Abstract
Small non-coding RNAs such as miRNAs, piRNAs and endo-siRNAs fine-tune gene expression through post-transcriptional regulation, modulating important processes in development, differentiation, homeostasis and regeneration. Using deep sequencing, we have profiled small non-coding RNAs in Hydra magnipapillata and investigated changes in small RNA expression pattern during head regeneration. Our results reveal a unique repertoire of small RNAs in hydra. We have identified 126 miRNA loci; 123 of these miRNAs are unique to hydra. Less than 50% are conserved across two different strains of Hydra vulgaris tested in this study, indicating a highly diverse nature of hydra miRNAs in contrast to bilaterian miRNAs. We also identified siRNAs derived from precursors with perfect stem–loop structure and that arise from inverted repeats. piRNAs were the most abundant small RNAs in hydra, mapping to transposable elements, the annotated transcriptome and unique non-coding regions on the genome. piRNAs that map to transposable elements and the annotated transcriptome display a ping–pong signature. Further, we have identified several miRNAs and piRNAs whose expression is regulated during hydra head regeneration. Our study defines different classes of small RNAs in this cnidarian model system, which may play a role in orchestrating gene expression essential for hydra regeneration.
INTRODUCTION
Small RNA-mediated silencing has emerged as an important mediator of gene regulation across all organisms, regulating diverse functions from defense against genomic pathogens in prokaryotes to regulation of self-renewal, differentiation, immune response, cell migration and cell cycle in eukaryotes (1–3). Gene regulation by small RNAs is mediated through degradation of target mRNAs, suppression of translation, DNA methylation, heterochromatin formation and programmed genome rearrangement. Based on their biogenesis and their associated proteins, regulatory small RNAs are classified into three types: microRNAs (miRNAs), endogenous silencing RNAs (endo-siRNAs) and piwi-associated RNAs (piRNAs) (4). miRNAs are 21–23-nucleotide (nt)-long RNAs that arise from hair-pin structures and mediate post-transcriptional gene regulation through mRNA degradation, translational repression and heterochromatin formation (1,4). Endo-siRNAs are synthesized through cleavage of long double-stranded RNAs, are 21–22 nt long and show perfect complementarity to their mRNA targets (5). siRNA-mediated silencing is evolutionarily conserved and is present in most of the eukaryotes. The least understood small RNAs, piRNAs, are 24–30 nt long and are expressed in germ cells at different developmental stages in Drosophila and mammals (6), whereas in Planarian Schmidtea mediterranea, they are expressed in neoblasts, which are the somatic adult pluripotent stem cells (7). piRNAs primarily mediate transposon silencing through heterochromatin formation and arise from intergenic repetitive elements. piRNAs also arise from protein coding genes such as the maf gene in Drosophila (8,9). In contrast to endo-siRNAs, miRNAs and piRNAs have co-evolved with metazoa and are expressed in all multicellular organisms including basal metazoa like sponges and cnidaria. They are also expressed in some, but not all, unicellular organisms (10–12). Interestingly, the number of miRNAs in an organism increases with increasing tissue complexity in metazoa (10), suggesting that miRNAs could have contributed to evolution of bilateria from basal metazoa by generating a more complex network of gene regulation from existing set of genes, for example, by regulating evolution of tissue identity (13).
Small RNAs also play an important role in development and regeneration by regulating cell proliferation and differentiation. Depletion of dicer, a key enzyme in miRNA biosynthesis, causes embryonic lethality in mice due to the lack of pluripotent stem cells (14). Depletion of DGCR8, another protein involved in miRNA biogenesis, in mouse embryonic stem cells (ESCs) leads to accumulation of ESCs in G1 phase due to a delayed G1-S phase transition (15). This defect in proliferation can be rescued by members of miR-290 family, implicating a role for miRNAs in G1-S phase transition in ESCs (16). miRNA biogenesis is also essential for caudal fin regeneration in zebrafish, where knock-down of dicer leads to regeneration defects and miRNA miR-203 regulates regeneration through down-regulation of Lef1, a transcription factor required for Wnt-dependent transcription (17). Recent studies on Planaria, a triploblastic metazoan with robust regeneration capability, also identified several classes of small RNAs and their pathway genes involved in stem cell function and regeneration (7,18).
Cnidaria is a sister clade of bilateria that is >500 million years old (19,20) and is classified into four distinct classes: anthozoa, hydrozoa, cubozoa and scyphozoa (21–23). These diploblastic radially symmetrical organisms usually alternate between two morphologically distinct forms—polyp form and a medusoid form. Unlike other diploblasts such as sponges and ctenophores, cnidarians have an axis and are organized into tissues. Cnidarians are the first multicellular organisms to use positional information for patterning and occupy an important position in the evolution of metazoa. Hydra is a fresh water cnidarian that exists exclusively in the polyp form and has been used as a model system for >250 years (24). Hydra has been used extensively to study regenerative biology, developmental biology and stem cell biology (25–27). Hydra has a remarkable ability to regenerate; when cut into pieces, each piece of body column, containing as little as 300 cells, can regenerate into a complete adult animal while maintaining the original polarity (28). It can also regenerate from a cluster of experimentally dissociated cells in which the axis has been disrupted and undergo de novo patterning (29); thus hydra stem cells have retained the ability to respond to morphogenetic signals and undergo patterning.
In this study, we profiled small non-coding RNAs in Hydra magnipapillata and specifically investigated the small RNA profile during head regeneration. Small RNAs have been profiled earlier from another cnidarian Nematostella vectensis (10). Seventeen miRNAs and few piRNA-like RNAs have also been identified in hydra during hydra genome sequencing (30). Our preliminary studies show that hydra expresses genes essential for small RNA biosynthesis such as drosha, two isoforms each of dicer, ago and piwi genes and RNA-dependent RNA polymerase (RDRP). Hydra appears to express functional RNAi machinery, and gene knock-down using siRNA or dsRNAs has been performed in hydra (31,32). Here, we profile small non-coding RNAs in hydra such as miRNAs, siRNAs and piRNAs and investigate their role in hydra regeneration. Small RNA libraries were generated from regenerating H. magnipapillata at different time points during head regeneration, and the libraries were subjected to deep sequencing and analysis to identify miRNAs, piRNAs and endo-siRNAs. We found 126 miRNA loci in hydra, of which only 14 have been reported earlier. Interestingly, 123 of 126 miRNAs are specific to hydra, suggesting either a highly diverse nature of hydra miRNAs or an independent evolution of hydra miRNA genes. We have also identified several small RNAs of size 18–24 nt whose precursors exhibit perfect stem–loop structures and are derived from inverted repeats. Interestingly, the most abundant class of small RNAs in hydra is piRNAs, which map to transposable elements, the annotated transcriptome and unique regions of the genome. A majority of the piRNAs that uniquely map to the genome lack ping–pong signature of piRNA biosynthesis, whereas a significant number of piRNAs that map to transposable elements and annotated transcriptome have ping–pong signature. Further, we also identified several miRNAs and piRNAs that are regulated during hydra head regeneration. In summary, our study reports 112 new miRNAs in hydra that might have distinct origins and also defines several different classes of small RNAs with a potential role in head regeneration.
MATERIALS AND METHODS
Hydra culture
All three strains of hydra, H. magnipapillata, H. vulgaris AEP and H. vulgaris Ind-Pune, were cultured in hydra medium (0.1 mM KCl, 1 mM CaCl2, 1 mM NaCl, 0.1 mM MgSO4, 1 mM Tris–Cl, pH 8.0) as described earlier at 18°C (33,34). Hydra were fed on alternate days with freshly hatched Artemia naupli but were starved 2–3 days before experiment. For regeneration experiment, H. magnipapillata were cut in the mid-gastric region and allowed to regenerate, the lower foot region regenerated head by 3 days. The foot region regenerating head was used for isolating RNA.
Small RNA library preparation
Hydra magnipapillata RNA was isolated from uncut hydra as control and from polyps regenerating head from foot. RNA from H. vulgaris AEP and H. vulgaris Ind-Pune was isolated from uncut controls only. RNA was isolated from ∼100 hydra regenerating head from foot region at different times, 3 h, 1 day, 2 day and 3 day using Trizol™ as per manufacturer’s instructions. Five micrograms of total RNA was used to generate small RNA libraries using TruSeq small RNA kit from Illumina as per manufacturer’s protocol. Briefly specific 5′ and 3′ adapters were ligated to RNA; ligated products were reverse-transcribed and amplified by polymerase chain reaction (PCR). PCR products were separated using polyacrylamide gel electrophoresis, and products corresponding to adapter-ligated 18–35-nt-long RNAs (140–160 bp) were eluted from the gel. The products were sequenced using Illumina HiSeq 1000. The entire experiment was performed twice to generate completely independent biological duplicates. The small RNA libraries for H. vulgaris AEP and H. vulgaris Ind-Pune were generated using Illumina Digital Gene Expression Small RNA Sample Prep kit (v1.5 protocol). The libraries were sequenced using Illumina GAIIx. The depth of sequencing for all libraries, whether sequenced using Illumina GAIIx or using HiSeq 1000, was comparable.
Bioinformatic analysis
About 10–20 million reads were obtained from each library, and the sequencing data from all 10 samples from H. magnipapillata (five samples each from two biological duplicates) were pooled for further analysis. To identify true miRNAs, 18–24 mer reads were mapped to hydra genome using Bowtie, and the reads mapping to hydra genome were further analysed. For each 18–24 nt read, 100 nt of genomic sequence flanking either side of the read was extracted, and its ability to form stem–loop structures, which are characteristic of pre-miRNAs, was analysed using RNAfold. The reads with stem–loop structures were further analysed using miRDeep 2 algorithm to search for miRNAs. Possible miRNAs were selected based on cut-off of −10 to 10. These putative miRNAs were then manually examined for the presence of 1-2 mismatches in the stem and loop sequence of 4–8 nucleotides, and only those exhibiting these characteristics were considered to be miRNAs.
To identify miRNAs that were differentially expressed in regenerating hydra as compared to the control, the reads from each sample were mapped to miRNAs identified by miRDeep2 and then were normalized to reads per million, to normalize for variations in the number of sequenced reads in each sample. We then calculated the correlation coefficient for two duplicates of each time point. For each time point of regeneration, the correlation coefficient was more than 0.95; we therefore did further analysis by taking average read numbers of the two duplicates. Averaged reads from all regenerates were individually compared to the control and expressed as fold-change over control. The small RNAs showing at least 2-fold variation for at least one time point were taken to be those showing differential expression in regeneration.
To identify piRNAs that map to transposable elements, all the reads of size 25–30 nt reads were aligned using Bowtie aligner allowing zero mismatches to transposable elements obtained from RepBase. piRNAs that map to unique regions of the genome were identified by mapping 25–30 nt reads to genome using Bowtie with option m1, which will retain the reads that map only once to the genome. To identify the piRNAs that map to annotated transcripts, all the reads of size 18–30 nt were aligned to annotated transcripts downloaded from NCBI using Bowtie aligner allowing no mismatches. The mapped files were converted to wiggle tracks, and the wiggle files were used as input for MATLAB to obtain density maps. The small RNA sequencing data have been deposited at the NCBI Sequence Read Archive (SRA) at http://www.ncbi.nlm.nih.gov/sra (accession number SRA050926).
Northern analysis
RNA was isolated from 2-day starved H. magnipapillata using Trizol™ as per manufacturer’s instructions. Forty micrograms of total RNA was fractionated on 12% denaturing polyacrylamide gel and transferred on Nylon N+ membrane. The blots were UV-cross-linked and baked at 80°C for 40 min. The probes were end-labelled with 32P using T4 kinase and purified on G-25 columns. The blots were incubated with hybridization solution (0.2 M NaH2PO4 and 7% SDS) alone for 2 h, followed by probe in hybridization solution at 40°C for 16–20 h. After hybridization, the blots were washed twice with 2× SSC + 0.1% SDS for 15 min at room temperature and exposed to Phosphorimager screen. The signal obtained was the same size as that obtained by performing Northern analysis of S. mediterranea total RNA using miR-71C probe (18), indicating that the signal obtained was of the expected size of 21–22 nt (data not shown). The probes used for Northern analysis are listed in Supplementary Table S6.
Quantitative RTPCR
Quantitative PCR was performed as reported for miRNAs earlier (35). miRNA-specific primers (MSPs) were designed for each miRNA for quantitative RTPCR. The primer for reverse transcription was designed with double-stranded stem–loop structure with universal reverse primer binding site at 5′ end and with last eight nucleotides at the 3′-end complementary to the 3′ -end of miRNA. PCR was carried out using miRNA-specific forward primers and universal primer with complementarity to the ‘stem’ sequence of the MSP. All primers used are listed in Supplementary Table S6. There is no suitable normalizing control for hydra miRNA PCRs; therefore, to normalize the variations that could occur during reverse transcription and PCR, all RNA samples were spiked with 200 ng of planarian RNA (sexual strain of S. mediterranea) before reverse transcription. The reverse transcription and PCR for miR-71-a1 was performed along with hydra-specific miRNA PCRs, and the expression levels of hydra miRNAs were normalized using miR71-a1. miR71-a1 expression was not detected in hydra during deep sequencing. Consistent with this observation, miR71-a1 primers did not amplify any product when used for reverse transcription and PCR using hydra RNA. Similarly, no significant amplification was observed when hydra miRNA-specific primers were used for reverse transcription and PCR using S. mediterranea RNA (data not shown). Reverse transcription was performed using SuperScript III RT (Invitrogen), and the quantitative PCR was carried out using SYBR Green (Applied Biosystems) as the fluorescent detector on an Applied Biosystems 7900HT machine. Each miRNA was analysed for expression profiles at different times of head regeneration, and the experiment was performed twice using with two sets of RNAs. Quantitative PCR for transcripts were performed using SYBR Green as the detector and actin as a normalizing control, and the probes used are listed in Supplementary Table S6.
In situ hybridization
Whole mount in situ hybridization was done using locked nucleic acid (LNA) miRCURY probes (EXIQON), using protocol for in situ hybridization as described previously for hydra (36). Probes designed were same as those used in Northern hybridization (Supplementary Table S6) except that they were tagged with digoxygenin at both 3′ and 5′ ends. Two-day starved hydra were relaxed with 2% urethane and fixed overnight with 4% PFA at 4°C. Animals were washed five times with 100% methanol followed by washes with 75% methanol + 25% PBT (phosphate buffered saline + 0.1% Tween-20), 50% methanol + 50% PBT, 25% methanol + 75% PBT and then thrice with PBT. Polyps were incubated with 5 μg/ml of Proteinase K for 10 min, and the reaction was stopped with two washes of 4 mg/ml glycine. Polyps were then washed with thrice with PBT, followed by two washes with 100 mM triethanolamine (TEA, pH 7.8). Hydra were then washed once with 0.25% acetic anhydride in 100 mM TEA (pH = 7.8) and then with 0.5% acetic anhydride. Animals were washed thrice with PBT and refixed in 4% PFA. Animals were washed thrice with PBT and treated with 0.2 M HCl for 30 min, followed by three washes with PBT. Animals were then incubated at 80°C for 30 min to inactivate endogenous alkaline phosphatase. Animals were washed thrice with PBT and then with 50% PBT + 50% Hybridization solution at 58°C for 10 min. The hybridization solution contained 70% deionized formamide, 5× SSC (diluted from 20× SSC: 3 M sodium chloride, 0.3 M sodium citrate, pH 7.0), 1× Denhardt’s Solution (made from 50× Stock: 1% Ficoll, 1% BSA, 1% polyvinyl pyrrolidone), 0.1% CHAPS, 0.1% Tween-20, Heparin 100 μg/ml and 0.2 mg/ml Yeast torula RNA. Animals were then washed with 100% hybridization solution at 58°C for 10 min. For pre-hybridization, animals were incubated with hybridization solution for 2 h at 58°C. After pre-hybridization, animals were incubated with 10 nM LNA probe in hybridization solution for 36 h at 58°C. Animals were then washed once each with hybridization solution, 75% hybridization solution + 25% 2× SSC, 50% hybridization solution + 50% 2× SSC, 25% hybridization solution + 75% 2× SSC and then with 100% 2× SSC. Animals were incubated with 20 μg/ml RNase A in 2× SSC + 0.1% CHAPS for 30 min at 37°C and then washed with 2× SSC + 0.1% CHAPS twice at hybridization temperature. This was followed by two washes with MAB (100 mM maleic acid, 150 mM sodium chloride, pH = 7.5) at room temperature for 10 min and with MAB-B (1% BSA in MAB) for 1 h. Animals were incubated with blocking solution (80% MAB-B + 20% Heat inactivated Horse Serum) at room temperature for 2 h and then overnight with pre-adsorbed 1:4000 anti-DIG alkaline phosphate conjugated antibody (Roche) at 4°C. Animals were then washed with MAB eight times and left overnight at 4°C in MAB. Animals were washed twice with NTMT (100 mM NaCl, 100 mM Tris–Cl pH 9.5, 50 mM MgCl2 + 0.1% Tween-20), incubated in 1 mM levamisole for 5 min and then incubated with BM-Purple (Roche) at 37°C till colour developed. Animals were washed with 100% methanol to remove non-specific staining and stored in PBS.
RESULTS
Deep sequencing of small RNA libraries from H. magnipapillata
Small RNAs in hydra have not been studied systematically, and their role in regeneration is entirely unexplored. The first genome sequencing report on H. magnipapillata identified 17 miRNAs and small RNAs that resembled piRNAs (30). In this study, we used next generation sequencing to comprehensively identify the small regulatory RNA repertoire in H. magnipapillata as well as to identify those small RNAs that are regulated during regeneration. Hydra has an enormous ability to regenerate after mid-gastric bisection, each piece regenerating into a complete functional organism in 3–4 days while maintaining the original polarity. Within 3 h post-bisection, the fusion of cut epithelial layers is complete (37), head activation begins (38,39) and Wnt3 is expressed, indicating initiation of axis formation (40). By 24 h, hypostome formation is observed, with some regenerates showing development of tentacle buds. By 2 days, tentacles emerge and by 3 days almost all hydra show well-formed tentacles (Supplementary Figure S1A) (41). In this study, we investigated the small RNA expression profile during head regeneration. Hydra polyps were cut in mid-gastric region, and the RNA was isolated from foot regions regenerating heads at 3 h, 1 day, 2 days and 3 days. RNA was isolated from duplicate regeneration experiments for analysis.
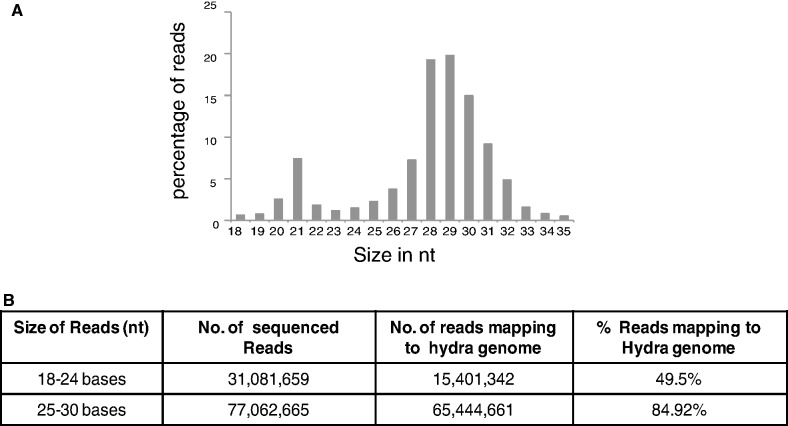
Total RNA isolated from uncut polyps and hydra regenerates at different times during head regeneration was ligated to adapters, amplified and size-selected for products corresponding to RNAs of size 18–35 nt on native PAGE. These small RNA libraries were then subjected to sequencing using an Illumina platform. About 10–20 million reads were obtained from each library, and sequencing data from all samples were pooled for further analysis. A total of 131 million reads corresponding to 18–35 nt size were obtained from these sequencing reactions. These reads were aligned to the hydra genome using Bowtie alignment (42), allowing no mismatch. Ninety-seven million reads mapped to the genome and of these 15.8% were 18–24 nt reads and 67.2% were 25–30 nt reads. Small RNA profiling of mapped reads revealed two distinct peaks, one at 21–22 nt position, corresponding to miRNA and endo-siRNA, and another peak at 27–29 nt position corresponding to piRNAs (Figure 1A).
Figure 1.
Small RNA profiling of H. magnipapillata. (A) Reads from deep sequencing of all libraries from H. magnipapillata were pooled and size distribution of reads was analysed. Reads between 18–35 nt show two peaks, one at size 21 nt and another at 28–29 nt. (B) Table shows the total number of reads of size 18–24 nt and 25–30 nt sequenced and the percentage of these reads mapping to hydra genome.
Intriguingly, we found that only 49.5% of the total 18–24 nt reads mapped to the genome (Figure 1B), which is less than expected. Potentially this could be due to the inadequate (6×) coverage in the published hydra genome (30). Inadvertent contamination is unlikely to account for this observation, as a high percentage (84.9%) of reads representing 25–30 nt sequences mapped to the genome (Figure 1B). It is unlikely that these reads arise from Artemia larvae that are fed to hydra, as the polyps were starved for two days before isolating RNA. Moreover when the unmapped reads (50.5% of total reads) were mapped to Artemia EST data set, only 6.2% of these reads mapped with no mismatch and 6.8% mapped when two mismatches were allowed. These reads also did not map to H. magnipapillata symbiont such as Curvibacter (30). But 8.6% of these unmapped reads did map to bacterial genomes that are available at http://www.genome.jp/kegg/, suggesting ∼4.3% of the total 18–24 nt reads are of bacterial origin, potentially from bacterial species that could be endosymbionts in hydra. No significant alignment was observed when these reads were mapped to Rfam database (which includes hydra non-coding RNA data set), genomic tRNA database at http://gtrnadb.ucsc.edu/, or small non-coding RNA database at http://biobases.ibch.poznan.pl/ncRNA/.
Interestingly, when the unmapped 18–24 nt reads were re-aligned to the hydra genome allowing one mismatch, 28% of these reads now mapped to the genome. Further increasing mismatches to two during alignment increased mapped reads to 58% (data not shown). In alignment carried out allowing two mismatches, 28% mismatches show A to G change, 13% show T to A change and 16% show T to C change. These small RNAs also show a preference for ‘A’ and ‘T’ at 5′-end, suggesting they are products of Dicer/Argonaute cleavage rather than degradation products (data not shown). The higher percentage of A-G mismatch among the small RNA reads (18–24 nt) suggests the possibility of RNA editing (43). Intriguingly, RNA editing alone does not explain the presence of mismatches such as T to A and T to C in the reads corresponding to 18–24 nt size, and reasons for these mismatches remain unclear.
Hydra expresses a unique set of miRNAs
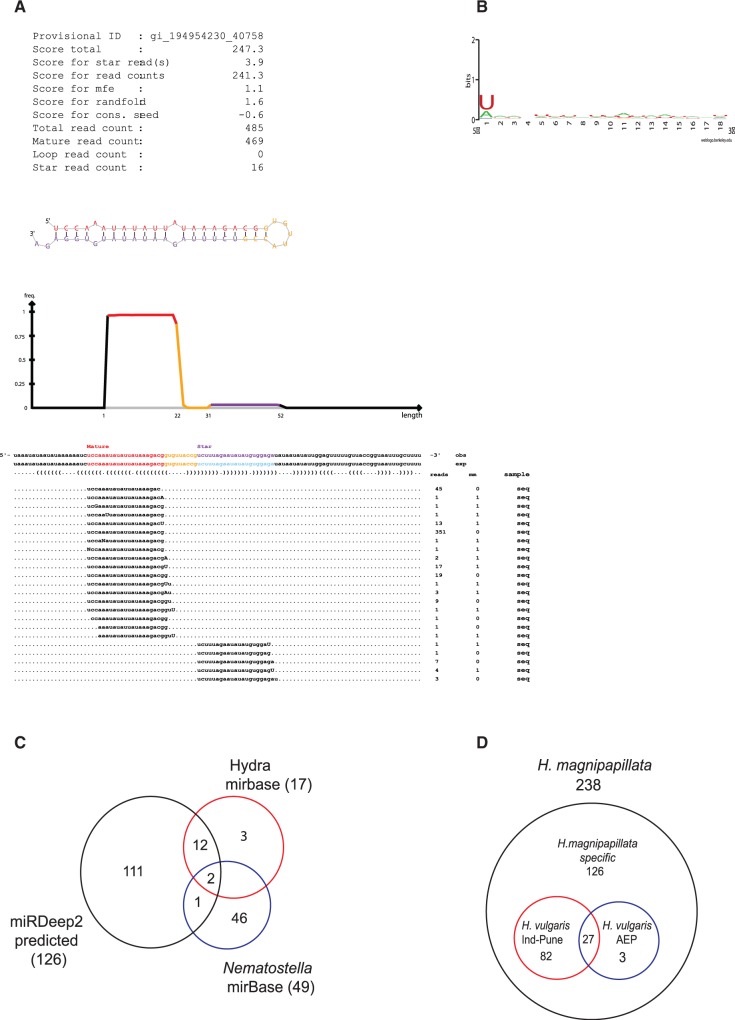
To identify novel miRNAs, we mapped all 18–24 bp reads to the hydra genome and predicted novel miRNAs using the program miRDeep2 (44). The miRDeep2 predicted list was filtered based on scores >+10 and randfold P-value <0.05. Precursor structures obtained after filtering were manually curated based on the presence of 1-2 mismatches in the stem region, a loop sequence of 4–8 nt, and the presence of mature sequence in the stem region (Figure 2A). Output from mirDeep2 consisted of loci from hydra genome to which 18–24 mers map allowing one mismatch. Using these criteria, miRDeep2 identified 128 miRNA loci in the H. magnipapillata genome. We reanalysed 18–24 mer reads that mapped to these miRNA loci, allowing no mismatch, and all 128 miRNA loci identified mapped to the genome without any mismatch (Supplementary Table S1). Of these 128 loci, two potential miRNAs, hma-mir-new2-1 and hma-mir-new2-2, were not considered for further analysis for reasons described below. Among the rest 126 loci, we found 112 with both mature and star strand with perfect alignment to genome (Supplementary Table S2). In case of 14 miRNAs, only the mature strand and not star strands showing perfect alignment to the hydra genome were identified (Supplementary Table S2).
Figure 2.
Identification of hydra miRNAs using miRDeep2. (A) Figure shows sequence of a typical miRNA (hma-mir-new46) identified by miRDeep2, showing mature strand and star strand and the frequency of the reads obtained for both strands during deep sequencing. (B) Base preference for predicted miRNAs. All predicted miRNAs show preference for U at 5′-end indicating processing by dicer. (C) Pie chart depicting miRNAs that are common between set of miRNAs identified in this study, hydra miRNAs present in miRBase and N. vectensis miRNAs present in miRBase. (D) 18–24 nt reads from H. vulgaris Ind-Pune and H. vulgaris AEP were mapped to miRNAs identified in H. magnipapillata allowing no mismatch and figure shows miRNAs conserved across these species of hydra.
To test the validity of miRDeep2 program, we checked for the presence of previously annotated hydra miRNAs from miRBase. We found 14 of 17 miRNAs previously predicted (30), underscoring the authenticity of the miRDeep2 program for prediction of hydra miRNAs (Figure 2C and Supplementary Table S1). The sequences corresponding to remaining three miRNAs were present in the list of 18–24 bp reads, but miRDeep2 failed to predict them as miRNA because of the absence of corresponding star strands from deep sequencing data. Further, we also analysed conservation of hydra miRNA loci in other metazoans. Only 3 of 126 miRNAs from hydra were conserved in another cnidarian, Nematostella vectensis (nve-miR-2022, nve-miR-2029 and nve-miR-2030) (10), as shown in Figure 2C, and none of these miRNAs were conserved in higher metazoa or in sponges. Interestingly, neither the highly conserved miRNA miR-100, which is expressed in most metazoa, including N. vectensis, nor miRNA let-7, which is conserved in bilateria (13), was identified in our sequencing analysis, suggesting a unique repertoire of hydra miRNAs.
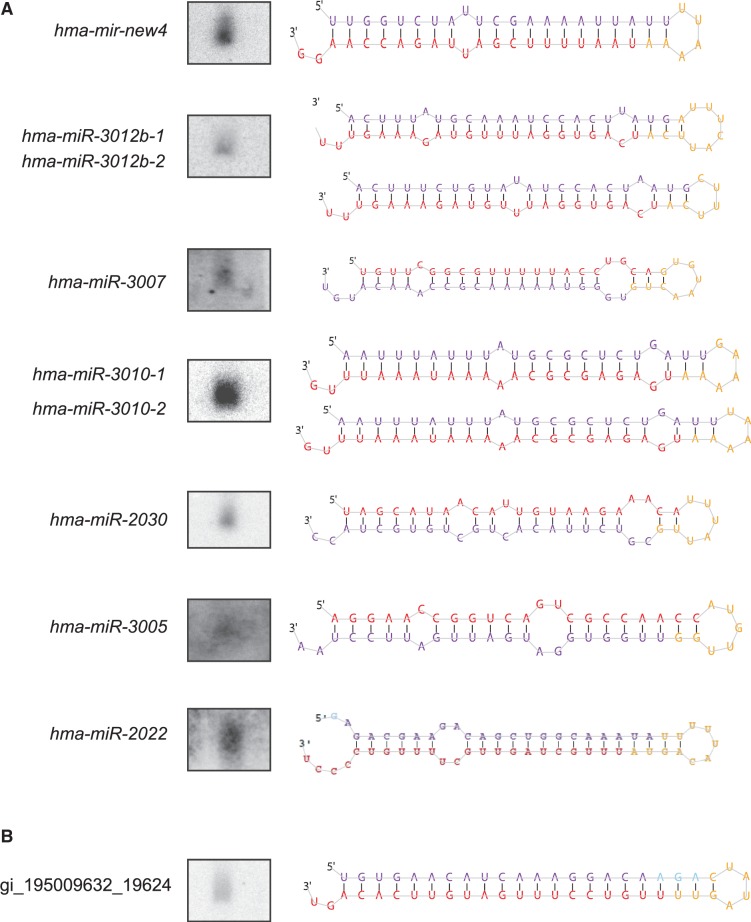
Most miRNAs range in size from 21–22 nt and show strong enrichment for ‘U’ or ‘A’ residue at 5′-end, a signature for potential Dicer cleavage products (Figure 2B). We found eight miRNAs with identical mature strand sequence but different precursor sequences (Supplementary Table S3). In general, many miRNAs in complex metazoans exist in a cluster and are derived from individual multi-cistronic transcripts. In hydra, we found a single miRNA cluster on contig ABRM01025207.1 with two miRNA loci (hma-mir-new22 and hma-mir-new63) separated by 500 bp (Supplementary Figure S1B and C). Interestingly, we also found another pair of miRNA loci (hma-mir-new66 and hma-mir-new11) that maps to the same genomic region but display an antisense orientation to each other on the contig ABRM01070152.1 (Supplementary Figure S1C). miRNAs are typically 21–22 nt in size and can be visualized by Northern hybridization using antisense probes against specific mature miRNAs. We analysed 13 of the miRDeep2-predicted miRNAs by Northern hybridization. As a control for Northern hybridization, we performed Northern hybridization with S. mediterranea RNA using miR71C probe as described (18). The signal obtained for hydra miRNAs was at the same position as miR71C (data not shown), confirming the expected size of 21–22 nt in 7 of 13 miRNAs investigated (Figure 3A). We further performed reverse transcription-polymerase chain reaction (RT-PCR) to confirm expression of new miRNAs that were identified in this study (35). The PCR products obtained were cloned and sequenced to confirm expression. We performed RT-PCR analysis on 13 newly identified miRNAs and were able to confirm expression of all 13 miRNAs by sequencing (Supplementary Table S2). Interestingly, two highly expressed small RNAs, hma-miR-new2-1 and hma-mir-new2-2, with different precursor sequences but same mature strand (Supplementary Table S1), could not be detected either by Northern hybridization or PCR, and hence have been removed from analysis. Interestingly, this small RNA was identified in libraries of all three strains of hydra sequenced in this study. The number of miRNAs reported in this study therefore is 126. We also performed in situ hybridization for miRNAs to investigate their localization in hydra as shown in Figure 4. We used planarian lin-4-3p miRNA as a negative control, as it is not expressed in hydra. All three miRNAs showed specific expression; hma-miR-3005 and hma-miR-2030 were expressed throughout the body column including ectoderm and endoderm, till the base of tentacles but not in tentacles, whereas hma-miR-3010 was expressed only in the endoderm and at the base of tentacles.
Figure 3.
Northern analysis of miRNAs identified by miRDeep2. (A) Northern analysis was performed on H. magnipapillata RNA using radiolabelled probe for mature sequence of miRNAs indicated, along with the stem–loop structure predicted by miRDeep2. There are two pairs of miRNAs (hma-miR-3012b-1 and hma-miR-3012b-2, hma-miR-3010-1 and hma-miR-3010-2) where two precursors have identical mature sequence. Stem–loop structures for both miRNAs from these pairs are depicted in the figure. (B) Figure shows Northern analysis done for endo-siRNA with perfect stem–loop structure, gi_195009632_19624.
Figure 4.
In situ hybridization showing expression pattern of hydra miRNAs. (A) In situ hybridization was performed using DIG-labelled locked nucleic acid probes for miRNAs hma-miR-2030, hma-miR-3005 and hma-miR-3010. lin-4-3p probe was used as a negative control, as it is not expressed in hydra. Inset shows expression pattern in ectoderm and endoderm at higher magnification. (B) Figure shows expression pattern of a perfect stem–loop structure gi_195009632_19624 seen after in situ hybridization.
Conservation of miRNAs across hydra strains
In general, the miRNA loci are highly conserved across all species of an organism. We therefore analysed for the sequence conservation of all 238 5p and 3p arms of 126 miRNA loci across two other strains of hydra, H. vulgaris AEP and H. vulgaris Ind-Pune, which are geographically separated from each other and from H. magnipapillata. H. magnipapillata was originally isolated from Japan and has been used for research over many years (30); the AEP strain of Hydra vulgaris was isolated from the USA (45) and is an important tool in hydra biology owing to its ability to readily undergo sexual reproduction. Hydra normally reproduces asexually by budding and sexual reproduction occurs under conditions of stress; the AEP strain can be readily induced to undergo sexual reproduction and has been used for generating transgenic lines of hydra (45). H. vulgaris Ind-Pune strain was isolated from Pune, India; it has been used for developmental studies and was recently classified as a strain of H. vulgaris (34,46). H. magnipapillata and H. vulgaris species are closely related and belong to the same monophyletic vulgaris group, although H. vulgaris AEP appears to be closer to H. carnea rather than rest of the vulgaris species (47).
Small RNA libraries were generated using total RNA from Hydra vulgaris AEP and H. vulgaris Ind-Pune and were sequenced using the Illumina platform. These reads were aligned to H. magnipapillata miRNA database consisting of 238 5p and 3p arm sequences, allowing no mismatches. Surprisingly the number of miRNAs conserved between different species of hydra is low, in contrast to bilateria where a higher percentage of miRNAs are conserved between different species, with only 27 miRNAs (5p and 3p) common between all three species. Out of 238 sequences, 109 were common in H. vulgaris Ind-Pune and H. magnipapillata, whereas 30 were common between H. magnipapillata and H. vulgaris AEP (Figure 2D and Supplementary Table S2). It is also possible that miRNAs expressed at very low levels in these two strains were perhaps not detected in this study. Our observations that H. vulgaris Ind-Pune shares more miRNAs with H. magnipapillata than H. vulgaris AEP can be attributed to the phylogenetic distance reported between these strains (47,48).
We also found several small RNAs that were identified as miRNAs by miRDeep2, but unlike miRNAs, they have perfect matches in the stem region. These small RNAs are highly AU rich and could have been derived from inverted repeats. They have 1-2 nt overhang at 3′-end of the precursor and constitute 19.6% of small RNAs identified as miRNAs by miRDeep2. We have classified these as endo-siRNAs, and they appear to be similar to the hairpin RNAs (hpRNAs) in the unicellular algae Chlamydomonas (12). Northern analysis for one such sequence showed hybridization to a 21 nt species confirming it as potential small RNA (Figure 3B). In situ hybridization of this small RNA revealed expression throughout the body column in both ectoderm and endoderm but not in the tentacles, with more expression in budding region than in the rest of the body column (Figure 4B).
Putative piRNAs in hydra
Our analysis also revealed a distinct class of small RNAs that were 27–30 nt in size. These are the most abundant class of small RNAs (67.2% of total reads) in hydra (Figure 1A) and might represent potential piRNAs, as they are of the same size as piRNAs and they possess sequence signatures seen in piRNAs from higher metazoa, as described below. Because we do not have direct evidence showing the binding of these small RNAs to PIWI proteins, we call these small RNAs putative piRNAs in this report. piRNAs are typically expressed in germ cells in Drosophila and mammals, and are important for germ line stem cell function (6). In planarian S. mediterranea, they are expressed in neoblasts, the adult pluripotent stem cells (7). piRNAs are also expressed in diploblastic animals like Nematostella and Amphimedon, but the expression pattern of these piRNAs is not known (10). In Drosophila, most piRNAs map to transposable elements and are responsible for transposon silencing (49,50). Drosophila piRNAs also to map to transcripts and may down-regulate them (8,9). In mammals, piRNAs are mostly expressed during spermatogenesis and are broadly divided into two categories: pre-pachytene piRNAs and pachytene piRNAs. Pre-pachytene piRNAs map to transposable elements and are involved in silencing of transposable elements (51). Pachytene piRNAs are expressed during late meiosis and map to unique regions in the mouse genome, but their function is not clearly understood (52,53). In the present work, small RNA libraries were made from the asexual form of hydra, which is devoid of gonads, and therefore putative piRNAs thus identified here are mostly present in somatic cells.
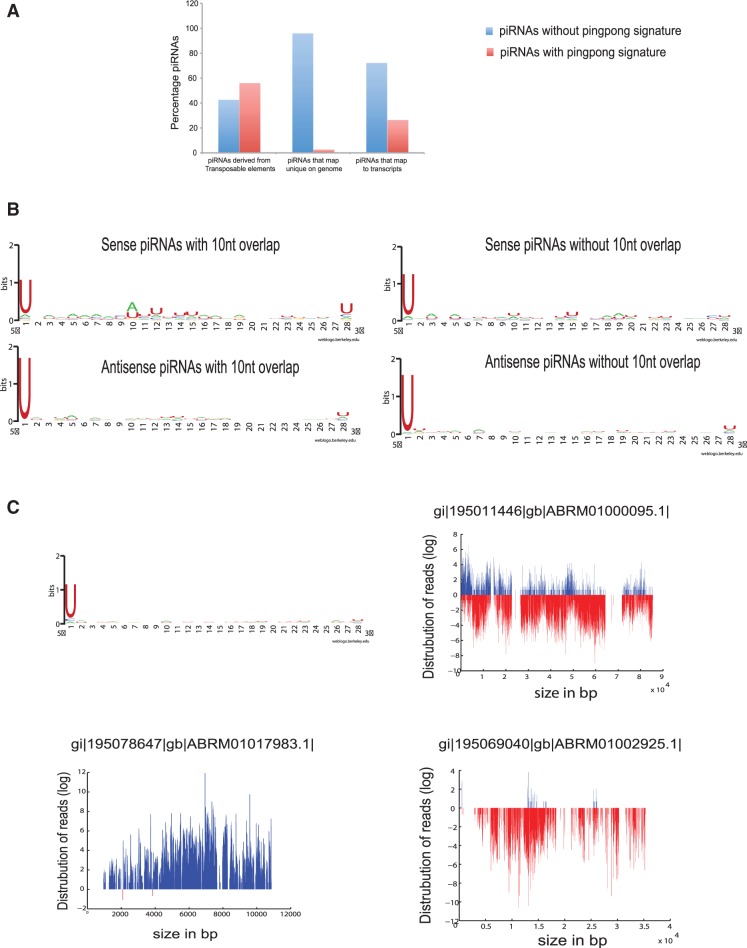
In Drosophila, piRNA biogenesis is proposed to follow two distinct models, which have been described as ‘ping–pong dependent’ and ‘ping–pong independent’ models (54). In the ping–pong model, piRNAs are bidirectional, with sense and antisense piRNAs showing 10 nt overlap. Further, sense strands are characterized by the presence of ‘A’ residue at 10th position and antisense strands have ‘U’ at the 5′-end. In ping–pong independent model, the sense and antisense strands could be bidirectional but do not have 10 nt overlap. A similar sequence analysis in mammals suggests that pre-pachytene piRNAs arise in ping–pong-dependent manner, whereas pachytene arise in ping–pong-independent manner. In order to find out the regions to which putative piRNAs map in hydra, we aligned all reads >24 nt to transposable elements of hydra from RepBase, to hydra genome and to the annotated transcriptome. We further analysed these putative piRNAs to predict the mechanism of biogenesis of these putative piRNAs. Interestingly, we find that in hydra 10% of reads map to transposable elements, 28.4% of reads map to unique regions on hydra genome and 13% of reads map to annotated transcriptome.
In all, 56.7% putative piRNAs that map to transposable elements have bidirectional orientation where sense strands have strong ping–pong signature with strong preference for ‘A’ residue at 10th nucleotide position, suggesting that the biogenesis of these putative piRNAs is likely to occur via ping–pong model (Figure 5A, left panel of Figure 5B, and Supplementary Table S7). The rest of the 43.2% bidirectional putative piRNAs that map to transposable elements do not have 10 nucleotide overlap, and the sense strands from these putative piRNAs do not have a preference for A at 10th nucleotide (Figure 5B, right panel and Supplementary Table S7). These putative piRNAs, however, have a strong preference for ‘U’ residue at the 5′-end, indicating they are products of processing by Argonaute family of proteins, rather than RNA degradation products. These sequence signatures suggest their biogenesis could occur via a ping–pong-independent mechanism rather than the absence of ping–pong partners owing to the lack of sequencing depth (Figure 5B, right panel).
Figure 5.
Putative piRNAs in hydra. (A) Graph showing distribution of ping–pong-dependent and ping–pong-independent putative piRNAs that map to repeat elements, unique regions on hydra genome and to transcripts. Putative piRNAs mapping to transcriptome and transposable elements show a significant number of piRNAs with ping–pong signature. (B) Base preference for piRNAs that map to transposable elements. The left panel shows base preference of sense strand from piRNAs with ping–pong signature (top) which shows preference for ‘A’ at 10th place and base preference for ‘U’ at the first nucleotide at 5′-end in the corresponding antisense piRNAs (bottom). The right panel shows base preference for the piRNAs that map to transposable elements but do not have ping–pong signature. The top right panel shows sense strand and the bottom right panel shows anti-sense strand. Both strands show preference for ‘U’ at 5′-end, indicating processing by dicer. (C) Figure shows distribution of piRNAs mapping to unique regions on genome. The graph on top left shows base preference for these piRNAs, which show enrichment for ‘U’ at 5′-end. The panel on the top right shows contig with bidirectional orientation of piRNAs. The lower panel shows orientation of unidirectional piRNAs on two contigs.
We found 28.4% of 25–30 nt reads mapping to unique non-coding regions on the genome, which resemble mammalian pachytene piRNAs. The numbers of reads that map to the contig vary from 1000–500 000 reads/contig. A total of 96.7% of these putative piRNAs do not have 10 nt overlap (Figure 5A) and have strong enrichment for ‘U’ at the 5′-end (Figure 5C, top panel and Supplementary Table S7), suggesting that they could arise in ping–pong-independent manner. With the exception of putative piRNAs that map to ABRM01000095.1 contig, which have bidirectional distribution (Figure 5C, top panel), most of the other major clusters (>100 000) map to either of the strands on the contigs (Figure 5C, bottom panel). It is, however, difficult to determine the exact number of such piRNA clusters on the genome owing to incomplete assembly of the reference hydra genome.
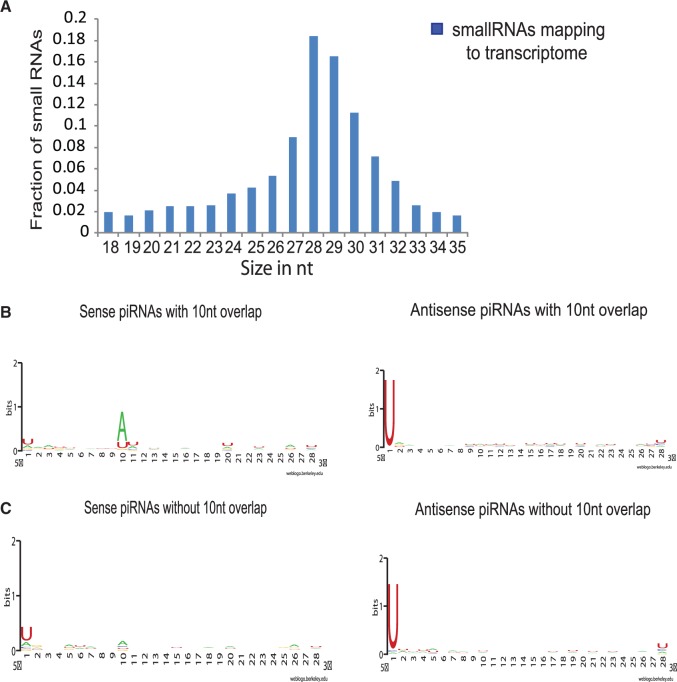
A significant number of small RNAs (13%) mapped to the annotated transcriptome. We selected all reads of size 18–30 nt and aligned them to the annotated transcriptome from NCBI. We found a strong enrichment for the reads of size 27–30 nt compared with 18–24 nt (Figure 6A). The small fraction (∼10%) of 18–24 bp reads that map to transcripts were evenly distributed across 18–24 nt without any specific enrichment for a particular size (Figure 6A). Further, 18–24 nt reads were mostly derived from the same regions of the transcript where 27–30 nt reads align, suggesting they are potential degradation products of 27–30 nt (data not shown). We then filtered the alignment file based on the density of reads that map to the transcripts. The density was the ratio of the number of reads that map to the transcript per kilobase of the transcript. For further analysis, transcripts with density of at least 100 reads of piRNAs mapping per kilobase of transcript were considered. We found 2141 transcripts with density of at least 1000, and 2265 transcripts with density of 100–999. Reads that map to these transcripts are both unidirectional and bidirectional. We found 27% of these reads have ping–pong signature (Figure 5A and Figure 6B). The remaining 73% of putative piRNAs do not have 10 nt overlap, but majority of antisense piRNAs have strong enrichment for U at the 5′-end, ruling out the possibility that these are degradation products (Figure 6C, right panel and Supplementary Table S7). However, most of the sense piRNAs that do not have 10 nt overlap lack base residue preferences at both the 5′-end and at 10 nt position, suggesting they are potential degradation products of transcripts and not piRNAs (Figure 6C, left panel and Supplementary Table S7). Thus, transcripts that have only sense orientation putative piRNAs mapping to them are removed from further analysis, and only those transcripts that either have putative piRNAs with bidirectional orientation or antisense orientation are taken into consideration.
Figure 6.
Putative piRNAs mapping to transcriptome. (A) Size distribution of small RNAs mapping to transcriptome. The figure shows fraction of 18–35 nt small RNAs mapping to transcriptome. Majority of the small RNAs are 28–29 nt long. (B) Base preference of putative piRNAs that map to transcriptome and show ping–pong signature. The left panel shows preference for ‘A’ at 10th nucleotide in sense strands and right panel shows preference for ‘U’ at the first nucleotide at 5′-end in the antisense strand. (C) Base preference of piRNAs that map to transcriptome but do not show ping–pong signature. The left panel show sense piRNAs that do not show any base preference, suggesting they are degradation products. The right panel shows antisense piRNAs with preference for ‘U’ at 5′-end.
Interestingly, among the transcripts to which piRNAs map, we find several histone genes. piRNAs on histones are distributed all across the length of the transcript. We mostly observed sense strand piRNAs mapping to the histone transcripts compared with the antisense strand (Supplementary Figure S2). We also looked for base residue preference and found strong enrichment for U residue at 5′-end, suggesting they are AGO/PIWI cleavage products.
Small RNAs in hydra regeneration
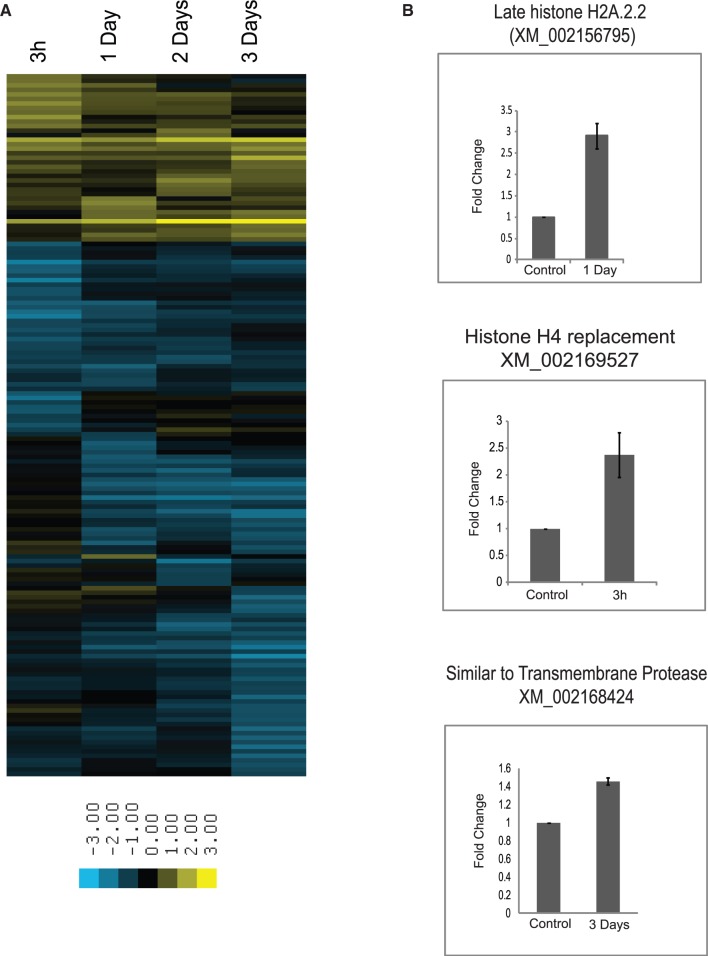
To investigate whether small RNAs uncovered in the sequence analysis play a role in regeneration, we investigated the regulation of small RNA expression in hydra regenerates. Sequencing was performed for two different sets of RNA isolated from regenerating heads at 3 h, 1 day, 2 days and 3 days after mid-gastric bisection. miRNA reads obtained from each head regenerate were first normalized to per million reads, and the correlation coefficient between the duplicates was calculated. There was a significant (>0.95) correlation between duplicates (Supplementary Figure S3), hence further analysis was done by averaging the number of reads per million for each duplicate. miRNAs with >10 raw reads were considered for further analysis for changes in expression levels. To investigate changes in the expression levels of miRNAs that were altered during regeneration, we calculated the fold difference in expression level of each miRNA, as compared to the control and identified miRNAs that showed at least two-fold difference in expression. Thus, we identified 10 miRNAs that showed differences in expression levels at different time points of hydra head regeneration; 2 miRNAs were up-regulated and 8 were down-regulated during regeneration (Figure 7A and Supplementary Table S4). We further validated the changes in expression for three miRNAs, hma-mir-new13-3p, hma-miR-2022-3p and hma-miR-new40-3p, by quantitative PCR (Figure 7B) (35). hma-miR-2022-3p showed >50% reduction in expression at 3 days of regeneration during deep sequencing, as compared to the control. Consistent with these observations, quantitative PCR showed a 60% reduction in the expression level of hma-miR-2022-3p. Similarly, miRNA hma-mir-new13-3p, which showed increased expression at 3 h by deep sequencing showed 80% up-regulation and hma-mir-new40-3p showed 4-fold up-regulation at 3 h and 2 days of regeneration by quantitative PCR.
Figure 7.
Changes in miRNA expression during head regeneration. (A) Heat map showing regulation of miRNAs during hydra head regeneration. The number of reads for each miRNA for each regeneration time point was normalized to per million reads; duplicates for each time point were averaged and then expressed as fold change over control expression levels. Log values of fold change to the base 2 were used for generating heat map. Only the miRNAs showing correlation coefficient of >0.95 and with at least 10 raw reads were considered for the analysis. (B) Quantitative PCR for miRNA hma-miR-2022-3p, hma-mir-new13-3p and hma-mir40-3p showing regulation of these miRNAs during head regeneration. hma-miR-2022-3p is down-regulated 3 days after mid-gastric cut, whereas hma-mir-new13-3p is up-regulated at 3 h after mid-gastric cut and hma-mir40-3p is up-regulated 3 h and 2 days after mid-gastric cut.
We performed similar analysis to investigate the changes in expression of putative piRNA clusters mapping to transcripts, genomic contigs and transposable elements. Although we did not find significant changes in expression of the putative piRNA cluster that map to transposable elements and unique regions on the contig, we identified 165 new putative piRNA clusters that map to various transcripts and show at least two-fold variation in expression levels (Supplementary Table S5). For this analysis, piRNAs mapping to 4406 transcripts as mentioned earlier under ‘putative piRNAs in hydra’ were considered. piRNA reads were normalized to per million of total reads mapping to transcriptome. Out of these filtered reads, reads showing at least two-fold difference in expression as compared to the control were selected as differentially expressed piRNAs. We identified 165 piRNA clusters that show either up-regulation or down-regulation during different stages of regeneration. Most of the putative piRNA clusters mapping to transcripts show decreased expression during hydra head regeneration. Interestingly, some of these putative piRNA clusters map to the genes encoding histones suggesting piRNAs might play a role in up-regulating histone gene expression during regeneration (Figure 8A, Supplementary Table S5). We also identified 38 putative piRNA clusters that map to transcripts that are up-regulated during hydra head regeneration. We further investigated whether the piRNAs could regulate the transcripts they map to in hydra. We performed quantitative PCRs to analyse whether the transcripts to which piRNAs map are differentially expressed. Transcripts for Late Histone H2A2.2, Histone H4 replacement and a transmembrane protease showed a small, but consistent, up-regulation at the time point when the corresponding piRNAs were down-regulated (Figure 8B). This suggests that piRNAs could potentially regulate expression of hydra transcripts during regeneration. Taken together, these results indicate a potential role for miRNAs and piRNAs in regulating gene expression during hydra regeneration.
Figure 8.
Changes in piRNA expression during head regeneration. (A) Heat map showing piRNAs that map to transcriptome and are regulated during hydra head regeneration. piRNA expression was first normalized to expression per million of total number of 25–30 nt reads that mapped to the transcriptome; duplicates for each time point of regeneration were averaged and normalized reads were expressed as fold change over control. Further only the piRNAs with >100 reads per kilobase of transcript were considered to check for changes in expression levels. (B) Quantitative PCR for transcripts to which piRNAs map and are differentially regulated during regeneration. Down-regulation in piRNA levels during regeneration is expected to lead to increased transcript levels. The figure shows increase in transcript levels of Late Histone H2A2.2, Histone H4 replacement and a transmembrane protease at time points when corresponding piRNAs are down-regulated as shown in Supplementary Table S5. Each qPCR was done with two independent biological replicates, with each PCR done in triplicate for each time point.
DISCUSSION
In this study, we report profiling of miRNAs and piRNAs from the cnidarian model system hydra by deep sequencing. Three new findings emerge from our analysis; first, we have identified 112 new miRNAs in hydra, adding substantially to the 17 miRNAs reported earlier (30). Second, hydra also expresses a number of endo-siRNAs which arise from perfect stem–loop structures, with 1-2 nt overhangs at the 3′-end of the precursor. Unexpectedly, the most abundant small RNAs in hydra are piRNAs that map to both repeat sequences and unique genomic regions, including transcripts. Sequence analysis of putative piRNAs suggests that these could be processed by both ping–pong-dependent and ping–pong-independent mechanisms. Finally, we show that several miRNAs and piRNAs are regulated during regeneration, indicating a role for small RNAs in hydra regeneration.
Our study has identified several novel miRNAs that were not previously identified (30), and this can be attributed to the improved depth of sequencing in this study. The most striking feature of hydra miRNAs identified here is their unique repertoire and diversity. Only three hydra miRNAs are conserved in another cnidarian, Nematostella, and none of the hydra miRNAs are conserved in any higher metazoan or in sponges. Surprisingly, hydra does not express two highly conserved miRNAs mir-100 and let-7. mir-100 is expressed in all bilaterians as well as Nematostella while let-7 is expressed in all bilateria (13). A search for sequences of let-7 or miR-100 yielded no genomic contigs that could potentially encode these miRNAs, suggesting that hydra genome does not encode these miRNAs. A similar observation has been made in the sponge Amphimedon, where none of the miRNAs identified were conserved in higher metazoans (10). It is interesting to note that miRNAs in bilateria are highly conserved, but those from diploblasts like Amphimedon and hydra appear to be unique to each organism. Conceivably, miRNA genes have independent evolutionary origins in the basal metazoa. Alternatively small RNAs in basal metazoa might be highly diverse owing to higher rate of evolution. Small RNA profiling of sponges and cnidaria like jellyfish, Hydractinia and Acropora can further explore this question.
Within the genus hydra, each species analysed expresses a large number of unique miRNAs, with only 11.3% miRNAs conserved between all three species analysed here. In all, 12.6% of miRNAs present in H. magnipapillata were identified in H. vulgaris AEP and 45.7% were identified in the H. vulgaris Ind-Pune. Whether this lack of detection is due to absence of expression of miRNAs or due to absence of miRNAs from the genome cannot be explored at this stage, as the genomic sequences of these two species are not available. Hydra miRNAs are perhaps under evolutionary pressure that necessitated evolution at a higher rate than higher metazoa, and therefore show less conservation across species. For example, each species of hydra has specific strains of bacteria as symbionts, and this pattern of association is maintained even after the species are cultured under laboratory conditions for several years (55). It is possible that the species-specific miRNAs evolved to regulate genes involved in response to such commensals, symbionts or parasites associated with each species. Alternatively, species-specific miRNAs could also target species-specific genes from these organisms. Several genome sequences analysed so far have reported around 10–20% genes that have no significant similarity to sequences in other organisms and these are termed as ‘taxonomically restricted genes’ (TRG) (56). It is possible that the species-specific miRNAs identified in this study regulate TRGs. Moreover, the hydra genome is itself highly dynamic, being rich in transposable elements, which contribute to 57% of the genome (30). The genome size within hydra species is also variable (57) and the genome organization is believed to have been influenced by bursts of transposable element expansion and horizontal gene transfer. These changes to genome organization and acquisition of genes through retrotransposition could have exerted evolutionary pressure leading to species-specific evolution of miRNAs to regulate these genes. Alternatively, it is also possible that evolution of miRNAs in turn could have led to speciation of hydra.
miRNAs are often found organized in clusters on genome in organisms like Drosophila and vertebrates, while the earliest triploblast Planarian S. mediterranea has seven such clusters (18). Such clusters of miRNAs are believed to be important for coordinated regulation of gene expression, and the numbers of miRNA clusters in a genome appear to correlate with organismal complexity. We found one miRNA cluster with two miRNA loci in the hydra genome (Supplementary Figure S1B and C). No clusters have been reported in Nematostella or Amphimedon (10), suggesting that coordinated regulation of miRNAs through organized clusters may not be a common mechanism of miRNA regulation in diploblasts.
We have also probed alterations in miRNA expression during hydra head regeneration. Ten miRNAs that are regulated at least by two-fold at various time points following bisection were identified, suggesting a role for miRNAs in the regulation of gene expression during regeneration. Most of these miRNAs (8 of 10) were down-regulated during head regeneration, indicating a potential role in suppression of genes that play a role in head formation and/or tentacle development. Interestingly, we observe that although at 3 days the regenerating hydra morphologically resembles a complete organism, none of the miRNAs that are regulated return to expression levels seen in unperturbed control organisms, suggesting that the process of regeneration is not complete at this time. Eight of these 10 miRNAs that were regulated during head regeneration are also conserved in H. vulgaris Ind-Pune. Interestingly we found only 2 out of 10 regulated miRNAs conserved in H. vulgaris AEP strain (Supplementary Table S4). Conceivably, the AEP strain, which is more closely related to H. carnea than other vulgaris species, might have evolved a set of novel miRNAs for regulation of regeneration.
As in Amphimedon and Nematostella (10), we find that piRNA-like small RNAs represent the most abundant class of small RNAs in hydra. These putative piRNA sequences were derived from the asexual form of hydra, suggesting they are not expressed in germ cells as in mammals, but as in Planarian S. mediterranea, may be expressed in stem cells (7). A transcriptome sequencing of different cell lineages of hydra shows that all three cell lineages in hydra, ectodermal cells, endodermal cells and interstitial cells, express piwi transcripts (58), which indicates that putative piRNAs could be present in the stem cells of all three cell lineages. In hydra, putative piRNAs mapped to both repeat elements as well as unique loci in the genome. A total of 57.2% putative piRNAs that map to repeat elements showed a signature of ping–pong-dependent biogenesis, confirming the earlier observations that this pathway of piRNA biogenesis evolved early (10). We also identified a large number of putative piRNAs without a ping–pong signature. At present, we cannot rule out the possibility that these are direct cleavage products of Argonaute proteins rather than PIWI proteins, which can only be resolved by knocking down the argonaute and piwi genes. We speculate that the putative piRNAs mapping to transposable elements would be involved in transposon silencing either via transposon degradation or heterochromatin formation, thus maintaining genome integrity. However, only 10% of putative piRNAs map to repeat elements and a larger number maps to unique regions on the genome, suggesting piRNAs in hydra may play a wider role than regulation of transposable elements. These putative piRNAs resemble pachytene piRNAs of mammals in terms of their genome organization. We found at least 12 clusters of piRNAs with >100 000 reads that map to different contigs. In the absence of genome assembly, it is difficult to predict the exact number of piRNA clusters and their location. In mammals, pachytene piRNA clusters are important for maintenance of germ line integrity (6) and the presence of similar piRNA clusters in an asexual strain of hydra makes it interesting to study their role in hydra biolog\y.
We identified several putative piRNA clusters that map to transcripts, suggesting that piRNAs may be important for gene regulation in hydra. A total of 27% of these piRNAs have ping–pong signature. The remaining 73% could either be associated with the ping–pong-independent pathway or could be Argonaute-processed small RNAs. These putative piRNA clusters could potentially regulate gene expression. Indeed, we find that a small, but significant, number of piRNAs that map to 165 transcripts are either up-regulated or down-regulated during head regeneration. Interestingly, some of these transcripts where piRNA clusters were down-regulated include various histone genes such as H2B, H1 and late histones H2A. Some of these genes such as Histone H2A and Histone H4 replacement show increase in expression levels at the time point where their corresponding piRNAs are down-regulated, indicating that these putative piRNAs could indeed regulate transcript levels. Recently, it has been shown that specific variants of histone H1 are expressed in human embryonic cells and induced pluripotent cells but not in differentiated cells (59). It is possible that a similar regulation of histone genes could be important for hydra regeneration. In Drosophila, piRNAs map to the 3′-UTR region of maf transcripts raising the possibility of regulation of expression of maf loci by piRNAs (8,9). piRNAs derived from the intronic region of the human melatonin receptor 1A (mtnr1a) also negatively regulate mtnr1a gene expression by binding to the genomic region (60). These isolated studies show that in addition to transposon regulation, piRNAs are likely to play alternate roles in regulating gene expression. It is interesting to note that putative piRNAs in hydra appear to regulate diverse genomic elements and that their role is not limited to regulation of gametogenesis as in higher metazoa. Similar to piRNAs in S. mediterranea, piRNAs in hydra could be important for adult stem cell function. It is possible that piRNAs that arose early in evolution played a wider role and their role eventually became more specialized for regulation of transposable elements in germ cells in higher organisms.
In summary, our study has identified a large number of novel miRNAs and piRNA-like small RNAs, some of which are regulated during head regeneration in H. magnipapillata. These findings serve as the platform from which to address more challenging task of defining the targets of miRNAs, expression of piRNAs in different cell types, biogenesis of piRNA-like small RNAs and functional roles played by these small RNAs in the unique biology of hydra.
ACCESSION NUMBERS
The small RNA sequencing data have been deposited at the NCBI Sequence Read Archive (SRA) at http://www.ncbi.nlm.nih.gov/sra (accession number SRA050926).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–7 and Supplementary Figures 1–3.
FUNDING
Institute for Stem Cell Biology and Regenerative Medicine (Government of India, Department of Biotechnology); Fast-Track fellowship from Department of Science and Technology, India [SERC/LS-0329/2008 to Y.G.]; DBT-Wellcome Trust India Alliance (to D.P.). Funding for open access charge: The publication charges will be covered by the Institute for Stem Cell Biology and Regenerative Medicine.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Prof Surendra Ghaskadbi for providing H. vulgaris Ind-Pune, Dr Girish Ratnaparkhi for providing H. magnipapillata and Prof Thomas Bosch for providing H. vulgaris AEP strains. Deep sequencing was performed at the Next Generation Sequencing facility at the Centre for Cellular and Molecular Platform (C-CAMP), Bangalore, supported by the Department of Biotechnology. A part of deep sequencing was also performed at Genotypic, Bangalore and Northern hybridizarion was performed at Anthem Biosciences, Bangalore. We also thank Aswin Seshasayee for critically reading the manuscript. The project was conceptualized by D.P., Y.G. and J.D. Experiments were performed by S.K. with help from A.N. and S.C. Bioinformatic analysis was performed by D. Poduval and D. Palakodeti. D.P. and Y.G. designed and supervised the project and prepared the manuscript.
REFERENCES
- 1.Liu Q, Paroo Z. Biochemical principles of small RNA pathways. Annu. Rev. Biochem. 2010;79:295–319. doi: 10.1146/annurev.biochem.052208.151733. [DOI] [PubMed] [Google Scholar]
- 2.Karginov FV, Hannon GJ. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol. Cell. 2010;37:7–19. doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 5.Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat. Rev. Mol. Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Juliano C, Wang J, Lin H. Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu. Rev. Genet. 2011;45:447–469. doi: 10.1146/annurev-genet-110410-132541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palakodeti D, Smielewska M, Lu YC, Yeo GW, Graveley BR. The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA. 2008;14:1174–1186. doi: 10.1261/rna.1085008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, Blower MD, Lai EC. A broadly conserved pathway generates 3′UTR-directed primary piRNAs. Curr. Biol. 2009;19:2066–2076. doi: 10.1016/j.cub.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- 10.Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao T, Li G, Mi S, Li S, Hannon GJ, Wang XJ, Qi Y. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 2007;21:1190–1203. doi: 10.1101/gad.1543507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molnar A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007;447:1126–1129. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- 13.Christodoulou F, Raible F, Tomer R, Simakov O, Trachana K, Klaus S, Snyman H, Hannon GJ, Bork P, Arendt D. Ancient animal microRNAs and the evolution of tissue identity. Nature. 2010;463:1084–1088. doi: 10.1038/nature08744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thatcher EJ, Paydar I, Anderson KK, Patton JG. Regulation of zebrafish fin regeneration by microRNAs. Proc. Natl Acad. Sci. USA. 2008;105:18384–18389. doi: 10.1073/pnas.0803713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palakodeti D, Smielewska M, Graveley BR. MicroRNAs from the Planarian Schmidtea mediterranea: a model system for stem cell biology. RNA. 2006;12:1640–1649. doi: 10.1261/rna.117206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, Seaver E, Rouse GW, Obst M, Edgecombe GD, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- 20.Philippe H, Derelle R, Lopez P, Pick K, Borchiellini C, Boury-Esnault N, Vacelet J, Renard E, Houliston E, Queinnec E, et al. Phylogenomics revives traditional views on deep animal relationships. Curr. Biol. 2009;19:706–712. doi: 10.1016/j.cub.2009.02.052. [DOI] [PubMed] [Google Scholar]
- 21.Bridge D, Cunningham CW, Schierwater B, DeSalle R, Buss LW. Class-level relationships in the phylum Cnidaria: evidence from mitochondrial genome structure. Proc. Natl Acad. Sci. USA. 1992;89:8750–8753. doi: 10.1073/pnas.89.18.8750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bridge D, Cunningham CW, DeSalle R, Buss LW. Class-level relationships in the phylum Cnidaria: molecular and morphological evidence. Mol. Biol. Evol. 1995;12:679–689. doi: 10.1093/oxfordjournals.molbev.a040246. [DOI] [PubMed] [Google Scholar]
- 23.Cartwright P, Halgedahl SL, Hendricks JR, Jarrard RD, Marques AC, Collins AG, Lieberman BS. Exceptionally preserved jellyfishes from the Middle Cambrian. PLoS One. 2007;2:e1121. doi: 10.1371/journal.pone.0001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bosch TC. Why polyps regenerate and we don't: towards a cellular and molecular framework for Hydra regeneration. Dev. Biol. 2007;303:421–433. doi: 10.1016/j.ydbio.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 25.Bosch TC, Anton-Erxleben F, Hemmrich G, Khalturin K. The Hydra polyp: nothing but an active stem cell community. Dev. Growth Differ. 2010;52:15–25. doi: 10.1111/j.1440-169X.2009.01143.x. [DOI] [PubMed] [Google Scholar]
- 26.Holstein TW, Hobmayer E, Technau U. Cnidarians: an evolutionarily conserved model system for regeneration? Dev. Dyn. 2003;226:257–267. doi: 10.1002/dvdy.10227. [DOI] [PubMed] [Google Scholar]
- 27.Bode H. Axis formation in hydra. Annu. Rev. Genet. 2011;45:105–117. doi: 10.1146/annurev-genet-102209-163540. [DOI] [PubMed] [Google Scholar]
- 28.Shimizu H, Sawada Y, Sugiyama T. Minimum tissue size required for hydra regeneration. Dev. Biol. 1993;155:287–296. doi: 10.1006/dbio.1993.1028. [DOI] [PubMed] [Google Scholar]
- 29.Gierer A, Berking S, Bode H, David CN, Flick K, Hansmann G, Schaller H, Trenkner E. Regeneration of hydra from reaggregated cells. Nat. New Biol. 1972;239:98–101. doi: 10.1038/newbio239098a0. [DOI] [PubMed] [Google Scholar]
- 30.Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, Rattei T, Balasubramanian PG, Borman J, Busam D, et al. The dynamic genome of Hydra. Nature. 2010;464:592–596. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lohmann JU, Endl I, Bosch TC. Silencing of developmental genes in Hydra. Dev. Biol. 1999;214:211–214. doi: 10.1006/dbio.1999.9407. [DOI] [PubMed] [Google Scholar]
- 32.Chera S, de Rosa R, Miljkovic-Licina M, Dobretz K, Ghila L, Kaloulis K, Galliot B. Silencing of the hydra serine protease inhibitor Kazal1 gene mimics the human SPINK1 pancreatic phenotype. J. Cell Sci. 2006;119:846–857. doi: 10.1242/jcs.02807. [DOI] [PubMed] [Google Scholar]
- 33.Horibata Y, Sakaguchi K, Okino N, Iida H, Inagaki M, Fujisawa T, Hama Y, Ito M. Unique catabolic pathway of glycosphingolipids in a hydrozoan, Hydra magnipapillata, involving endoglycoceramidase. J. Biol. Chem. 2004;279:33379–33389. doi: 10.1074/jbc.M401460200. [DOI] [PubMed] [Google Scholar]
- 34.Chandramore K, Ito Y, Takahashi S, Asashima M, Ghaskadbi S. Cloning of noggin gene from hydra and analysis of its functional conservation using Xenopus laevis embryos. Evol. Dev. 2010;12:267–274. doi: 10.1111/j.1525-142X.2010.00412.x. [DOI] [PubMed] [Google Scholar]
- 35.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bode H, Lengfeld T, Hobmayer B, Holstein TW. Detection of expression patterns in hydra pattern formation. In: Vincan E, editor. Methods in Molecular Biology. New York: Humana Press; 2008. Vol. 469, pp. 69–84. [DOI] [PubMed] [Google Scholar]
- 37.Shimizu H, Zhang X, Zhang J, Leontovich A, Fei K, Yan L, Sarras MP., Jr Epithelial morphogenesis in hydra requires de novo expression of extracellular matrix components and matrix metalloproteinases. Development. 2002;129:1521–1532. doi: 10.1242/dev.129.6.1521. [DOI] [PubMed] [Google Scholar]
- 38.MacWilliams HK. Hydra transplantation phenomena and the mechanism of hydra head regeneration. I. Properties of the head inhibition. Dev. Biol. 1983;96:217–238. doi: 10.1016/0012-1606(83)90324-x. [DOI] [PubMed] [Google Scholar]
- 39.MacWilliams HK. Hydra transplantation phenomena and the mechanism of Hydra head regeneration. II. Properties of the head activation. Dev. Biol. 1983;96:239–257. doi: 10.1016/0012-1606(83)90325-1. [DOI] [PubMed] [Google Scholar]
- 40.Hobmayer B, Rentzsch F, Kuhn K, Happel CM, von Laue CC, Snyder P, Rothbacher U, Holstein TW. WNT signalling molecules act in axis formation in the diploblastic metazoan Hydra. Nature. 2000;407:186–189. doi: 10.1038/35025063. [DOI] [PubMed] [Google Scholar]
- 41.Bode HR. Head regeneration in Hydra. Dev. Dyn. 2003;226:225–236. doi: 10.1002/dvdy.10225. [DOI] [PubMed] [Google Scholar]
- 42.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farajollahi S, Maas S. Molecular diversity through RNA editing: a balancing act. Trends Genet. 2010;26:221–230. doi: 10.1016/j.tig.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wittlieb J, Khalturin K, Lohmann JU, Anton-Erxleben F, Bosch TC. Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc. Natl Acad. Sci. USA. 2006;103:6208–6211. doi: 10.1073/pnas.0510163103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reddy PC, Bidaye SS, Ghaskadbi S. Genome-wide screening reveals the emergence and divergence of RTK homologues in basal Metazoan Hydra magnipapillata. J. Biosci. 2011;36:289–296. doi: 10.1007/s12038-011-9065-6. [DOI] [PubMed] [Google Scholar]
- 47.Kawaida H, Shimizu H, Fujisawa T, Tachida H, Kobayakawa Y. Molecular phylogenetic study in genus Hydra. Gene. 2010;468:30–40. doi: 10.1016/j.gene.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 48.Reddy PC, Barve A, Ghaskadbi S. Description and phylogenetic characterization of common hydra from India. Curr. Sci. 2011;101:736–738. [Google Scholar]
- 49.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 50.Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007;35:5430–5438. doi: 10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 52.Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 55.Fraune S, Bosch TC. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc. Natl Acad. Sci. USA. 2007;104:13146–13151. doi: 10.1073/pnas.0703375104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TC. More than just orphans: are taxonomically-restricted genes important in evolution? Trends Genet. 2009;25:404–413. doi: 10.1016/j.tig.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 57.Zacharias H, Anokhin B, Khalturin K, Bosch TC. Genome sizes and chromosomes in the basal metazoan Hydra. Zoology (Jena) 2004;107:219–227. doi: 10.1016/j.zool.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 58.Hemmrich G, Khalturin K, Boehm AM, Puchert M, Anton-Erxleben F, Wittlieb J, Klostermeier UC, Rosenstiel P, Oberg HH, Domazet-Loso T, et al. Molecular signatures of the three stem cell lineages in Hydra and the emergence of stem cell function at the base of multicellularity. Mol Biol Evol. 2012;29:3267–3280. doi: 10.1093/molbev/mss134. [DOI] [PubMed] [Google Scholar]
- 59.Yang L, Duff MO, Graveley BR, Carmichael GG, Chen LL. Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 2011;12:R16. doi: 10.1186/gb-2011-12-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Esposito T, Magliocca S, Formicola D, Gianfrancesco F. piR_015520 belongs to Piwi-associated RNAs regulates expression of the human melatonin receptor 1A gene. PLoS One. 2011;6:e22727. doi: 10.1371/journal.pone.0022727. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.