Abstract
RNA sequencing has become an important method to perform hypothesis-free characterization of global gene expression. One of the limitations of RNA sequencing is that most sequence reads represent highly expressed transcripts, whereas low level transcripts are challenging to detect. To combine the benefits of traditional expression arrays with the advantages of RNA sequencing, we have used whole exome enrichment prior to sequencing of total RNA. We show that whole exome capture can be successfully applied to cDNA to study the transcriptional landscape in human tissues. By introducing the exome enrichment step, we are able to identify transcripts present at very low levels, which are below the level of detection in conventional RNA sequencing. Although the enrichment increases the ability to detect presence of transcripts, it also lowers the accuracy of quantification of expression levels. Our results yield a large number of novel exons and splice isoforms, suggesting that conventional RNA sequencing methods only detect a small fraction of the full transcript diversity. We propose that whole exome enrichment of RNA is a suitable strategy for genome-wide discovery of novel transcripts, alternative splice variants and fusion genes.
INTRODUCTION
The introduction of high-throughput genome wide approaches to study transcription has revealed a significant diversity and complexity of transcription. Tiling arrays were used to discover that a large fraction of the human genome is transcribed at low levels (1-4) . Arrays targeted at splice junctions have provided further insight into the diversity of transcripts and indicate that many isoforms are specific to certain cell types or developmental stages. Recently, the ability to use high-throughput technologies to sequence RNA (RNA-seq) has further expanded our understanding of the human transcriptome (1,4,5). The unprecedented levels of sensitivity and low background of deep RNA-seq compared with other methods (6,7), enable the identification and characterization of previously unannotated gene structures, exons and alternative splice isoforms (8–12). At the same time, the apparent differences between expression array results and RNA-seq data have initiated a discussion regarding the true nature and function of low level pervasive transcription (13,14). To a large extent, these differences may be explained by the fact that arrays are targeting specific subsets of the total pool of RNA, whereas RNA-seq targets all transcripts. Despite the fact that the introduction of RNA-seq has offered a deeper insight into the complexity of the transcriptome, the catalogue of all expressed transcripts is still far from complete. Several studies have highlighted that while large amounts of reads map to intergenic and intronic regions (15,16), potentially indicative of new exons or functional RNA, a large fraction of sequence reads are also used up by highly expressed transcripts, thereby lowering the ability to detect other transcripts present at low levels (15,16).
One way to increase the ability to detect rare transcripts is to use target enrichment. This approach has previously been used on RNA for specific subsets of genes (17) and was recently combined with deep sequencing to reveal a large number of novel transcripts for select regions of the genome (18). In this study, we aimed to utilize the increased sensitivity of targeted enrichment in combination with genome wide assessment of transcription by using whole exome capture of RNA followed by massively parallel sequencing, hypothesizing that we may improve detection of low-level transcripts genome wide. We compare our results with conventional RNA sequencing of the same samples. Our data support that exome RNA capture sequencing (ExomeRNAseq) improves detection of splice junctions and rare transcripts, but is less quantitative, as compared with total RNA sequencing (TotalRNAseq). Our data support that ExomeRNAseq is an advantageous strategy for RNA based genome-wide transcript discovery and may prove to be an efficient strategy for RNA-based clinical diagnostics.
MATERIALS AND METHODS
Preparation of cDNA
Total RNA for all samples was purchased from Biochain. Starting with 1 µg of total RNA, cDNA was synthesized with the SMARTer Pico PCR cDNA Synthesis kit (Clontech) according to manufacturer’s recommendations. The resulting first strand cDNA from each RNA sample was then amplified with the Advantage2 PCR kit (Clontech) using 18 polymerase chain reaction (PCR) cycles. 3.5 µg of double-stranded cDNA was used for library preparation.
Capture and sequencing
The cDNA was sheared using a Covaris instrument (Covaris, Inc.). Fragment libraries were created from the sheared samples using AB Library Builder System and captured using the Agilent SureSelect 50 Mb exome enrichment kit, according to the manufacturer’s protocols. Exome capture was conducted by hybridizing the cDNA libraries with biotinylated RNA baits for 24 hr followed by extraction using streptavidin-coated magnetic beads. Captured cDNA was then amplified followed by emulsion PCR using EZ Bead System and sequenced using SOLiD4. Each sample was sequenced on 1/3 of a slide, producing between 98 261 580 and 148 056 068 fifty base-pair reads per library. Total RNA sequencing for the same samples was performed as part of a previous study (16).
Sequencing and mapping
Mapping of the reads from all five cDNA libraries and the five total RNA libraries were conducted using version 1.3.1 of Bioscope (Applied Biosystems) and version 1.3.3 of TopHat (19), using default settings for color space reads. All reads were aligned to the hg19 assembly version (GRCh37) of the human genome and the prebuilt color-space index of the hg19 genome assembly (TopHat) was acquired from the TopHat homepage (http://tophat.cbcb.umd.edu/). The poly(A) dataset (SRA accession SRX056683) was downloaded from the Short Read Archive (SRA) and mapped using TopHat.
Gene expression
Gene expression was estimated based on reads per kilobase per million mapped reads (RPKM) (20) over RefSeq transcripts (downloaded from the UCSC Genome Browser). The RPKM was calculated for the exonic regions of each transcript. To establish a cut-off for expressed transcripts, RefSeq annotations were compared with background RPKM values as follows. To create the distribution of expression representing background, regions of a length between 120 and 240 bp were randomly distributed in regions of the genome not covered by any RefSeq transcripts, UCSC known genes, ENSEMBL gene predictions or UCSC spliced expressed sequence tags (ESTs). Centromeric and telomeric regions were also excluded. For each sample and each transcript in RefSeq a random transcript was created using as many random regions as there were exons in the transcript. The RPKM was then calculated for each of these random transcripts. The random transcript distribution constructed for each sample was then compared with the RefSeq transcript distribution of that sample. The cutoff was then set so that 98% of the random transcripts (representing background) were removed. To correct for difference in read count between the TotalRNAseq and ExomeRNAseq samples, aligned reads were randomly drawn from the ExomeRNAseq sample to a create dataset with equal number of aligned reads as in the corresponding TotalRNAseq dataset.
Identification of differentially expressed transcripts
Cufflinks Cuffcompare version 1.1.0 (http://cufflinks.cbcb.umd.edu) was used to calculate the difference in expression between all samples in a pair wise comparison. Corresponding ExomeRNAseq and TotalRNAseq sample pairs were then compared as follows. Transcripts determined not to be expressed in either sample were excluded from all four samples. The remaining transcripts were then tested for differential expression using Cuffcompare and transcripts determined to be differentially expressed were counted for each pair. Presented results correspond to comparison between frontal cortex and liver.
Splice junction analysis
TopHat was used to calculate the number of splice junctions in each tissue (TotalRNAseq and ExomeRNAseq), as well as for the poly(A) dataset. Novel junctions were defined by comparison with the UCSC ‘known genes’ (downloaded from UCSC Genome Browser) and each detected junction not annotated among the known genes were considered novel. To estimate the false positive rate of novel junctions, they were compared with the spliced ESTs track (downloaded from the UCSC Genome Browser). The junctions corresponding to an annotation in the spliced ESTs track were counted as a validation of a novel junction. Further categorization of the splice junctions was made using in-house Perl scripts, defining each junction-end not overlapping an exon as a marker for a novel exon. A subset of novel junctions was then chosen for validation on the basis that they were located in sequences amenable to standard PCR primer design.
PCR and sequencing
Novel exons and splice isoforms were validated using PCR. The PCR was performed with initial denaturation at 95°C for 10 min followed by 35 cycles of (95°C for 15 s, 60°C for 30 s and 72°C for 30 s). The reaction contained 5 ng human fetal frontal cortex single stranded cDNA, 0.4 μM for each primer (Supplementary Table S1) and 12.5 μl of Maxima Hot Start Taq DNA Polymerase (Fermentas) in a total volume of 25 μl. The PCR products were subsequently analysed on 2% agarose gel. The products were confirmed by Sanger sequenced using standard protocols. Single-stranded cDNA, used in the PCR and quantitative real-time PCR (qRTPCR) reactions, was synthesized from 1 μg of total RNA using SMARTer Pico PCR cDNA Synthesis Kit (Clontech) according to the manufacturer recommendations.
Quantitative real-time PCR
qRTPCR was used to measure the relative expression of the newly identified alternatively spliced exons in human fetal frontal cortex cDNA. The qRTPCR was performed with Stratagene Mx3000P in 96-well plates. The reaction was carried out with initial denaturation at 95°C for 10 min followed by 40 cycles of denaturation at 95°C for 15 s, primer annealing at 58°C for 30 s and extension at 72°C for 30 s followed by dissociation curves step. The qRTPCR contained 12.5 ng single stranded cDNA, 0.4 μM for each primer (Supplementary Table S1) and 12.5 µl Maxima SYBR Green/ROX qPCR Master Mix (Fermentas) in 25 μl reactions. All samples were amplified in triplicate and the mean values were used to calculate the expression level of each target. The relative expression levels in the samples were determined using the corresponding standard curve for each primer pair. Expression levels were normalized to the level of β-actin. Raw data were analysed using MxPro software (Stratagene).
RESULTS
Sequencing and expression analysis
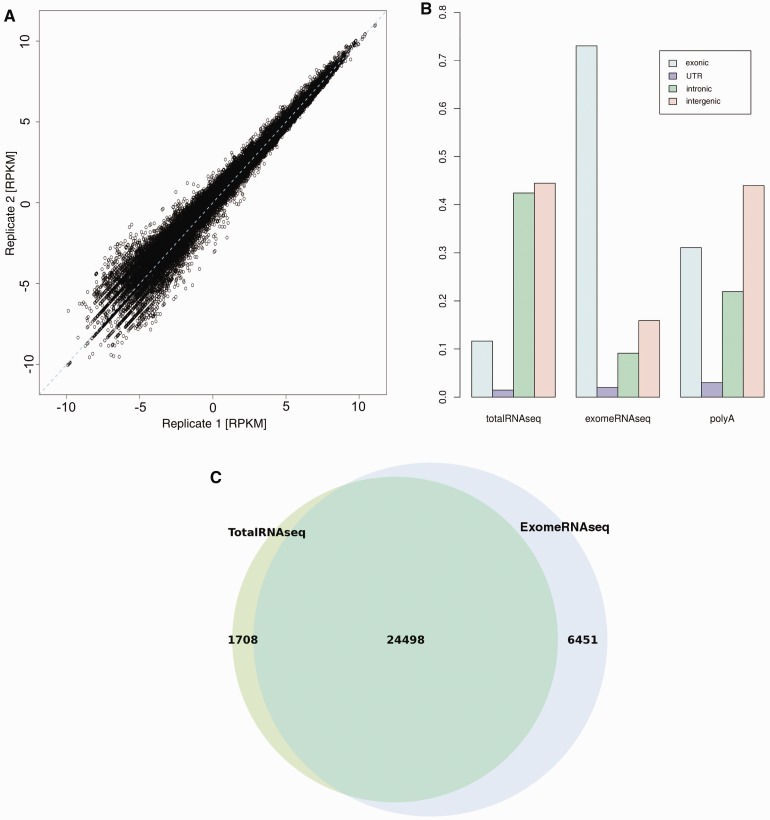
Target enrichment of exome sequences was performed in solution with the Agilent SureSelect 50 Mb kit to capture cDNA targets. RNA from four different human tissues was used for the experiments (adult liver, adult cortex, fetal liver and fetal cortex), with fetal cortex prepared and run in duplicate (independent capture, library and sequencing) to evaluate reproducibility. Captured targets were sequenced using SOLiD4, generating on average 73 million mapped reads per tissue. For evaluating the results, we used the RefSeq transcripts and calculated the RPKM (20) for each transcript in each tissue. We first tested the reproducibility of the method by comparing the RPKM values for corresponding transcripts in the replicate experiment (Figure 1A). The results are highly correlated (Pearson correlation = 0.99), indicating that the experimental procedure is consistent and reproducible. When comparing RPKM values for corresponding transcripts between ExomeRNAseq and TotalRNAseq from the same RNA sample only a weak correlation could be seen (Supplementary Figure S1A). We further investigated these differences by exploring the sequence read distributions. In addition to ExomeRNAseq and TotalRNAseq, a Poly(A) selected RNA-seq dataset produced using the same sequencing method was downloaded from the Short Read Archive and included in the comparison. We plotted the fraction of reads mapping to exonic, UTR, intronic and intergenic regions to explore the distribution of coverage of the genome. The result shows that a significantly larger fraction of the reads fall into exons in the ExomeRNAseq samples, as compared with the two other RNA-seq approaches (Figure 1B).
Figure 1.
Sequencing and expression analysis. (A) Plot showing the correlation of RPKM values for each transcript in the two independent replicate experiments (adult cortex) from the same starting material. The results show a very high correlation (R = 0.99), indicating that the protocol is stable and results are reproducible. (B) A bar graph showing the fraction of the reads mapping to intronic, exonic, intergenic and UTR regions for each RNA-seq dataset. The results indicate that ExomeRNAseq have the highest fraction of reads mapped to exonic regions. (C) A Venn diagram showing the number of genes expressed in adult cortex tissue using ExomeRNAseq (blue) and TotalRNAseq (green). The overlap of the circles represents genes shown to be expressed using both sequencing approaches. (D) Graphs showing the expression measured by TotalRNAseq, ExomeRNAseq and qRTPCR. For clarity, the values of each technology were normalized using the highest values measured by that approach.
To determine a cut-off to define expressed genes, we created a random transcript distribution by creating exon-sized regions corresponding to each RefSeq transcript, which were then distributed in non-coding parts of the genome (representing background). We then defined all genes that had an RPKM higher than the 98th percentile of the random transcript distribution as expressed (Supplementary Figure S1B). The results show that ExomeRNAseq yields a substantially larger number of expressed transcripts than TotalRNAseq (30 949 vs. 26 206 transcripts, see Figure 1C), indicating that many transcripts expressed at a low level can be readily detected using the capture strategy. The results are similar in all tissues and remain highly significant after correcting for different read counts in the datasets (Supplementary Figure S2). We also compared the distribution of the 20% lowest expressed transcripts in TotalRNAseq with the expression of the same transcripts in ExomeRNAseq, and vice versa, and conclude that transcripts with low expression levels in TotalRNAseq generally show higher expression in the ExomeRNAseq data (Supplementary Figure S1C). A smaller number of transcripts are detected in total RNA but not in the capture data (n = 1708), and these represent transcripts that lack capture probes (14%), have a low number of probes, or regions where no target was captured despite the presence of probes (something that also evident for some probes in standard DNA-based exome sequencing). To further investigate the quantitative properties of ExomeRNAseq, we used Cufflinks (21) to identify RefSeq transcripts that were differentially expressed between tissues. We find that the fraction of differentially expressed transcripts is similar in ExomeRNAseq and TotalRNAseq, but with a larger absolute number of differentially expressed genes identified in the capture data (Supplementary Figure S3).
To further test the quantitative ability of ExomeRNAseq, we performed qRTPCR on fetal frontal cortex single stranded cDNA to measure the relative expression of exonic regions selected from twelve different genes. Five of the selected regions exhibited differential expression between ExomeRNAseq and TotalRNAseq and seven were chosen in an unbiased manner. The qRTPCR showed a better correlation with expression values from TotalRNAseq, and we conclude that this may be partially explained by differential capture efficiency between probes in the exome capture (Figure 1D).
Splice junction discovery
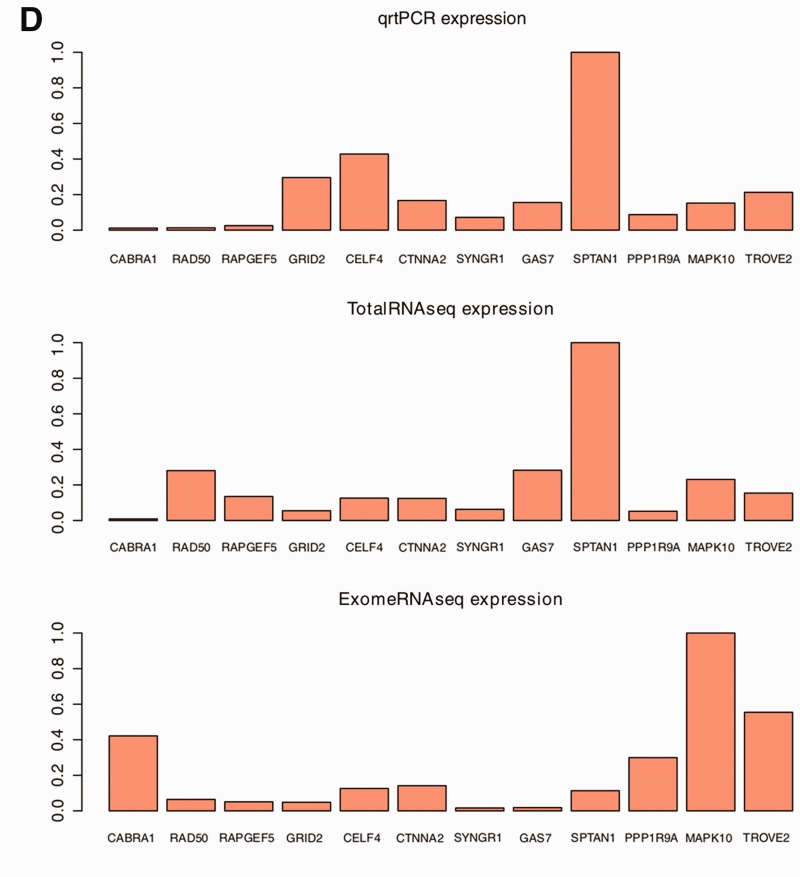
Discovery and detection of novel transcript isoforms and alternative splicing events are vital for expanding our knowledge of the transcriptome. As our result shows that a higher fraction of the ExomeRNAseq reads map to exons, we next sought to identify splice junctions and novel exons in the capture data. We used TopHat (19) and SplitSeek (22) on our sequencing results to identify splice junctions in each sample and found similar results with both algorithms (data not shown). On average, 100 000 junctions were identified in the ExomeRNAseq samples, compared with an average of 25 000 junctions in the TotalRNAseq samples. To account for all relevant technologies, we investigated a dataset based on poly(A)-RNAseq (SOLiD4 sequencing of neural progenitor cells). The poly(A) dataset yielded more junctions (∼44 000) than TotalRNAseq, but substantially fewer than our ExomeRNAseq approach (Figure 2A). As the number of mapped reads was slightly different between samples, the fraction of reads spanning splice junctions was calculated to compare samples (Figure 2B). In accordance with the significantly higher fraction of reads mapping to exonic regions (Figure 1B) and better ability to detect low level transcripts, ExomeRNAseq results show a significantly larger fraction of spliced reads than any other investigated method.
Figure 2.
Splice junction discovery (A) Total number of splice junctions identified in each sample in ExomeRNAseq and TotalRNAseq. For comparison, the number of splice junctions identified in a publicly available poly(A)-RNAseq performed with the same sequencing platform is included. (B) Bar graph showing the fraction of splice junction reads. The two bars plotted for each tissue shows the fraction (in orange) of spliced reads detected in ExomeRNAseq and TotalRNAseq, respectively. (C) The number and distribution of novel splice junctions in the data. The distributions plotted show different types of novel splice junctions including those bridging known exons in same transcripts (Novel Junctions), those connecting a known and a novel exon (NovelExons), those connecting two non-exon regions (Novel/Novel), and junctions linking independent transcripts (Transcript/Transcript).
To investigate the nature and novelty of the splice junctions identified, we downloaded annotations for known genes from UCSC (23) (containing more annotated splice junctions than RefSeq) and compared these with the junctions identified in each of the samples. The junctions were divided into two different categories, known and novel, where those assigned as ‘known’ match a splice junction reported in the known genes track. As expected, the ExomeRNAseq had a greater abundance of novel splice junctions compared with the other RNA-seq approaches. To assess the impact of sequencing depth on splice junction discovery in TotalRNAseq, we resequenced the fetal frontal cortex sample, producing four times more reads than the previous TotalRNAseq datasets. However, the deeper sequencing of TotalRNAseq led to a marginal increase in splice junction detection (totaling 25 575 junctions), indicating that an extreme depth of coverage would be required to have similar power to detect splice junctions as we show with ExomeRNAseq.
Novel splice junction detection
In total, we identify an average of 22 432 and 3036 novel splice junctions in the ExomeRNAseq and TotalRNAseq data, respectively. Our definition of “novel” is that the splice junctions are not part of current mRNA annotations in the “known genes” track at UCSC. All novel splice junctions were further categorized into those bridging known exons in the same transcripts, connecting a known and a novel exon, connecting two non-exon regions, or linking independent transcripts (Figure 2C). The novel isoforms identified by the different RNA-seq approaches show a similar distribution between these categories.
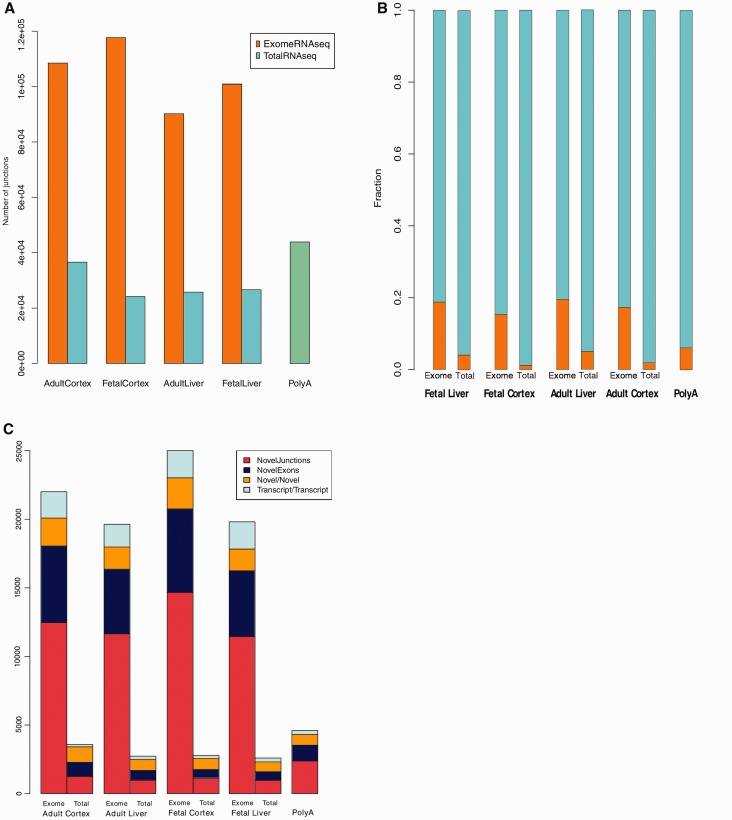
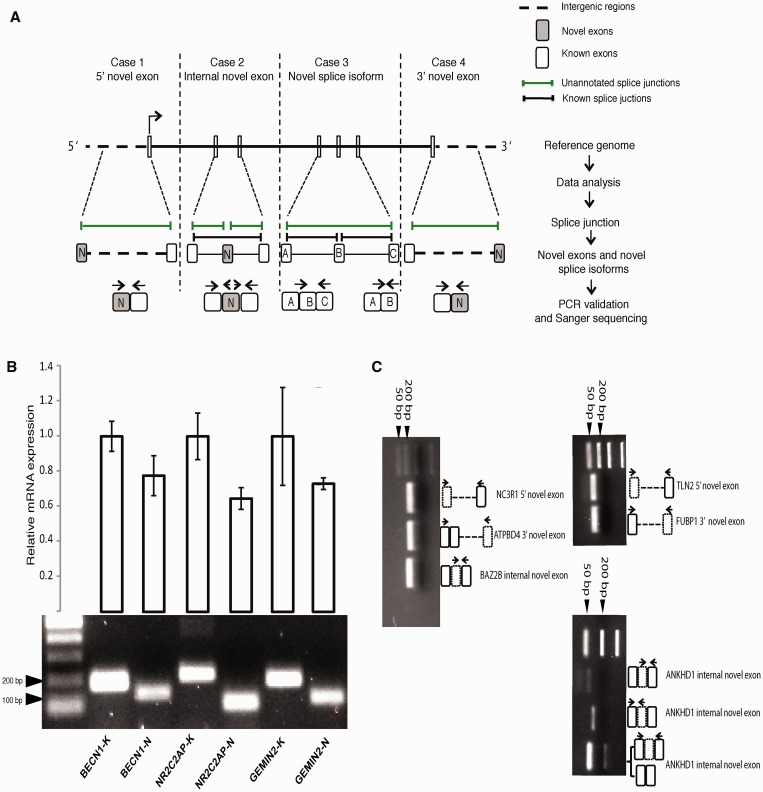
The large number of novel splice junctions identified could be due either to a large number of false positive junctions or that there are indeed a considerable number of hitherto unannotated isoforms in the human transcriptome. To further assess the false positive rate, we determined the number of novel splice isoforms that match junctions reported in the EST data (downloaded from UCSC). Even though the ExomeRNAseq had a substantially larger number of novel splice junctions compared with TotalRNAseq, we find the fraction of junctions overlapping spliced ESTs to be similar, indicating that there is no increase in false positive splice junctions in the capture data (Supplementary Figure S4). To experimentally validate novel splice junctions, we grouped our findings into four categories: novel alternative splice isoforms, novel 5′ exons, novel exons located within genes and novel 3′ exons (Figure 3A). Three splice junctions and six novel exons were selected from different genes for validation using PCR. All PCRs were analysed by gel electrophoresis, and further confirmed by Sanger sequencing. The three splice variants were also investigated using qRTPCR and our data show that the expression levels of the novel isoforms were all lower as compared with established alternative transcripts for the same gene (Figure 3B and C). These data further support the increased power of ExomeRNAseq to detect and identify transcripts present at low levels that may escape detection by conventional RNA-seq approaches.
Figure 3.
Identification and validation of novel exons and splice junctions. (A) A schematic illustration for the workflow used for identification and validation of novel exons and splice isoforms. Examples from each category including novel 5’ exon (case 1), novel internal exon (case 2), novel splice isoform (case 3) and novel 3′ exon (case 4) were chosen for experimental validation. Novel exons and alternative transcripts were identified using TopHat, and validated using PCR and Sanger sequencing. The horizontal arrows indicate the primers used for experimental validation. (B) Novel alternative splice isoforms were validated for five different genes. The lower panel shows the expected PCR products of the transcripts from frontal cortex cDNA. The top panel shows (qRTPCR) results representing relative RNA levels of the known (K) and novel (N) splice isoforms. The results indicate that the novel splice isoforms are expressed at lower level than the previously known isoforms for each transcript, highlighting the strength of ExomeRNAseq to detect rare transcripts. The qRTPCR values are based on three independent experiments (error bars show ± sd). qRTPCR values were normalized to the level of β-actin. (C) PCR was used to validate cases of novel exons. In total, six exons were amplified (two located upstream, two downstream and two within genes) from fetal frontal cortex cDNA. Amplicons corresponding to the expected size of the novel exons were detected on 2% agarose gel. The schematic illustrations on the right side of the gel pictures show the name of the validated genes (italics), location of the novel exons in the transcripts (dashed rectangles) and the location of the primers used in the PCR (arrows). Dashed lines represent intergenic regions surrounding the transcripts.
Tissue-specific splice isoforms
Many alternative splice isoforms are specific to certain tissues or developmental stages. To further investigate tissue-specific expression and splicing, the number of shared splice junctions in pair-wise comparison of the tissues was calculated. The results show that ∼65–70% of the splice junctions identified are shared between at least two tissues. As might be expected, the fetal and adult frontal cortex show the highest number of shared splice junctions (Supplementary Figure S5). Surprisingly, the same pattern was not found for the two liver samples sequenced. The result showed instead that adult liver had the fewest number of shared junctions with the other sequenced tissues, reflecting that this was the tissue where the smallest number of junctions was found. When comparing the novel splice junctions in the same way, the fetal and adult cortex still shared the highest number of junctions.
SNP discovery
To evaluate the possibility of using ExomeRNAseq as a method for identification of coding variation, we compared single nucleotide variant (SNV) calling in the data from ExomeRNAseq and TotalRNAseq. The number of coding SNVs found in the ExomeRNAseq data (average 9106 variants per sample) greatly exceeded the number identified in TotalRNAseq data (average 1919 SNVs per sample) (Supplementary Figure S6A). To further investigate the accuracy of the SNV calls, the results were compared with known variants reported in NCBI’s database dbSNP (Supplementary Figure S6B). The results show that while approximately four times as many variants were identified in the ExomeRNAseq, the overlap with dbSNP is higher in the TotalRNAseq (average 91% vs. 83% in the capture data). There are several potential explanations for a higher accuracy in SNV calls from the TotalRNAseq data. One reason may be that the distribution of read coverage over exons may be different after target enrichment, with lower coverage at the edges of exons as compared with TotalRNAseq. To explore the possibility of a position bias in the ExomeRNAseq data, the relative exonic position of each SNV was plotted (See Supplementary Figure S6C). The result indicates that there is no strong tendency for non-dbSNP variants to accumulate near edges of exons.
DISCUSSION
We show for the first time that whole exome capture can be applied to cDNA for efficient detection of transcripts and alternative splice variants. Our data support and extends on results of more targeted studies (18), showing a significantly increased power to detect transcripts present at very low level. There are several potential research and diagnostic applications for whole exome capture. One advantage of the approach is the ability to discover new exons. We find evidence for multiple previously unannotated exons, with the largest number identified in fetal frontal cortex. The majority of these map within existing gene annotations (n = 4515) making them by far the most over-represented group, as compared with novel 5′ or 3′ exons (n = 495 and 477, respectively). Previous studies have shown that a large fraction of sequence reads from poly(A)-RNAseq map to intronic regions (15,16). The fact that so many of the novel exons we find map to introns may explain a significant portion of those intronic reads. A consequence of novel exons that extend existing gene models is that a larger fraction of the genome will be covered by genes than previously thought. Again, this may explain a significant portion of sequence reads mapping to intergenic regions.
We find a large number of previously unannotated alternative splice variants, based on splice junctions between known exons (on average 13 032 per tissue). We also note that many genes have multiple alternative 3′- UTRs, although many of these occur at different places within existing 3′-UTR annotations. This seems to be a common feature of many genes that is currently not well annotated. Our validation data show that the majority of new alternative splice isoforms that we detect are expressed at low levels, explaining why they have not been reported previously. Overall, recent data from us and others (15,16) indicate that there are many low level transcripts that are not captured by current array or sequence-based methods.
There are some limitations to the ExomeRNAseq approach. Not all transcripts are represented in the existing commercial exome enrichment kits, and not all probes are efficient at capturing their targets. In addition, a drawback of RNA capture is that the ability to accurately quantify gene expression is significantly lowered compared with conventional RNA-seq strategies. This is mainly due to the differential efficiency of capture probes to hybridize to their targets. We attempted to correct for this by using results from DNA exome capture based on the same probes, where we know that the number of targets is two for most probes. Normalizing against the DNA coverage of the same probes seems to work well for some genes, but has only minor effect on the global correlation between TotalRNAseq and ExomeRNAseq (data not shown). One problem with this approach is that it normalizes also for other factors that affect coverage, such as mappability, and because the normalization is applied only to ExomeRNAseq and not TotalRNAseq, it may in some cases skew the results further. It is possible that additional correction that includes mappability scores could help improve the situation further, but the main conclusion from our results is that quantification accuracy is inevitably lowered after target enrichment.
There are several applications for which ExomeRNAseq may be beneficial compared with current approaches. The ability to detect junctions present at low levels opens up for the possibility to use ExomeRNAseq for genome wide discovery of translocations causing novel fusion transcripts in solid tumors. Translocations are challenging to detect in DNA tumor samples, and detection is further complicated by the fact that samples often represent a mix of tumor and normal cells so that only a fraction of the cells carry the rearrangement. We propose that RNA capture may be a way to approach these challenges.
Based on the fact that we identify a large number of new exons, we propose that ExomeRNAseq may be an excellent approach for cross-species comparisons. It was recently shown that exome capture on DNA can efficiently be used to map variation across primates (24,25), and it should work equally well for RNA based capture. Since we show that we can find a large number of coding variants in the data, exome enrichment at the level of RNA can be used both for annotation of gene models and identification of variation.
Our data support previous findings that our understanding of transcription and post-transcriptional regulation is limited, and that current approaches are only finding a fraction of the transcript diversity. Our results suggest that very deep sequencing of captured or enriched portions of the transcriptome may be the best way towards uncovering the complete spectrum of transcript diversity in any given cell type or tissue.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–6 and Supplementary Table 1.
FUNDING
The Swedish Foundation for Strategic Research; the Swedish Medical Research Council; the Kjell and Märta Beijer Foundation (to L.F.). Funding for open access charge: Swedish Foundation for Strategic Research.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the Uppsala Genome Center for technical support with the library preparations and sequencing experiments. The alignment and variant calling were performed on resources provided by SNIC through the Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX). They are grateful for access to the poly(A) RNA-seq data produced as part of the International Human Epigenome Consortium.
REFERENCES
- 1.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 3.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 4.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okoniewski MJ, Miller CJ. Hybridization interactions between probesets in short oligo microarrays lead to spurious correlations. BMC Bioinformatics. 2006;7:276. doi: 10.1186/1471-2105-7-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilhelm BT, Landry JR. RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods. 2009;48:249–257. doi: 10.1016/j.ymeth.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 8.Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 9.Bainbridge MN, Warren RL, Hirst M, Romanuik T, Zeng T, Go A, Delaney A, Griffith M, Hickenbotham M, Magrini V, et al. Analysis of the prostate cancer cell line LNCaP transcriptome using a sequencing-by-synthesis approach. BMC Genomics. 2006;7:246. doi: 10.1186/1471-2164-7-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torres TT, Metta M, Ottenwalder B, Schlotterer C. Gene expression profiling by massively parallel sequencing. Genome Res. 2008;18:172–177. doi: 10.1101/gr.6984908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blekhman R, Marioni JC, Zumbo P, Stephens M, Gilad Y. Sex-specific and lineage-specific alternative splicing in primates. Genome Res. 2010;20:180–189. doi: 10.1101/gr.099226.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson R. Dark matter transcripts: sound and fury, signifying nothing? PLoS Biol. 2010;8:e1000370. doi: 10.1371/journal.pbio.1000370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson M. The evolution of gene regulation, the RNA universe, and the vexed questions of artefact and noise. BMC Biol. 2010;8:97. doi: 10.1186/1741-7007-8-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most “dark matter” transcripts are associated with known genes. PLoS Biol. 2010;8:e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ameur A, Zaghlool A, Halvardson J, Wetterbom A, Gyllensten U, Cavelier L, Feuk L. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat. Struct. Mol. Biol. 2011;18:1435–1440. doi: 10.1038/nsmb.2143. [DOI] [PubMed] [Google Scholar]
- 17.Levin JZ, Berger MF, Adiconis X, Rogov P, Melnikov A, Fennell T, Nusbaum C, Garraway LA, Gnirke A. Targeted next-generation sequencing of a cancer transcriptome enhances detection of sequence variants and novel fusion transcripts. Genome Biol. 2009;10:R115. doi: 10.1186/gb-2009-10-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, Mattick JS, Rinn JL. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat. Biotechnol. 2011;30:99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 21.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010; 28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ameur A, Wetterbom A, Feuk L, Gyllensten U. Global and unbiased detection of splice junctions from RNA-seq data. Genome Biol. 2010;11:R34. doi: 10.1186/gb-2010-11-3-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC known genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 24.George RD, McVicker G, Diederich R, Ng SB, MacKenzie AP, Swanson WJ, Shendure J, Thomas JH. Trans genomic capture and sequencing of primate exomes reveals new targets of positive selection. Genome Res. 2011;21:1686–1694. doi: 10.1101/gr.121327.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vallender EJ. Expanding whole exome resequencing into non-human primates. Genome Biol. 2011;12:R87. doi: 10.1186/gb-2011-12-9-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.