Abstract
Ribosomes, after one round of translation, must be recycled so that the next round of translation can occur. Complete disassembly of post-termination ribosomal complex (PoTC) in yeast for the recycling consists of three reactions: release of tRNA, release of mRNA and splitting of ribosomes, catalyzed by eukaryotic elongation factor 3 (eEF3) and ATP. Here, we show that translocation inhibitors cycloheximide and lactimidomycin inhibited all three reactions. Cycloheximide is a non-competitive inhibitor of both eEF3 and ATP. The inhibition was observed regardless of the way PoTC was prepared with either release factors or puromycin. Paromomycin not only inhibited all three reactions but also re-associated yeast ribosomal subunits. On the other hand, sordarin or fusidic acid, when applied together with eEF2/GTP, specifically inhibited ribosome splitting without blocking of tRNA/mRNA release. From these inhibitor studies, we propose that, in accordance with eEF3’s known function in elongation, the release of tRNA via exit site occurs first, then mRNA is released, followed by the splitting of ribosomes during the disassembly of post-termination complexes catalyzed by eEF3 and ATP.
INTRODUCTION
Protein synthesis consists of four phases: initiation, elongation, termination and ribosome recycling (1). The elongation phase involves the movement of tRNA (translocation) through the three tRNA-binding sites (A, P and E sites corresponding to aminoacyl, peptidyl and exit sites, respectively) on the ribosome. Translocation by eukaryotic elongation factor 2 (eEF2) and GTP shifts the peptidyl-tRNA from the A site to the P site and the deacylated tRNA from the P to the E site. The eukaryotic translocation step is widely accepted as the specific target of cycloheximide (CHX) and related compounds such as lactimidomycin (LTM) (2). In yeast and other fungi, in addition to eEF2 and eEF1A, eEF3 is believed to be essential for the peptide elongation step (3) and is indispensable for yeast (4). eEF3 has been shown to facilitate the exchange of labelled E-site-bound tRNA with added deacylated tRNA and cause the release of the E-site-bound tRNA upon addition of eEF1A/GTP/aminoacyl-tRNA (5). Furthermore, eEF3 interacts with eEF1A (6). For the termination step, the release factor eRF1, bound at the A-site with the termination codon, hydrolyzes the peptidyl-tRNA ester bond with the help of eRF3 and GTP, forming the post-termination complex (PoTC) (7,8). The ribosome of PoTC needs to be recycled to initiate the next round of translation.
As originally the term was coined (9), ‘ribosome recycling’ was meant to represent the reaction to recycle the spent ribosome for the next round of translation of new mRNA. We define this reaction as disassembly of PoTC involving release of mRNA and tRNA from the ribosome accompanied by splitting of the ribosome into subunits. In yeast, we reported that ribosome recycling is catalysed by eEF3/ATP. The reaction is inhibited by aminoglycosides such as neomycin and hygromycin. It is clearly energy-dependent because a non-hydrolysable analogue of ATP did not replace ATP (10). In this system, PoTC was created from puromycin-treated polysomes assuming that the behaviour of ribosomes in the naturally occurring PoTC is identical to that of this model PoTC. In addition to this system, ABCE1 (Rli1 in yeast) and ATP have been reported to catalyse the splitting of yeast PoTC into mRNA/40S subunit complex [Figure 5A of (11)]. Although the scheme they presented indicates that tRNA is released (step 6 of Figure 7 of their article), no data for the tRNA release were presented (11). In their experiment, PoTC with three-codon ORF containing tRNAPhe at the P-site and UAA at the A-site was used. Despite the fact that there has not been any description of yeast factors responsible for the release of mRNA and tRNA from the complex of 40S subunits formed by Rli1, we assume that the complete disassembly of PoTC occurs after the action of Rli1 by some unknown means. Available data will be dealt with in the discussion section regarding the possibility that yeast Rli1 and eEF3 function in the ribosome recycling in vivo.
ABCE1 is also suggested to be involved in Archaea ribosome recycling on the basis of its ribosome splitting activity of the mRNA/ribosome complex [Figure 4 of (12)]. Possible structural basis of this action was presented using crystal structure of ABCE1. However, it is not clear whether ABCE1 is acting on the PoTC or some other ribosomal complex in their system. Although no data on the release of tRNA or mRNA were presented, their scheme (Figure 6 of their article) indicates that both mRNA and tRNA are still on the subunits after splitting of ribosomes by ABCE1, suggesting that additional steps must take place to complete ribosome recycling (12). As for higher eukaryotes, ABCE1 splits rabbit reticulocytes PoTC leaving mRNA and tRNA on the 40S subunits (13). An additional factor called Ligatin (14) or a combination of initiation factors eIF3/eIF1/eIF1A/eIF3j (13) is required to remove mRNA and tRNA to complete the disassembly of PoTC. In addition to these ABCE1/Ligatin and ABCE1/eIF3/eIF1/eIF1A/eIF3j pathways, an earlier report suggested that eIF3, eIF1, eIF1A and eIF3j, without ABCE1, disassemble PoTC in 2.5 mM MgCl2/1 mM GTP (15). This was confirmed in nucleotide-unbound MgCl2 concentration of 1 mM and 1 mM ATP/0.4 mM GTP (13). It is unlikely that Ligatin (called Tma64 in yeast) is involved in yeast ribosome recycling because Tma64 is not essential for yeast (16) while the ribosome recycling is an essential step for life (9).
In this article, as reported previously, we deal with the case where the components of PoTC including mRNA and tRNA are completely released from the complex by eEF3 and ATP. The time course of the complete disassembly measured by the conventional method suggested that the three reactions—tRNA release, mRNA release and ribosome splitting—may take place almost simultaneously. However, we show here that CHX, LTM, paromomycin (PAR), eEF2/GTP/sordarin (SOR) and eEF2/GTP/fusidic acid (FA) inhibit the eEF3-dependent recycling of PoTC in a fashion that suggests that the disassembly begins with tRNA release followed by the release of mRNA, then the splitting of ribosomes.
MATERIALS AND METHODS
Buffers
Buffer X/Y contains 20 mM Hepes-KOH (pH 7.6), X mM MgCl2, Y mM KCl and 2 mM DTT.
Preparation of PoTC polysome, elongation factors, termination factors and 32P-labelled 60S subunits
The polysomes consisting of model PoTC strung together with mRNA were prepared as described (10). Briefly, Saccharomyces cerevisiae strain WY344 was grown at 30°C in 4.8 l of yeast extract/peptone/dextrose medium with shaking (190 rpm) for 1–1.5 days, until the culture reached a density of OD600 = 1.6. Cells were immediately cooled by the addition of crushed ice and were centrifuged at 3000g for 10 min at 4°C. One cell volume (about 12 ml) of buffer 10/50 containing 250 mM sucrose, 0.2 mg/ml heparin and 0.2 mM PMSF was added together with 12 ml of acid-washed glass beads (Sigma, 425–600 μm). The cells were disrupted by vortexing five times for 30 s with 1-min breaks on ice between each vortexing. The disrupted cells were centrifuged as above to remove intact cells and glass beads. The supernatant was re-centrifuged at 17 000g for 10 min at 4°C to remove debris, and the lysate obtained (15 ml containing the polysomes) was adjusted to high salt buffer 25/500 and left standing for 5 min on ice. This process stops movement of ribosomes on the mRNA. The extract was then subjected to sucrose density gradient centrifugation (SDGC), and the fractions containing only polysomes were collected. The high-salt wash removes most of the non-ribosomal proteins and the E-site-bound tRNA (17). Run-off ribosomes that might have been produced during the above process were removed by this procedure. Thus, in the experiments, polysome run-off can be avoided in the absence of CHX. The polysomes were incubated with eEF2 and GTP to translocate peptidyl-tRNA from the A to the P site, followed by a second high-salt wash to remove added eEF2 and E-site-bound tRNA. Puromycin (PUR) was then added to make model PoTC. The model PoTC consists of a ribosome with tRNA on the P-site and any triplet on the A-site. Each model PoTC is strung together with mRNA thus keeping the form of a polysome. Elongation factors were prepared as described (10).
32P-labelled 60S subunits were prepared as described (15) using casein kinase (18). Briefly, 0.56 µM 60S subunits were prepared as described (19) and were phosphorylated with 500 U/µl casein kinase II (New England BioLabs) and 50 µCi of [γ-32P]ATP (6000 Ci/mmol, Perkin Elmer) in 110 µl of the reaction mixture containing buffer 10/50 for 30 min at 30°C. The mixture was loaded onto 4.5 ml of 5–30% (w/v) sucrose gradient prepared in buffer 5/50 and sedimented for 7 h at 4°C (Beckman SW50.1 rotor, 49 000g). The sedimentation behaviour was monitored using an ISCO UA-6 spectrophotometer at 254 nm, and the fractions containing the [32P]60S subunits (2 ∼ 4 × 104 dpm/pmol) were pooled (0.6 ml) and stored at −80°C. Termination factors were prepared as described in the Supplementary Methods.
Disassembly of the PoTC
The complete disassembly of the PoTC was analysed as previously described (10) with some modifications detailed below. To detect the release of tRNA from the PoTC (Figure 1G, 4A, 5A and Table 1), polysomes (3 A260 units) were disassembled in buffer 3/150 (600 μl) with 1 mM PUR, 50 μM ATP (potassium salt) and 0.5 μM eEF3. The reaction mixture was filtered through a Millipore filter, and tRNA, which had been released from the ribosomes, was measured in the filtrate by charging it with a mixture of [14C]amino acids and aminoacyl-tRNA synthetases. To detect the release of mRNA from the PoTC (Figure 1A–C and H, 2, 4B and 5B), polysomes (0.75 A260 units, 15 pmol) were disassembled in buffer 3/150 (150 μl) by the addition of PUR, ATP and eEF3 as described above. The reaction was stopped by the addition of MgCl2 to 25 mM. The mixture was loaded onto 4.5 ml of 15–45% sucrose gradient prepared in buffer 25/150 and sedimented for 75 min at 4°C (Beckman SW50.1 rotor, 150 000g). To detect the ribosome splitting of the PoTC (Figure 1D–F and I, 4C, 5C–D and 7), the disassembly reaction was performed in the same way as the mRNA release assay and stopped by 4% (v/v) formaldehyde. The mixture was loaded onto 4.5 ml of 5–30% sucrose gradient prepared in buffer 3/150 and sedimented for 128 min at 4°C (SW50.1 rotor, 150 000g). The sedimentation behaviour of polysomes and the ribosomes was monitored using an ISCO UA-6 spectrophotometer at 254 nm. In all experiments, inhibitors were added at time zero, and the reaction was allowed to proceed for 1 min. The percent inhibition was calculated from the extent of the reaction with each concentration of inhibitors compared with that without antibiotic.
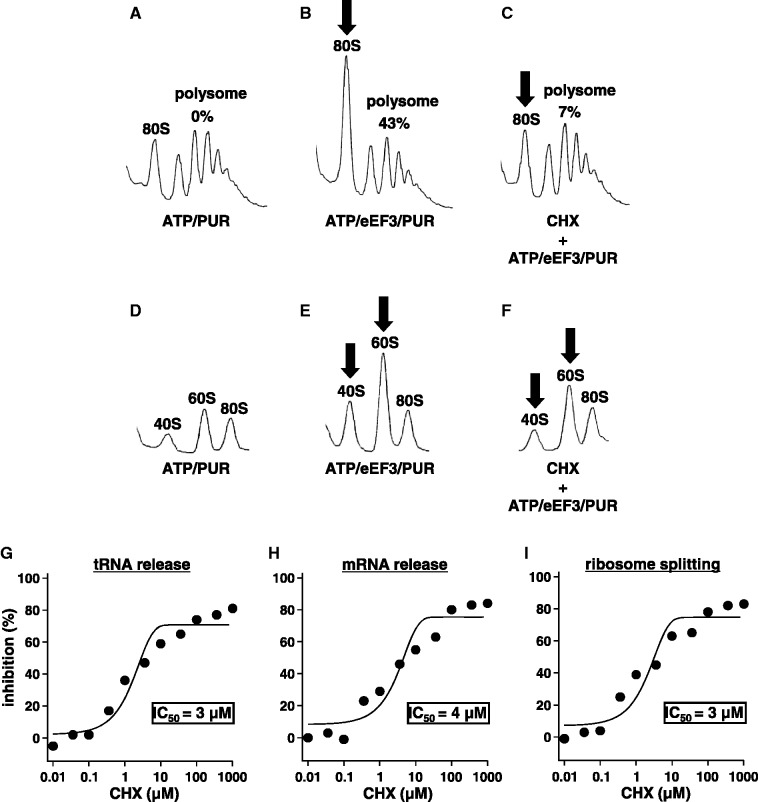
Figure 1.
Inhibitory effect of CHX on the disassembly of the PoTC by eEF3/ATP. (A–C) CHX inhibits the release of mRNA from PoTC. Sedimentation profiles of polysomes after the disassembly reaction of PoTC by eEF3/ATP under various conditions are shown. Final concentrations of each component are as follows: 50 µM ATP, 0.5 µM eEF3, 1 mM PUR, 360 µM CHX. The standard reactions for the disassembly of PoTC were stopped by buffer 25/150. This buffer converts the formed subunits to 80S ribosomes. The mixtures were subjected to SDGC with 150 000g for 75 min. (A) Only ATP/PUR was added. (B) Complete reaction mixture. (C) B in the presence of 360 µM CHX. Incubation was for 1 min at 30°C. The decrease of polysome area is accompanied by increase of the 80S ribosome (the arrows). Numbers on top of the polysome region indicate the percentage decrease of the area under each experimental condition. Note that in (C) (in the presence of CHX) the decrease of PoTC is significantly lower (43% versus 7%), and the increase of 80S ribosomes is less. (D–F) CHX inhibits the ribosome splitting of PoTC. For D–F, the reaction mixtures were fixed by formaldehyde to preserve subunits formed during the disassembly (10). They were centrifuged for 128 min at 150 000g to observe subunits. Remaining PoTC was pelleted and not observed under these sedimentation conditions. (D) Polysomes with ATP and PUR. (E) Complete reaction mixture. (F) E in the presence of 360 µM CHX. Note that subunits (arrows) were much less in F than E, indicating that the splitting of PoTC is inhibited by CHX. (G–I) Determination of IC50 of CHX on the three disassembly reactions of the PoTC by eEF3/ATP. Various amounts of CHX were added just before the disassembly reaction. IC50s of tRNA release (G), mRNA release (H) and the ribosome splitting (I) are shown.
Table 1.
eEF2/GTP/SOR or eEF2/GTP/FA preferentially inhibits ribosome splitting by eEF3/ATP
| Compound added | Release of |
Ribosome splitting |
||||
|---|---|---|---|---|---|---|
| tRNA, cpm | Inhibition, % | mRNA, % | Inhibition, % | Splitting, AU | Inhibition, % | |
| None | 693 | 0 | 40 | 0 | 1488 | 0 |
| eEF2/GTP/SOR | 535 | 23 | 31 | 22 | 109 | 93 |
| eEF2/GTP/FA | 570 | 18 | 36 | 11 | 41 | 97 |
The reaction mixture and experimental conditions were as in Figure 1 without CHX, except for the presence of eEF2/GTP/SOR or eEF2/GTP/FA where indicated. Percentages of mRNA release (z) were calculated: z = 100 × (1 − y/w), where (y) is the polysome area remaining after the reaction and (w) is that without any factors.
Ribosome splitting was calculated as follows. Splitting (arbitrary units, AU) = (area of subunits after the reaction) − (initial area of subunits). The percent inhibition was calculated from the extent of the reaction with inhibitor mix compared with that without inhibitor mix.
In Figure 3B and C, the disassembly of PoTC prepared with eRF1 and eRF3 was performed and the effect of CHX was studied. For this experiment, the purified polysomes (15 pmol) were incubated in 45 µl buffer 5/50 with 0.75 µM eRF1, 0.75 µM eRF3 and 0.33 mM GTP for 30 min at 30°C. Next, MgCl2, KCl, Hepes-KOH (pH 7.5), DTT and water were added to the reaction mixture so that it was in 150 µl buffer 3/150. Then, 0.5 µM eEF3 and 50 µM ATP were added to the reaction mixture, followed by 3-min incubation at 30°C. The reaction was stopped by the addition of 25 mM MgCl2 and analyzed by 15–45% SDGC (SW50.1 rotor, 150 000g, 75 min at 4°C) in buffer 25/150. In Figure 3C, various concentrations of CHX were added at time zero, and the disassembly of PoTC prepared with eRF1 and eRF3 was allowed to run for 1 min.
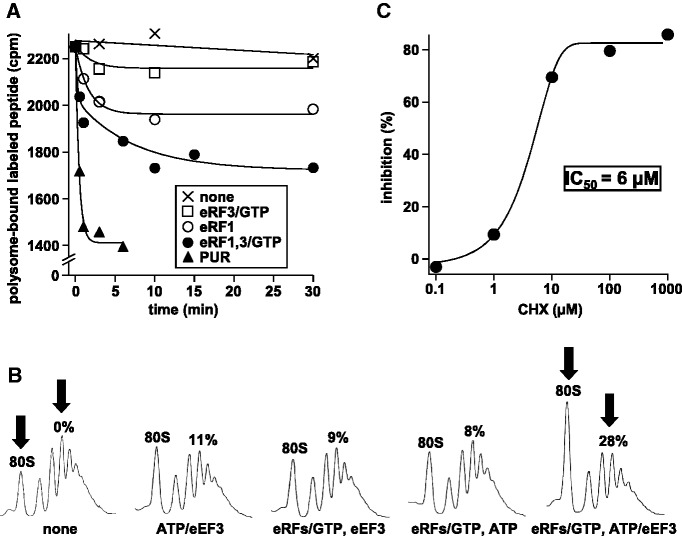
Figure 3.
PoTC created by eRF1 and eRF3 is disassembled by eEF3. This reaction is also sensitive to CHX. (A) eRF1/eRF3/GTP releases 35S-labelled nascent peptides from the polysomes. The complete reaction mixture (60 µl) contained 20 pmol of labelled polysomes, 30 pmol of eRF1, 30 pmol of eRF3 and 20 nmol of GTP. Components of each reaction tube are as indicated in the inset. The reaction was carried out at 30°C and stopped by 1 mM anisomycin at the indicated times. The mixture was subjected to SDGC, and the radioactivity remaining on the polysomes was counted. (B) eEF3/ATP disassembles the PoTC formed by eRFs/GTP. Non-labelled polysomes were incubated with eRFs/GTP for 30 min to release peptide as in A. eEF3/ATP was then added, incubated for 3 min at 30°C to disassemble the formed PoTC. The reaction was stopped by 25 mM MgCl2, which also converts formed subunits to 80S ribosomes. The percentage of mRNA release was estimated from the reduction of the polysome and is indicated above the polysome region. Note the decrease (28% less) in polysome area and the increase of 80S peak between the extreme left and right panels (arrows). (C) CHX inhibits the disassembly of the PoTC formed by eRFs/GTP. Experimental conditions were as in B, except that various amounts of CHX were added just before the disassembly reaction, which was carried out for 1 min. Inhibitory effect of CHX (percent inhibition of mRNA release) is plotted against concentration of CHX.
To verify that the predominant 80S ribosomes observed in Figure 7 in the presence of SOR or FA were indeed derived from polysomes (PoTCs strung together with mRNA), we conducted an experiment shown in Figure 8A–E. In this experiment, the disassembly of the PoTC was carried out for 1 min as described above for the mRNA release assay in the presence or absence of 0.5 µM eEF2, 10 µM GTP and 100 µM SOR or 1 mM FA. The reaction was stopped by 4% formaldehyde, and the disassembly was measured by subjecting the reaction mixture to 15–50% SDGC (SW50.1 rotor, 150 000g, 90 min at 4°C) in buffer 3/150.
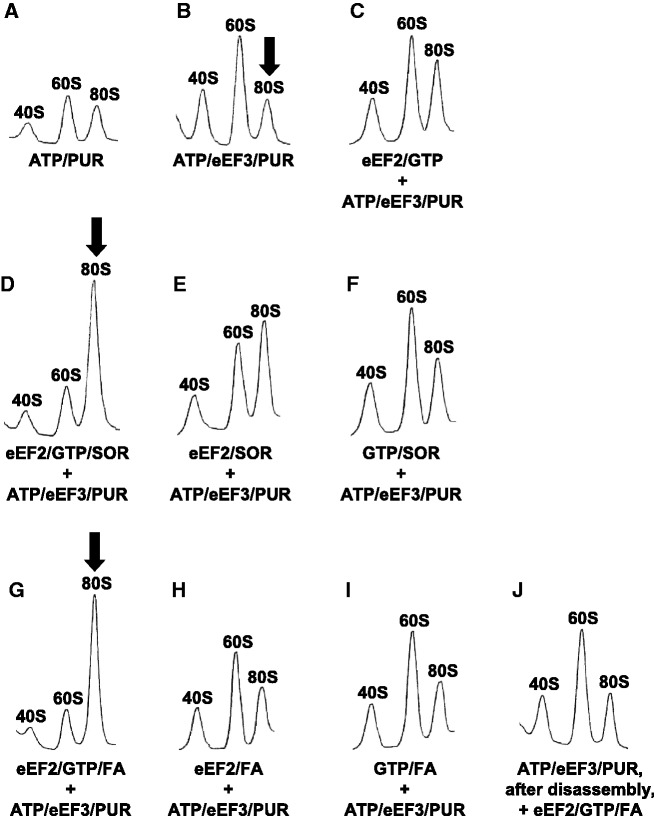
Figure 7.
eEF2/GTP/SOR or eEF2/GTP/FA specifically inhibits eEF3/ATP-dependent splitting of 80S ribosomes of the PoTC. Sedimentation profiles of ribosomes after the disassembly reaction of PoTC by eEF3/ATP under various conditions are shown. Final concentrations of each component are as follows: 50 µM ATP, 0.5 µM eEF3, 1 mM PUR, 0.5 µM eEF2, 10 µM GTP, 100 µM SOR, 1 mM FA. An inhibitor mix (i.e. eEF2/GTP/SOR or eEF2/GTP/FA) was added before the disassembly reaction unless otherwise indicated. The reaction was stopped at 1 min by formaldehyde and centrifuged through 5–30% SDGC as in Figure 1D–F. (A) Only ATP/PUR was added. These ribosomal peaks represent background values. (B) Complete disassembly reaction. Note the increase of subunits. (C) Lack of effect of eEF2/GTP alone. (D) Complete reaction with inhibitor mix (eEF2/GTP/SOR). Note the increase of 80S peak and the decrease of subunits. (E) D without GTP. (F) D without eEF2. (G), (H) and (I) correspond to D, E and F, respectively, except that FA was added instead of SOR. Note the large peak of 80 S in D and G (arrows), presumably the complex with eEF2/inhibitors. (J) corresponds to G except that eEF2/GTP/FA was added after disassembly into subunits took place (1 min). Note that no increase of 80S ribosome was observed in this case.
Figure 8.
eEF2/GTP/SOR or eEF2/GTP/FA inhibits ribosome splitting but not release of mRNA. (A–E) eEF3/ATP releases 80S ribosomes from the PoTC in the presence of eEF2/GTP/SOR or FA. (A) No additions. (B) ATP/PUR was added. (C) Complete reaction mix for disassembly. (D) Inhibitor mix (eEF2/GTP/SOR) was added to C. (E) Inhibitor mix (eEF2/GTP/FA) was added to C. Note the large peak of 80S in D and E (arrows), but the amount of polysomes are about the same for these three conditions. (F and G) Lack of incorporation of externally added labelled 60S subunits into 80S ribosomes formed from PoTC by eEF3/ATP in the presence of eEF2/GTP/SOR or FA. The reaction shown in Table 1 with 15 pmol of PoTC and other components was performed in the presence of 32P-labelled 60S subunits (0.25 pmol) with eEF2/GTP/SOR (F) or FA (G). The mixture was subjected to 5–30% SDGC, and both UV absorbance and radioactivity were measured for each fraction.
In Figure 8F and G, the disassembly reaction was carried out in the presence of 32P-labelled 60S subunits (2 ∼ 4 × 104 dpm/pmol) and eEF2/GTP/SOR or FA as described above. The reaction was stopped by 4% formaldehyde. The mixture was subjected to 5–30% SDGC (SW50.1 rotor for 128 min at 150 000g), and both the UV absorbance and radioactivity of each fraction were measured.
Peptide release assay
To establish the hydrolysis of ribosome-bound peptidyl-tRNA by eRF1 and eRF3 in the absence of a termination codon at the A-site, we prepared 35S-labelled peptidyl-tRNA bound to the polysomes as described below. The yeast lysates (4.8 ml) prepared as described above were incubated with 25.2 nM [35S]methionine (1.175 Ci/mmol, Perkin Elmer), 1 mM ATP, 0.5 mM GTP, 20 mM creatine phosphate and 40 µg/ml creatine kinase in buffer 3/150 for 10 min at 30°C to label the nascent peptides. After the incubation, MgCl2, KCl, Hepes-KOH (pH 7.5), DTT and water were added so that the reaction mixture was in buffer 25/500. The labelled polysomes (9.3 ml) were subjected to SDGC (6 ml of 15–45% sucrose gradient layered on 1.13 ml of 80% sucrose cushion prepared in buffer 10/50) for 5 h at 37 500 rpm (174 000g) in Beckman rotor SW41. The isolated polysomes (4.4 nmol) were then translocated with 14 nmol of eEF2 in 7 ml of buffer 5.5/150 containing 1.4 µmol of GTP. The translocated labelled polysomes were again subjected to SDGC (as described above) to further purify the polysomes.
35S-labelled polysomes thus prepared (20 pmol, containing [35S]peptidyl-tRNA (113 cpm/pmol) at the P-site) were incubated as shown in Figure 3A in 60 µl buffer 5/50 with 0.5 µM eRF1, 0.5 µM eRF3 and 0.33 mM GTP at 30°C. The reaction was stopped by 1 mM anisomycin (20), followed by the addition of MgCl2 to 25 mM to stabilize the complex. The mixture was loaded onto 4.5 ml of 15–45% sucrose gradient prepared in buffer 25/50 and sedimented for 75 min at 4°C (SW50.1 rotor, 150 000g). The fractions containing polysomes were manually collected, and cold trichloroacetic acid (5%) insoluble radioactivity of the polysomes was counted. Decrease of polysome-associated radioactivity was regarded as hydrolysis of peptidyl-tRNA by eRF1 and eRF3.
ATP hydrolysis assay
In the ATPase experiment shown in Figure 6, a standard reaction mixture (15 µl) contained 0.1 µM polysomes, 0.5 µM eEF3, 1 mM PUR and 50 µM [γ-32P]ATP (specific activity 1.32 mCi/µmol), as well as various concentrations of an inhibitor in 3/150 buffer. Incubation was carried out at 30°C for 1 min. The release of inorganic phosphate from [γ-32P]ATP upon hydrolysis was analysed by monitoring ammonium molybdate-soluble radioactivity as described (21) except that the reaction volumes were reduced by 25%.
Figure 6.
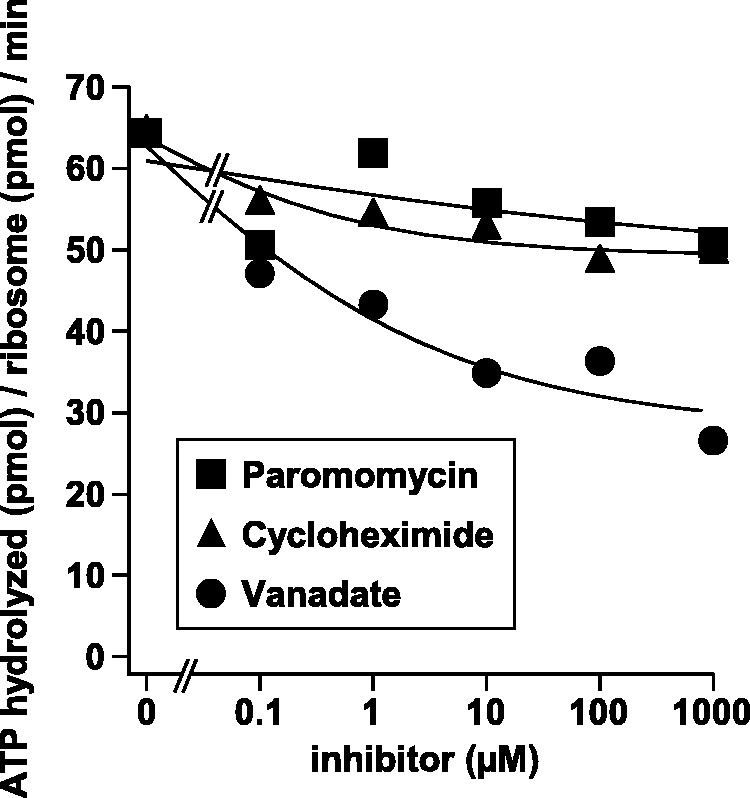
Effects of CHX or PAR on ATP hydrolysis during the eEF3-catalysed disassembly of PoTC. The reaction mixture (15 µl) contained 0.1 µM PoTC, 0.5 µM eEF3, 50 µM [γ-32P]ATP, and various concentrations of inhibitors in buffer 3/150. Incubation was carried out at 30°C for 1 min, and the hydrolysis of ATP was plotted against the concentrations of inhibitors. Vanadate [an inhibitor of ATPase (32)] is a positive control and indicates the partial inhibition of the ATPase of eEF3/ribosome.
RESULTS
CHX inhibits disassembly of the PoTC by eEF3/ATP
Preliminary studies suggested that CHX inhibited eEF3-dependent release of mRNA from PoTC (10). Here, we examined the mechanism of the CHX action. In the experiment described in Figure 1, we prepared model PoTC by removing nascent peptide chains from the peptidyl-tRNA by PUR on the poly-ribosomes isolated from growing yeast. Before the puromycin reaction, peptidyl-tRNA bound at the A-site was translocated to the P-site by eEF2/GTP. The complex thus prepared consists of several PoTCs strung together with mRNA keeping the form of polysome except for the absence of the nascent peptide. The release of deacylated tRNA from the PoTC was assayed by charging the released tRNA with [14C]amino acids (see Materials and Methods). The release of mRNA from the PoTC was detected by the decrease of the polysomes and simultaneous appearance of 80 S ribosomes as shown in Figure 1A–C. The reason why we observe 80S ribosomes instead of subunits is that we added Mg2+ at the end of the reaction to convert subunits to 80S ribosomes for the ease of analysis. It is clear from these figures that eEF3 created a large peak of 80S ribosomes, with the sacrifice of polysomes (Figure 1B). When CHX was added, the conversion was mostly stopped (Figure 1C). In Figure 1D–F, the addition of Mg2+ was omitted, and the mixture was sedimented longer to observe the subunits. There were some pre-existing subunits (Figure 1D), but the amount of subunits (Figure 1E, arrows) was significantly more when eEF3 was added. In the presence of CHX, the increase of subunits was effectively prevented (compare E with F).
Figure 1G–I show the dose–response curves of CHX on the three reactions of the complete disassembly (tRNA release, mRNA release and ribosome splitting). The concentrations of CHX required to cause 50% inhibition (IC50) of the three reactions were very similar (3–4 μM). Furthermore, those IC50 values were comparable with the IC50 for the inhibition of translocation [1.2 μM for CHX (22)], suggesting that the binding constant of CHX to PoTC is virtually the same as for binding to elongating ribosomes.
CHX is a non-competitive inhibitor of both eEF3 and ATP
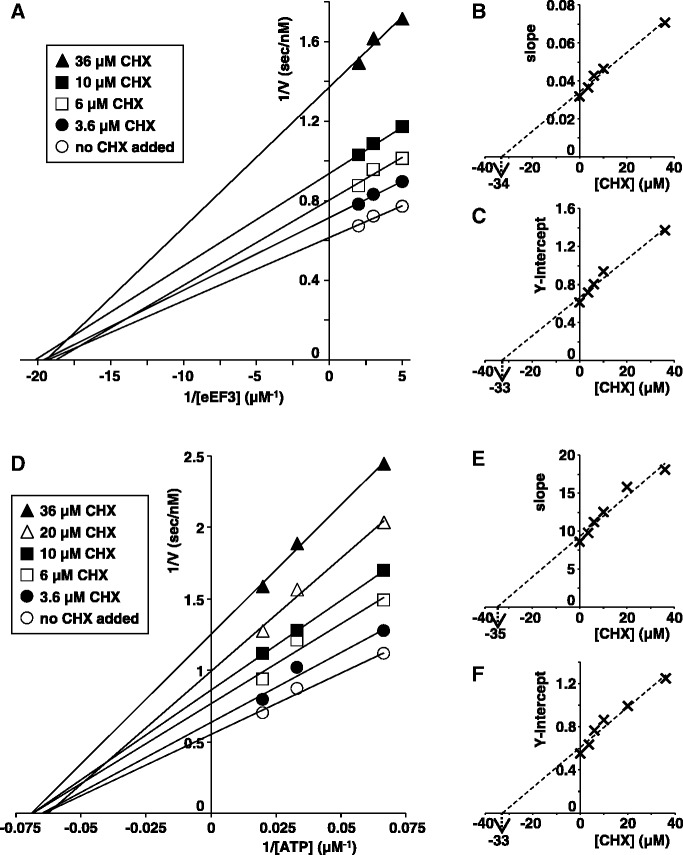
Kinetic analyses show that CHX is a non-competitive inhibitor of eEF3 (Figure 2A–C). This is because the ribosomal binding site of CHX is different from that of eEF3. Thus, CHX binds to rabbit ribosomes at the site surrounded by 28S rRNA (corresponds to 25S rRNA in yeast), L27a (yeast L28) and L36a (yeast L42) in the 60S subunit (2,23), while eEF3 binds to 5S rRNA, L5 and L11 in the 60S subunit as well as the 40S subunit (24). The Ki and Ki’ values of CHX were about 34 and 33 μM, respectively, indicating that the affinity of CHX is not influenced by eEF3. These values are comparable with the Kd value of CHX with vacant ribosomes (15 μM) (2). CHX was also a non-competitive inhibitor of ATP (Figure 2D–F), probably because CHX binds to the ribosome while ATP binds to eEF3 (24).
Figure 2.
CHX is a non-competitive inhibitor of both eEF3 and ATP. (A and D) Lineweaver–Burk plots of the disassembly reaction (mRNA release) inhibited by CHX at various concentrations of eEF3 (A) or ATP (D). (B and C) The slope (B) or Y intercept (C) of A was plotted against the concentrations of CHX. Ki and Ki’ of CHX were estimated to be 34 µM and 33 µM from B and C, respectively, where Ki and Ki’ are the dissociation constants for the enzyme–inhibitor (EI) complex or the enzyme–substrate–inhibitor (ESI) complex, respectively. (E and F) The slope (E) or Y intercept (F) of D was plotted against the concentrations of CHX. Ki and Ki’ of CHX were estimated to be 35 µM and 33 µM from E and F, respectively.
CHX inhibits disassembly of PoTC created by eRF1 and eRF3
It has been reported that ABCE1 dissociates the 60S subunit from rabbit PoTC prepared with eRF1 (and eRF3), but not the PoTC formed by PUR (13). Since our yeast PoTC was prepared with PUR, we determined whether PoTC prepared with eRF1 and eRF3 would behave identically to that prepared with PUR. Because our substrate was derived from polysomes isolated from growing yeast, most of the ribosomes on the mRNA do not have a termination codon at the A-site. Therefore, eRF1 would not be expected to release the peptide. However, yeast eRF1 is known to release the peptides slowly from P-site-bound peptidyl-tRNA without a termination codon at the A-site in vitro (25). Prokaryotic release factors (RF1 and RF2) are proposed to hydrolyse the ester bond of peptidyl-tRNA in vivo even when the A-site does not have a termination codon in stalled ribosomes (26). In accordance with these findings, Figure 3A indicates that eRF1/eRF3/GTP ternary complex released 35S-labelled nascent peptides from the polysomes, resulting in the model PoTC. eEF3/ATP released 28% of the mRNA from the PoTC formed by termination factors (Figure 3B) though the efficiency was modest compared with what we showed previously for PoTC prepared with PUR (59%) (10). This is probably due to smaller amount of PoTC when prepared with eRF1/eRF3 as shown in Figure 3A. We should point out that we never removed the added eRF1 and eRF3 from the resulting PoTC, suggesting that these factors are still bound to the PoTC as the case of higher eukaryotes (13). Possible role of eRF1/eRF3 and eEF3 in the rescue of stalled ribosomes based on these data is discussed in the Supplementary data. We further demonstrate the inhibitory effect of CHX on the mRNA release from that PoTC created by eRF1/eRF3 as shown in Figure 3C. This figure indicates that the IC50 (6 µM) of CHX with this substrate was comparable with IC50 with the PoTC obtained by PUR (compare with IC50 shown in Figure 1H).
Inhibitory effect of LTM on the disassembly of the PoTC by eEF3/ATP
LTM was recently shown to bind to the CHX-binding site and inhibited the translocation step (2). As shown in Figure 4, LTM also inhibited all three reactions of the disassembly equally. The observed IC50 values (15 ∼ 18 μM) were about five times higher than those of CHX, and the maximum inhibition was obtained at 100 μM. This value is similar to the concentration (200 μM LTM) which inhibits translocation almost completely (2). These data, together with the data shown in the preceding section, suggest that there is a reaction common to recycling and translocation despite that they are catalysed by different factors.
Figure 4.
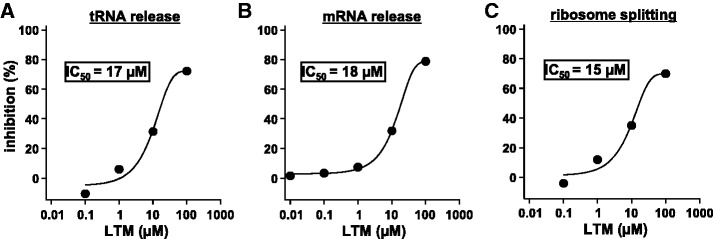
Inhibitory effect of LTM on the disassembly of the PoTC by eEF3/ATP. The experimental conditions are the same as in Figure 1 except that various concentrations of LTM were added. Transfer RNA release (A) and mRNA release (B) from the PoTC and the ribosome splitting of the PoTC (C) are shown.
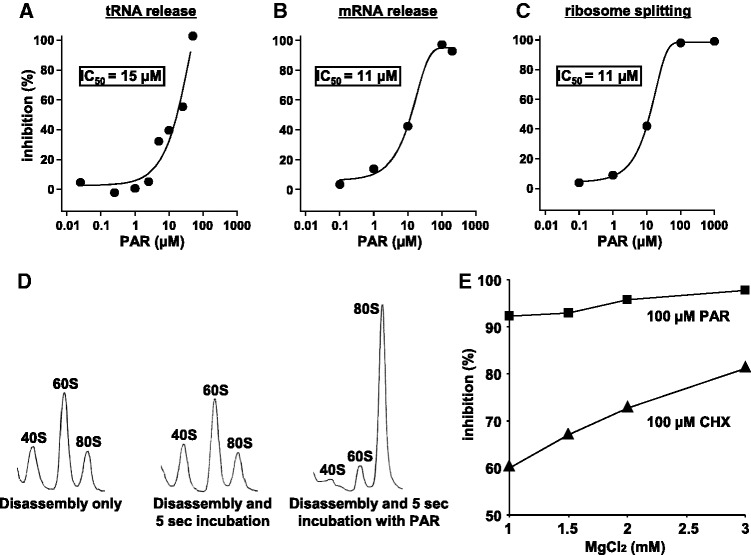
Dual effects of PAR: inhibition of PoTC disassembly and induction of subunit association
Aminoglycosides, such as streptomycin (27) and PAR (28), are known to inhibit translocation as well as inducing miscoding (29). Since we found that PAR is a potent inhibitor of ribosome recycling (10,30), we investigated this compound further to gain insights into the mechanism of the recycling in yeast. As shown in Figure 5, PAR has dual actions. PAR inhibited all three reactions of the disassembly process (Figure 5A–C). In addition, it re-associated yeast ribosomal subunits (Figure 5D) as it did the bacterial ribosomal subunits (31). This suggested that it acts like Mg2+, but this was disproven by the experiment shown in Figure 5E, demonstrating that the inhibitory effect of PAR on the disassembly reaction was not significantly influenced by Mg2+. These data imply that PAR inhibits ribosome recycling not by its association activity of ribosomal subunits but by some other mechanism(s). In contrast, Mg2+ has a remarkable stimulatory effect on the inhibition by CHX. Significance of these observations will be described in the discussion section.
Figure 5.
Dual effects of PAR. (A–C) Inhibition of PoTC disassembly. The experimental conditions are the same as in Figure 1 except that various concentrations of PAR were added. (D) Induction of subunit association by PAR. PoTC was disassembled into subunits by eEF3/ATP (left panel). The disassembled ribosomes were further incubated for 5 s at 30°C without PAR (middle panel) while the mixture shown in the right panel was incubated with 1 mM PAR for 5 s at 30°C. Then, the mixtures were subjected to SDGC, and sedimentation behaviours of the ribosomes are shown. (E) Effect of Mg2+ on the inhibitory effect of CHX and PAR. The disassembly of PoTC was carried out as in Figure 1, except that the reaction mixture contained 100 µM CHX (▴) or PAR (▪) and various concentrations of MgCl2 as indicated. The percent inhibition was calculated from the extent of the release of mRNA with antibiotic compared with that without antibiotic at each concentration of MgCl2.
CHX or PAR inhibits the ATP hydrolysis and the disassembly reaction
The eEF3-dependent disassembly of PoTC requires ATP (10), but how the ATP energy is used for the disassembly reaction remains obscure. As shown in Figure 6, ∼20% of the ATP hydrolysis by eEF3/ribosomes was inhibited by CHX or PAR at a high antibiotic concentration (1 mM). At this concentration, CHX or PAR gives almost complete maximum inhibition of eEF3-dependent disassembly reaction (Figure 1G–I and 5A–C). The amount of ATP hydrolysis (97 pmol) accompanying the recycling is far greater than the molar amount of the recycled PoTC (1.5 pmol). The amount of ATPase inhibited (22 pmol) was also far greater than the amount of recycled PoTC. We could not estimate how many molecules of ATP hydrolysis were necessary for the recycling of one molecule of PoTC. We can conclude, however, that the recycling inhibitors inhibit both recycling and ATP hydrolysis, consistent with the notion that ATP hydrolysis is necessary for the recycling. On the other hand, vanadate, an ATPase inhibitor (32), inhibited ATP hydrolysis by eEF3/ribosomes about 50% (52 pmol) without inhibiting the disassembly of PoTC (10), suggesting that the ATP energy consumption with eEF3 consists of two parts, one important for the disassembly of PoTC (vanadate-insensitive and CHX-sensitive) and the other not important (vanadate-sensitive and CHX-insensitive).
eEF2/GTP/SOR or eEF2/GTP/FA inhibits ribosome splitting but not release of tRNA or mRNA
Next, we explored the effect of SOR and FA, which are well-known translocation inhibitors in yeast, but only slightly inhibit the eEF3-dependent release of mRNA from PoTC (10). To our surprise, however, upon combination with eEF2 and GTP, these translocation inhibitors preferentially inhibited the splitting of ribosomes, with much less inhibition of the two other reactions, the release of tRNA and mRNA (Table 1, details of experimental conditions are described below).
In Figure 7, we analysed the ribosome splitting of PoTC by eEF3 and ATP. It is clear that the major reaction products were 40S and 60S subunits (Figure 7B). When the reaction was carried out in the presence of eEF2/GTP/SOR, the major products were 80S ribosomes (Figure 7D), showing that the splitting was preferentially inhibited. Similar results were obtained with eEF2/GTP/FA (Figure 7G). Other panels indicate that these three components (eEF2/GTP/SOR or eEF2/GTP/FA) are essential to produce significant accumulation of 80S ribosomes. As shown in Table 1, in contrast to the clear-cut inhibition of the subunits formation, the release of tRNA measured by the amount of 14C-aminoacyl tRNA formed was not significantly inhibited (Supplementary Table S1). Similarly, the release of mRNA measured by the decrease of the polysome was not significantly inhibited (Figure 8, compare C with D or with E). These results show that the inhibitory effect is specific on the ribosome splitting.
However, the data described above could mean that, in the presence of inhibitor complexes, 80S ribosomes were released from PoTC as such or that re-association of the subunits occurred after the ribosomes of PoTC were split by eEF3/ATP. The latter possibility was ruled out by the inability of the inhibitor complexes to produce 80S ribosomes when added after the completion of the splitting reaction (Figure 7J). We conclude that eEF2/GTP/FA does not stimulate re-association of split subunits but keeps subunits together on the PoTC and allows the release of mRNA. To substantiate this conclusion further, the eEF3/ATP-dependent reaction on PoTC was performed in the presence of added 32P-labelled 60S subunits (Figure 8F and G). It is clear that an insignificant portion of 32P-labelled 60S subunits was converted into 32P-80S ribosomes. This reveals that the 80S ribosomes observed in Figure 7D and G or Figure 8D and E were not formed after the ribosomes of PoTC were split into subunits. As a control, we established that the 32P-labelled 60S subunits used in the experiment were capable of forming 80S ribosomes with the 40S subunits under identical ionic conditions but in the presence of PAR, which is found to associate subunits (Supplementary Figure S1). We thus conclude that eEF2/GTP/SOR or FA preferentially inhibited the ribosome splitting reaction by eEF3/ATP.
DISCUSSION
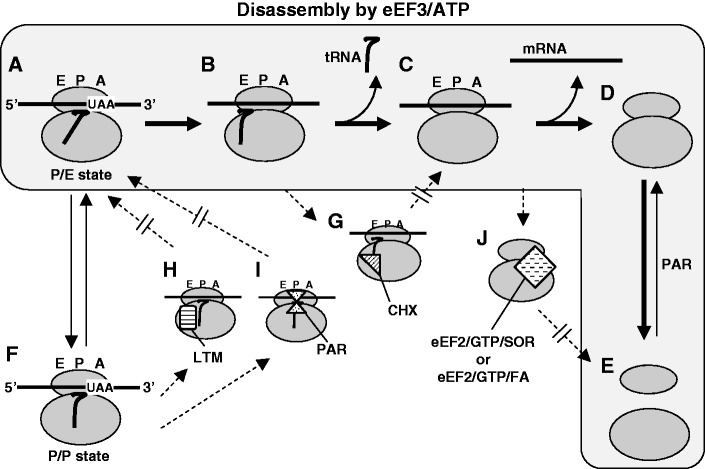
Based on these results, known characteristics of the inhibitors used in this study and an analogy to the ribosome recycling steps in E. coli, we propose the sequential steps of the ribosome recycling by eEF3 and ATP as shown in Figure 9A–E. The substrate for eEF3/ATP is PoTC with deacylated tRNA at the P/E site (complex A), which is in equilibrium with PoTC with deacylated tRNA at P/P site (complex F) (33–35). It has been suggested that CHX prevents translocation by fixing tRNA at the E-site (36) while LTM inhibits translocation by binding to the E-site, which prevents tRNA from coming to the E-site (2). On these bases, we propose that CHX and LTM inhibit disassembly of PoTC by not allowing tRNA to be released from the E-site (complex G and H). We suggest that eEF3 moves tRNA from P/E site to E/E site (complex A to B) and releases tRNA from the ribosomal E site (complex B to C). This is consistent with our observation that the inhibitory effect of CHX on the disassembly reaction was enhanced at high Mg2+ concentration (Figure 5E). This is probably because Mg2+ increases the affinity of deacylated tRNA to the E site (37) and fits well with the hypothesis that CHX traps tRNA on the E site of the ribosomes (36). We propose that tRNA is released from PoTC first as suggested from the ribosome recycling in E. coli (30). CHX and LTM prevent this first step by two different mechanisms as mentioned above (Figure 1G and 4A) (Figure 9, complexes A through C). It has been shown that PAR prevents the exchange of P-site-bound tRNA with free tRNA (38). The binding site of PAR on the large subunit overlaps with the P-site (39). Recently, other aminoglycoside kanamycin (2-deoxystreptamine-like PAR) was shown to stabilize P/P-site-bound tRNA (40). These findings are consistent with the possibility that PAR fixes tRNA on the P-site (Figure 9, complex I), resulting in the inhibition of tRNA release (Figure 5A).
Figure 9.
Proposed steps of the eEF3/ATP-dependent complete disassembly of PoTC. A and F are in equilibrium. A through E show the stepwise disassembly of PoTC (in the area coloured with pale grey). G through J show the presumed complexes of PoTC or partially disassembled PoTC with the inhibitors.
After PoTC loses tRNA, the stability of bound mRNA is significantly weakened (41), causing mRNA release (Figure 9, complex C to D). We should hasten to add that this does not necessarily mean that the release of mRNA is spontaneous after the release of tRNA. Since the inhibition of ribosome splitting by eEF2/GTP/SOR or by eEF2/GTP/FA gave 80S ribosomes rather than poly-80S without tRNA (Figure 8 and Table 1), we propose that the ribosome splitting occurs after the release of mRNA (Figure 9, complex D to E). The order of events is further supported by the fact that we never observed polysomes with 40S subunits as an intermediate during the disassembly reaction by eEF3/ATP (compare polysome region of Figure 1A with B). The ribosome splitting is inhibited by eEF2/GTP/SOR or eEF2/GTP/FA because these complexes would bind to the 80S ribosome forming complex J shown in Figure 9 (42,43). Given that eEF2 binds to helix 44 of the 40S subunit (h44) and to helix 69 of the 60S subunit (H69) (43), it is conceivable that the eEF2/GTP/SOR or FA stabilizes the subunits’ bridge resulting in the 80S ribosomes (Table 1). PAR induces the association of subunits (Figure 5D) in addition to the inhibitory effect on the tRNA release as discussed above. PAR is known to inhibit splitting of 70S ribosomes in E. coli (31) probably because PAR binds to h44 and H69 (39). However, in contrast to eEF2/GTP/SOR or FA, PAR does not inhibit ribosome splitting without inhibiting tRNA/mRNA release in the disassembly reaction (Figure 5A–C). This is in accordance with the hypothesis that the complete disassembly by eEF3 must proceed according to the order described in Figure 9. Since PAR inhibits the first reaction, the effect on any of the reactions, such as splitting, after the first reaction cannot be seen. The lack of Mg2+ effect on the inhibitory activity of PAR (Figure 5E) is consistent with this scheme because Mg2+ stimulates ribosome subunits association. The stepwise disassembly proposed in this figure should be further confirmed using fast kinetics with mRNA of known sequence.
Regarding the possible role of ABCE1 and eEF3 in yeast ribosome recycling in vivo, ABCE1 (44,45) and eEF3 mutants (6) are of interest. The in vivo inactivation of ABCE1 resulted in accumulation of monosomes (44,46) and reduction of 40S-bound eIF1 and eIF2 (44). On this basis, the authors suggested that ABCE1 may function in the initiation process. In addition, inactivation of ABCE1 caused read-through of the termination codon, leading the authors to conclude that it functions in the termination process (45). Recent in vitro experiments suggest that ABCE1 is involved in the termination process (11). In contrast, when eEF3 is inactivated in vivo, polysomes accumulate (6). Since eEF3 was believed to be necessary only for the elongation process before our finding (10), this was interpreted as a result of the halt of the ribosomes during the peptide chain elongation. However, in vivo inactivation of eEF1A (which is known to be involved in the elongation only) gave much less accumulation of polysome compared with that accumulated upon inactivation of eEF3 [Figure 3 of (6)]. These observations strongly suggest that the conspicuous accumulation of polysomes upon in vivo loss of eEF3 function is mostly the result of inactivation of the recycling activity of eEF3. It is understandable that inhibition of the release of ribosomes from mRNA at the termination site leaves more ribosomes on the mRNA, causing the increase of polysomes. Taken together, we suggest that eEF3, rather than ABCE1, may play the major role for the ribosome recycling step at the end of most of ORF in yeast. Since eEF3 is involved in the peptide chain elongation by removing the E-site-bound tRNA, this suggestion is consistent with the present finding that there is common mechanism between elongation and recycling in yeast. We further propose that the recycling mechanism involving ABCE1 described for Archaea and mammals, and thus presumably primordial, has been supplanted in yeast with an eEF3-catalysed reaction, even though ABCE1 is conserved and essential in yeast. The reason why ABCE1 is essential for yeast may be because it is important for initiation, termination and other functions such as ribosome biogenesis (47), but may not be due to its possible function in ribosome recycling.
We are aware of the possibility that the use of model PoTC that does not have a termination codon at the A-site may invalidate our conclusions in this article. Furthermore, most of our model PoTC was created by PUR instead of eRF1 and eRF3. However, we justify the use of model PoTC because this led us to the discovery of ribosome recycling factor (RRF) which is recognized to recycle ribosomes in prokaryotes (48,49). In addition, everything discovered with the model PoTC was confirmed by the in vitro experiments using naturally occurring ORF which harbours seven codons (50–53). Moreover, these in vitro results are consistent with the results of in vivo experiments (54,55). In this article, some of PoTC was prepared with eRF1 and eRF3 rather than with PUR. They gave similar results as those prepared with PUR. No in vitro ribosome recycling with long ORF has been reported. Studies are currently in progress to achieve in vitro disassembly of natural PoTC with long mRNA in yeast. Further evidence that LTM binds to the E site as shown in Figure 9 has recently been published in (56).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figure 1, Supplementary Methods and Supplementary References.
FUNDING
National Institutes of Health [CA106150 to B.S.]; Creative Biomedical Research Institute (to A.K., H.K.). Funding for open access charge: Creative Biomedical Research Institute.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Eric Wickstrom and Mr. Jamison Leid for critical reading and linguistic help to the manuscript. We also thank Drs. Roland Beckmann and Thomas Becker for a gift of the strain for production of eEF3, and Dr. Terri G. Kinzy for supplying the strain for eEF2.
REFERENCES
- 1.Hirokawa G, Demeshkina N, Iwakura N, Kaji H, Kaji A. The ribosome-recycling step: consensus or controversy? Trends Biochem. Sci. 2006;31:143–149. doi: 10.1016/j.tibs.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, Liu JO. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 2010;6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skogerson L, Wakatama E. A ribosome-dependent GTPase from yeast distinct from elongation factor 2. Proc. Natl Acad. Sci. USA. 1976;73:73–76. doi: 10.1073/pnas.73.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin SL, Xie AG, Bonato MC, McLaughlin CS. Sequence analysis of the translational elongation factor 3 from Saccharomyces cerevisiae. J. Biol. Chem. 1990;265:1903–1912. [PubMed] [Google Scholar]
- 5.Triana-Alonso FJ, Chakraburtty K, Nierhaus KH. The elongation factor 3 unique in higher fungi and essential for protein biosynthesis is an E site factor. J. Biol. Chem. 1995;270:20473–20478. doi: 10.1074/jbc.270.35.20473. [DOI] [PubMed] [Google Scholar]
- 6.Anand M, Chakraburtty K, Marton MJ, Hinnebusch AG, Kinzy TG. Functional interactions between yeast translation eukaryotic elongation factor (eEF) 1A and eEF3. J. Biol. Chem. 2003;278:6985–6991. doi: 10.1074/jbc.M209224200. [DOI] [PubMed] [Google Scholar]
- 7.Frolova L, Goff XL, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni AL, Celis JE, Philippe M, et al. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- 8.Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov S, Kisselev L, Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janosi L, Shimizu I, Kaji A. Ribosome recycling factor (ribosome releasing factor) is essential for bacterial growth. Proc. Natl Acad. Sci. USA. 1994;91:4249–4253. doi: 10.1073/pnas.91.10.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurata S, Nielsen KH, Mitchell SF, Lorsch JR, Kaji A, Kaji H. Ribosome recycling step in yeast cytoplasmic protein synthesis is catalyzed by eEF3 and ATP. Proc. Natl Acad. Sci. USA. 2010;107:10854–10859. doi: 10.1073/pnas.1006247107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shoemaker CJ, Green R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc. Natl Acad. Sci. USA. 2011;108:E1392–E1398. doi: 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barthelme D, Dinkelaker S, Albers SV, Londei P, Ermler U, Tampe R. Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. Proc. Natl Acad. Sci. USA. 2011;108:3228–3233. doi: 10.1073/pnas.1015953108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol. Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24:1787–1801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pisarev AV, Hellen CU, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleischer TC, Weaver CM, McAfee KJ, Jennings JL, Link AJ. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 2006;20:1294–1307. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beckmann R, Spahn CM, Eswar N, Helmers J, Penczek PA, Sali A, Frank J, Blobel G. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell. 2001;107:361–372. doi: 10.1016/s0092-8674(01)00541-4. [DOI] [PubMed] [Google Scholar]
- 18.Hasler P, Brot N, Weissbach H, Parnassa AP, Elkon KB. Ribosomal proteins P0, P1, and P2 are phosphorylated by casein kinase II at their conserved carboxyl termini. J. Biol. Chem. 1991;266:13815–13820. [PubMed] [Google Scholar]
- 19.Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR. Reconstitution of yeast translation initiation. Methods Enzymol. 2007;430:111–145. doi: 10.1016/S0076-6879(07)30006-2. [DOI] [PubMed] [Google Scholar]
- 20.Barbacid M, Vazquez D. (3H)anisomycin binding to eukaryotic ribosomes. J. Mol. Biol. 1974;84:603–623. doi: 10.1016/0022-2836(74)90119-3. [DOI] [PubMed] [Google Scholar]
- 21.Raj VS, Kaji H, Kaji A. Interaction of RRF and EF-G from E. coli and T. thermophilus with ribosomes from both origins-insight into the mechanism of the ribosome recycling step. RNA. 2005;11:275–284. doi: 10.1261/rna.7201805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dresios J, Panopoulos P, Suzuki K, Synetos D. A dispensable yeast ribosomal protein optimizes peptidyltransferase activity and affects translocation. J. Biol. Chem. 2003;278:3314–3322. doi: 10.1074/jbc.M207533200. [DOI] [PubMed] [Google Scholar]
- 23.Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334:941–948. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- 24.Andersen CB, Becker T, Blau M, Anand M, Halic M, Balar B, Mielke T, Boesen T, Pedersen JS, Spahn CM, et al. Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature. 2006;443:663–668. doi: 10.1038/nature05126. [DOI] [PubMed] [Google Scholar]
- 25.Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop off to initiate no-go decay. Science. 2010;330:369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vivanco-Dominguez S, Bueno-Martinez J, Leon-Avila G, Iwakura N, Kaji A, Kaji H, Guarneros G. Protein synthesis factors (RF1, RF2, RF3, RRF, and tmRNA) and peptidyl-tRNA hydrolase rescue stalled ribosomes at sense codons. J. Mol. Biol. 2012;417:425–439. doi: 10.1016/j.jmb.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 27.Igarashi K, Ishitsuka H, Kaji A. Comparative studies on the mechanism of action of lincomycin, streptomycin and erythromycin. Biochem. Biophys. Res. Commun. 1969;37:499–504. doi: 10.1016/0006-291x(69)90943-7. [DOI] [PubMed] [Google Scholar]
- 28.Eustice DC, Wilhelm JM. Mechanisms of action of aminoglycoside antibiotics in eucaryotic protein synthesis. Antimicrob. Agents Chemother. 1984;26:53–60. doi: 10.1128/aac.26.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies J, Gorini L, Davis BD. Misreading of RNA codewords induced by aminoglycoside antibiotics. Mol. Pharmacol. 1965;1:93–106. [PubMed] [Google Scholar]
- 30.Hirokawa G, Kiel MC, Muto A, Selmer M, Raj VS, Liljas A, Igarashi K, Kaji H, Kaji A. Post-termination complex disassembly by ribosome recycling factor, a functional tRNA mimic. EMBO J. 2002;21:2272–2281. doi: 10.1093/emboj/21.9.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirokawa G, Kaji H, Kaji A. Inhibition of anti-association activity of translation initiation factor 3 by paromomycin. Antimicrob. Agents Chemother. 2007;51:175–180. doi: 10.1128/AAC.01096-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uritani M, Miyazaki M. Characterization of the ATPase and GTPase activities of elongation factor 3 (EF-3) purified from yeasts. J. Biochem. 1988;103:522–530. doi: 10.1093/oxfordjournals.jbchem.a122302. [DOI] [PubMed] [Google Scholar]
- 33.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol. Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol. Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 35.Cornish PV, Ermolenko DN, Staple DW, Hoang L, Hickerson RP, Noller HF, Ha T. Following movement of the L1 stalk between three functional states in single ribosomes. Proc. Natl Acad. Sci. USA. 2009;106:2571–2576. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pestova TV, Hellen CU. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lill R, Robertson JM, Wintermeyer W. Affinities of tRNA binding sites of ribosomes from Escherichia coli. Biochemistry. 1986;25:3245–3255. doi: 10.1021/bi00359a025. [DOI] [PubMed] [Google Scholar]
- 38.Shoji S, Walker SE, Fredrick K. Reverse translocation of tRNA in the ribosome. Mol. Cell. 2006;24:931–942. doi: 10.1016/j.molcel.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borovinskaya MA, Pai RD, Zhang W, Schuwirth BS, Holton JM, Hirokawa G, Kaji H, Kaji A, Cate JH. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat. Struct. Mol. Biol. 2007;14:727–732. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- 40.Wang L, Pulk A, Wasserman MR, Feldman MB, Altman RB, Doudna Cate JH, Blanchard SC. Allosteric control of the ribosome by small-molecule antibiotics. Nat. Struct. Mol. Biol. 2012;19:957–963. doi: 10.1038/nsmb.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uemura S, Dorywalska M, Lee TH, Kim HD, Puglisi JD, Chu S. Peptide bond formation destabilizes Shine-Dalgarno interaction on the ribosome. Nature. 2007;446:454–457. doi: 10.1038/nature05625. [DOI] [PubMed] [Google Scholar]
- 42.Bodley JW, Lin L, Salas ML, Tao M. Studies on translocation V: fusidic acid stabilization of a eukaryotic ribosome-translocation factor-GDP complex. FEBS Lett. 1970;11:153–156. doi: 10.1016/0014-5793(70)80516-6. [DOI] [PubMed] [Google Scholar]
- 43.Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong J, Lai R, Nielsen K, Fekete CA, Qiu H, Hinnebusch AG. The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J. Biol. Chem. 2004;279:42157–42168. doi: 10.1074/jbc.M404502200. [DOI] [PubMed] [Google Scholar]
- 45.Khoshnevis S, Gross T, Rotte C, Baierlein C, Ficner R, Krebber H. The iron-sulphur protein RNase L inhibitor functions in translation termination. EMBO Rep. 2010;11:214–219. doi: 10.1038/embor.2009.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen ZQ, Dong J, Ishimura A, Daar I, Hinnebusch AG, Dean M. The essential vertebrate ABCE1 protein interacts with eukaryotic initiation factors. J. Biol. Chem. 2006;281:7452–7457. doi: 10.1074/jbc.M510603200. [DOI] [PubMed] [Google Scholar]
- 47.Yarunin A, Panse VG, Petfalski E, Dez C, Tollervey D, Hurt EC. Functional link between ribosome formation and biogenesis of iron-sulfur proteins. EMBO J. 2005;24:580–588. doi: 10.1038/sj.emboj.7600540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirashima A, Kaji A. Factor-dependent release of ribosomes from messenger RNA. Requirement for two heat-stable factors. J. Mol. Biol. 1972;65:43–58. doi: 10.1016/0022-2836(72)90490-1. [DOI] [PubMed] [Google Scholar]
- 49.Hirashima A, Kaji A. Purification and properties of ribosome-releasing factor. Biochemistry. 1972;11:4037–4044. doi: 10.1021/bi00772a005. [DOI] [PubMed] [Google Scholar]
- 50.Ryoji M, Berland R, Kaji A. Reinitiation of translation from the triplet next to the amber termination codon in the absence of ribosome-releasing factor. Proc. Natl Acad. Sci. USA. 1981;78:5973–5977. doi: 10.1073/pnas.78.10.5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryoji M, Hsia K, Kaji A. Read-through translation. Trends Biochem. Sci. 1983;8:88–90. [Google Scholar]
- 52.Ryoji M, Karpen JW, Kaji A. Further characterization of ribosome releasing factor and evidence that it prevents ribosomes from reading through a termination codon. J. Biol. Chem. 1981;256:5798–5801. [PubMed] [Google Scholar]
- 53.Ogawa K, Kaji A. Requirement for ribosome-releasing factor for the release of ribosomes at the termination codon. Eur. J. Biochem. 1975;58:411–419. doi: 10.1111/j.1432-1033.1975.tb02388.x. [DOI] [PubMed] [Google Scholar]
- 54.Janosi L, Mottagui-Tabar S, Isaksson LA, Sekine Y, Ohtsubo E, Zhang S, Goon S, Nelken S, Shuda M, Kaji A. Evidence for in vivo ribosome recycling, the fourth step in protein biosynthesis. EMBO J. 1998;17:1141–1151. doi: 10.1093/emboj/17.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Inokuchi Y, Hirashima A, Sekine Y, Janosi L, Kaji A. Role of ribosome recycling factor (RRF) in translational coupling. EMBO J. 2000;19:3788–3798. doi: 10.1093/emboj/19.14.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee S, Liu B, Lee S, Huang SX, Shen B, Qian SB. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc. Natl Acad. Sci. USA. 2012;109:E2424–E2432. doi: 10.1073/pnas.1207846109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.