly correlated with gene expression (4). Genome-wide hypomethylation is involved with aging and cellular differentiation (5–7). Aberrant DNA methylation patterns of gene promoter CpG islands have been widely observed in many kinds of human cancers (8–10). Laurent et al. (11) found that differentially methylated patterns between exons and introns in gene bodies could have epigenetic roles in transcript splicing. Low methylation levels in gene promoters and high methylation levels in gene bodies are associated with highly expressed genes (12).
Over the past 20 years, however, studies of DNA methylation mainly focused on CpG-rich regions, such as CpG islands or gene promoter regions, because of the limitations of DNA methylation analysis technologies, such as high cost, low resolution and sequence-specific basis (13). Bisulfite conversion sequencing remains the most standard and accurate method for examining the methylation status of cytosines at single-base resolution. Several technologies for measuring DNA methylation (BC-seq, MethyC-seq, BSPP, RRBS, WGBS and Methyl-MAPS) have been developed based on bisulfite conversion and next-generation sequencing methods, which have been widely applied to measure genome-wide methylation maps of human, mouse, Arabidopsis and other 17 eukaryotic genomes (7,11,12,14–18). Several alignment tools of short reads, such as Bowtie, Maq, SOAP and SOAP II, may be used to pre-process methylation data from short sequencing reads by aligning DNA methylation sequencing reads to a reference genome (19–22). Furthermore, several special programs [e.g. Bismark (23), BS Seeker (24), BSMAP (25), MethTools (26), QUMA (27), BISMA (28) and BiQ Analyzer HT (29)] have been developed to map high-throughput bisulfite sequencing data into reference genomes. A database (NGSmethDB) has been devised for the storage and retrieval of single-base resolution methylation data (30), which provides the actual methylation landscape of various cell types or tissues in eukaryotic genomes. These methods primarily relate to the determination of the methylation status or level of CpGs that takes the first step to further identify and analyze the genomic DNA methylation patterns from high-throughput bisulfite sequencing data.
In plant and animal genomes, the identification for DNA methylation patterns of genomic regions has been widely studied (31,32). The methylated regions could be associated with gene silencing and heterochromatin structures(11,33), while the unmethylated regions, such as promoter regions or CpG islands, are strongly associated with transcriptional activity (via RNA polymerase II) and active chromatin modification H3K4me3 (3). Studies on the methylation patterns of genomic regions have generally involved ‘interesting’ genomic regions (such as CpG islands, gene promoters, transposons, exons and introns). The average methylation levels of CpGs in the pre-defined genomic regions are computed to determine the methylation patterns of genomic regions (34,35). However, many of pre-defined genomic regions are arbitrary, such as ‘2-kb upstream of gene transcriptional start sites’, and the methylation patterns of shorter regions within the genomic regions could be masked by the average methylation level of all CpGs in the genomic regions. Therefore, the identification and analysis of methylation patterns of genomic regions from dozens of millions of CpG methylation data will be invaluable in revealing the biological function of genomic methylation patterns.
In previous studies, two main approaches have been used to identify the unmethylated regions: (i) computational methods for the identification of CpG-rich regions by DNA sequence features (36–39) and (ii) experimental methods for physically unmethylated regions by ChIP–chip (40) or CXXC affinity purification (41). However, both computational and experimental methods are biased toward the CpG-rich regions, and it is inevitably difficult to determine accurately the boundaries of these potentially unmethylated regions. In addition, the methylated regions may be identified from genome-wide methylation data measured by MeDIP-seq or MBD-seq based on affinity enrichment (42,43). To our knowledge, however, no specific bioinformatics tool may be used to identify the unmethylated and methylated regions from high-throughput bisulfite sequencing data. In the present study, we proposed an algorithm to identify the unmethylated and methylated regions by measuring the methylation status of cytosines based on the reliable bisulfite sequencing data, without the limitation of sequence features. It only depends on the methylation status of neighboring CpGs determined by bisulfite sequencing data, thus it overcomes the limitation of pre-defined regions and sequence features. It can accurately identify the boundaries of unmethylated and methylated regions. The genome-scale determination of unmethylated and methylated regions makes it possible to efficiently assess the biological function of genome-wide methylation patterns.
Differentially methylated regions (DMRs) have been widely identified among tissues, developmental cells and cancer types as being involved in tissue-, cell- or cancer-specific gene expression (11,44–47). Therefore, the identification and analysis of DMRs for paired or multiple samples would be of wide interest. Several methods for identification of DMRs among different samples (cell types or tissues) have been proposed, based on the statistical methods of the t-test for paired samples (48), the Kruskall–Wallis test and analysis of variance (ANOVA) for multiple samples (49,50). These traditional statistical methods are unfit for the identifications of DMRs because DNA methylation data are in general bimodal distribution rather than normal distribution. Especially, the determination of methylation patterns for DMRs among the multiple samples (≥3) remains a challenge for statistical methods (50,51). For example, the average methylation levels of the pre-defined regions in three genome regions 0.05, 0.15 and 0.2 could show significant difference by ANOVA. However, the genomic regions actually preserve the unmethylated pattern and the difference among the three samples could be caused by the experimental errors or sequencing depth. Recently, we developed a method of QDMR based on the Shannon entropy independent of the DNA methylation distribution, which may be used to quantitatively identify DMRs based on the average methylation level of CpGs in the pre-defined regions (52). However, QDMR could not directly identify the DMRs from bisulfite sequencing data at single-base resolution rather than methylation levels of pre-defined regions. Sliding window is a traditional method for pre-defined regions that are arbitrarily chosen and not taken the actual methylation status of CpGs into consideration. Here, we developed a combinatorial algorithm to determine the potentially genomic regions with different methylation patterns based on the bisulfite sequencing data across the paired or multiple samples. The combinatorial algorithm may qualitatively identify conservatively methylated regions (CMRs) and DMRs by determining whether the methylation patterns of genomic regions change among paired or multiple samples. Based on average methylation levels of CpGs in the potentially functional regions among paired or multiple samples, we further used the method of Shannon entropy to quantitatively assess the consistency or difference in the identified regions.
CpG_MPs was applied to the bisulfite sequencing data for five human cell types during stem-cell differentiation. Genome-wide unmethylated and methylated regions were identified for each of the five human cell types. And a large number of CMRs and DMRs were identified among the five human cell types that show significant sequence bias and cell-specific methylation patterns of genomic regions. To facilitate the users to effectively identify and analyze the potentially functional regions with different DNA methylation patterns from the bisulfite sequencing reads of CpGs at single-base resolution, the comprehensive tool of CpG_MPs provides the functions of data normalization, sequence features and visualization of genomic regions. A free version of the CpG_MPs software is available at http://bioinfo.hrbmu.edu.cn/CpG_MPs. The identification and analysis of potentially functional regions from paired or multiple samples could provide a new perspective, revealing the stability or dynamic changes of chromatin, epigenetic inheritance and regulation during cellular differentiation.
MATERIALS AND METHODS
Database
The human reference sequences and Refseq gene annotation were downloaded from the UCSC Genome Browser (53). Promoter regions were defined as being the 2-kb upstream and downstream of transcription start sites. The high-throughput bisulfite sequencing data of five human cell types during cellular differentiation were from two experiment laboratories. The data were from human embryonic stem cells (H1) and fetal lung fibroblasts (IMR90) downloaded from http://neomorph.salk.edu/human_methylome/ (7), human embryonic stem cells (H9), a fibroblastic differentiated derivative of the human embryonic stem cell (H9_fibro) and neonatal foreskin fibroblasts (Neonatal_fibro) from NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) (11). These raw bisulfite sequencing data were converted into the number of methylated reads and covered reads of cytosines (including unmethylated/methylated reads) by aligning them to the human reference genome (hg18) using the software of Bismark for bisulfite sequence alignment (23). The 14 318 physical unmethylated regions in human blood cells, based on the method of unmethylated CpG affinity chromatography, were obtained from Illingworth et al. (41).
Overview of software
A comprehensive tool, CpG_MPs, is introduced for identification and analysis of genomic regions with unmethylated and methylated patterns from bisulfite sequencing data, and to identify CMRs and DMRs for paired or multiple samples. It comprises four modules: (i) data normalization of the sequencing reads of CpGs; (ii) identification of unmethylated and methylated regions; (iii) identification of CMRs and DMRs across paired or multiple samples; and (iv) extraction of sequence features and visualization of the genomic regions with various methylation patterns. The pipeline of the software is summarized in Figure 1 and the detailed information as follows.
Figure 1.
Workflow of the software CpG_MPs for the identification and analysis of genomic regions with different methylation patterns from bisulfite sequencing data. (A) Input data of CpG_MPs include two sections: The bisulfite sequencing data need to be processed by converting raw bisulfite sequences into the numbers of methylated reads and covered reads for each cytosine, using the short-read alignment tool of Bismark. The reference sequence data of an organism may be downloaded directly from UCSC, NCBI or ENSEMBL. (B) Data normalization of CpG_MPs includes three functions of incorporating sequencing reads of the complementary positions of CpGs in sense and antisense strands, quality control of sequencing depth and calculation of methylation level of CpGs. (C) Identification of unmethylated regions and methylated regions based on the method of hotspot extension from the normalized methylation level of CpGs at single-base resolution. (D) Identification of CMRs and DMRs deduced from the determination of unmethylated and methylated regions of each sample. (E) Analysis of sequencing features and visualization for the genomic regions identified by CpG_MPs.
Data input
The input data of CpG_MPs consist of high-throughput bisulfite sequencing data of CpGs at single-base resolution. The bisulfite sequencing data are the sequencing reads of cytosines at single-base resolution in the TXT format, which needs to be processed by converting raw bisulfite sequences into the numbers of methylated reads and covered reads for each cytosine, using a specific tool of Bismark (23) for short-read alignment of high-throughput bisulfite sequencing. The input data are sorted automatically according to the order of their coordinates in the reference genome.
Normalization of the sequencing reads of CpGs
To determine accurately and efficiently the methylation patterns of genomic regions from the bisulfite sequencing data at single-base resolution, CpG_MPs first normalizes the sequencing reads of CpGs into the methylation level of CpGs in the unit interval of [0,1]. In the module of data normalization, CpG_MPs provides three basic procedures: incorporating sequencing reads, quality control and calculation of methylation level of CpGs. CpG_MPs facilitates the above procedures with a user-friendly interface to convert the sequencing reads into the standard methylation level of CpGs. The detailed procedures are as follows.
Incorporating sequencing reads in sense and antisense strands
Sequencing technologies for measuring DNA methylation may provide sequencing reads of single-base CpGs in sense and antisense strands. CpG_MPs provides a procedure that incorporates sequencing reads of symmetrical CpGs in sense and antisense strands. If the users wish to investigate CpG methylation in a single strand or compare CpG methylation statuses between sense and antisense strands, they only need to input methylation data of a single strand. And the procedure of incorporating sequencing reads of both strands is automatically skipped.
Quality control
To obtain accurate CpG methylation levels from high-throughput bisulfite sequencing technologies, CpG_MPs sets sequencing depth as an important parameter to measure the methylation level of CpGs. Recently, Laurent and colleagues performed saturated analysis of bisulfite sequencing depth and showed that at a sequencing depth ≥3, the accuracy of determining the methylation status of CpGs was high (79%) (11). To obtain more accurate methylation statuses of CpGs, the default parameter of sequencing depths of CpGs in CpG_MPs is set as 5, which may be adjusted by the users as necessary. All the CpGs with low sequencing depths are filtered.
Calculation of the methylation level of CpGs
The sequencing data of CpGs in each chromosome are aligned to the reference genome. The methylation level of each CpG is defined by the sequencing reads of CpGs as follows:
| (1) |
where reads (mCpG) represents the number of methylated CpG reads at the CpG dinucleotides and reads (mCpG) represents the total number of cover reads comprising unmethylated and methylated reads at the CpG dinucleotides. By this definition, the methylation level of CpG is normalized into the unit interval [0, 1].
Identification of the unmethylated and methylated regions using hotspot extension algorithm
To identify the genomic regions with unmethylated and methylated patterns, the methylation status for each CpG was divided into four categories, based on the normalized methylation levels of CpGs: (i) unmethylated CpGs with methylation levels ≤0.3, (ii) partially unmethylated CpGs ranging from 0.3 to 0.5, (iii) partially methylated CpGs ranging from 0.5 to 0.7 and (iv) methylated CpGs whose methylation levels ≥0.7. We devised an algorithm, termed as hotspot extension, to identify the genomic regions with the unmethylated and methylated patterns including two main steps: searching for hotspots and the extension of hotspots. The detailed algorithm is shown as follows.
Step 1: Convert the normalized methylation level of CpGs into the methylation status of CpGs.
Step 2: Scan CpGs from a 5′- to 3′-direction to extract the genomic regions including at least
 successively unmethylated (methylated) CpGs as unmethylated (methylated) hotspots.
successively unmethylated (methylated) CpGs as unmethylated (methylated) hotspots.Step 3: Extend the unmethylated (methylated) hotspots upstream and downstream to incorporate unmethylated (methylated) or partially unmethylated (methylated) CpGs into the hotspots as unmethylated regions, until methylated (unmethylated) or partially methylated (unmethylated) CpGs are met. The method allows for at most one CpG with different methylation statuses during the extension of hotspots.
Step 4: Combine two neighboring genomic regions with the same methylation pattern together if their distance is <200 bp.
Step 5: Compute the mean value and standard deviation of methylation level of CpGs in each unmethylated/methylated region.
The thresholds of  were determined by comparing the distributions of successively unmethylated/methylated CpGs between experimental samples of bisulfite sequencing data and computationally generated control samples. The default values of the parameters within the software may be adjusted for users from the setup options of CpG_MPs.
were determined by comparing the distributions of successively unmethylated/methylated CpGs between experimental samples of bisulfite sequencing data and computationally generated control samples. The default values of the parameters within the software may be adjusted for users from the setup options of CpG_MPs.
Identification of CMRs and DMRs
Based on the identification of methylation patterns of genomic regions in each sample by the second module of CpG_MPs, a method for the identification of conservatively unmethylated regions (CUMRs), CMRs and DMRs was devised by combining the combinatorial algorithm for determination of potentially functional regions with the method of Shannon entropy for quantitatively assessing consistency or difference of the identified regions. The schematic figure for identification of CMRs and DMRs is shown in Figure 2.
Figure 2.
The schematic figure for identification of CMRs and DMRs by combining the combinatorial algorithm with Shannon entropy.
First, the unmethylated/methylated regions of each sample identified by the second module of CpG_MPs are mapped into reference genome and the sample-methylation patterns of overlapping regions (ORs) in the reference genome are recorded (Figure 2A). To effectively mark the DNA methylation patterns of genomic regions, the unmethylated pattern is labeled as ‘−1’ and the methylated pattern as ‘1’. For  (
( ) samples, the two mathematical measures to determine the methylation patterns of ORs (conservatively or differentially methylated patterns) are defined as follows:
) samples, the two mathematical measures to determine the methylation patterns of ORs (conservatively or differentially methylated patterns) are defined as follows:
 |
(2) |
where  represents the number of samples with methylated pattern (‘1’) in the ORs and
represents the number of samples with methylated pattern (‘1’) in the ORs and  represents the number of samples with unmethylated pattern (‘−1’) in the ORs (Figure 2B). The measure
represents the number of samples with unmethylated pattern (‘−1’) in the ORs (Figure 2B). The measure  is defined to determine the methylation patterns of ORs across multiple samples, that is
is defined to determine the methylation patterns of ORs across multiple samples, that is
 |
(3) |
Since the genomic regions with different methylation patterns among multiple samples by CpG_MPs are dynamical regions rather than pre-defined regions, the number of samples in the ORs is unfixed. Therefore, we defined the measure  to assess the overlapping ratio of the number of samples with determined methylation patterns to the total number samples in ORs. Obviously, the larger
to assess the overlapping ratio of the number of samples with determined methylation patterns to the total number samples in ORs. Obviously, the larger  values (
values ( ) of ORs are, the more robust the determined methylation patterns of ORs across multiple samples are. Therefore, the methylation patterns and reliabilities of ORs may be marked by the
) of ORs are, the more robust the determined methylation patterns of ORs across multiple samples are. Therefore, the methylation patterns and reliabilities of ORs may be marked by the  values and
values and  values of ORs, respectively.
values of ORs, respectively.
Next, a strategy of hotspot extension is used to merge the neighboring ORs with the same methylation pattern across multiple samples, because many neighboring ORs with the same methylation patterns are split by their different  values caused by sequencing depth of raw data or the dynamic algorithm for the identification of unmethylated and methylated regions in each sample using CpG_MPs (Figure 2C). The strategy of hotspot extension was devised as follows. First, the ORs are ranked from the largest to the smallest according to their
values caused by sequencing depth of raw data or the dynamic algorithm for the identification of unmethylated and methylated regions in each sample using CpG_MPs (Figure 2C). The strategy of hotspot extension was devised as follows. First, the ORs are ranked from the largest to the smallest according to their  values that is used to assess the reliability of methylation patterns of ORs. The ORs with the maximum
values that is used to assess the reliability of methylation patterns of ORs. The ORs with the maximum  values are optimized and chosen as the hotspots. Then, the hotspots are extended to merge the neighboring ORs with the same methylation pattern when the distances between hotspots and neighboring ORs are <200 bp and
values are optimized and chosen as the hotspots. Then, the hotspots are extended to merge the neighboring ORs with the same methylation pattern when the distances between hotspots and neighboring ORs are <200 bp and  values of neighboring ORs <0.5. The rule for merging the hotspots and neighboring ORs with same methylation pattern into new hotspots is devised as follows:
values of neighboring ORs <0.5. The rule for merging the hotspots and neighboring ORs with same methylation pattern into new hotspots is devised as follows:
 |
(4) |
where  and
and  represent, respectively, the
represent, respectively, the  value of hotspots and its neighboring ORs. This algorithm is repeated until
value of hotspots and its neighboring ORs. This algorithm is repeated until  values of all the remaining ORs are <0.5. Finally, the new hotspots containing at least four CpGs are identified as the CMRs, CUMRs or DMRs. In this study,
values of all the remaining ORs are <0.5. Finally, the new hotspots containing at least four CpGs are identified as the CMRs, CUMRs or DMRs. In this study,  = 0.5 is used as a cutoff to assess the reliability of methylation patterns of ORs, that is the methylation patterns of at least half of samples are determined in ORs. And the threshold for
= 0.5 is used as a cutoff to assess the reliability of methylation patterns of ORs, that is the methylation patterns of at least half of samples are determined in ORs. And the threshold for  and the number of CpGs included in the identified regions may be adjusted by users as necessary in the software of CpG_MPs.
and the number of CpGs included in the identified regions may be adjusted by users as necessary in the software of CpG_MPs.
Furthermore, the  values of DMRs may be used to classify DMRs into two methylation patterns as shown in Equation (5).
values of DMRs may be used to classify DMRs into two methylation patterns as shown in Equation (5).
| (5) |
Therefore, the  values of DMRs may further be used to extract sample-specific DMRs. For example, DMRs are sample-specific, if the absolute value of
values of DMRs may further be used to extract sample-specific DMRs. For example, DMRs are sample-specific, if the absolute value of  for the DMRs across
for the DMRs across  (
( ) samples. The software of CpG_MPs provides the function to extract the sample-specific DMRs that may be regarded as the DNA methylation markers of the sample in contrast to other samples.
) samples. The software of CpG_MPs provides the function to extract the sample-specific DMRs that may be regarded as the DNA methylation markers of the sample in contrast to other samples.
Finally, a method of modified Shannon entropy was used to quantitatively assess the identified regions across paired or multiple samples. The average methylation levels of CpGs in the identified regions were computed. Then, the entropy values of the identified regions were computed based on the average methylation level of each identified region in paired or multiple samples by modified Shannon entropy. The detailed formulas and algorithms of determined thresholds for modified Shannon entropy were shown in our previous study for the quantitative identification of DMRs (52). The larger the entropy values are, the more consistent the determined CUMRs/CMRs among multiple samples are, while the lower the entropy values are, the larger methylation changes of the determined DMRs among multiple samples are. The significant CUMRs/CMRs or DMRs were extracted based on the corresponding thresholds of entropy for different number of samples.
Sequence features of genomic regions of different methylation patterns
The sequence features of genomic regions identified by CpG_MPs from high-throughput bisulfite sequencing data include length, GC content and CpG ratio.
Length is equal to the number of the nucleotides in a genomic region.
| (6) |
| (7) |
where 
 and
and  are the number of C, G and CpG nucleotides in a genomic region, respectively.
are the number of C, G and CpG nucleotides in a genomic region, respectively.
Identification of DMRs by Fisher’s exact test based on sliding window
As the description of Lister et al., we developed a program to identify the DMRs from the bisulfite sequencing data of CpGs between the H1 and IMR90 cells by the method of Fisher’s exact test based on sliding window (FET_SW) with 1-kb window size and 1000-bp sliding step (7). Total 491 de novo methylated regions and 75 378 demethylated regions were identified by FET_SW from H1 to IMR90 cell, respectively.
Gene ontology annotation
The functional annotation analysis of the genes with de novo methylated regions among the five human samples was performed by the DAVID Bioinformatics Resources 6.7 website of http://david.abcc.ncifcrf.gov (54). The human NCBI gene list was used as the reference genome. The P-value modified by Benjamini correction for significance was set at 10 × e−3for Gene Ontology (GO) analysis.
RESULTS AND DISCUSSION
Data normalization of bisulfite sequencing data
To quantitatively measure the methylation level of CpGs from bisulfite sequencing data, we devised a module of data normalization to convert the sequence reads of CpGs into the methylation level of CpGs in the unit interval [0, 1]. Two main factors are taken into consideration: sequencing information of both strands and sequencing depth of CpGs at single-base resolution. CpG dinucleotides are known to be symmetrical in sense and antisense strands, based on the principle of complementary base pairing. Earlier studies have shown that methyltransferase DNMT1 maintains the symmetric structure of CpG methylation in both strands after DNA replication, because DNMT1 has high preference for hemimethylated target sites during DNA replication (55,56). Therefore, CpG_MPs provides a simple procedure that merges sequencing reads of symmetrical CpGs in both strands, which may double their sequencing depths to improve the saturation of sequencing depths without any extra cost. On the other hand, several studies have reported that a few regions of strand-biased DNA methylation were observed in animal and plant genomes (especially in centromeric regions) associated with heterochromation structure and transcriptional silencing during DNA replication (57,58).Therefore, the procedure of incorporating sequencing reads of both strands may be automatically skipped if the users take the strand-biased CpG methylation into consideration (detailed information in ‘Materials and Methods’ section). In addition, the users may further identify the strand-specific methylation regions through setting the methylation information of sense strand and antisense strand as two samples. The assessment of methylation level of CpGs is related to the sequencing depth of the DNA methylation mapping technologies. The saturation of sequencing depth may give rise to accurate DNA methylation level of CpG-specific loci (42). Therefore, the sequencing depth of CpGs is regarded as an important parameter of quality control in the module of data normalization. Finally, the conventional method is used to convert the sequencing read into the standard methylation level of CpG by Equation (1) as shown in ‘Material and Methods’ section.
The module of data normalization of CpG_MPs was applied to determine the genome-wide methylomes of five human cell types: H1, H9, H9_fibro, Neonatal_fibro and IMR90 as shown in ‘Materials and Methods’ section. In this study, the sequencing information of both strands is merged together and the threshold of sequence depth methylation level of CpGs is set as 5. These five standard methylomes of CpGs for different cell types in human genome have been depicted that may be downloaded freely from our website. The basic statistics of measured CpGs by bisulfite sequencing data are shown in the Supplementary Table S1. The results show high genome-wide coverage ratio (>0.75) of measured CpGs by the module of data normalization of CpG_MPs for the five different types of human genome that can be used to further investigate the methylation patterns of genomic regions.
Identification of unmethylated and methylated regions from the normalized methylation level of CpGs
Inspired by the tendency of neighboring CpGs sharing the same methylation status (50,51,59,60), we devised a searching algorithm of hotspot extension to identify the unmethylated and methylated regions by searching for successive CpGs with the same methylation status from a single sample (cell type or tissue). The accurate methylation status of CpGs is regarded as the ‘switch’ of genomic methylation patterns.
To verify the potential epigenetic mechanism of neighboring CpGs sharing uniform methylation pattern genome-wide, we first computed the distribution of the four kinds of methylation statuses of CpGs in five human experimental samples (cell types) (see ‘Materials and Methods’ section). The average proportions of CpGs for each methylation status in the five cell types were regarded as the distribution of methylation statuses of CpGs for the human genome. The mean proportions of CpGs with the four kinds of methylation statuses were computed (see Supplementary Figure S1). We observed that most CpGs (65–95%) were methylated or partially methylated in the five cell types of human genome during differentiation, which was consistent with the previous findings of global DNA methylation of human genome (2). However, the distributions of methylation statuses of CpGs were significantly different among the five cell types, which are consistent with dynamic changes of human methylomes during cellular differentiation (7,11).
Next, the average proportions of CpGs with the four methylation statuses in the five cell types were regarded as the distribution of methylation statuses of CpGs in human experimental samples. Based on the distribution of methylation statuses of CpGs in human experimental samples, we computationally generated 1000 control samples with random methylation status of CpGs as follows. The random sequence comprised 1 000 000 CpGs, and each CpG was randomly assigned one of four kinds of methylation statuses. The occurrence probabilities for the four kinds of methylation statuses of CpGs in the random sequence follow their corresponding distribution in human genome, as determined by the five bisulfite sequencing data as mentioned above. The process was repeated 1000 times. The 1000 random sequences with CpGs of random methylation statuses were generated as control samples. We searched the genomic regions for successive CpGs with the same methylation status in the experimental samples and control samples. The cumulative distribution functions of CpGs for the number of successive CpGs with the four methylation statuses were computed in the experimental samples and the control samples, respectively (Figure 3). We observed that the cumulative distribution functions of CpGs of unmethylated and methylated status in the experimental samples were significantly lower than those in the randomly generated control samples (Figure 3A). For example, <5% of CpGs of unmethylated status continuously appeared at least three times in the control samples, yet those in experimental samples accounted for >60% of the total CpGs. These results indicate that the unmethylated CpGs gather into significant clusters to form genomic regions with the unmethylated pattern in human methylomes, rather being generated randomly. The methylated CpGs have a similar trend to form genomic regions with the methylated pattern (Figure 3D). On the other hand, the cumulative distribution functions of successive CpGs with a partially methylated status and a partially unmethylated status in the experimental samples are consistent with those in the control samples (Figure 3B and C). These results suggest that the unmethylated/methylated CpGs significantly gather into clusters to form genomic regions with unmethylated/methylated pattern, while partially unmethylated/methylated CpGs may not gather into clusters to form genomic regions with the partially unmethylated/methylated pattern. Therefore, the methylation patterns of human genomic regions may be divided into two classes: unmethylated and methylated patterns. The genomic regions with successive unmethylated (methylated) CpGs are regarded as hotspots of the unmethylated (methylated) pattern, respectively. The threshold of the amount of successive unmethylated CpG sites was set as 3 to identify statistically significant unmethylated hotspots using the probability (P-value < 0.05) of CpGs for successive three unmethylated CpGs in the total CpGs in random samples (Figure 3A).
Figure 3.
Distributions of successive CpGs with the same methylation status. For the four kinds of methylation statues of CpGs, the red solid lines represent the mean-fit lines of the cumulative distribution functions of CpGs for the number of successive CpGs in five experimental samples, and the blue dashed lines are for the 1000 control samples generated randomly.
The unmethylated and methylated hotspots were identified from the standard methylomes in five experiential samples. However, we found that the average length of the unmethylated hotspots was only 704 bp and that of methylated hotspots was 1068 bp, which were shorter than that the expected length of unmethylated/methylated regions (Table 1). We observed that the methylation levels for the unmethylated CpGs and partially unmethylated CpGs were similar; these marginal CpGs of partially unmethylated status could be excluded, and neighboring unmethylated hotspots were divided by the strict thresholds or missed values for the identification of unmethylated hotspots. The same trend was observed in the methylated hotspots. Therefore, the shorter lengths of unmethylated/methylated hotspots could be caused by the strict thresholds used in determining the methylation status or sequencing depth by the measuring technologies.
Table 1.
The basic statistics of unmethylated/methylated hotspots and extended unmethylated/methylated regions in five human cell types
| Unmethylated hotspots | Unmethylated regions | Methylated hotspots | Methylated regions | |
|---|---|---|---|---|
| Number | 855 009 | 594 795 | 6 651 980 | 1 187 182 |
| Length ± SDa | 704 ± 33 058 | 2169 ± 41 691 | 1068 ± 19209 | 9274 ± 64 097 |
| CpG number ± SDa | 13 ± 32 | 25 ± 46 | 10 ± 12 | 77 ± 156 |
| Methylation level ± SDa | 0.11 ± 0.06 | 0.2 ± 0.10 | 0.88 ± 0.05 | 0.78 ± 0.07 |
aMethylation level represents the average methylation level of all CpGs in the identified regions.
To confirm the validation of hotspot extension to preserve the integrity of genomic methylation patterns, we identified the unmethylated and methylated regions using hotspot extension in five experimental samples. As shown in Table 1, we found that the average length and number of CpGs in unmethylated/methylated regions extended by hotspots increased by >3- (or 9-) fold compared to those identified using strict thresholds. Meanwhile, the total number of unmethylated and methylated regions was reduced by >70%. These results suggest that many neighboring hotspots with the same methylation patterns were split by the strict threshold, yet the extension of hotspots could combine them into a complete region. The average methylation level and standard deviation of CpGs in the unmethylated/methylated regions extended by hotspots were computed to quantitatively evaluate the mean methylation level and variation of CpGs in the identified regions. The results show that the average methylation level of unmethylated regions is still low (0.20), while that of methylated regions keeps high average methylation level (0.78). Meanwhile, the average standard deviations of unmethylated/methylated regions keep the low level (<0.10) (Table 1). These results indicate that the extended unmethylated /methylated regions still preserve their hypomethylated/hypermethylated level, even when the threshold of methylation status is loosened.
Based on the above observations, we devised an algorithm of hotspot extension to identify the genomic regions with unmethylated/methylated pattern using two main steps. First, we marked the genomic unmethylated/methylated regions with the stringent rule, based on the clusters of successive unmethylated/methylated CpGs that guarantee the accurate positioning of the unmethylated/methylated regions. Then, we loosened the condition for determining the methylation status and allowed one CpG with different methylation statuses during the extension of hotspots that may reduce the effect of the strict categories of methylation status and the sequencing depth. The CpGs with similar methylation status in their flanking regions were incorporated into the hotspots as genomic unmethylated/methylated regions. The detailed algorithm is shown in the ‘Materials and Methods’ section. The method of hotspot extension may not only accurately determinate the locations of unmethylated/methylated regions by strict thresholds but also could maintain the integrity of unmethylated/methylated regions through hotspot extension. Overcoming the limitation of average methylation level of window size or pre-defined regions, CpG_MPs becomes feasible for discovering dynamic regions depending on the methylation status of CpGs at single-base resolution from bisulfite sequencing data. Therefore, it could identify the accurate boundary of the genomic regions with different methylation patterns, because a linear searching strategy of CpG sites is used to search the neighboring CpGs with the same methylation status rather than the average methylation level of CpGs in specific regions.
The second module of CpG_MPs is further used to identify the unmethylated and methylated regions for each of the five samples as mentioned above. The numbers and coverage ratios of methylated regions are significantly greater than those of unmethylated regions consistent with the global methylation of human genome (2,61) (Supplementary Table S2). The average methylation level and standard deviation of CpGs in each unmethylated/methylated regions were computed by CpG_MPs as two quantitative indicators of methylation patterns of genomic regions. As shown in Figure 4A, we found that the unmethylated regions and methylated regions show the distinct distributions of the average methylation levels among the five cell types. Almost all of unmethylated regions keep the hypomethylated average methylation levels (<0.3), while >85% of methylated regions keep the hypermethylated average methylation levels (>0.7). On the other side, the methylation levels of CpGs in unmethylated and methylated regions maintain the low standard deviation (<0.2) (Figure 4B). These results indicate that CpG_MPs may effectively identify unmethylated and methylated regions and the CpGs in the identified regions preserve the consistent methylation status.
Figure 4.
Comparison of average methylation levels and standard deviations of CpGs in genomic regions with different methylation patterns among five human cell types. Box plots in (A) and (B) show, respectively, the genome-wide distributions of average methylation level and standard deviation of CpGs in five cell types of H1, H9, H9_fibro, Neonatal_fibro and IMR90.
To further confirm the reliability of the unmethylated regions identified by CpG_MPs, we used the genome-wide 14 318 physically unmethylated regions identified by Illingworth et al. as the test set of unmethylated regions (see ‘Materials and Methods’ section). These physically unmethylated regions from human blood cells were biased toward CpG-enriched regions and limitation of CXXC affinity chromatography techniques (41); therefore, the amount of physically unmethylated regions is relative lower than one identified by CpG_MPs. Therefore, we only computed the overlapping numbers of the physical unmethylated regions with the unmethylated regions identified by CpG_MPs from each of five cell types (Supplementary Figure S2). The high overlapping ratios of physically unmethylated regions with the identified unmethylated regions from five cell types were observed: 92% in H1, 94% in H9, 93% in H9_fibro, 96% in Neonatal_fibro and 96% in IMR90. This result indicated that almost all the physically unmethylated regions could be identified by our methods in the five different cells. On the other hand, the numbers of unmethylated regions identified by CpG_MPs from the five cells are 2- to 16-fold of physical unmethylated regions. These results indicate that CpG_MPs could identify accurately unmethylated regions, and many new unmethylated regions identified by CpG_MPs from bisulfite sequencing data of the different cell types or tissues, may be regarded as physical unmethylated regions of human cell- or tissue-specific genomes. Although the systematic investigation of the methylated regions has not been investigated to date, the genome-wide methylated regions identified by CpG_MPs could provide a new perspective in the study of the function of DNA methylation and the dynamic structures of heterochromatin in different cell types or tissues.
Identification of CUMRs, CMRs and DMRs across paired or multiple samples
The accurate identification of DMRs is key requisite to investigate the regulatory functions of DNA methylation across paired or multiple samples. Here, we developed a novel method to identify DMRs, CUMRs and CMRs across paired or multiple (≥3) samples, by combining the combinatorial algorithm to identify the genomic regions with different methylation patterns, with a method of Shannon entropy to quantitatively evaluate the difference or consistency of the identified regions. It includes two main steps and the detailed algorithm is shown in ‘Materials and Methods’ section.
First, a combinatorial algorithm was devised to qualitatively identify DMRs, CUMRs and CMRs by a combinatorial approach for determining the methylation patterns of ORs across paired or multiple samples. However, the combinatorial approach faces two main problems. The determination of methylation patterns of ORs is a complex problem, because there are in theory  kinds of combinatorial methylation patterns of ORs for
kinds of combinatorial methylation patterns of ORs for  samples. The kinds of methylation patterns of genomic regions across multiple samples rise exponentially along with the number of samples. Additionally, the locations of unmethylated/methylated regions identified by CpG_MPs are unfixed among different samples that cause the complex information of methylation patterns and shorter lengths of ORs in the process of combinatorial iteration when the number of samples is larger (Figure 2A). To resolve two problems, we defined two mathematical measures
samples. The kinds of methylation patterns of genomic regions across multiple samples rise exponentially along with the number of samples. Additionally, the locations of unmethylated/methylated regions identified by CpG_MPs are unfixed among different samples that cause the complex information of methylation patterns and shorter lengths of ORs in the process of combinatorial iteration when the number of samples is larger (Figure 2A). To resolve two problems, we defined two mathematical measures  and
and  to determine the methylation patterns and reliabilities of ORs as shown in Equation (2). The measure
to determine the methylation patterns and reliabilities of ORs as shown in Equation (2). The measure  is used to determine three methylation patterns of ORs (DMRs, CUMRs/CMRs) by Equation (3), which overcomes the complex problem for
is used to determine three methylation patterns of ORs (DMRs, CUMRs/CMRs) by Equation (3), which overcomes the complex problem for  kinds of combinatorial methylation patterns of ORs across
kinds of combinatorial methylation patterns of ORs across  samples in theory. However, the algorithm omits the detailed information of methylation patterns in each sample of ORs. To resolve the defect, CpG_MPs provides the information of average methylation level of the identified ORs in each sample that is convenient for the users to further investigate the dynamic mechanism of methylation patterns across multiple samples. Next, the strategy of hotspot extension was devised to merge neighboring ORs with the same methylation patterns by Equation (4), which may reduce the effect of shorter and scattered ORs caused by dynamic locations of genomic regions in different samples. In addition, the
samples in theory. However, the algorithm omits the detailed information of methylation patterns in each sample of ORs. To resolve the defect, CpG_MPs provides the information of average methylation level of the identified ORs in each sample that is convenient for the users to further investigate the dynamic mechanism of methylation patterns across multiple samples. Next, the strategy of hotspot extension was devised to merge neighboring ORs with the same methylation patterns by Equation (4), which may reduce the effect of shorter and scattered ORs caused by dynamic locations of genomic regions in different samples. In addition, the  values may be further determined the tendency of the methylation patterns of DMRs by Equation (5), which may be used to identify the sample-specific DMRs. Second, a method of Shannon entropy has been used to quantitatively assess the consistency of the identified CUMRs/CMRs and difference of the identified DMRs across paired or multiple samples, respectively. The significant CUMRs/CMRs or DMRs were excavated across paired multiple samples according to the corresponding thresholds of entropy that may reduce the false positive of the genomic regions with different methylation patterns qualitatively identified by the combinatorial algorithm.
values may be further determined the tendency of the methylation patterns of DMRs by Equation (5), which may be used to identify the sample-specific DMRs. Second, a method of Shannon entropy has been used to quantitatively assess the consistency of the identified CUMRs/CMRs and difference of the identified DMRs across paired or multiple samples, respectively. The significant CUMRs/CMRs or DMRs were excavated across paired multiple samples according to the corresponding thresholds of entropy that may reduce the false positive of the genomic regions with different methylation patterns qualitatively identified by the combinatorial algorithm.
The third module of CpG_MPs was applied to identify the CUMRs/CMRs and DMRs between H1 and IMR90 cell, whose bisulfite sequencing data were obtained from one experimental laboratory (7). For paired samples, the DMRs may be further classified into de novo methylated regions and demethylated methylation regions according to the orientation of methylation change from H1 to IMR90 cells. As shown in Table 2, the 27 459 CUMRs, 4654 de novo methylated regions, 199 213 demethylated regions and 353 209 CMRs were identified from H1 to IMR90. The number of demethylated regions is significantly higher (>75-fold) than that of de novo methylated regions. These results suggest that the methylated regions could be more instability than the unmethylated regions during cellular differentiation.
Table 2.
DNA methylation patterns and sequence features of CMRs and DMRs between H1 and IMR90
| Regions | Number | Length ± SD | GC Content ± SD | CpG Ratio ± SD |
|---|---|---|---|---|
| CUMRs | 27 459 | 1263 ± 1498 | 0.58 ± 0.09 | 0.64 ± 0.22 |
| De novo methylated regions | 4654 | 597 ± 643 | 0.52 ± .1 | 0.49 ± 0.28 |
| Demethylated regions | 199 213 | 2384 ± 68 906 | 0.42 ± 0.08 | 0.25 ± 1.58 |
| CMRs | 353 209 | 4624 ± 31 721 | 0.45 ± 0.08 | 0.33 ± 0.26 |
CUMRs represent the conservatively unmethylated region in both H1 and IMR90; de novo methylated regions represent unmethylated in H1 and methylated in IMR90; Demethylated regions represent methylated in H1 and unmethylated in IMR90; and CMRs is conservatively methylated region in both H1 and IMR90.
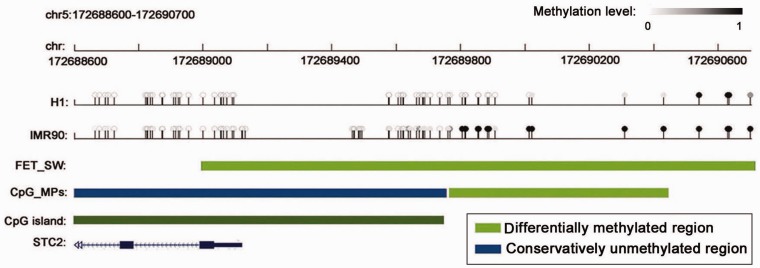
Compared with the method of FET_SW based on sliding window for identification of DMRs proposed by Lister et al. (see ‘Materials and Methods’ section), CpG_MPs identified a large number of new and short DMRs consisting of de novo methylated regions and demethylated regions from H1 to IMR90 cell. The number (4654) of de novo methylated regions by CpG_MPs is >9-fold of one (491) by FET_SW and >2-fold for the demethylated regions as shown in Figure 5 and Supplementary Table S3. The absolute difference of average methylation levels of CpGs in each DMR was computed; the results show that both de novo methylated regions and demethylated regions by CpG_MPs show the larger difference of DNA methylation than those by FET_SW from H1 to IMR90 cell (Figure 5). CpG_MPs identified 3831 new de novo methylated regions from H1 to IMR90, where 903 out of these regions located in the promoter regions of 827 genes (Supplementary Table S4). Many short de novo methylated regions were observed in the gene promoters. For example, CpG_MPs identified the shortest de novo methylated region with only 11 bp in the promoter region of FASLG gene that plays important roles in human stem-cell differentiation and cancer development (62,63). GO annotation analysis was performed for these target genes whose promoter regions contain the new de novo methylated regions. The results show that they are closely associated with positive regulation of gene transcription, RNA metabolic process, nucleobase, nucleoside, nucleotide and nucleic acid metabolic process, embryonic morphogenesis, embryonic organ development, skeletal system development and cell morphogenesis involved in differentiation as shown in Table 3. It suggests that CpG_MPs may identify many new de novo methylated regions and their target genes are involved in the regulation of gene transcription during embryonic development. As for the DMRs identified by FET_SW, 97% (478) of de novo methylated regions and >81% (61 194) of demethylated regions may overlap with the corresponding DMRs by CpG_MPs. What’s more, CpG_MPs may accurately identify the boundaries of DMRs. For example, both CpG_MPs and FET_SW may identify the DMRs in promoter region of gene SCT2 (Figure 6). Our algorithm of CpG_MPs may accurately identify the boundaries of DMR. However, the DMR identified by FET_SW include many non-differentially methylated CpGs in CpG islands. In fact, these CpGs in the CpG island keep low methylation level and are identified as a CUMR by CpG_MPs. The actual DMR is in the flanking regions of CpG islands rather than in CpG islands.
Figure 5.
Comparison of DNA methylation difference of DMRs identified by CpG_MPs and FET_SW from H1 to IMR90. Absolute difference of average methylation levels in DMRs (de novo methylated regions and demethylated regions) represents the absolute difference of average methylation levels of CpGs in DMRs between H1 and IMR90. The basic statistic is shown in the Supplementary Table S3.
Table 3.
GO annotation analysis of 827 genes containing new de novo methylated regions in their promoter regions between H1 and IMR90
| GO terms | Count | Benjamini P-value | GO terms | Count | Benjamini P-value |
|---|---|---|---|---|---|
| Positive regulation of transcription | 55 | 2.97 × e−06 | Embryonic organ development | 24 | 8.26 × e−05 |
| Embryonic morphogenesis | 38 | 5.40 × e−06 | Positive regulation of biosynthetic process | 57 | 8.94 × e−05 |
| Positive regulation of gene expression | 55 | 5.60 × e−06 | Regulation of transcription from RNA polymerase II promoter | 59 | 8.98 × e−05 |
| Positive regulation of transcription, DNA dependent | 47 | 1.59 × e−05 | Skeletal system development | 34 | 1.11 × e−04 |
| Positive regulation of RNA metabolic process | 47 | 1.72 × e−05 | Positive regulation of transcription from RNA polymerase II promoter | 37 | 1.51 × e−04 |
| Positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 56 | 1.93 × e−05 | Cellular component morphogenesis | 38 | 2.63 × e−04 |
| Embryonic organ morphogenesis | 22 | 2.60 × e−05 | positive regulation of macromolecule metabolic process | 64 | 2.85 × e−04 |
| Positive regulation of nitrogen compound metabolic process | 56 | 2.79 × e−05 | Embryonic skeletal system development | 15 | 2.92 × e−04 |
| Positive regulation of macromolecule biosynthetic process | 56 | 4.16 × e−05 | Cell morphogenesis | 35 | 3.42 × e−04 |
| Positive regulation of cellular biosynthetic process | 57 | 7.29 × e−05 | Cell morphogenesis involved in differentiation | 27 | 7.12 × e−04 |
The annotations of GO terms with Benjamini P-value < 10 × e−03 are listed.
Figure 6.
A promoter region of STC2 gene in human chromosome 5 overlapped with the de novo methylated regions identified by both methods of FET_SW and CpG_MPs from the bisulfite sequencing data of H1 and IMR90. The CpG island in promoter regions of STC2 gene was obtained from UCSC Genome Browser (53).
CpG_MPs may also be applied to identify the CMRs and DMRs among multiple samples (see ‘Materials and Methods’ section). It identified 25 009 CUMRs, 411 756 CMRs and 245 685 DMRs across the five cell types related to embryo stem-cell differentiation (Table 4). As for these CUMRs, we found that they overlapped with >88% of the14 318 physical unmethylated regions in human blood cell as shown in ‘Materials and Methods’ section. It indicates that the 25 009 CUMRs identified by CpG_MPs were reliable unmethylated regions in different cell types. In addition, we found these CUMRs obtained high GC contents and CpG ratios as shown in Table 4. The 25 009 CUMRs satisfy the two key criterions of unmethylated regions and CpG-rich regions for identification of CpG islands. Therefore, they may be regarded as the reliable ‘CpG islands’ in human genome. Among the 245 685 DMRs among the five cell types, 63 951cell-specific DMRs were identified by CpG_MPs and may be regarded as the ‘DNA methylation fingerprint’ to distinguish the cell types as shown in Table 4 (64,65). The basic statistics of the methylation patterns of cell-specific regions are shown in Supplementary Table S5. The results show that the number (53 086) of cell-specific unmethylated regions is >5-fold of the number (10 865) of methylated regions. And most of cell-specific regions of human embryonic stem cells (H1) are methylated regions, yet the most of cell-specific regions for Neonatal_fibro and IMR90 of newborn human fibroblasts are unmethylated regions. These results indicate that the methylation patterns of cell-specific regions in five cell types possess significant preference and a portion of genomic regions demethylated from embryonic stem cells to human fibroblasts.
Table 4.
DNA methylation patterns and sequence features of CMRs and DMRs for five human samples
| Regions | Number | Length ± SD | GC content ± SD | CpG ratio ± SD |
|---|---|---|---|---|
| CMRs | 411 756 | 3742 ± 5740 | 0.45 ± 0.07 | 0.30 ± 1.31 |
| CUMRs | 25 009 | 1214 ± 1256 | 0.59 ± 0.08 | 0.64 ± 0.22 |
| DMRs | 245 685 | 1526 ± 9123 | 0.45 ± 0.09 | 0.27 ± 1.09 |
| Sample-specific DMRsa | 63 951 | 1154 ± 17 606 | 0.47 ± 0.10 | 0.34 ± 2.06 |
aSample-specific DMRs of each cell of H1, H9, H9_fibro, Neonatal_fibro and IMR90 are shown in Supplementary Table S5.
Sequence features and visualization of genomic regions of different methylation patterns
DNA methylation patterns of genomic regions are associated with the sequence features of their DNA context (32,66). The tissue-specific and cancer-specific methylation patterns of genomic regions are associated with regions that are the flanking sequences of CpG islands (4,46). However, deconstructing the function of the CpG distribution for DNA methylation patterns remains challenging because of the distributions of both CpG sites and their methylation statuses are uneven throughout the whole genome. Therefore, CpG_MPs provides the fourth module to analyze sequence features (length, GC content, CpG ratio) and visualize genomic regions of different methylation patterns. The detailed equations for the sequence features are shown in ‘Materials and Methods’ section. It is useful for users to further explore the potential effect of DNA sequence features on the DNA methylation patterns of the genomic regions.
The fourth module of CpG_MPs was used to compute the sequence features of CUMRs, CMRs and DMRs of paired or multiple samples from the five human cell types as shown in Tables 2 and 4. We found that the genomic regions with different methylation patterns showed preferences for GC contents and CpG ratios: high in CUMRs, moderate in DMRs and low in CMRs consistent with the previous studies (47,67). All of the genomic regions identified by CpG_MPs include at least four CpGs, which guarantees the stability of the methylation patterns of the genomic regions. Thus, sequence features analysis of genomic regions with different methylation patterns facilitates the identification of the epigenetic mechanism of changing methylation.
In addition, the genomic regions with different methylation patterns identified by CpG_MPs from bisulfite sequencing data may be output as a table or a graph. To facilitate taking advantage of other genetic and epigenetic information (such as genes, SNPs, CpG islands, DNA methylation and histone modifications) in the relevant biological databases, it provides a directly linked server from the genomic methylation regions out to the bioinformatics secondary databases of UCSC (53), MethyCancer (68) and HHMD (69). Users may rapidly and efficiently obtain the relevant information for the genomic regions with different methylation patterns.
The detailed information of identified genomic regions by the four modules of CpG_MPs from the five cell types can be downloaded from http://bioinfo.hrbmu.edu.cn/CpG_MPs.
CONCLUSION
CpG_MPs provides an efficient and comprehensive tool for the identification and analysis of genomic regions with different methylation patterns from high-throughput bisulfite sequencing data. CpG_MPs includes four modules to standardize methylation level of CpGs, identifies genomic regions of different methylation patterns, analyzes sequence features and visualizes the identified regions. It may accurately identify and analyze the unmethylated and methylated regions based on the methylation status of CpGs at single-base resolution from the bisulfite sequencing data without the limitation of fixed-length regions. The mixed model combining qualitative identification of a combinatorial algorithm and quantitative assessment of Shannon entropy may accurate identify the CMRs and DMRs without the limitation of number of samples. The user-friendly interface and adjustable threshold of parameters of CpG_MPs facilitate the efficient identification and analysis of potentially functional regions with different methylation patterns from high-throughput bisulfite sequencing data.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5 and Supplementary Figures 1–2.
FUNDING
Funding for open access charge: The National Natural Science Foundation of China [61075023, 30971645 and 31171383]; Science Foundation of Heilongjiang Province [C201012 and QC2011C061]; Scientific Research Fund of Heilongjiang Provincial Education Department [12511272].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors like to thank Dr Shengqiang Liu and Jingyuan Fu for reviewing the manuscript.
REFERENCES
- 1.Bird A, Taggart M, Frommer M, Miller OJ, Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985;40:91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 3.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 4.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Issa JP. CpG-island methylation in aging and cancer. Curr. Top. Microbiol. Immunol. 2000;249:101–118. doi: 10.1007/978-3-642-59696-4_7. [DOI] [PubMed] [Google Scholar]
- 6.Fuke C, Shimabukuro M, Petronis A, Sugimoto J, Oda T, Miura K, Miyazaki T, Ogura C, Okazaki Y, Jinno Y. Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: an HPLC-based study. Ann. Hum. Genet. 2004;68:196–204. doi: 10.1046/j.1529-8817.2004.00081.x. [DOI] [PubMed] [Google Scholar]
- 7.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum. Mol. Genet. 2001;10:687–692. doi: 10.1093/hmg/10.7.687. [DOI] [PubMed] [Google Scholar]
- 9.Esteller M, Fraga MF, Guo M, Garcia-Foncillas J, Hedenfalk I, Godwin AK, Trojan J, Vaurs-Barriere C, Bignon YJ, Ramus S, et al. DNA methylation patterns in hereditary human cancers mimic sporadic tumorigenesis. Hum. Mol. Genet. 2001;10:3001–3007. doi: 10.1093/hmg/10.26.3001. [DOI] [PubMed] [Google Scholar]
- 10.Liu H, Su J, Li J, Lv J, Li B, Qiao H, Zhang Y. Prioritizing cancer-related genes with aberrant methylation based on a weighted protein-protein interaction network. BMC Syst. Biol. 2011;5:158. doi: 10.1186/1752-0509-5-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nature Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat. Rev. Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 14.Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiang H, Zhu J, Chen Q, Dai F, Li X, Li M, Zhang H, Zhang G, Li D, Dong Y, et al. Single base-resolution methylome of the silkworm reveals a sparse epigenomic map. Nature Biotechnol. 2010;28:516–520. doi: 10.1038/nbt.1626. [DOI] [PubMed] [Google Scholar]
- 16.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 18.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 22.Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 23.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen PY, Cokus SJ, Pellegrini M. BS Seeker: precise mapping for bisulfite sequencing. BMC Bioinformatics. 2010;11:203. doi: 10.1186/1471-2105-11-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grunau C, Schattevoy R, Mache N, Rosenthal A. MethTools–a toolbox to visualize and analyze DNA methylation data. Nucleic Acids Res. 2000;28:1053–1058. doi: 10.1093/nar/28.5.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumaki Y, Oda M, Okano M. QUMA: quantification tool for methylation analysis. Nucleic Acids Res. 2008;36:W170–W175. doi: 10.1093/nar/gkn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rohde C, Zhang Y, Reinhardt R, Jeltsch A. BISMA–fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC Bioinformatics. 2010;11:230. doi: 10.1186/1471-2105-11-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lutsik P, Feuerbach L, Arand J, Lengauer T, Walter J, Bock C. BiQ Analyzer HT: locus-specific analysis of DNA methylation by high-throughput bisulfite sequencing. Nucleic Acids Res. 2011;39:W551–W556. doi: 10.1093/nar/gkr312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hackenberg M, Barturen G, Oliver JL. NGSmethDB: a database for next-generation sequencing single-cytosine-resolution DNA methylation data. Nucleic Acids Res. 2011;39:D75–D79. doi: 10.1093/nar/gkq942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 33.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 34.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O'Malley R, Castanon R, Klugman S, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bock C, Halachev K, Buch J, Lengauer T. EpiGRAPH: user-friendly software for statistical analysis and prediction of (epi)genomic data. Genome Biol. 2009;10:R14. doi: 10.1186/gb-2009-10-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ponger L, Mouchiroud D. CpGProD: identifying CpG islands associated with transcription start sites in large genomic mammalian sequences. Bioinformatics. 2002;18:631–633. doi: 10.1093/bioinformatics/18.4.631. [DOI] [PubMed] [Google Scholar]
- 37.Su J, Zhang Y, Lv J, Liu H, Tang X, Wang F, Qi Y, Feng Y, Li X. CpG_MI: a novel approach for identifying functional CpG islands in mammalian genomes. Nucleic Acids Res. 2010;38:e6. doi: 10.1093/nar/gkp882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hackenberg M, Previti C, Luque-Escamilla PL, Carpena P, Martinez-Aroza J, Oliver JL. CpGcluster: a distance-based algorithm for CpG-island detection. BMC Bioinformatics. 2006;7:446. doi: 10.1186/1471-2105-7-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sujuan Y, Asaithambi A, Liu Y. CpGIF: an algorithm for the identification of CpG islands. Bioinformation. 2008;2:335–338. doi: 10.6026/97320630002335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heisler LE, Torti D, Boutros PC, Watson J, Chan C, Winegarden N, Takahashi M, Yau P, Huang TH, Farnham PJ, et al. CpG island microarray probe sequences derived from a physical library are representative of CpG Islands annotated on the human genome. Nucleic Acids Res. 2005;33:2952–2961. doi: 10.1093/nar/gki582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Illingworth R, Kerr A, Desousa D, Jorgensen H, Ellis P, Stalker J, Jackson D, Clee C, Plumb R, Rogers J, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS biology. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bock C, Tomazou EM, Brinkman AB, Muller F, Simmer F, Gu H, Jager N, Gnirke A, Stunnenberg HG, Meissner A. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nature Biotechnol. 2010;28:1106–1114. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nature Biotechnol. 2010;28:1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Esteller M. Epigenetics in cancer. N. Eng. J. Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 45.Ooi SK, Wolf D, Hartung O, Agarwal S, Daley GQ, Goff SP, Bestor TH. Dynamic instability of genomic methylation patterns in pluripotent stem cells. Epigenet. Chromatin. 2010;3:17. doi: 10.1186/1756-8935-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Su J, Shao X, Liu H, Liu S, Wu Q, Zhang Y. Genome-wide dynamic changes of DNA methylation of repetitive elements in human embryonic stem cells and fetal fibroblasts. Genomics. 2012;99:10–17. doi: 10.1016/j.ygeno.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 48.Bibikova M, Chudin E, Wu B, Zhou L, Garcia EW, Liu Y, Shin S, Plaia TW, Auerbach JM, Arking DE, et al. Human embryonic stem cells have a unique epigenetic signature. Genome Res. 2006;16:1075–1083. doi: 10.1101/gr.5319906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Byun HM, Siegmund KD, Pan F, Weisenberger DJ, Kanel G, Laird PW, Yang AS. Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Hum. Mol. Genet. 2009;18:4808–4817. doi: 10.1093/hmg/ddp445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bock C, Walter J, Paulsen M, Lengauer T. Inter-individual variation of DNA methylation and its implications for large-scale epigenome mapping. Nucleic Acids Res. 2008;36:e55. doi: 10.1093/nar/gkn122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, Liu H, Lv J, Xiao X, Zhu J, Liu X, Su J, Li X, Wu Q, Wang F, et al. QDMR: a quantitative method for identification of differentially methylated regions by entropy. Nucleic Acids Res. 2011;39:e58. doi: 10.1093/nar/gkr053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 55.Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem. 2004;279:48350–48359. doi: 10.1074/jbc.M403427200. [DOI] [PubMed] [Google Scholar]
- 56.Bock C, Lengauer T. Computational epigenetics. Bioinformatics. 2008;24:1–10. doi: 10.1093/bioinformatics/btm546. [DOI] [PubMed] [Google Scholar]
- 57.Vilkaitis G, Suetake I, Klimasauskas S, Tajima S. Processive methylation of hemimethylated CpG sites by mouse Dnmt1 DNA methyltransferase. J. Biol. Chem. 2005;280:64–72. doi: 10.1074/jbc.M411126200. [DOI] [PubMed] [Google Scholar]
- 58.Luo S, Preuss D. Strand-biased DNA methylation associated with centromeric regions in Arabidopsis. Proc. Natl Acad. Sci. USA. 2003;100:11133–11138. doi: 10.1073/pnas.1831011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shoemaker R, Deng J, Wang W, Zhang K. Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res. 2010;20:883–889. doi: 10.1101/gr.104695.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nautiyal S, Carlton VE, Lu Y, Ireland JS, Flaucher D, Moorhead M, Gray JW, Spellman P, Mindrinos M, Berg P, et al. High-throughput method for analyzing methylation of CpGs in targeted genomic regions. Proc. Natl Acad. Sci. USA. 2010;107:12587–12592. doi: 10.1073/pnas.1005173107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl Acad. Sci. USA. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brunlid G, Pruszak J, Holmes B, Isacson O, Sonntag KC. Immature and neurally differentiated mouse embryonic stem cells do not express a functional Fas/Fas ligand system. Stem Cells. 2007;25:2551–2558. doi: 10.1634/stemcells.2006-0745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Y, Wen QJ, Yin Y, Lu XT, Pu SH, Tian HP, Lou YF, Tang YN, Jiang X, Lu GS, et al. FASLG polymorphism is associated with cancer risk. Eur. J. Cancer. 2009;45:2574–2578. doi: 10.1016/j.ejca.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 64.Fernandez AF, Assenov Y, Martin-Subero JI, Balint B, Siebert R, Taniguchi H, Yamamoto H, Hidalgo M, Tan AC, Galm O, et al. A DNA methylation fingerprint of 1628 human samples. Genome Res. 2012;22:407–419. doi: 10.1101/gr.119867.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brunner AL, Johnson DS, Kim SW, Valouev A, Reddy TE, Neff NF, Anton E, Medina C, Nguyen L, Chiao E, et al. Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res. 2009;19:1044–1056. doi: 10.1101/gr.088773.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 67.Edwards JR, O‘Donnell AH, Rollins RA, Peckham HE, Lee C, Milekic MH, Chanrion B, Fu Y, Su T, Hibshoosh H, et al. Chromatin and sequence features that define the fine and gross structure of genomic methylation patterns. Genome Res. 2010;20:972–980. doi: 10.1101/gr.101535.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He X, Chang S, Zhang J, Zhao Q, Xiang H, Kusonmano K, Yang L, Sun ZS, Yang H, Wang J. MethyCancer: the database of human DNA methylation and cancer. Nucleic Acids Res. 2008;36:D836–D841. doi: 10.1093/nar/gkm730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y, Lv J, Liu H, Zhu J, Su J, Wu Q, Qi Y, Wang F, Li X. HHMD: the human histone modification database. Nucleic Acids Res. 2010;38:D149–D154. doi: 10.1093/nar/gkp968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.