Abstract
microRNAs (miRNAs) regulate gene expression at multiple levels by repressing translation, stimulating deadenylation and inducing the premature decay of target messenger RNAs (mRNAs). Although the mechanism by which miRNAs repress translation has been widely studied, the precise step targeted and the molecular insights of such repression are still evasive. Here, we have used our newly designed in vitro system, which allows to study miRNA effect on translation independently of deadenylation. By using specific inhibitors of various stages of protein synthesis, we first show that miRNAs target exclusively the early steps of translation with no effect on 60S ribosomal subunit joining, elongation or termination. Then, by using viral proteases and IRES-driven mRNA constructs, we found that translational inhibition takes place during 43S ribosomal scanning and requires both the poly(A) binding protein and eIF4G independently from their physical interaction.
INTRODUCTION
microRNAs (miRNAs) are small non-coding RNAs that participate in many cellular processes as essential gene regulators. miRNAs act as guides for the RNA-induced silencing complex (RISC) to bind messenger RNAs (mRNAs) and to repress their translation and/or decrease their stability. Usually, miRNAs bind to their target mRNAs at the 3′-untranslated region (3′-UTR) through partial base pairing (1). As a consequence, miRNAs can potentially interact with numerous target mRNAs. In agreement with this, 60% of all mammalian mRNAs have been reported to contain conserved miRNA target sequences (2).
Many mechanisms have been proposed to explain how miRNAs could regulate gene expression including translational repression, mRNA deadenylation and accelerated decay, which are non exclusive but rather sequential. In fact, recent data suggest that translational repression is the first mechanism of miRNA-induced gene repression, followed by mRNA deadenylation and eventually its degradation (3–7). Concerning translational repression, miRNAs were first reported to regulate translation at post-initiation steps (8–12), but recent data strongly suggest that repression takes place at the initiation stage (7,13–20).
However, there is still some controversy about the stage at which translation initiation could be repressed. Although many reports point to the 5′ cap structure as an essential factor necessary for translational repression (7,14,16–19), the need for other cis-acting factors such as the poly(A) tail is less clear with some data indicating an essential role for the poly(A) tail (14,19,21,22), while others report that its removal or replacement by the 3′ stem-loop tail of histone transcripts does not affect miRNA activity (17,23,24). In addition, PABP has been recently implicated in miRNA effect by interacting with the C-terminal domain of GW182 to promote translational repression and deadenylation (6,25–28).
Moreover, studies have failed to converge regarding the actual stage of translation initiation regulated by miRNAs. Some reports state that miRNAs act by targeting cap recognition and recruitment of the 43S complex (16,17,29), while others describe repression at the level of 60S ribosomal subunit joining (13,20). Thus, the precise molecular mechanisms by which miRNAs mediate translational repression remain a matter of debate.
Recent data strongly suggest that translational repression occurs prior to transcript deadenylation and degradation (3,5,30,31). This fits well with the rabbit reticulocyte lysate (RRL) model that we have previously described, which contains endogenous miRNAs that are fully functional to repress translation and to induce an siRNA response (22). One of the main advantages of this system is that repression occurs only at the level of translation with no effect on transcript degradation or deadenylation (22). Thus, it allows to focus only on the molecular mechanism used by miRNAs to interfere with protein synthesis. We have exploited the advantage of the RRL to assess the impact of miRNA repression on each individual step of protein synthesis (e.g. initiation, ribosomal subunit joining, elongation, termination and peptide release). Our results first show that miRNAs interfere only with translation initiation. Using a combination of viral proteases together with reporter genes containing cellular 5′-UTR with different structure or IRESes from different viral families, we could show that repression takes place at the level of 43S ribosomal scanning. Moreover, we show evidence that both PABP and eIF4G are necessary for miRNA-mediated translation inhibition, but this requirement is independent from the physical interaction between these two proteins.
MATERIALS AND METHODS
DNA constructs and in vitro transcription
Plasmids containing target sites for miR-451 (Luc, Luc-451X6, Luc-451mut) were already described (22). 5′-UTRs were obtained by PCR on cDNA obtained from Hela-cells total RNAs (BCL3, GAPDH, Cyclin D2, Line-1 and Hsp70-1), pEMCV-renilla and pHCV-renilla plasmids (32), pXLPV and pEMCV-PV plasmids (33), pXLCSFV 1–423.NS plasmid (34) avian encephalomyocarditis viruses (AEVs) plasmid (35), and Seneca Valley virus (SVV)+55 construct (36) using specific primers containing EcoRV restriction site and T7 promoter (for sense primers) and BamHI restriction site (for antisense primers). PCR products were digested and cloned in Luc and Luc-451X6 vectors previously digested by PvuII and BamHI restriction enzymes.
Plasmids were digested using EcoRI (polyadenylated RNAs), NaeI [internalized poly(A)] or XbaI (non polyadenylated RNAs) restriction enzymes. RNAs were obtained by using 1 mg linearized plasmid, 10 U T7 RNA polymerase (Promega Co., Madison, WI, USA), 20 U of RNAsin (Promega Co, Madison, WI, USA), 10 mM of rATP, rUTP and rCTP, 0.48 mM rGTP, 3 mM DTT and 1.3 mM m7GpppG (capped RNAs) or ApppG (uncapped RNAs) cap analog (New England Biolabs) in transcription buffer [40 mM Tris–HCl (pH 7.9), 6 mM MgCl2, 2 mM spermidine and 10 mM NaCl]. The transcription reaction was carried out at 37°C for 2 h, and the RNAs were treated with RQ1 DNAse (Promega Co., Madison, WI, USA) and precipitated with Ammonium Acetate at 2.5 M final concentration. The integrity of the RNAs was checked by electrophoresis on non-denaturating agarose gels and their concentration was quantified by spectrophotometry at 260 nm using Nanodrop (NanoDropTechnologies, Wilmington, DE, USA). For radiolabeled RNAs the same protocol was used except that rUTP was omitted and replaced by 20 mCi of aP32 rUTP.
Western blotting
To test for initiation factor integrity, 3 µl of each reaction was recovered after translation and resolved on a 10% SDS–PAGE. Proteins were then transferred to a PVDF membrane by electroblotting and incubated with antibodies against PABP or eIF4G (kind gifts from Dr Morley).
Preparation of untreated RRL and in vitro translation assays
Untreated RRL was prepared essentially as previously described (32). Translation reactions were performed in a final volume of 30 µl consisting of 20 µl of untreated RRL, 0.46 fmol of heat denatured mRNAs, in the presence of KCl (100 mM), MgCl2 (0.5 mM) and amino acids mixture (20 µM each). RRL under full translational condition was incubated together with the heat denatured mRNA for 1 h at 10°C, followed by 2 min at 20°C, 2 min at 25°C and 30 or 60 min at 30°C. The reaction was then stopped by the addition of 50 µl of luciferase lysis buffer to 10 µl of the translation reaction.
Renilla activity was measured and normalized to an internal Firefly luciferase mRNA for all experiments that do not involve the addition of a translational inhibitor (Figures 1, 2, 4 and 5).
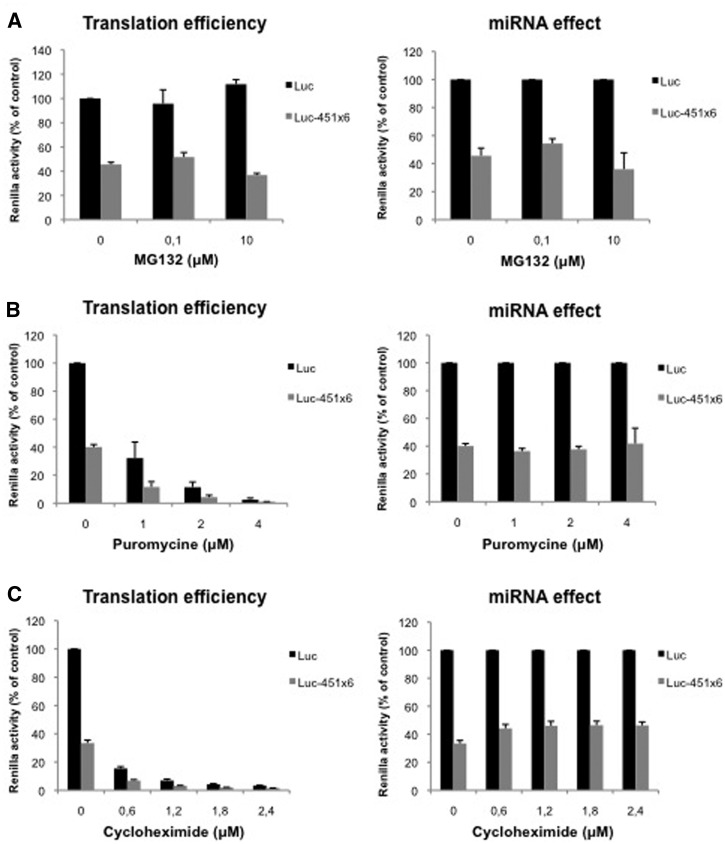
Figure 1.
miRNAs do not target translation elongation nor degradation of nascent peptides through the proteasome. (A) Translation of Luc and Luc-451X6 RNAs was carried out in untreated RRL in presence of indicated concentration of MG132. (B) Translation of Luc and Luc-451X6 RNAs was carried out in untreated RRL in presence of indicated concentration of puromycine. (C) Translation of Luc and Luc-451X6 RNAs was carried out in untreated RRL in presence of indicated concentration of cycloheximide. Results are shown as translation efficiency (left panels) and miRNA effect (right panels), as described in ‘Materials and Methods’ section. Error bars correspond to SD obtained from three independent experiments.
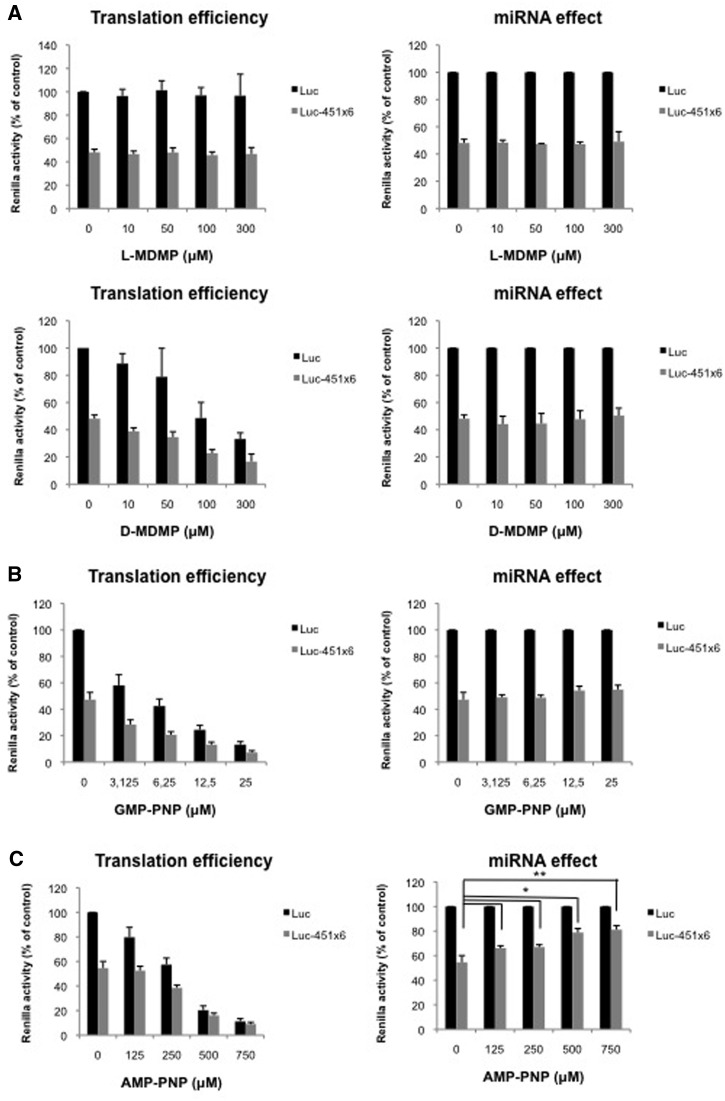
Figure 2.
60S ribosomal joining is not regulated by miRNAs. (A) Translation of Luc and Luc-451X6 RNAs was carried out in untreated RRL in presence of indicated concentration of l-MDMP (top panels) or d-MDMP (bottom panels). (B) Translation of Luc and Luc-451X6 RNAs was carried out in untreated RRL in presence of indicated concentration of GMP-PNP. (C) Translation of Luc and Luc-451X6 RNAs was carried out in untreated RRL in presence of indicated concentration of AMP-PNP. Results are shown as translation efficiency (left panels) and miRNA effect (right panels), as described in ‘Materials and Methods’ section. Error bars correspond to SD obtained from three independent experiments. * corresponds to a P-value <0.05; ** corresponds to a P-value <0.01; (non directional t-test).
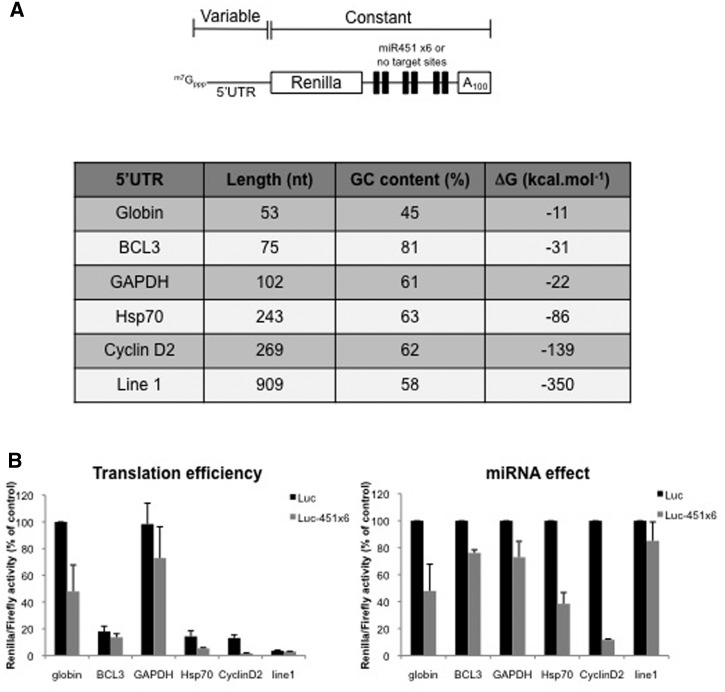
Figure 4.
miRNA-mediated translation inhibition is affected by 5′-UTR composition. (A) Schematic representation of the Renilla luciferase RNA used, which contains either no target sites (Luc) or six target sites for miR-451 (Luc-451) at the 3′-end. Expression of this construct was driven by various 5′-UTR that were derived from cellular transcripts as shown on the table (stability of the 5′-UTR was predicted using the mfold program). (B) Translation of Luc and Luc-451X6 RNAs driven by different 5′-UTR was carried out in untreated RRL. Results are shown as translation efficiency (left panel) and miRNA effect (right panel), as described in ‘Materials and Methods’ section. Error bars correspond to SD obtained from three independent experiments.
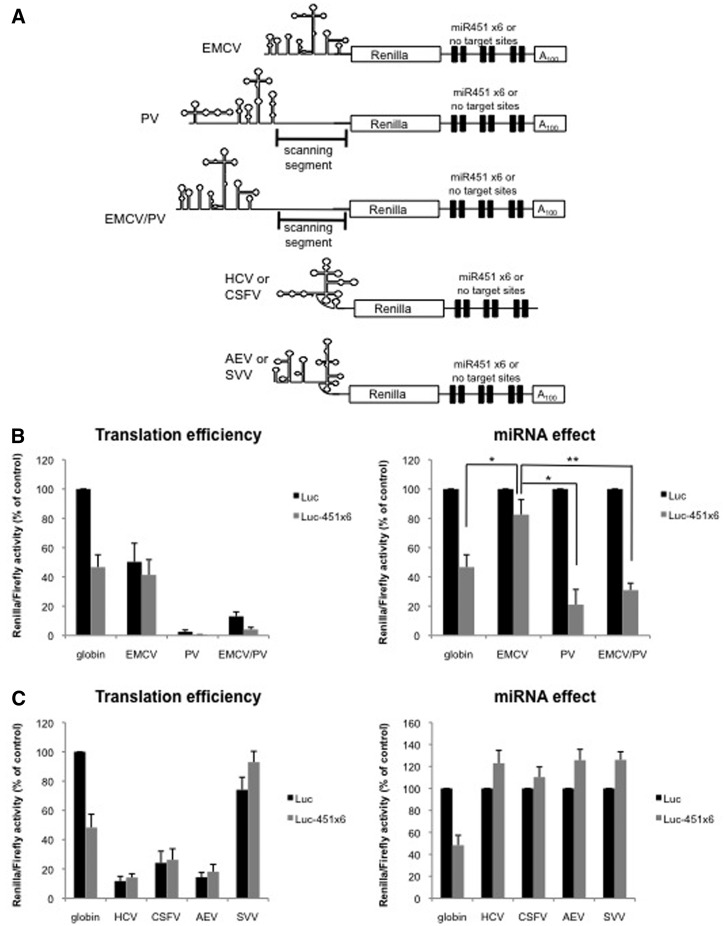
Figure 5.
Ribosomal scanning is required for miRNA-mediated inhibition. (A) Schematic representation of the Renilla luciferase RNAs in which translation initiation was driven by EMCV, PV, the EMCV/PV chimera, HCV, CSFV, SVV or AEV IRES. (B) Translation of Luc or Luc-451X6 constructs containing the EMCV, PV, EMCV/PV IRES or globin 5′-UTR as a control, in the untreated RRL. (C) Translation of Luc or Luc-451X6 constructs containing the HCV, CSFV, SVV, AEV IRES or globin 5′-UTR as a control in the untreated RRL. Results are shown as translation efficiency (left panels) and miRNA effect (right panels), as described in ‘Materials and Methods’ section. Error bars correspond to SD obtained from three independent experiments; * corresponds to a P-value <0.05; ** corresponds to a P-value <0.01; (non directional t-test).
For all the experiments, we express translation efficiency as the percentage of luciferase activity compared to the control Luc mRNA (set at 100%); for miRNA effect, in each condition, the luciferase activity from the Luc-451X6 mRNA is expressed as a percentage of its control Luc mRNA (set to 100%).
For analysis of radiolabeled RNA integrity, RNA were translated as described and extracted at indicated times with Tris-reagent following manufacturer conditions. Total RNAs were run on a 2% agarose gel and analyzed by autoradiography on a phosphorimager.
Preparation of viral proteases
Commercial 3C protease from human rhinovirus (HRV) was obtained from Novagen (Madison, WI, USA). The l-protease from the foot-and-mouth disease virus (FMDV) was produced by in vitro translation using nuclease-treated RRL as previously described (32) and 2 μl were added prior to translation. The human immunodeficiency virus type-2 (HIV-2) protease was obtained from the NIH and 2 μl were added prior to translation. For rescue experiment, translation reactions were treated 10 min with 2 μl HIV-2 protease, cleavage was then blocked with 10 μM palinavir and translation were carried during 1 h in presence of dialysis buffer or recombinant PABP or eIF4G (kind gift of C.S. Fraser).
Preparation of PABP recombinant protein
E scherichia coli BL21 cells expressing the pET15b-PABP vector (kindly provided by Martin Bushell) were grown until A600 reached 0.6–0.8 and then, induced overnight at 30°C with IPTG 0.5 mM. Bacterial pellets were resuspended in native lysis buffer (50 mM NaH2PO4 [pH 8.0]; 300 mM NaCl and 10 mM imidazole) supplemented with 1 mg/ml lysozyme (Sigma) and cells were lysed by sonication. Supernatant was recovered and incubated with Ni-NTA resin (Qiagen) (previously equilibrated in lysis buffer) at 4°C for 2 h under gentle shaking. The resin was then washed three times with five volumes of washing buffer [50 mM NaH2PO4 [pH 8.0]; 300 mM NaCl and 20 mM imidazole and protease inhibitor cocktail (Roche)] and protein was then eluted with elution buffer (50 mM NaH2PO4 [pH 8.0]; 300 mM NaCl; 500 mM Imidazole and protease inhibitor cocktail). The eluted protein was desalted and concentrated with dialysis buffer (20 mM HEPES [pH 7.5], 100 mM KCl, 2 mM DTT and protease inhibitor cocktail) using Spin-X UF Concentrators (Corning).
RESULTS
miRNAs repress mRNA translation independently of deadenylation and decay of target mRNAs
In this study, we have used the untreated RRL as a model in vitro system to study the effects of endogenous miRNAs on translation of exogenous reporter transcripts. For this, we used a Renilla luciferase reporter construct that harbors, unless specified, a globin 5′-UTR and are followed by a 3′-UTR containing either six target-sites for miR-451 (namely ‘Luc-451X6’) or lacking miRNA target sites (namely ‘Luc’). We deliberately chose to use miR-451 as a model miRNA for this study as we previously showed that it is highly expressed in the reticulocyte lysate and recapitulates all aspects of the miRNA response (22). Protein synthesis was quantified by measuring luciferase activity and both translation efficiency and miRNA effect are quantified and plotted on individual bar graphs (see ‘Materials and Methods’ section).
In our previous published work, we had shown that translational repression occurred in the absence of any deadenylation of the target mRNA. This was demonstrated by checking the integrity of radiolabeled RNAs after translation and by real-time quantitative PCR (22). However, because the extent of deadenylation could be difficult to assess, we have used another experimental strategy to show evidence that inhibition of gene expression in our system was not due to shortening of the poly(A) tail. In order to do this, we have adapted the method recently described by Tomari and colleagues (37) which consists of internalizing the poly(A) tail by adding a stretch of non specific residues at the 3′ end of the poly(A) tail (Supplementary Figure S1A). In the present case, 280 nt were added and the integrity of the radiolabeled transcripts that contain 6 miR-451 target sites or 6 mutated sequences was checked on agarose gel (Supplementary Figure S1B). We did not observe any deadenylation or degradation of the mRNAs at the early time points (0, 0.5, 1 and 2 h). However, at 3 h, we detected a decrease in the amount of both control and targeted mRNA, which suggests that some decay has occurred (Supplementary Figure S1B). We then tested the effect of poly(A) tail internalization on miRNA-mediated repression. Interestingly, RNAs with normal and internalized poly(A) were inhibited by miR-451 to a similar level (50%), even after 6 h of incubation (Supplementary Figure S1C). As an additional control, we have also investigated whether mutations in the seed region of the target gene could reverse the effects of miR-451. Supplementary Figure 1D shows the translational efficiency and the miR-451 effects over a 6 hours period of time.
Altogether, these data confirm the results that we had obtained in our previous published work and further validates that inhibition of gene expression takes place at the level of translation in the RRL.
miRNAs do not target translation elongation nor degradation of nascent peptides through the proteasome
Taking advantage of our system that allows translational repression in the absence of deadenylation and mRNA decay, we have assessed the impact of miRNA repression in the presence of a large spectrum of effectors that are known to block translation at each of the various steps of protein synthesis (initiation, ribosomal subunit joining, elongation, termination and peptide release).
We initially started with the proteasome inhibitor MG132 as miRNAs have been suggested to regulate translation elongation through the proteolytic cleavage of nascent peptides (11) and was already used to study miRNA activity in living cells (17). As observed (Figure 1A, left panel), addition of MG132 did not significantly affect luciferase production from both Luc and Luc-451X6 mRNAs. Furthermore, translational repression mediated by miR-451 was not affected thus suggesting that miRNAs do not target the proteolytic degradation of nascent peptides, at least not through a proteasome-induced process (Figure 1A, right panel).
We then tested whether miR-451 could cause the premature drop-off of elongating ribosomes as previously suggested for the artificial miRNA CXCR4 (12). For this, we added increasing amounts of puromycin to our translation reactions and then monitored translation of Luc and Luc-451X6 mRNAs (Figure 1B). Puromycin causes the premature termination of elongating ribosomes thus inducing ribosome drop-off. As observed (Figure 1B, left panel), addition of puromycin to translation extracts led to a dose-dependent inhibition of luciferase expression from both RNAs reaching a 60-fold inhibition at the highest concentration. Strikingly, the relative level of miRNA-mediated repression remained constant to ∼60% of the control in all conditions tested (Figure 1B, right panel). Interestingly, similar results were obtained when translation elongation was inhibited by the addition of cycloheximide, which stalls elongating 80S ribosomes on the mRNA without inducing their drop-off (Figure 1C). Indeed, as observed for puromycin, addition of increasing amounts of cycloheximide strongly inhibited luciferase expression without inducing further effects on miRNA-451 repression (Figure 1C, right panel).
Taken together, these results suggest that miRNAs do not interfere with translation elongation or proteosomal degradation of nascent peptides.
60S ribosomal joining is not regulated by miRNAs
We next tested whether the ribosomal subunits joining step could be affected by miR-451. This was suggested by a report showing that let-7 miRNAs could regulate this stage of translation with RISC being able to associate with 60S ribosomes through eIF6 to interfere with the formation of the 80S ribosome on targeted mRNAs (13); such an hypothesis has also been challenged by other researchers (38). A similar mechanism was also proposed in the nuclease-treated RRL using the artificial CXR4 miRNA system showing poor association of the 60S subunit to the 40S ribosomal subunit (20).
In an attempt to reduce formation of 80S ribosomes, we took advantage of MDMP, a chemical compound that specifically impairs the association of 60S and 40S ribosomal subunits (39,40). As observed, addition of increasing amounts of d-MDMP (the biologically active stereoisomer) led to a dose-dependent inhibition of both Luc and Luc-451X6 mRNAs translation (Figure 2A, bottom left panel) whereas addition of the inactive l-stereoisomer did not affect translation (Figure 2A, top left panel). Interestingly, despite the inhibition of translational rates, miRNA-mediated repression was not changed upon addition of d-MDMP (Figure 2A, bottom right panel) suggesting that miRNAs did not interfere with the joining of the 60S ribosomal subunit.
Ribosomal subunit joining is also regulated by the release of initiation factors from the 40S ribosome upon recognition of the AUG initiation codon. This stage is mediated by hydrolysis of the GTP molecule associated to eIF2 and eIF5B (41). Interestingly, since Argonaute 2 was first characterized to be a protein that regulates eIF2 recycling both in RRL and wheat germ extracts (42), it was of interest to test whether it could affect 60S ribosome joining by inhibiting eIF2 release. For this, we used GMP-PNP, a non-hydrolysable GTP analog that stalls the 48S complex at the AUG start codon and prevents 60S ribosome binding. As observed (Figure 2B, left panel), addition of GMP-PNP led to a dose-dependent inhibition of Luc and Luc-451X6 mRNAs translation reaching a 7-fold inhibition at the highest concentration. However, as observed for MDMP, translational repression Luc-451X6 mRNA was not affected therefore suggesting that RISC did not interfere with the hydrolysis of eIF2-bound GTP (Figure 2B, right panel).
As discussed earlier, translation initiation requires two molecules of GTP to proceed but is also dependent on ATP hydrolysis through the use of various DEAD-Box RNA helicases that are required for translation initiation, among which is eIF4A (43). The latter is an ATP dependent RNA helicase which is required both for initial ribosome binding to the mRNA and linear scanning of the ribosome from the binding site to the AUG initiation codon (44). In fact, eIF4A was shown to ‘prepare’ a landing pad for ribosomes on the mRNA and then unwinds the RNA secondary structures encountered during ribosomal scanning (44). In order to test whether ATP hydrolysis can be targeted by miRNA repression, we used AMP-PNP, a non-hydrolysable analogue of ATP. As observed (Figure 2C, left panel), addition of AMP-PNP to translation extracts led to a dose-dependent inhibition of Luc translation reaching up to 10-fold repression. Interestingly, Luc-451X6 translation appeared to be relatively more resistant to AMP-PNP than Luc translation, especially at low amounts of AMP-PNP (Figure 2C, left panel). This is particularly striking when Luc-451X6 translation was normalized against that of Luc RNA (Figure 2C, right panel) and we could observe a derepression of miRNA activity following the addition of AMP-PNP suggesting that miR-451 interferes with ATP hydrolysis. However, although they are indicative of a link between ATP hydrolysis and miRNA repression, these data must be interpreted with care due to the involvement of ATP in many biological processes including tRNA aminoacylation or even formation of the RISC complex (45).
Taken together, data obtained with various inhibitors suggest that miRNA repression does not take place at the level of 60S subunit joining, nor elongation, nor termination thus suggesting a potential role at the level of initiation.
PABP and eIF4G are required for miRNA-mediated repression independently from their physical interaction
miRNA-mediated repression at the level of translation initiation has been suggested by other studies in cell free system to primarily depend on the involvement of PABP and eIF4F (6,16). This prompted us to further assess the role of these factors in our in vitro system. For this, we used a combination of different viral proteases that are able to cleave PABP, eIF4G or both.
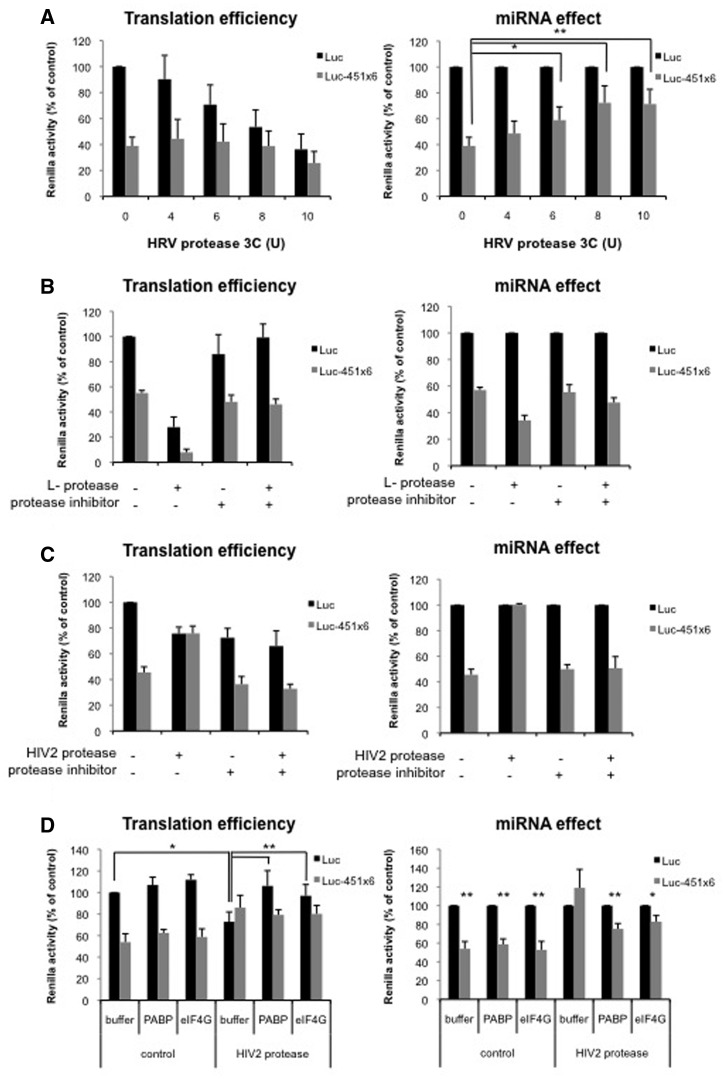
The protease 3C from Human Rhinovirus is known to inhibit translation initiation through cleavage of PABP without affecting the composition of the eIF4F complex (46). We thus used this enzyme to block PABP function prior to translation of Luc or Luc-451X6 mRNAs. As shown on western blotting, PABP was efficiently cleaved at the highest concentration of the protease added (Supplementary Figure S2A) and this resulted in translation inhibition (Figure 3A, left panel). However, the relative effect of miRNA-mediated translation inhibition was significantly relieved, from 50% to 20% (Figure 3A, right panel) suggesting that PABP is required for the miRNA response in the RRL.
Figure 3.
PABP and eIF4G are required for miRNA-mediated repression independently from their physical interaction. (A) Translation of Luc and Luc-451X6 RNAs was carried out in untreated RRL after addition of HRV 3C protease as indicated. (B) Translation of Luc and Luc-451X6 RNAs was carried out in untreated RRL after addition of 2 μl in vitro translated l-protease, E64 protease inhibitor (10 μM) or both as indicated. (C) Translation of Luc and Luc-451X6 RNAs was carried out in untreated RRL after addition of 2 μl recombinant HIV-2 protease, the palinavir protease inhibitor (10 μM) or both as indicated. (D) Translation of Luc and Luc-451X6 RNAs was carried out in RRL and treated with 2 μl HIV-2 protease for 10 min. The reaction was then stopped by addition of the palinavir protease inhibitor (10 μM) and the recombinant PABP (500 ng) or eIF4G (1 μg) were added to the mixture for a further 50 min before luciferase analysis. Results are shown as translation efficiency (left panels) and miRNA effect (right panels), as described in ‘Materials and Methods’ section. Error bars correspond to SD obtained from three independent experiments; * corresponds to a P-value <0.05; ** corresponds to a P-value <0.01; (non directional t-test).
We then used the l-protease of FMDV which destabilizes the eIF4F complex by processing eIF4G in a N-terminus fragment that contains binding sites for eIF4E and PABP and a C-terminus domain which harbors the binding sites for eIF3 and eIF4A (47). Thus, in vitro translated l-protease was added to the untreated RRL prior to translation of Luc and Luc-451X6 RNAs. As visualized by western blot, eIF4G was cleaved upon addition of l-protease (Supplementary Figure S2B) and this resulted in translation inhibition (Figure 3B, left panel, compare Control and l-protease lanes). Addition of the protease inhibitor E64 restored wild-type levels of translation indicating that translational repression was merely the result of the cleavage of eIF4G. Interestingly, miRNA-mediated repression was not affected and, if anything, rather slightly stimulated by the addition of l-protease (Figure 3B, right panel). This indicates that neither the integrity of eIF4F nor the N-terminal domain of eIF4G is necessary for miRNA function and suggests that RISC does not target the eIF4G-PABP physical interaction.
Finally, we have used the viral protease from the HIV-2 to measure its effect on translation of Luc and Luc-451X6 mRNAs. This enzyme has been shown to cleave eIF4G at two different sites yielding three different fragments (48,49). Interestingly, it also hydrolyses PABP at two different sites and proteolysis of both factors contributes to translation inhibition (50). Cleavage of eIF4G was shown to be complete upon addition of the HIV-2 protease in the RRL as visualized by western blotting (Supplementary Figure S2C). Remarkably, the consequence of this cleavage resulted in the complete loss of miR-451 effect. This was dependent on proteolytic activity since addition of specific inhibitors was sufficient to restore the miRNA response (Figure 3C, right panel). To test whether the specific loss of PABP and eIF4G cleavage was responsible for the effect on the miRNA-mediated inhibition, we have supplemented the HIV-2 protease treated lysate with the intact proteins (Figure 3D). Interestingly, both proteins were able to restore the miRNA effect to a partial, but significant, level. Addition of both in the same reaction did not significantly increase the rescue (data not shown). This nicely confirms the role for PABP in translation inhibition caused by miRNAs. However, the effect of eIF4G proteolysis seems to be, a priori, not in agreement with our data obtained with the L protease (see above) and showing no change upon eIF4G cleavage. But it is important to remember that those two virally encoded enzymes do not cleave the eIF4G molecule at the same sites. As a result, the consequences of the proteolytic action of these two virally encoded proteases are very different on translation initiation (49). We have previously shown that the HIV-2 protease removes a short region of 40 amino acids from the central domain of eIF4G that was shown to exhibit a strong RNA binding activity and this event results in the specific inhibition of ribosomal scanning by a, yet, unknown mechanism (48,49).
Taken together, these data suggest that both PABP and the carboxy-terminal domain of eIF4G are required for efficient miRNA repression and this effect seems to be linked to ribosomal scanning.
miRNA-mediated repression is modulated by the composition of the 5′-UTR
Ribosomal scanning is a difficult process to study, as it involves many factors and it is hard to distinguish from other translational steps, such as ribosomal binding or AUG recognition (51). Nevertheless, both 5′-UTR length and structure have been shown to modulate ribosomal scanning rate. We thus decided to investigate the influence of 5′-UTR composition on miRNA effect. For this we cloned different human 5′-UTR in our Luc and Luc-451X6 vectors (Figure 4A). In addition to the 5′-UTR of β-globin which was used as a positive control in the system, we have deliberately chosen genes harboring complex and structured 5′-UTRs such as BCL3 (75 nt., 81% GC rich), GAPDH (102 nt., 61% GC rich), Hsp70 (243 nt., 63% GC rich), Cyclin D2 (269 nt., 62% GC rich) and Line-1 (909 nt., 58% GC rich) (Figure 4A). As expected (Figure 4B, left panel), the efficiency of translation strongly differed between the different 5′-UTR tested, with β-globin and GAPDH driving translation at similar rates, while translation driven by BCL3, Hsp70, Cyclin D2 and Line-1 5′-UTRs was significantly less important. Interestingly, the nature of the 5′-UTR considerably influenced the level of miRNA-mediated inhibition (Figure 4B, right panel). However, we were surprised to observe that we could not draw any linear correlation between the complexity of the 5′-UTR and the level of miRNA repression (Line-1 mRNA being only slightly affected by miRNAs although its 5′-UTR corresponds to the longest tested). Nevertheless, it is noteworthy that no linear correlation could be drawn either between the complexity of the 5′-UTR structure and the overall translational efficiency (compare translational efficiency for GAPDH with the others).
A possible explanation for these rather puzzling results is to consider that the structure of the 5′-UTR does not only influence ribosomal scanning but also initial binding of the ribosome. Thus, we may have combined two effects, binding and scanning, that could explain this lack of correlation. Alternatively, it has recently been shown that a specific subset of mRNAs with complex RNA structures can recruit additional RNA helicases such as DHX29 and DDX3 to facilitate the progression of the 43S complex (52,53). Therefore, complex-structured 5′-UTR may be better translated because they have the ability to recruit additional RNA helicases. Taken together these data suggest that the structure of the 5′-UTR plays a role in the magnitude of the miRNA response but the mechanism by which this occurs needs to be investigated further.
Ribosomal scanning is required for miRNA-mediated inhibition
As stated earlier, the 5′-UTR corresponds to the region where ribosomes initially contact the mRNA (via a large number of eIFs) and is also the place where linear scanning of the 43S pre-initiation complex takes place. Therefore, to distinguish which one of these two mechanisms (binding or scanning) is targeted by miRNAs, we have used the well-characterized IRESes from the encephalomyelocarditis virus (EMCV) and poliovirus (PV) viruses. These two IRESes are known to recruit the 40S ribosomal subunit in a similar way with entry being some 25 nt downstream the conserved polypyrimidine rich tract at the vicinity of an AUG codon (54). However, once bound to the EMCV mRNA, the 40S ribosome recognizes the AUG codon and initiate translation exclusively from this point with limited ribosome scanning, whereas, in the case of PV, the pre-initiation complex scans some 150 nt before it finds and recognizes the authentic AUG codon (54). Thus, we have exploited these differences to test whether miR-451 repression interferes with ribosome scanning. These IRESes have been inserted in front of the luciferase reporter gene (see Figure 5A) and reporter constructs were transcribed to create uncapped and polyadenylated RNAs as they are naturally found during the viral replication cycle. As observed (Figure 5B), translation mediated by these viral IRESes exhibited a different sensitivity to miRNA repression with the PV construct being repressed whereas function of the EMCV IRES was only slightly affected. Although a lack of effect of miRNAs on the EMCV IRES has been reported previously (14,16,17,22,45), the ability of miRNAs to repress translation driven by the PV IRES had never been tested so far. This most probably reflects differences in ribosomal movement and further supports the finding that miRNAs repress translation by interfering with ribosomal scanning with no, or little, effect on ribosomal attachment.
As it could be argued that the differences between PV and EMCV IRESes could also be explained by their relative different translational efficiency in the RRL, we have used the L protease to enhance translation of the PV IRES as previously reported (55). This is presented in Supplementary Figure S3 and nicely shows that the magnitude of miRNA repression does not vary upon stimulation of the PV IRES by the L protease.
To further confirm these data, we have used a chimeric IRES made between EMCV and PV (Figure 5A); interestingly, it has been shown that switching the 3′IRES boundaries between PV and EMCV was sufficient to induce ribosomal scanning on EMCV (33). We therefore tested the impact of miRNA activity on this EMCV/PV chimera (Figure 5B). Interestingly, translation of the EMCV/PV chimeric IRES which contains the EMCV core and the PV ‘scanning segment’ became sensitive to miRNA binding (see EMCV/PV) further suggesting that ribosomal scanning is the step of translation that is regulated by miRNAs.
miRNAs have no effect on ‘non scanning’ mRNAs such as HCV and HCV-like IRESes
Data presented in this article suggest so far that translation inhibition induced by miR-451 requires ribosomal scanning. Thus, given these data, we made the assumption that a mRNA that could initiate translation independently from ribosomal scanning would not be affected by miR-451. Such an mRNA exists in the form of HCV and CSFV IRESes that are naturally uncapped and non-polyadenylated (56). Thus, HCV- and CSFV-luciferase were translated in their natural conformation with or without miR-451 target sites as shown in Figure 5A. As it was anticipated and previously shown for HCV (17,21,22), translation driven by both IRESes was not repressed by miR-451 (Figure 5C). We have recently shown that the presence of the poly(A) tail was absolutely required for miRNA-mediated translation repression (22). Thus, from the experiment carried out with HCV/CSFV, it could be argued that the lack of miRNA effect could be due to the physical absence of a poly(A) tail on these two mRNAs. To circumvent this problem, we next used the IRESes of two members of the Picornaviridae family that were shown to initiate translation in a way that is closely related to HCV and CSFV. These picornavirus are namely the AEV and SVV (35,36), for which the viral RNA is uncapped but it contains a poly(A) tail (Figure 5A); however, it does not functionally initiate translation in a poly(A)/PABP dependent manner but rather rely on an HCV- and CSFV-like mechanism for ribosome recruitment. Interestingly, translation of these constructs in our system was not inhibited at all by the binding of miR-451 and showed a very similar and comparable behavior to that obtained with the HCV and CSFV IRESes (Figure 5C). Thus, it confirms that miRNA repression cannot occur when translation initiation operates in the absence of ribosomal scanning even if the mRNA target harbors a poly(A) tail.
DISCUSSION
miRNA repression affects several stages of gene expression including translation (7,9,12–17,19,20), deadenylation and decay of target mRNAs (6,7,23,24). Although it has been shown that miRNA-mediated deadenylation is translation independent (6,57), recent data suggest that miRNA repression first occurs at the translational stage before it can undergo deadenylation (3,5,30,31). Differences in the mechanism by which miRNAs repress gene expression are likely to reflect the fact that several overlapping mechanisms are at use or that differences in experimental design may introduce bias. To support this latter hypothesis, important variations in the magnitude of miRNA repression have been reported with different methods of cell transfection such as cationic lipids versus electroporation or DNA versus RNA (58), but also in the composition of the intrinsic promoter (SV40 versus TK) (59) or even in the sequence of the miRNA itself (60). In order to minimize any experimental bias, we have chosen to study the effect of the most abundant endogenous miRNA (miR-451) contained in the RRL, which was shown to faithfully recapitulate translational repression without inducing deadenylation nor RNA degradation (22). This was further verified in this present article by using an internalized poly(A) tail and we could show that this did not change significantly the level of repression induced by miRNAs (Supplementary Figure S1). Using this system, our initial goal was to find which stage of translation was repressed. This is an important unresolved issue as several conflicting reports showed that it could occur at initiation or elongation steps (7,9,12–17,19,20) or even by inducing proteolytic cleavage of nascent peptides through a, yet, uncharacterized machinery (11).
Our experimental strategy consisted of using a spectrum of natural or chemical molecules that are specifically targeting different stages of translation. By using puromycin and cycloheximide, we showed that the relative level of miRNA repression was not affected suggesting a mode of action upstream from the elongation step (Figure 1). This result is consistent with type I miRNA-repressed mRNAs, defined by Kong and coworkers, for which miRNA repression takes place at translation initiation and are not affected by the addition of low amounts of cycloheximide (59). Furthermore, it also confirms that miRNA-targeted mRNAs are undergoing active translocation of elongating ribosomes as previously suggested (12,59). Addition of the proteasome inhibitor MG132 showed that miRNA did not induce neo-synthesized peptide degradation (Figure 1) and the use of both MDMP and GMP-PNP ruled out any involvement of miRNAs in the control of 60S ribosome subunit joining (Figure 2). This suggests that repression induced by miR-451 happens at an early stage of translation, pointing out to an effect on the initiation stage of protein synthesis. Translational inhibition by miRNAs has already been shown to require both eIF4F and PABP in cell extracts (6,16). We thus wanted to test whether this is also the case in RRL. To this end, we tested a combination of proteins that cleave either PABP or eIF4G or both (Figure 3). We found that PABP and eIF4G are both required but this is not due to their physical interaction. Rather we observed that an RNA binding motif of eIF4G implicated in scanning may be important. We thus tested whether the composition of the 5′-UTR, which is known to modulate ribosomal scanning, could also exert an effect on RISC activity (Figure 4). Although we did not find any linear correlation between 5′-UTR structure and the magnitude of miRNA repression, we observed strong variations among the different 5′-UTR tested. This simply indicates a strong relationship between the composition of the 5′-UTR and the level of repression by miRNAs. Since the 5′-UTR controls translation initiation in eukaryotes both by modulating ribosomal binding and scanning (61), we then wanted to investigate further which of these two steps was involved in the repression mediated by miRNAs.
For this, we used an experimental approach consisting of testing the EMCV and PV IRESes that are known to bind and recruit the ribosome in a similar manner but are very different in terms of ribosome scanning (62). Interestingly, we found that miR-451 massively repressed PV-driven translation with only marginal effects on EMCV (Figure 5), suggesting that interference did not occur at the ribosome binding step but rather during ribosome scanning. Finally, the use of pestivirus and pestivirus-like IRESes confirmed the lack of miRNA repression in the absence of ribosomal scanning despite the physical presence of the poly (A) tail (Figure 6).
Although our results show the implication of ribosomal scanning and the involvement of both PABP and eIF4G in this mechanism, we are still lacking the exact molecular mechanistic. In a recent report by Zekri et al. (28), Drosophila GW182 (a RISC associated factor) was shown to bind PABP and to impair its interaction with eIF4G therefore disrupting the closed-loop structure and inducing translation inhibition. While this work was in progress, Fukaya and Tomari (37) showed by using a Drosophila based in vitro system that the PABP was neither required for translational repression nor deadenylation. Our data show that, both eIF4G and PABP are required for miRNA-mediated inhibition (Figure 3) but this would not be due to their physical interaction. Consistent with this is the fact that PABP cleavage mediated by 3C or HIV-2 protease does not separate the eIF4G binding domain from poly(A) binding activity (46). Indeed, both eIF4G and poly(A) interact specifically with the N-terminal part of PABP, while proteases rather cleave the C-terminal domain (46). Interestingly, PABP cleavage by HRV 3C protease has been shown to eliminate its interaction with other factors implicated in translation, such as Paip1, Paip2, eRF3 and eIF4B (46,63,64). Strikingly, PAM2 domain of GW182 has also been shown to interact with the C-terminal domain of PABP (6,28,65). GW182 may thus interfere with PABP association with one of those important translation factors. The understanding of how miRNAs and GW182 can interfere with PABP function and ribosomal scanning will be a very interesting challenge for future work.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3.
FUNDING
ANRS; ANR; Ministère de la Recherche et de la Technologie (to E.P.R.); Fondation pour la Recherche Medicale (FRM) fellowships (to E.P.R. and T.L.); Région Rhône-Alpes fellowship (to T.L.); Conicyt-Chile fellowship (to R.S.R.); Agence Nationale de Recherche sur le SIDA et les Hepatites Virales fellowship (to R.S.R.); Becas Chile/Conicyt fellowship (to P.S.R.). Funding for open access charge: INSERM.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank S. Morley for PABP and eIF4G antibodies. We thank the NIH for the HIV-2 protease. We thank Dr Lisa O. Roberts for the kind gift of the AEV and SVV IRES constructs, Dr Martin Bushell for pET15b-PABP plasmid and Dr C. Fraser for the gift of eIF4G. We also thank Dr H. Ougham (Aberystwyth University, UK) for providing l- and d-MDMP. Finally, we thank Pr Melissa Moore for critical reading of the article.
REFERENCES
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bethune J, Artus-Revel CG, Filipowicz W. Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep. 2012;13:716–723. doi: 10.1038/embor.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol. Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zdanowicz A, Thermann R, Kowalska J, Jemielity J, Duncan K, Preiss T, Darzynkiewicz E, Hentze MW. Drosophila miR2 primarily targets the m7GpppN cap structure for translational repression. Mol. Cell. 2009;35:881–888. doi: 10.1016/j.molcel.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM, Ruvkun G. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc. Natl Acad. Sci. USA. 2004;101:360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maroney PA, Yu Y, Fisher J, Nilsen TW. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat. Struct. Mol. Biol. 2006;13:1102–1107. doi: 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- 10.Nelson PT, Hatzigeorgiou AG, Mourelatos Z. miRNP: mRNA association in polyribosomes in a human neuronal cell line. RNA. 2004;10:387–394. doi: 10.1261/rna.5181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nottrott S, Simard MJ, Richter JD. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat Struct. Mol. Biol. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- 12.Petersen CP, Bordeleau M-E, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol. Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 13.Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- 14.Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc. Natl Acad. Sci. USA. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwasaki S, Kawamata T, Tomari Y. Drosophila argonaute1 and argonaute2 employ distinct mechanisms for translational repression. Mol. Cell. 2009;34:58–67. doi: 10.1016/j.molcel.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 16.Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 17.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 18.Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- 19.Wang B, Love TM, Call ME, Doench JG, Novina CD. Recapitulation of short RNA-directed translational gene silencing in vitro. Mol. Cell. 2006;22:553–560. doi: 10.1016/j.molcel.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 20.Wang B, Yanez A, Novina CD. MicroRNA-repressed mRNAs contain 40S but not 60S components. Proc. Natl Acad. Sci. USA. 2008;105:5343–5348. doi: 10.1073/pnas.0801102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walters RW, Bradrick SS, Gromeier M. Poly(A)-binding protein modulates mRNA susceptibility to cap-dependent miRNA-mediated repression. RNA. 2010;16:239–250. doi: 10.1261/rna.1795410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ricci EP, Limousin T, Soto-Rifo R, Allison R, Poyry T, Decimo D, Jackson RJ, Ohlmann T. Activation of a microRNA response in trans reveals a new role for poly(A) in translational repression. Nucleic Acids Res. 2011;39:5215–5231. doi: 10.1093/nar/gkr086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl Acad. Sci. USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braun JE, Huntzinger E, Fauser M, Izaurralde E. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol. Cell. 2011;44:120–133. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Huntzinger E, Braun JE, Heimstadt S, Zekri L, Izaurralde E. Two PABPC1-binding sites in GW182 proteins promote miRNA-mediated gene silencing. EMBO J. 2010;29:4146–4160. doi: 10.1038/emboj.2010.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jinek M, Fabian MR, Coyle SM, Sonenberg N, Doudna JA. Structural insights into the human GW182-PABC interaction in microRNA-mediated deadenylation. Nat. Struct. Mol. Biol. 2010;17:238–240. doi: 10.1038/nsmb.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zekri L, Huntzinger E, Heimstadt S, Izaurralde E. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol. Cell. Biol. 2009;29:6220–6231. doi: 10.1128/MCB.01081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141–1151. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 30.Mishima Y, Fukao A, Kishimoto T, Sakamoto H, Fujiwara T, Inoue K. Translational inhibition by deadenylation-independent mechanisms is central to microRNA-mediated silencing in zebrafish. Proc. Natl Acad. Sci. USA. 2012;109:1104–1109. doi: 10.1073/pnas.1113350109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moretti F, Kaiser C, Zdanowicz-Specht A, Hentze MW. PABP and the poly(A) tail augment microRNA repression by facilitated miRISC binding. Nat. Struct. Mol. Biol. 2012;19:603–608. doi: 10.1038/nsmb.2309. [DOI] [PubMed] [Google Scholar]
- 32.Soto Rifo R, Ricci EP, Décimo D, Moncorgé O, Ohlmann T. Back to basics: the untreated rabbit reticulocyte lysate as a competitive system to recapitulate cap/poly(A) synergy and the selective advantage of IRES-driven translation. Nucleic Acids Res. 2007;35:e121. doi: 10.1093/nar/gkm682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohlmann T, Jackson RJ. The properties of chimeric picornavirus IRESes show that discrimination between internal translation initiation sites is influenced by the identity of the IRES and not just the context of the AUG codon. RNA. 1999;5:764–778. doi: 10.1017/s1355838299982158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fletcher SP, Jackson RJ. Pestivirus internal ribosome entry site (IRES) structure and function: elements in the 5′ untranslated region important for IRES function. J. Virol. 2002;76:5024–5033. doi: 10.1128/JVI.76.10.5024-5033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bakhshesh M, Groppelli E, Willcocks MM, Royall E, Belsham GJ, Roberts LO. The picornavirus avian encephalomyelitis virus possesses a hepatitis C virus-like internal ribosome entry site element. J. Virol. 2008;82:1993–2003. doi: 10.1128/JVI.01957-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willcocks MM, Locker N, Gomwalk Z, Royall E, Bakhshesh M, Belsham GJ, Idamakanti N, Burroughs KD, Reddy PS, Hallenbeck PL, et al. Structural features of the Seneca Valley virus internal ribosome entry site (IRES) element: a picornavirus with a pestivirus-like IRES. J. Virol. 2011;85:4452–4461. doi: 10.1128/JVI.01107-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fukaya T, Tomari Y. PABP is not essential for microRNA-mediated translational repression and deadenylation in vitro. EMBO J. 2011;30:4998–5009. doi: 10.1038/emboj.2011.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 2008;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- 39.Baxter R, Knell VC, Somerville HJ, Swain HM, Weeks DP. Effect of MDMP on protein synthesis in wheat and bacteria. Nat. New Biol. 1973;243:139–142. doi: 10.1038/newbio243139a0. [DOI] [PubMed] [Google Scholar]
- 40.Gay A, Thomas H, Roca M, James C, Taylor J, Rowland J, Ougham H. Nondestructive analysis of senescence in mesophyll cells by spectral resolution of protein synthesis-dependent pigment metabolism. New Phytol. 2008;179:663–674. doi: 10.1111/j.1469-8137.2008.02412.x. [DOI] [PubMed] [Google Scholar]
- 41.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chakravarty I, Bagchi MK, Roy R, Banerjee AC, Gupta NK. Protein synthesis in rabbit reticulocytes. Purification and properties of an Mr 80,000 polypeptide (Co-eIF-2A80) with Co-eIF-2A activity. J. Biol. Chem. 1985;260:6945–6949. [PubMed] [Google Scholar]
- 43.Parsyan A, Svitkin Y, Shahbazian D, Gkogkas C, Lasko P, Merrick WC, Sonenberg N. mRNA helicases: the tacticians of translational control. Nat. Rev. Mol. Cell Biol. 2011;12:235–245. doi: 10.1038/nrm3083. [DOI] [PubMed] [Google Scholar]
- 44.Pause A, Methot N, Svitkin Y, Merrick WC, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S, Liu Q, Tomari Y. ATP-dependent human RISC assembly pathways. Nat. Struct. Mol. Biol. 2009;17:17–23. doi: 10.1038/nsmb.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bushell M, Wood W, Carpenter G, Pain VM, Morley SJ, Clemens MJ. Disruption of the interaction of mammalian protein synthesis eukaryotic initiation factor 4B with the poly(A)-binding protein by caspase- and viral protease-mediated cleavages. J. Biol. Chem. 2001;276:23922–23928. doi: 10.1074/jbc.M100384200. [DOI] [PubMed] [Google Scholar]
- 47.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohlmann T, Prevot D, Decimo D, Roux F, Garin J, Morley SJ, Darlix JL. In vitro cleavage of eIF4GI but not eIF4GII by HIV-1 protease and its effects on translation in the rabbit reticulocyte lysate system. J. Mol. Biol. 2002;318:9–20. doi: 10.1016/S0022-2836(02)00070-0. [DOI] [PubMed] [Google Scholar]
- 49.Prevot D, Decimo D, Herbreteau CH, Roux F, Garin J, Darlix JL, Ohlmann T. Characterization of a novel RNA-binding region of eIF4GI critical for ribosomal scanning. EMBO J. 2003;22:1909–1921. doi: 10.1093/emboj/cdg175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Castello A, Franco D, Moral-Lopez P, Berlanga JJ, Alvarez E, Wimmer E, Carrasco L. HIV-1 protease inhibits Cap- and poly(A)-dependent translation upon eIF4GI and PABP cleavage. PLoS One. 2009;4:e7997. doi: 10.1371/journal.pone.0007997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hinnebusch AG. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 2011;75:434–467. doi: 10.1128/MMBR.00008-11. first page of table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pisareva VP, Pisarev AV, Komar AA, Hellen CU, Pestova TV. Translation initiation on mammalian mRNAs with structured 5′UTRs requires DExH-box protein DHX29. Cell. 2008;135:1237–1250. doi: 10.1016/j.cell.2008.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soto-Rifo R, Rubilar PS, Limousin T, de Breyne S, Decimo D, Ohlmann T. DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J. 2012;31:3745–3756. doi: 10.1038/emboj.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jackson RJ. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem. Soc. Trans. 2005;33:1231–1241. doi: 10.1042/BST0331231. [DOI] [PubMed] [Google Scholar]
- 55.Ziegler E, Borman AM, Kirchweger R, Skern T, Kean KM. Foot-and-mouth disease virus Lb proteinase can stimulate rhinovirus and enterovirus IRES-driven translation and cleave several proteins of cellular and viral origin. J. Virol. 1995;69:3465–3474. doi: 10.1128/jvi.69.6.3465-3474.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev. 2007;21:1857–1862. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′UTR as in the 3′ UTR. Proc. Natl Acad. Sci. USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kong YW, Cannell IG, de Moor CH, Hill K, Garside PG, Hamilton TL, Meijer HA, Dobbyn HC, Stoneley M, Spriggs KA, et al. The mechanism of micro-RNA-mediated translation repression is determined by the promoter of the target gene. Proc. Natl Acad. Sci. USA. 2008;105:8866–8871. doi: 10.1073/pnas.0800650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu E, Thivierge C, Flamand M, Mathonnet G, Vashisht AA, Wohlschlegel J, Fabian MR, Sonenberg N, Duchaine TF. Pervasive and cooperative deadenylation of 3′UTRs by embryonic microRNA families. Mol. Cell. 2010;40:558–570. doi: 10.1016/j.molcel.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jackson RJ, Wickens M. Translational controls impinging on the 5′-untranslated region and initiation factor proteins. Curr. Opin. Genet. Dev. 1997;7:233–241. doi: 10.1016/s0959-437x(97)80133-5. [DOI] [PubMed] [Google Scholar]
- 62.Jackson RJ, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 63.Craig AW, Haghighat A, Yu AT, Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 64.Khaleghpour K, Svitkin YV, Craig AW, DeMaria CT, Deo RC, Burley SK, Sonenberg N. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol. Cell. 2001;7:205–216. doi: 10.1016/s1097-2765(01)00168-x. [DOI] [PubMed] [Google Scholar]
- 65.Kozlov G, Safaee N, Rosenauer A, Gehring K. Structural basis of binding of P-body-associated proteins GW182 and ataxin-2 by the Mlle domain of poly(A)-binding protein. J. Biol. Chem. 2010;285:13599–13606. doi: 10.1074/jbc.M109.089540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.