Abstract
Influenzavirus non-structural protein NS1 is involved in several steps of the virus replication cycle. It counteracts the interferon response, and also exhibits other activities towards viral and cellular RNAs. NS1 is known to bind non-specifically to double-stranded RNA (dsRNA) as well as to viral and cellular RNAs. We set out to search whether NS1 could preferentially bind sequence-specific RNA patterns, and performed an in vitro selection (SELEX) to isolate NS1-specific aptamers from a pool of 80-nucleotide(nt)-long RNAs. Among the 63 aptamers characterized, two families were found to harbour a sequence that is strictly conserved at the 5′ terminus of all positive-strand RNAs of influenzaviruses A. We found a second virus-specific motif, a 9 nucleotide sequence located 15 nucleotides downstream from NS1’s stop codon. In addition, a majority of aptamers had one or two symmetrically positioned copies of the 5′-GUAAC / 3′-CUUAG double-stranded motif, which closely resembles the canonical 5′-splice site. Through an in-depth analysis of the interaction combining fluorimetry and gel-shift assays, we showed that NS1’s RNA-binding domain (RBD) specifically recognizes sequence patterns in a structure-dependent manner, resulting in an intimate interaction with high affinity (low nanomolar to subnanomolar KD values) that leads to oligomerization of the RBD on its RNA ligands.
INTRODUCTION
Influenza A viruses are enveloped viruses with a segmented RNA genome consisting of eight single-stranded RNAs of negative polarity. Transcription and replication in the nucleus of infected cells generate 8 complementary RNAs and 10 or more messenger RNAs, which collectively encode 13 proteins, including the surface glycoproteins haemagglutinin (HA) and neuraminidase (NA) (1–3). Non-structural protein 1 (NS1) is encoded by the smallest RNA fragment that also encodes nuclear export protein (NEP)/NS2, and harbours determinants of pathogenicity. There are two alleles of NS1, and several variant proteotypes within each allele (4,5). Allele A is by far the most frequent, whereas allele B is found only in some viruses of avian origin. NS1, which is absent from the virion, is a ∼230-residue protein that exhibits several pro-viral activities (6). (i) Through binding to the 30 kDa subunit of Cleavage-and-Polyadenylation Specificity Factor (CPSF30), to the poly-A-binding protein and to several components of the mRNA export machinery (7–9), it prevents the maturation and nucleocytoplasmic export of cellular mRNAs, thus participating in the host shut-off. (ii) Reciprocally, it regulates the splicing of the two viral mRNAs encoding M1 and NS1 (10), and also enhances translation of the viral mRNAs (11). (iii) Through direct and indirect interactions with several proteins involved in the interferon response, it prevents the triggering of the interferon system and also blocks many of the interferon-mediated antiviral activities (12,13). Through its N-terminal RNA-binding domain (RBD, which is a dimer of amino-acids 1–73), NS1 interacts with many types of viral and cellular RNAs, while its C-terminal effector domain (ED) is involved in most of its interactions with proteins. Whereas NS1 is necessary for normal growth of the virus, natural isolates harbouring premature stop codons in NS1 open reading frame have been found, and viruses with even shorter NS1, or no NS1 at all, have been constructed by reverse genetics (14–16). However, these viruses are severely impaired in their replication, and they grow only in interferon-deficient systems (14–17). Single amino-acid substitutions that abolish the functionality of the RBD have the same effects as truncations or deletions of NS1 (18–20), thus emphasizing the critical role of RNA binding in the multiple activities of NS1. Other less deleterious substitutions that have a major effect on virulence have also been described in natural isolates (19,21).
NS1 has been shown to bind various types of RNAs: viral genomic RNAs (22,23), viral mRNAs (11), U6 and U6atac small nuclear RNAs (24–26), double-stranded RNAs (27–29). The diversity of RNA ligands that have been described, together with the different modes of interactions that have been proposed (30–32), prompted us to re-examine the molecular patterns that favour the interaction of an RNA with NS1. To this end, we used a SELEX method (Systematic Evolution of Ligands by Exponential Enrichment) (33) to identify RNA motifs that bind recombinant protein NS1. We isolated and sequenced 63 clones, and further characterized one of the most frequently isolated candidates. Importantly, two motifs were shared by two or more families of aptamers: AGCAAAAG, which is invariably present in all the messenger and complementary RNAs of all influenza A viruses, as well as two symmetrically positioned double-stranded RNA (dsRNA) motifs containing the canonical 5′-splice site of introns. Another virus-specific pattern corresponded to a motif that is found in NS1 mRNA, 15 nucleotides downstream of its stop codon. To delineate the patterns that are specifically recognized by NS1, interaction of aptamers and their derivatives with NS1 was assayed in vitro, using fluorimetry-based methods and gel-shift experiments. We showed that NS1’s RBD specifically recognizes sequence patterns, provided these are displayed within a proper RNA structure.
MATERIALS AND METHODS
General methods
Current molecular biology methods were performed according to Sambrook and Russell (34).
Recombinant NS1 proteins
The NS1 proteins of the two viruses A/HK/213/03 (H5N1) and A/turkey/Italy/977/1999 (H7N1), corresponding to the alleles A and B of the protein, respectively, were PCR amplified from their cloned cDNA (a kind gift from William Dundon and Ilaria Capua) with appropriate primers containing NdeI and NotI restriction sites (primer sequences can be provided on request). The digested PCR products were ligated to the NdeI–NotI digested pET22b(+) DNA (Novagen), generating plasmids pNS1-A and pNS1-B, which encoded a full-length NS1 with an added C-terminal extension DPGGGLNDIFEAQKIEWHEDAAALEHHHHHH containing both a biotin-acceptor peptide (35) and a C-terminal His6 tag. Both proteins were overexpressed as inclusion bodies in BL21-(DE3) bacteria, then purified as described (36). A 1:1 mixture of these two recNS1 was used in the selection process. Subsequently, for the interaction studies, the allele A-full-length protein and its truncated variants [1–73] and [80–230], corresponding to the RBD and ED, respectively, were all produced with only the C-terminal His tag peptide AAALEHHHHHH. Using the QuikChange kit (Stratagene), substitutions were introduced by PCR in the RBD. Soluble recombinant His-tagged proteins were overexpressed in BL21-(DE3) bacteria, RNase-treated as described (37), then added with urea up to 2 M and purified through cobalt affinity. They were then equilibrated in 10 mM Hepes pH 7.5, 150 mM NaCl, 5% glycerol, 0.005% Tween-20 through passage onto a desalting column (Bio-Gel P6 Desalting Gel, Biorad). Purified proteins were distributed in small aliquots that were snap-frozen into liquid nitrogen, then stored at −80°C until use.
Aptamer construction and transcription
An initial pool of 80-nt RNAs with a central random sequence of 40 nucleotides flanked by two 20-nt regions of determined sequence was prepared as described (38). DM01 transcription template was mutagenized by PCR to produce mutated derivatives of aptamer DM01. Transcription templates for the small hairpin RNAs were obtained by annealing and elongation of overlapping complementary DNA oligonucleotides. Fluorescein-labelled RNAs were in vitro transcribed using the T7 Megashortscript kit (Ambion), in the presence of 1 mM fluorescein-12-UTP (Roche), together with 2 mM of unlabelled UTP. The same Megashortscript kit was used to transcribe 32P-labelled RNAs: the concentration of ATP, CTP and GTP was lowered to 0.5 mM (instead of 7.5), while both unlabelled and [alpha 32P]-UTP were present (10 µM each). The concentrations of enzyme and template DNA were lowered, and the reaction was incubated for 10 min at 37°C. RNAs were separated from free nucleotides through Sephadex G25 filtration.
In vitro selection
A microplate selection method was used, based on that described by Drolet et al. (39). Duplicate wells of a Nunc Maxisorp 96-well microplate were coated (2 h at room temperature) with 0.15–2.5 µg of recNS1 in 70 µL of phosphate-buffered saline (PBS), then washed three times with 200 µl of binding buffer (BB: 50 mM Tris-Cl, 100 mM NaCl, 2 mM MgCl2, 0.05% Tween 20, pH 7.5). Before applying the RNA pool, wells were saturated (90 min, room temperature) with 5 µM of yeast tRNA in 45 µl of BB. To minimize the selection of aptamers with affinity to the plasticware, candidate RNAs were first incubated (1 h at room temperature) at their final concentrations in several uncoated wells (25 µl per well) in BB. Subtracted RNAs from the previous step were added to the coated wells already containing the saturating yeast tRNA. After a 30- to 60-min incubation at 37°C, wells were washed three times with 200 µl of BB, and three times with 200 µl of prewarmed BB. Amounts per well of the protein used to coat the well, as well as respective concentrations of the candidate RNAs and competing tRNAs were varied through the SELEX in order to gradually increase the selection pressure. These ranged from 90 pmoles of NS1 per well / 2 µM candidate RNA / 2 µM tRNA in the first cycles, to 2 pmol NS1 per well / 0.6 µM candidate RNA / 5 µM tRNA in the last (11th) cycle.
Reverse transcription, PCR amplifications, cloning and sequence analysis
At the end of each SELEX cycle, selected RNAs were reverse transcribed, and a new pool of transcription templates was prepared as described (38). At the end of the final SELEX cycle, PCR products were cloned into vector PCR-2 (TOPO-TA cloning kit, Invitrogen) and sequenced as described (38). RNA folding predictions were performed with Mfold 3.2 (40). Motifs searches were performed, either directly on the influenza resource database, or using “fuzznuc” in the Emboss software suite (41).
Electrophoretic mobility shift assays
RNA [150 nanograms (6 pmol)] was mixed with increasing amounts of NS1-RBD (4–100 pmol of the dimer) in a 10 µl reaction containing 50 mM Tris-borate, 1 mM EDTA, pH 8.2 (TBE). After a 15-min incubation on ice, RNA and RNA–protein complexes were resolved through migration in a 10% non-denaturing polyacrylamide gel (150 min, 70 volts, 4°C), using a Mini-Protean II system (Biorad). Gels were soaked in SyBr-Gold (Molecular Probes) and visualized on a UV-transilluminator. The same protocol was used for gel-shift assays with 32P-labelled RNAs, except for the presence of yeast tRNA in the reaction mixture as a non-specific competitor (30 µg/ml). Gels were dried, then exposed to a Kodak Biomax MR film, with or without an intensifying screen. The intensity of the bands was not quantified because the signal was likely not linear.
Fluorescence anisotropy (FA)
Fluorescein-labelled RNAs were diluted to 5–7 nM in FA buffer (8 mM HEPES, 80 mM NaCl, 20 mM KCl, 2 mM MgCl2, 5% glycerol, 0.025% tween-20 and 30 µg/ml yeast tRNA, representing a ∼200-fold molar excess of the non-specific tRNA). Fluorescence measurements were performed at 18°C in a PTI-Quantamaster 40 (Photon Technologies International) in a T configuration, allowing the simultaneous measure of the fluorescence intensities in the parallel (IVV) and perpendicular (IVH) polarization planes. Anisotropy was calculated using the equation r = (Ivv − G × Ivh) / (Ivv + 2 × G × Ivh), where the G-factor corrects for the relative efficiency of the two optical paths VV and VH. Protein was first added at its maximal concentration in the reaction mixture (3 ml in a quartz cuvette containing a magnetic stirrer), and a series of decreasing concentrations was achieved by 20–25 successive 2/3 dilutions. Each dilution was followed by 2 min of magnetic stirring and 2.5 min of equilibration before fluorescence measurements. Fluorescence intensities (IVV and IVH) and anisotropy were measured in triplicate at each dilution point (λexc = 495, λemi = 515, slitwidth = 6 nm, each measure was averaged over 20 s). Total fluorescence intensity was calculated as IVV + 2 × G × IVH (42), taking into account the G-factor. For the competition assay, the instrument’s sensitivity was augmented by increasing the slitwidth, and the concentration of the labelled RNA was lowered to 0.5 nM.
Fluorescence spectra of the RBD
RBD was diluted to 100 nM in 0.6 ml of fluorescence buffer (8 mM HEPES, 80 mM NaCl, 20 mM KCl, 2 mM MgCl2), either alone or in the presence of varying amounts of RNAs DM01 or SB01. Tryptophan fluorescence was recorded in a 0.5 ml quartz cuvette using the “Synchronous scan” function of the spectroflurimeter: fluorescence intensity was measured from 280 to 400 nm, while the excitation wavelength was simultaneously varied from 260 to 380 nm. Blank spectra were recorded with buffer alone, or with buffer containing RNA (50 or 100 nM), and were subtracted from the RBD, SB01 and DM01 spectra containing the corresponding amounts of RNA. Each scan is the average of two successive scans on the same sample.
Data analysis
Anisotropy and fluorescence intensity values were analysed using the Prism GraphPad software. When appropriate, the fraction of bound RNA was calculated using the equation
Fbound = (r − rfree) / [Q(rbound − r) + (r − rfree)], where r is the anisotropy and Q is the quantum yield enhancement (43). Data were fitted to the appropriate model through non-linear regression.
RESULTS
In vitro selection identifies virus-specific RNA sequences
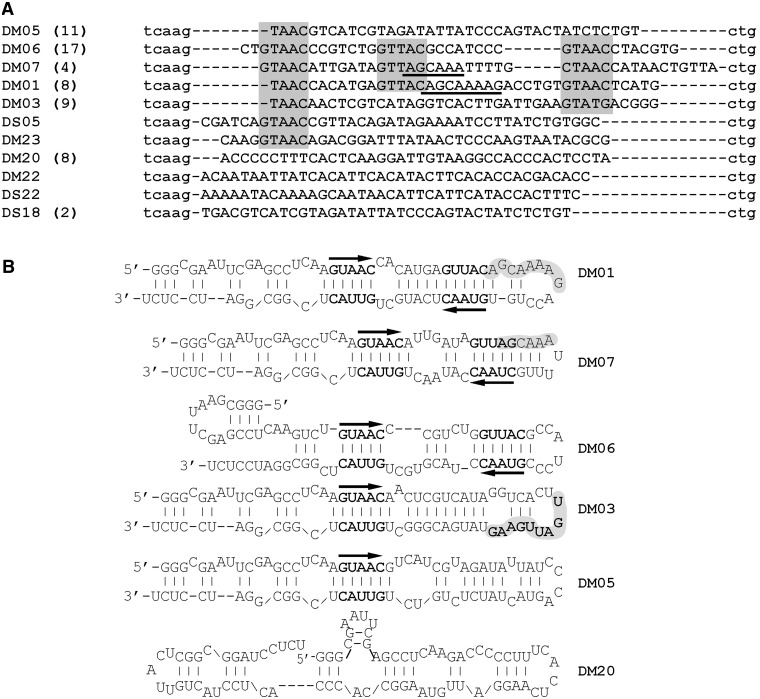
Two recombinant NS1 proteins of viruses A/HK/213/03 (H5N1) and A/turkey/Italy/977/1999 (H7N1), corresponding respectively to the alleles A and B of the protein, were purified from overexpressing BL21(DE3) Escherichia coli. To isolate RNA aptamers that bind NS1, we performed the SELEX method, using a 1:1 mixture of the two proteins and a pool of in vitro transcribed 80-nt RNAs harbouring a central 40-nt random region. We chose to perform the selection with both alleles because we did not want to miss potential candidates that may have bound to only one allele of NS1. At the end of the iterative selection process (11th cycle), RNAs were reverse-transcribed and cloned. In all, 63 sequences were obtained, which were then compared and grouped by similarity in six distinct families of sequences and a few unique sequences (Figure 1). Strikingly, two families of sequences (DM01 and DM07), altogether comprising twelve of the 63 sequences, harboured the 5′-part of a 12-nucleotide-long motif that is invariably present at the 5′ end of all positive-strand RNAs of all influenzaviruses A: AGCAAAAG or AGCAAA (44). There is at least a second virus-specific motif, which we did not further explore: UGAUUGAAG, which is found in the nine members of the DM03 family (Figure 1B), is almost invariably present in NS1 mRNA at position 735, 15 nucleotides downstream of NS1 stop codon. More than 84% of the ∼20 000 segment 8 sequences in Genbank harbour the motif URAUUGAAG at this position (see Table 1). Interestingly, among the ∼3000 NS1 sequences lacking this motif are the ∼1700 sequences corresponding to the allele B of NS1: these harbour a slightly different motif at that position: most frequently it is UAAUUGCAG, instead of UGAUUGAAG. Furthermore, the UGAUUGAAG and UAAUUGAAG motifs are also present in a significant fraction of the (+)-strand RNAs corresponding to viral polymerase subunits PA and PB2 (see Table 1). Less conspicuous is the ACGUGCU (in DM06), which is found at position 723 in 30% of the PB2 sequence (in most cases in the form of ACGUGCUG). Finally, a fourth pattern was even more frequent than these virus-specific motifs: 49 of the 63 aptamers, including the 12 aptamers from families DM01 and DM07, harboured one (n = 20) or two copies (n = 29) of the double-stranded motif 5′-GUAAC-3′ / 3′-CAUUG-5′ (Figure 1A and B). Beside these motifs, we noticed that the Mfold-predicted structure of all these aptamers consisted of a long unbranched dsRNA structure with bulges (Figure 1B). Noteworthily, in all the 29 aptamers that harboured two copies of the double-stranded motif 5′-GUAAC-3′ / 3′-CAUUG-5′ (families DM01, DM06 and DM07 in Figure 1B), these two motifs were symmetrically positioned, about 7 bp apart, in the double-stranded structure.
Figure 1.
Sequences and predicted structures of the NS1-aptamers. (A) Sequences of the 63 aptamers were aligned and grouped according to their similarity. Lower case letters at the 5′ and 3′ end are part of the constant sequences, whereas nucleotides from the N40 central region are in upper case letters. Alignment was manually edited to emphasize the nucleotide motifs that are present in several independent sequences, such as the AGCAAA(AG) (underlined) and GUAAC/GUUAC motifs (shaded boxes). The number of sequences in each family is indicated in parentheses. (B) Structures of the most frequently isolated aptamers were predicted with Mfold. Bold letters represent the conserved double-stranded motifs 5′-GUAAC-3′ / 3′-CAUUG-5′, the orientation of which is shown by the arrows. Shaded areas highlight the virus-specific sequences AGCAAA(AG) and UGAUUGAAG.
Table 1.
The URAUUGAAG and ACGUGCU motifs in influenza virus positive-strand RNAs
| NS1 | PA | PB2 | Total number in (+) strand | Total number in (−) strand | |
|---|---|---|---|---|---|
| UGAUUGAAG | 10191 / 20583 (735) | 7302 / 19350 (1808) | 17583 / 205947 | 687 | |
| UAAUUGAAG | 7265 / 20583 (735) | 10914 / 19350 (257) | 3456 / 19554 (2051) | 21696 | 87 |
| ACGUGCU | 6243 / 19554 (723) | 6508 | 562 |
The values indicate number of sequences harbouring the given motif / total number of sequences. The number in parentheses indicates the position of the motif in the (+)RNA. Last column shows the total number of each motif in negative-strand RNAs. Search was performed on 26 September 2012.
RBD strongly binds aptamers in gel-shift experiments
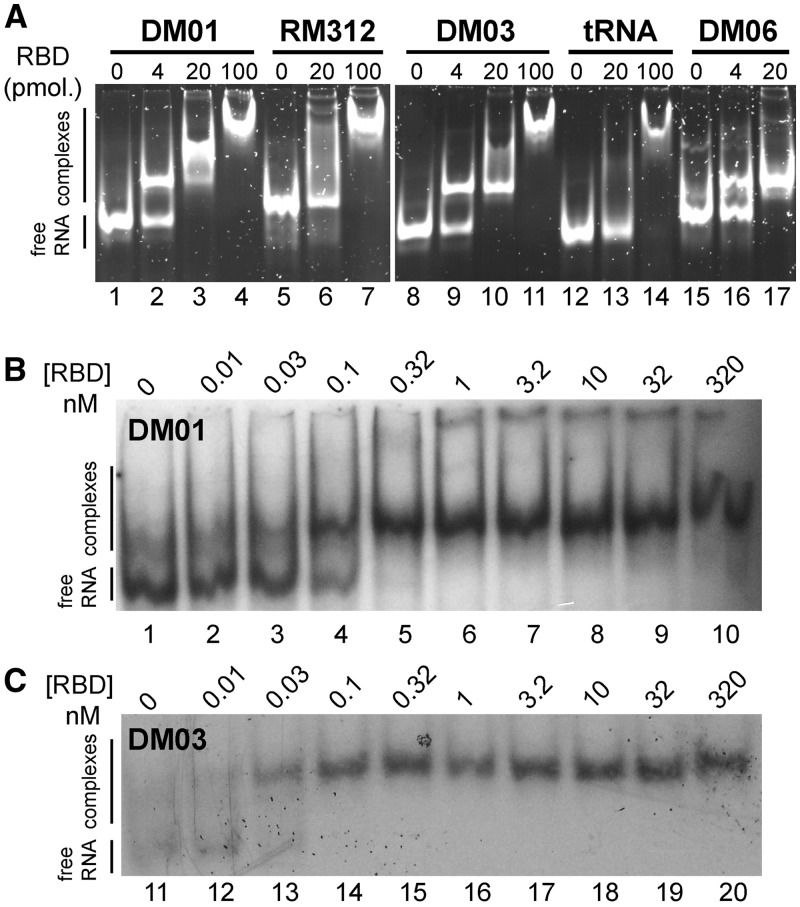
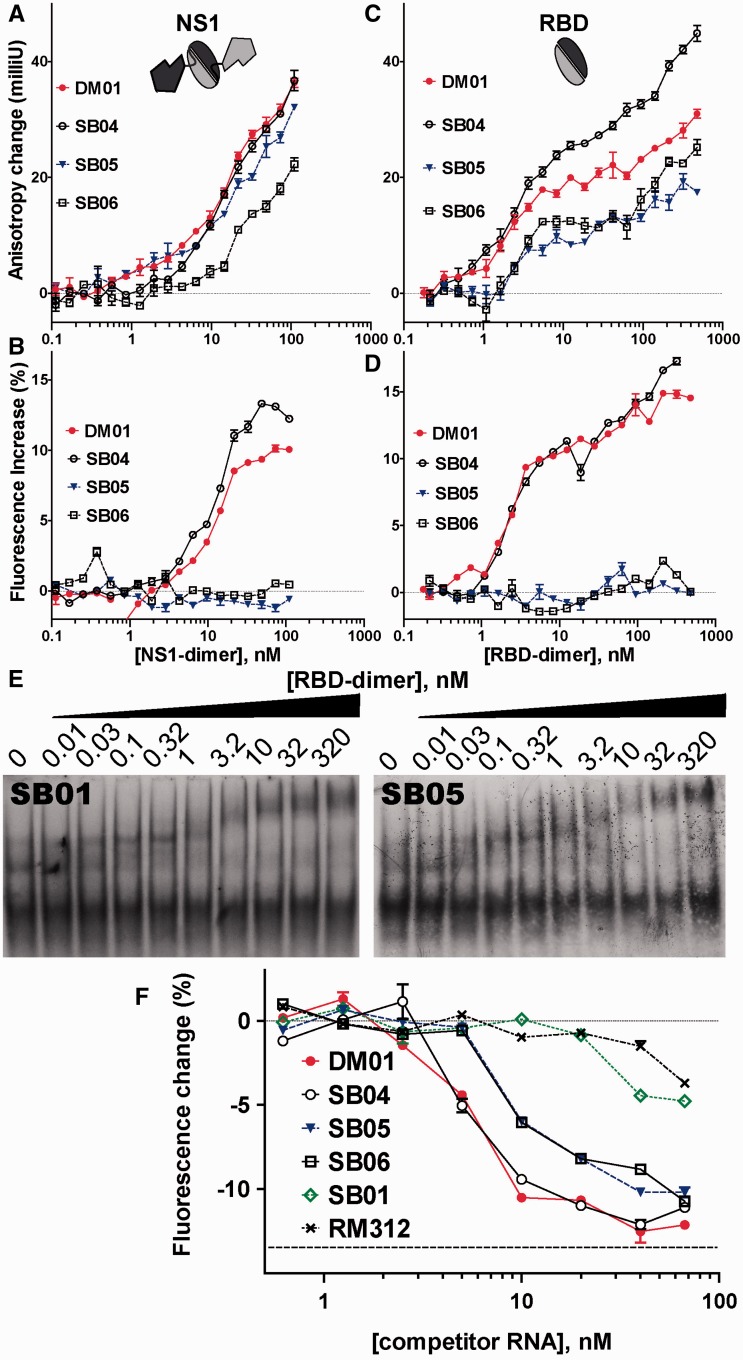
We then chose to study the interaction of these aptamers only with allele A of NS1 because the latter is far more prevalent than allele B, which is found only in some viruses of avian origin. To confirm their interaction with NS1, aptamers were assayed through an electrophoretic mobility shift assay (EMSA). Full-length NS1 could not be used in such assays, however, because it perturbed the electrophoresis of all RNAs, probably as a result of its known propensity to aggregate (45). Instead, RNAs were incubated with the RBD, then resolved through a non-denaturing polyacrylamide gel. As shown in Figure 2A, aptamers DM01, DM03 and DM06 (as well as DM05 and DM07, not shown) readily formed complexes with the RBD. By contrast, with RM312, an aptamer with the same construction that was raised against the ovine prion protein (38), as well as with tRNAs that were used as controls, only high amounts of RBD resulted in the formation of complexes (Figure 2A, lanes 7 and 14). Furthermore, formation of the RBD–aptamer complexes was robust with respect to the various buffer conditions that were assayed (TBE buffer, as well as 50–100 mM Tris-Cl buffers containing 100–150 mM of NaCl, 0–40 mM of KCl, 1–4 mM of MgCl2) (not shown). Gel-shift experiments conducted with radiolabelled RNAs DM01 and DM03 (Figure 2B and C) indicated a very low, subnanomolar KD value (all the RNA was bound at RBD concentrations close to 300 pM or even lower).
Figure 2.
In vitro interaction of RBD with aptamers (EMSA). (A) The indicated RNAs (150 ng each, ∼6 pmol) were incubated with increasing amounts of the RBD (4–100 pmol of dimer). RNA–protein complexes were separated through electrophoresis in a 10% non-denaturing polyacrylamide gel, then visualized after SyBr-Gold staining. (B and C) Gel-shift experiments were further performed with 32P-labelled DM01 and DM03.
Evaluating the sequence and structural requirements by deconstruction of DM01
Examination of the sequences and predicted structures of the aptamers (Figure 1B) revealed at least three important features: (i) a pronounced double-stranded character, (ii) the presence of a virus-specific sequence (AGCAAAAG or AGCAAA), and (iii) the frequent presence of the GUAAC sequence and complementary GUUAC, which in most aptamers was present as two symmetrical copies. We set out to examine the importance of these features in the NS1–RNA interaction, choosing as a model aptamer DM01, which harbours these three main features. The RNAs and proteins that were used throughout this work are shown in Figure 3.
Figure 3.
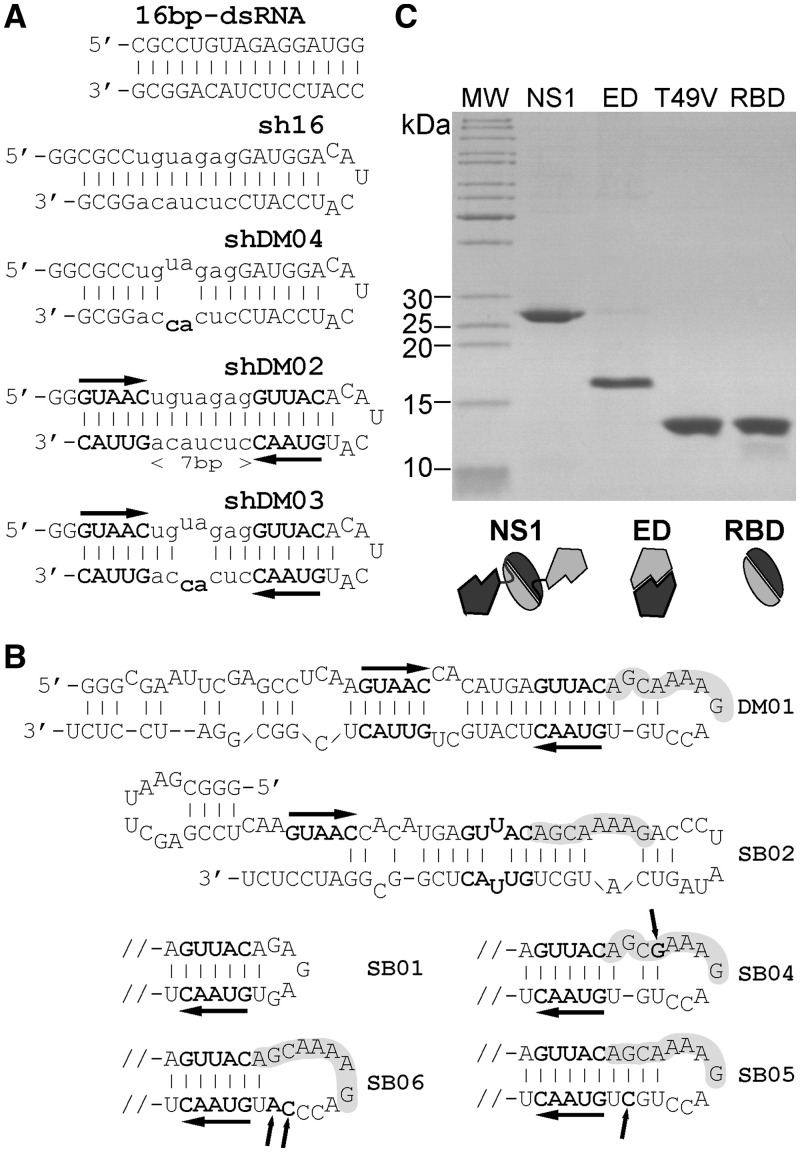
RNAs and proteins used in the interaction studies. (A) Structure of the short RNAs of the sh series. Sh16 is a derivative of the 16 bp-dsRNA model that was shown to bind NS1 (30,32). Both shDM02 and shDM03 harbour the two symmetrical motifs 5′-GUAAC-3′ / 3′-CAUUG-5′, with an intervening spacer consisting of a 7 bp helix that is fully double stranded in shDM02, or interrupted by a symmetric internal loop in shDM03. Two nucleotide substitutions, creating a symmetric internal loop, were introduced in sh16 to produce shDM04. (B) Mfold-predicted structures of mutated aptamers SB01-06. Horizontal arrows highlight the GUUAC sequence, and the AGCAAAAG motif is shaded. Only the terminal hairpin is shown for SB01 and SB04-06. Arrows point to modified or added nucleotides in SB04-06. (C) Recombinant proteins (2 µg of the RBDs and 1 µg of NS1 and ED) were analysed by SDS-PAGE and revealed by Coomassie staining.
RBD specifically recognizes the two double-stranded motifs 5′-GUAAC-3′ / 3′-CAUUG-5′
First we remarked that the central symmetry formed by the two GUAAC double-stranded motifs in the RNA structure can be superimposed onto that of the RBD dimer, which was shown to bind a model 16-bp RNA duplex in a defined crystallographic structure (32). Therefore we first chose to investigate the importance of these two 5′-GUAAC-3′ / 3′-CAUUG-5′ motifs. To that end, we produced a series of short hairpin (sh) RNAs that were derived from the model 16-bp dsRNA mentioned above. Short RNA sh16 was designed in order to mimic this 16-bp dsRNA. Two Gs at the 5′-terminus, as well as a small loop connecting the two strands, were added so that it could be produced as a single molecule by in vitro transcription (Figure 3A). Next, the two symmetrical 5′-GUAAC-3′ / 3′-CAUUG-5′ motifs were introduced into that backbone, with an intervening spacer consisting of a 7 bp helix that was either fully double stranded (shDM02) or interrupted by a symmetric internal loop (shDM03). In addition, shDM04 was designed essentially as an sh16, with an internal loop identical to that of shDM03 (see Figure 3A).
RBD binds to sequence-specific dsRNA with low nanomolar KD value
Gel-shift experiments with unlabelled RNAs (not shown) repeatedly showed that shDM03 stood out as the best RBD ligand among the short RNAs of the sh series. However, the interaction was difficult to quantify, and for a better quantification, the interaction of these small RNAs with RBD was measured in solution using fluorescence anisotropy (FA). An increased anisotropy of fluorescence reflects a reduced rotational mobility of the emitting fluorophore, generally resulting from the formation of complexes that increase the inertia of the fluorescent molecule. In addition, the computation of the bound fraction from FA takes into account the quantum yield enhancement (see methods), therefore the fluorescence intensity values IVV and IVH were also used to record the total fluorescence intensity. Interaction with a protein can increase the emission of a fluorescent dye in a manner that is very sensitive to the proximity of the dye to the protein (46,47), therefore we also used the intensity change to estimate the intimacy of the interaction. Fluorescein-labelled RNAs were synthesized through in vitro T7-transcription, then submitted to FA in the presence of RBD. FA was first measured in 3 ml of RNA containing the highest amount of protein (usually 1–1.5 micromolar). Then successive dilutions of the protein were made by replacing 1 ml of the reaction mixture with 1 ml of free RNA.
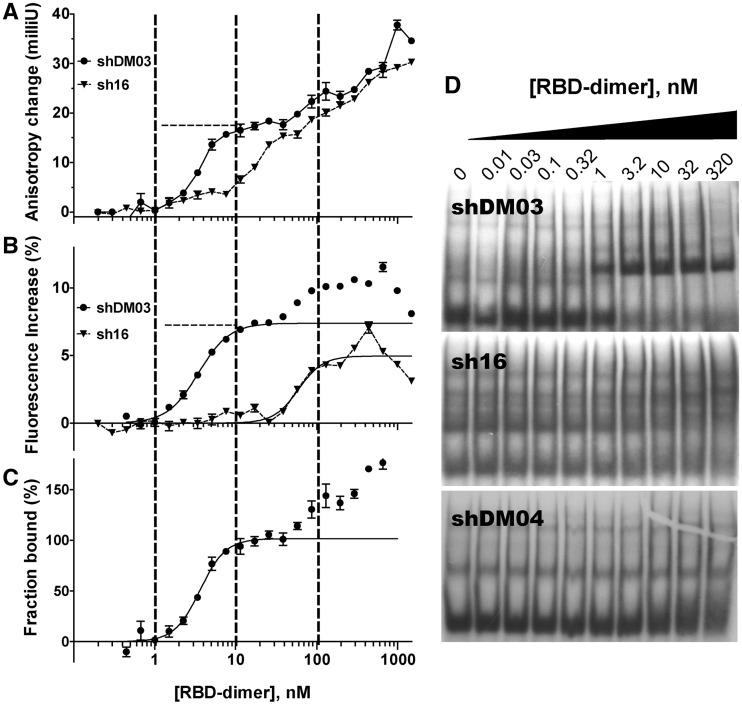
As shown in Figure 4A, increasing amounts of the RBD resulted in an increased anisotropy of shDM03 (under our experimental conditions, anisotropy of the free state for the RNAs of the sh series was close to 80 milliunits). Anisotropy augmented and reached a first plateau as soon as the RBD concentration was about 10 nM. Then it increased further with higher concentrations of RBD. The anisotropy of small RNA sh16 also gradually increased with increasing amounts of added RBD, but no such plateau was seen as was observed with shDM03 (shDM04 yielded results similar to those of sh16). Several repetitions of this comparison (of which Figure 4 is only one representative experiment) always gave the same results. In addition, varying the amount of RBD through successive additions rather than successive dilutions also yielded similar results; however, the dilution series repeatedly yielded better curves, probably resulting from a better homogenization of the reaction mixture (43). Furthermore, essentially the same results were obtained when in vitro-transcribed shDM03 was replaced by its chemically synthesized counterpart carrying a fluorescein moiety attached to its 5′-end.
Figure 4.
Interaction of RBD with sh16, shDM03 and shDM04. Anisotropy change (A) and change in fluorescence intensity (B) were recorded for shDM03 (filled circles) and sh16 (triangles), when mixed with increasing amounts of RBD (see methods). (C) The bound fraction deduced from the anisotropy was calculated for shDM03, using two fixed parameters that are indicated by dashed lines in panels A and B: rbound = 17.5 milliunits, and fluorescence increase of the bound state = 7.4% (i.e. Q = 1.074). This was fit to the model ‘specific binding with Hill slope’ (for data points below 50 nM) (mean ± SEM, n = 3 for each point). (D) The three 32P-labelled RNAs were further subjected to gel-shift experiments.
Interestingly, the fluorescence intensity of shDM03 increased by as much as 10% on binding to RBD (Figure 4B), and furthermore the shape of the intensity curve clearly paralleled that of the anisotropy curve, reaching a first plateau at about 10 nM of RBD. By contrast, the relative intensity of sh16 began to increase at much higher concentrations of RBD, and reached a maximum increase of ∼5% only. The quantum yield enhancement for 100% of bound shDM03 was estimated from the first plateau (about 7.4% in Figure 4B, dashed line, therefore Q = 1.074), and was used together with the anisotropy values to compute the bound fraction of shDM03 (see methods), which is displayed in Figure 4C. We assumed that after the first plateau, the increase in both anisotropy and fluorescence intensity was due to the formation of higher order complexes in which several RBD dimers bound the fluorescent RNA, resulting in values higher than 100% in Figure 4C. These large complexes are probably comparable with those observed with aptamer DM01 in gel-shift experiments (Figure 2A, lanes 3 and 4). The binding model which best fitted the steepness of our experimental binding isotherm was that of a specific binding with Hill slope, in which one RNA molecule can specifically bind more than one RBD dimer [corresponding to the model: RNA + n RBD -> RNA(RBD)n]. Applying this model to the bound fraction of shDM03 (Figure 4C) yielded a KD value of 3.63 nM [95% confidence interval (CI) = 3.19–4.07], with an apparent Hill coefficient (h) of 2.75 (R2 = 0.96, 95% CI = 1.97–3.54). Similar values were obtained with shDM02 (not shown). Interestingly, fitting the same model to the fluorescence increase (Figure 4B) yielded very similar KD and h values of 3.46 nM and 2.27, respectively (R2 = 0.98). We feel that our data do not allow us to provide a more precise KD value, mainly because the accuracy of our calculation is inherently limited by the RNA concentration (∼5–7 nM), which is too high to reliably determine a subnanomolar value of KD (see for example http://structbio.vanderbilt.edu/chazin/wisdom/kdcalc.htm). Indeed, when the concentration of shDM03 was varied in several repetitions of this assay, we remarked that the KD was always close to half the concentration of the labelled RNA. Therefore the KD value we determined should rather be viewed as the upper bound of its range. Gel-shift experiments (Figure 4D) indeed were consistent with a KD close to 1 nM for shDM03, while virtually no RNA–RBD complexes were observed for sh16 and shDM04 (slowly migrating bands in the sh16 and shDM04 gels probably correspond to RNA complexes resulting from intermolecular annealing of the RNAs, favoured by their internal symmetry). Furthermore, whereas RNA–RBD complexes were observed with sh16 and shDM04 in solution (as reflected by the increase in anisotropy), these were probably disrupted during electrophoresis. Altogether, our data clearly show that RBD interacts strongly and specifically with small RNAs containing the symmetrical motif 5′-GUAAC-3′ / 3′-CAUUG-5′, with a KD close to 1 nM.
RBD specifically recognizes the virus-specific pattern AGCAAAAG
We next investigated the importance of the viral sequence AGCAAAAG in the interaction of NS1 with RNAs. To that end, a second series of RNAs (SB01 to SB06) was designed, derived from aptamer DM01 (Figure 3B). Although we are aware that Mfold-predicted secondary structures (Figure 1B) are biased towards double-stranded conformations and only represent the most energetically favourable ones, we emphasize that the alternate less favourable structures of DM01 (not shown) all contain the viral sequence AGCAAAAG in the same environment. Instead of attempting to chemically or enzymatically probe the structure of DM01, we preferred to introduce point mutations that would most likely result in either alterations of the structure, or only in substitutions within an unchanged structure. The terminal loop of DM01 containing the AGCAAAAG motif was deleted and replaced by a GAGAG loop in SB01, whose predicted structure is otherwise identical to that of DM01. SB02 still retained the viral sequence, but the immediately downstream sequence ACCUGUGUAAC, which is involved in the double-stranded structure 5′-GUAAC-3′ / 3′-CAUUG-5′, was modified to ACCCUAUAG. Because of this modification, one of the two double-stranded motifs can no longer form, which dramatically alters the overall structure and the environment of the viral sequence. SB04 differed from DM01 by a single G to A substitution, which changed the AGCAAAAG motif to AGCGAAAG. SB05 and SB06 were designed in order to modify the structural context of the sequence AGCAAAAG. In SB05, an additional C was introduced to base-pair with the bulged G, resulting in an uninterrupted 14-bp helix, whereas, to the contrary, replacement of a UG dinucleotide by CA in SB06 lengthens the terminal loop containing the AGCAAAAG motif.
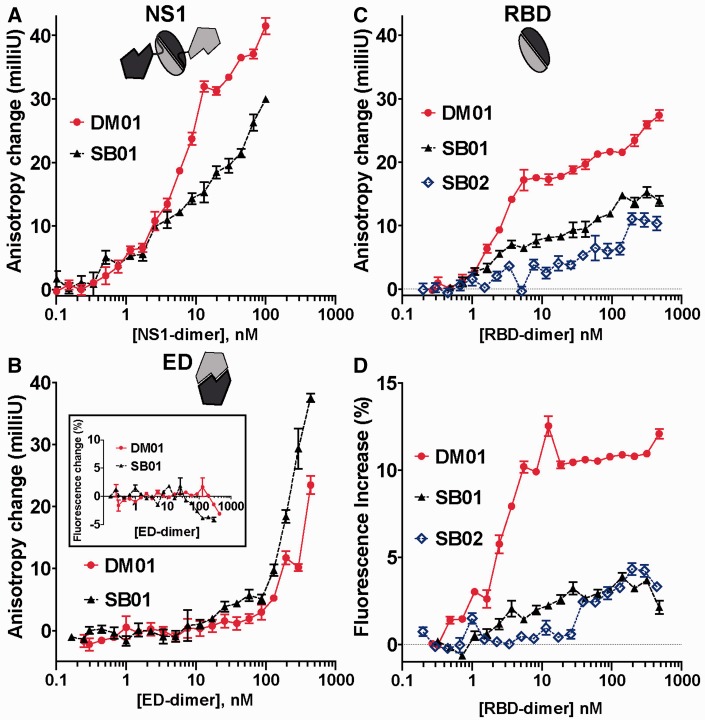
Because we showed that the RBD interacts specifically with the two symmetrical 5′-GUAAC-3′ / 3′-CAUUG-5′ motifs, we first thought that the recognition of the AGCAAAAG motif would likely require the participation of the ED. Therefore, we first measured the interaction of DM01 and SB01 with both the full-length NS1 and the ED. As judged by the difference between the anisotropy curves of the two RNAs, DM01 binds much better than SB01 to full-length NS1 (Figure 5A). By contrast, when assaying the ED, only high concentrations of the protein (>200 nM) could significantly increase the anisotropy of both RNAs (Figure 5B) (for these ∼80-nt RNAs, the anisotropy of the free molecule was close to 100 milliunits). This anisotropy did not reach a plateau, and furthermore the fluorescence intensity of both SB01 and DM01 remained flat in the presence of high amounts of ED (Figure 5B, inset). Therefore these two observations indicate that the ED does not significantly interact with either DM01 or SB01, and our interpretation is that high amounts of ED create a ‘crowded environment’, in which frequent stochastic collisions with the protein reduce the rotational mobility of the fluorescent RNAs, hence the increased anisotropy but unchanged fluorescence intensity. Anisotropy of DM01, SB01 and SB02 with the RBD (Figure 5C) revealed that DM01 strongly binds RBD with a low nM KD value, producing a first plateau similar to that observed previously in the shDM03–RBD interaction (compare with Figure 4A). By contrast, interaction of SB01 and SB02 with RBD was much reduced, as judged by the absence of the first anisotropy plateau (Figure 5C). This is further supported by the increased fluorescence intensity (Figure 5D) that was observed with DM01 only, and not with SB01 or SB02. Finally, gel-shift experiments clearly showed that DM01 bound to the RBD with a KD value close to 100 pM (see above, Figure 2B), while only low amounts of gel-shifted RNA–RBD complexes were observed when SB01 was mixed with large amounts of the RBD (see below, Figure 6E). Altogether, these data demonstrate that the RBD strongly interacts with DM01, which harbours the AGCAAAAG motif, while removing this motif (in SB01) or altering its structural presentation (in SB02) are sufficient to dramatically reduce the interaction. Thus the RBD binds the AGCAAAAG motif with high affinity, provided this motif is displayed in the structural context of DM01.
Figure 5.
Interaction of DM01, SB01 and SB02 with full-length NS1, ED and RBD. Anisotropy change of DM01 (red dots) and SB01 (filled triangles) was recorded when mixed with increasing amounts of full-length NS1 (A), ED (B) or RBD (C). SB02 was also included in (C). (D) and (B, inset) also show the change in fluorescence intensity of the RNAs when mixed with ED (B, inset) or with RBD (D) (mean ± SEM, n = 3 for each point).
Figure 6.
Interaction of DM01, SB01, SB04, SB05 and SB06 with full-length NS1 and RBD. Anisotropy change of DM01 (red dots), SB04 (open circles), SB05 (blue triangles) and SB06 (open squares) was recorded when mixed with increasing amounts of full-length NS1 (A) or RBD (C). (B and D) Relative change in fluorescence intensity of the RNAs when mixed with full-length NS1 (B) or with RBD (D) (mean ± SEM, n = 3 for each point). (E) 32P-labelled RNAs SB01 and SB05 were further subjected to gel-shift experiments. (F) Fluorescein-labelled DM01 (0.5 nM) was mixed with RBD (3 nM) in the presence of varying amounts of unlabelled competitor RNAs. Fluorescence intensity was recorded, and the relative change was expressed in % of the maximum intensity (mean ± SEM, n = 3 for each point). Horizontal dashed line represents the fluorescence intensity of unbound DM01 (∼−13.5% relative to its bound counterpart).
Binding of RBD to AGCAAAAG is structure dependent
We next asked whether more subtle modifications to the sequence or to the structural context of the AGCAAAAG motif could alter the interaction. DM01, SB04, SB05 and SB06 were assayed for their interaction with both full-length NS1 and the RBD. Interaction with full-length NS1 increased the fluorescence anisotropy of the four RNAs (Figure 6A). However, there was no clear-cut difference, except for the fact that the increase was less pronounced for SB06 than for SB04 and DM01, while SB05 gave an intermediate curve (Figure 6A). We surmise that most of this increase resulted not from genuine interactions, but rather from stochastic collisions between the RNAs and the protein. Full-length NS1 is probably prone to stochastic collisions, as it can be viewed as a compact RBD-dimer loosely linked to two ED monomers (see schematic drawing in Figure 6A), which in turn tend to associate independently with other EDs (45). Indeed, fluorescence intensity curves (Figure 6B) showed a net increase in fluorescence for DM01 and SB04 only, while fluorescence intensity remained flat for both SB05 and SB06. With the RBD (Figure 6C), the anisotropy increase for both DM01 and SB04 reached a first plateau at 20 milliunits, and then increased further, as was previously observed in the shDM03–RBD interaction. The anisotropy increase was lower with both SB05 and SB06, and furthermore the fluorescence intensity curves confirmed the strong interaction of RBD with both DM01 and SB04, whereas the flat curves observed for both SB05 and SB06 indicated a weak interaction (Figure 6D). Non-linear fitting of the bound fraction for both DM01 and SB04 yielded similar values of KD and Hill coefficients (∼2 nM and h = 1.7, respectively). Gel-shift experiments with SB05 (Figure 6E) and SB06 (not shown) yielded results similar to those obtained with SB01: only low amounts of gel-shifted RNA–RBD complexes were observed at high concentrations of the RBD. These may result from binding of the RBD to the two double-stranded motifs 5′-GUAAC-3′ / 3′-CAUUG-5′, although this interaction seems less intense than in the context of small RNA shDM03 (see Figure 4D). Therefore, only subtle alterations to the structure of the AGCAAAAG motif are sufficient to abolish its interaction with the RBD.
In an attempt to further explore the specificity of the RBD towards the structural presentation of the AGCAAAAG motif, we compared DM01, SB04, SB05 and SB06 in a competition experiment. Fluorescein-labelled DM01 (0.5 nM) was mixed with RBD (3 nM) in the presence of varying concentrations of unlaballed RNAs. Increasing amounts of competitor RNAs readily reduced the fluorescence anisotropy of DM01, the best competitor being DM01. However, anisotropy alone was not sufficient to clearly distinguish SB04, -05 and -06 (not shown), while differences were clearly observed through the decreased fluorescence intensity of DM01, reflecting its unbound state (Figure 6F). Although SB01 was only slightly better than the control aptamer RM312, SB04 was as efficient as DM01 as a competitor. SB05 and SB06, which both harbour the AGCAAAAG motif in a modified structure, were indistinguishable, being of intermediate potency as competitors. Therefore, these data show that RBD binds equally well the AGCAAAAG (DM01) and AGCGAAAG (SB04) motifs. Furthermore, this competition assay shows that SB05 and SB06, which both harbour the AGCAAAAG motif in a sub-optimal presentation, still retain a partial efficiency as competitors, compared with SB01 which lacks the virus-specific sequence. This suggests that the RBD still recognizes the AGCAAAAG motif in a modified structure, albeit with much less affinity than the same motif in a proper structural context (i.e., that of DM01).
Finally, we also observed that both the fluorescence intensity and the anisotropy rose more slowly with full-length NS1 than with RBD (compare Figure 6A and B with Figure 6C and D, respectively), which might suggest that the ED not only does not favour but also somewhat hinders the interaction.
T49V substitution in the RBD prevents its oligomerization on the RNA
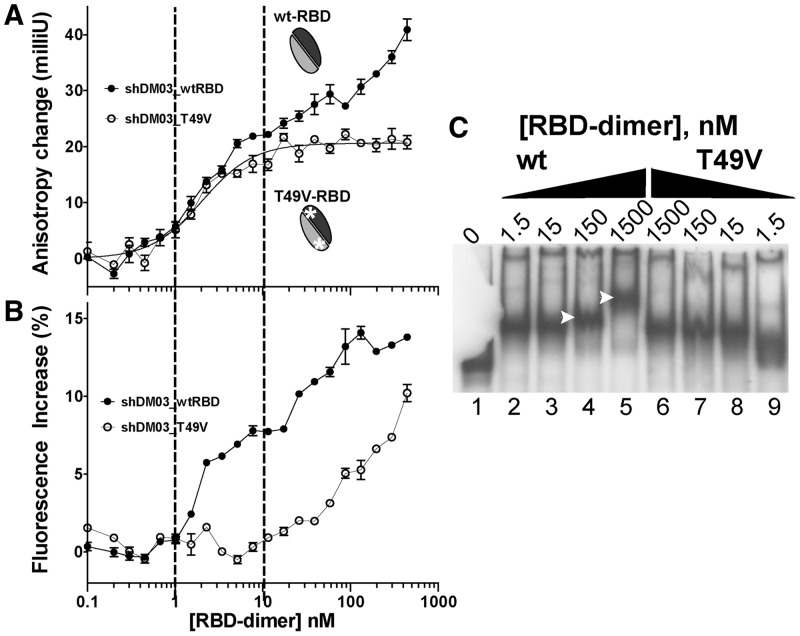
In a first attempt to more accurately determine by anisotropy the binding parameters of the RBD with the small RNAs of the sh series (data not shown), we had constructed one RBD variant by substituting threonine 49 with valine, based on the fact that the conserved T49 had previously been shown to be involved in the RBD–RNA interaction (48). T49 lies at the end of helix 2, which harbours the amino acids involved in RNA binding (i.e. R38 and K41), and the T49V exchange results in the mere substitution of a single hydroxyl group by a methyl group. Interestingly, differences were regularly observed between the two RBD variants (i.e. wt and T49V) on interaction with the small RNA shDM03. As shown in Figure 7A, the anisotropy of shDM03, when mixed with T49V-RBD, rose rapidly to a plateau that was ∼20 milliunits above that of the free RNA, but 5–8 milliunits below the first plateau observed with the wild-type RBD. Moreover, there was no further increase in anisotropy once this plateau was reached, contrary to what occurred with the wild-type RBD. This suggested that, contrary to its wild-type counterpart, T49V-RBD could not oligomerize on its RNA substrate. Further, the fluorescence increase of shDM03 (Figure 7B) was much lower with T49V-RBD than with wt-RBD, suggesting that the interaction of T49V-RBD with shDM03 was less intimate than it was with wt-RBD. Similar anisotropy and fluorescence curves were obtained with shDM02 (not shown). Furthermore, a similar observation was made when comparing the interaction of SB04 with wt- and T49V-RBD (data not shown).This peculiar behaviour of T49V-RBD was confirmed in gel-shift experiments using radiolabelled RNA DM01 (Figure 7C): the major band of RNA–RBD complex increased in size with higher amounts of the wt-RBD (Figure 7C, lanes 4 and 5, arrows). Slightly shifted RBD–RNA complexes with increasing amounts of wt-RBD were also observed in other gel-shift experiments (Figure 2B, lane 10; Figure 2C, lane 20; Figure 2A, lane 3). To the contrary, this was not observed with T49V-RBD, even at the highest concentration (Figure 7C, lane 6).
Figure 7.
T49V-RBD does not form oligomers. (A, B) Anisotropy changes and fluorescence increase of shDM03 on binding to wt-RBD (filled circles) or to T49V-RBD (open circles). Curve on top graph represents the fitted bound fraction for the interaction of shDM03 with T49V-RBD (plateauing at 100%). (C) Gel-shift experiment with 32P-labelled DM01 and increasing amounts of either wt-RBD or T49V-RBD (the gel was inadvertently broken, but the equivalent of lane 1 can be seen in Figure 2B, lane 1).
RNA binding induces a conformational change in the RBD
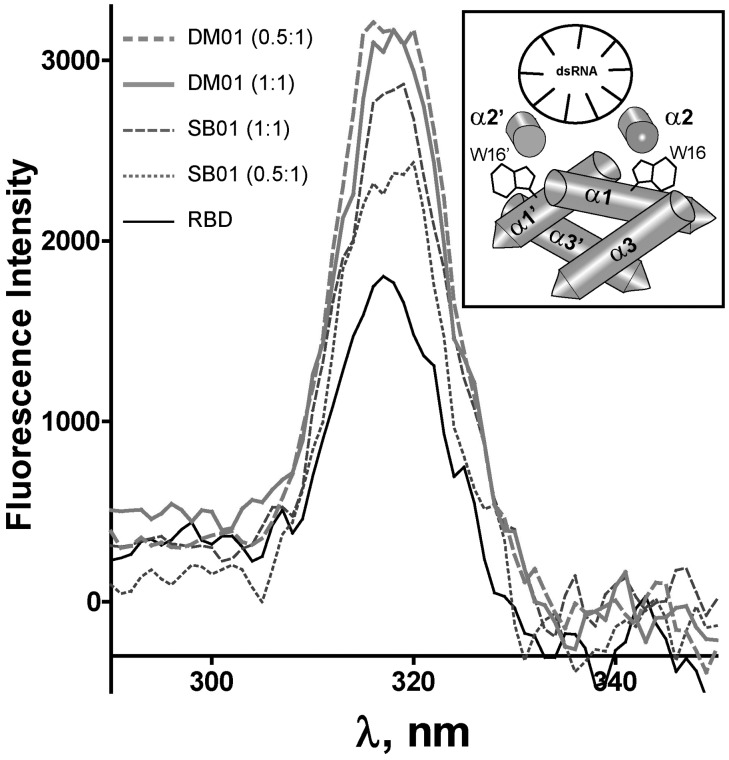
We next asked whether binding to its RNA ligands could induce structural changes in the RBD. The RBD dimer contains two absolutely conserved tryptophan residues (W16 and W16′), which are located in the alpha helices α1 and α1′ and are buried deeply in the RBD structure. We reasoned that interaction of the RBD with RNAs might induce structural changes that could modify the environment of these tryptophan residues and modify their fluorescence properties. We recorded the fluorescence spectrum of the RBD, either alone or in the presence of RNAs DM01 or SB01. As shown in Figure 8, the fluorescence spectrum of RBD clearly showed a tryptophan-specific peak at 320 nm. The intensity of this peak was proportional to the concentration of RBD (not shown), and it was increased in the presence of both SB01 and DM01, the latter repeatedly producing a stronger effect, especially at a low RNA:protein ratio. Graphs obtained with non-specific aptamer RM312 (not shown) were intermediate between those obtained with SB01 (1:1 ratio) and DM01 (1:1 ratio).
Figure 8.
RNAs induce structural modifications in the RBD. The fluorescence spectrum of the tryptophan residues was recorded on the RBD (100 nM), which was either alone in buffer (solid thin line) or mixed with 50 nM or 100 nM of DM01 or SB01, as indicated (0.5:1 or 1:1 ratios). Each point represents the fluorescence intensity at a given wavelength on excitation at (λemi—20 nm), using the ‘synchronous scan’ function of the Quantamaster instrument. Inset shows the structure of the RBD bound to a model 21 bp-dsRNA, drawn after the structure 2ZKO deposited in the Protein Data Bank (48). Tryptophan residues W16 and W16′, located in the alpha helices α1 and α1′, are depicted. The axes of the antiparallel helices α2 and α2′, and of the dsRNA helix, are perpendicular to the image plane.
DISCUSSION
Influenzavirus protein NS1 is generally thought to bind double-stranded RNAs with no structure or sequence specificity. Using the SELEX method, we performed an in vitro selection of small RNAs that bind NS1, in an attempt to identify sequential or structural motifs that may favour their interaction with NS1. Our rationale was that we may thus identify RNA patterns with which NS1 preferentially interacts, and thereby establish functional relationships between the biophysical properties of NS1 in vitro and its activities in vivo. Although high-throughput sequencing combined to a less stringent SELEX (49) or to a crosslinking method (50) would probably yield broader sets of ligands, our set of 63 sequences contained a number of patterns that match well with the known functions of NS1. Firstly, and consistent with the dsRNA-binding property of NS1, their Mfold-predicted structures (Figure 1) were all characterized by an unbranched double-stranded conformation, which is also reminiscent of the structure of miRNA precursors (51). Secondly, at least two conspicuous virus-specific sequences were identified in a significant fraction of our NS1-aptamers: first, AGCAAAAG is invariably present at the 5′end of all influenzavirus RNAs of positive polarity (although RNAs corresponding to the three polymerase subunits bear the slightly different sequence AGCGAAAG) (44). The second viral pattern consists of the 9-nt motif UGAUUGAAG, which is almost invariably present in NS1 mRNA at position 735, 15 nucleotides downstream of NS1 stop codon. In addition to these patterns, our set of aptamers may contain yet other virus-specific motifs that we have not explored, like the ACGUGCU motif identified in DM06 (Table 1). Noteworthily, the two virus-specific sequences in our set of aptamers were always optimally exposed, close to the terminal loop of their predicted structure. Thirdly, the majority of our aptamers had at least one copy of the double-stranded motif (5′-GUAAC-3′/3′-CAUUG-5′).
Using aptamer DM01 as a model, we evaluated the importance of the AGCAAAAG sequence and of the double-stranded motif (5′-GUAAC-3′/3′-CAUUG-5′) by deconstructing DM01 in two series of modified RNAs. Gel-shift experiments, as well as fluorescence anisotropy and protein-induced fluorescence enhancement (46), were used to quantify the interaction.
The RBD binds a short double-stranded motif with high affinity
Previous studies had shown that RBD bound in vitro to a model 16-bp RNA duplex (32). The short hairpin RNA derived from this model RNA effectively behaved as a non-specific RBD ligand, as judged from both fluorimetry and gel-shift assays (Figure 4). Introducing the two copies of the dsRNA motif 5′-GUAAC-3′/3′-CAUUG-5′ into this model RNA converted it to a specific and high affinity ligand of the RBD. The KD value calculated from FA for this specific binding was in the low nanomolar range, consistent with the ∼1 nM value estimated through gel-shift. This low nM KD value is about three orders of magnitude less than that previously measured by a sedimentation equilibrium method for the model 16-bp RNA (30). High-affinity binding to this double motif probably results from the perfect match between the symmetry of the RNA and that of the RBD. Indeed, in all the aptamers harbouring the two double-stranded motifs, these were symmetrically positioned, 7 bp apart in the double-stranded structure. Assuming the structural geometry that was previously described for the dsRNA–RBD dimer complex (32,48), the central symmetry in the RNA structure can be superimposed onto that of the RBD dimer, and each of the GUAAC/CAUUG motifs would establish symmetrical interactions with the RBD in the vicinity of residues S42-R46′ and S42′-R46, respectively.
The RBD recognizes at least two virus-specific patterns
NS1 specifically bound the AGCAAAAG motif. The ED is not involved in this interaction, which relies solely on the RBD, as revealed by anisotropy, fluorescence intensity and gel-shifts (Figures 5A, B and 6E). Binding of the AGCAAAAG sequence to the RBD is also structure dependent: changing its structural environment (SB02 in Figure 5C and D), even through minor alterations (SB05 and SB06 in Figure 6), is sufficient to dramatically reduce the interaction. On the contrary, changing the AGCAAAAG motif in DM01 to AGCGAAAG does not seem to alter its interaction with either full-length NS1 or the RBD. Although we did not investigate the other virus-specific motif UGAUUGAAG, this pattern is also specifically recognized by the RBD, as judged by the strong binding of aptamer DM03 in gel-shift experiments. Therefore the RBD can specifically recognize at least three distinct unrelated sequence patterns, i.e. the two virus-specific sequences and the double-stranded motif. The low KD values of the specific ligands compares well with the nanomolar to subnanomolar KD values that have been determined by various methods for other RNA-binding proteins (52,53). How RBD, a small domain composed of two 73-residue polypeptides, can specifically recognize such distinct targets? This ‘chameleon-like’ behaviour has already been observed for other RNA-binding proteins (52,54–56), and can also be paralleled with the observation that a single transcription factor can bind several distinct DNA targets (57). This chamelonism of the RBD is probably favoured by both the flexibility of its structure and the number of arginine and lysine residues. On the other hand, whether distinct target RNAs are recognized by the same molecule, or by different structural conformations of NS1, remains to be established. Such a conformational plasticity could arise from alternate oligomerization modes of NS1. Indeed, elucidation of its crystal structure has revealed that NS1 can form either single dimeric units or long oligomeric structures of alternating RBD and ED dimers, resulting from the fact that each of the two domains of NS1 (RBD and ED) can dimerize separately (45). It is therefore possible that NS1, depending on its oligomerization status, could bind distinct RNA patterns and exert distinct activities. Reciprocally, different RNA ligands could promote distinct oligomerization modes and thereby modulate the various activities of NS1.
Cooperative binding and oligomerization
As indicated by the steepness of our binding isotherms, specific binding is cooperative, i.e. once a RBD is bound, binding of other RBDs to the same RNA molecule is facilitated, corresponding to the model: RNA + n RBD -> RNA(RBD)n. How could an RNA as short as shDM03 (Figure 4) bind two or three RBDs? First, the symmetry of this small RNA could favour its binding to two RBDs, symmetrically with respect to the dsRNA axis. However, it is even more likely that at least three RBDs could bind, forming a short tunnel around the dsRNA. Indeed, elucidation of the crystal structure of full-length NS1 has shown that it can form long tubular structures with an internal diameter of 20 angstroms (45), which is wide enough to accommodate a dsRNA molecule. Cooperative binding of the RBD likely promotes its oligomerization on its target RNA template. Oligomerization was abolished by the T49V substitution in the RBD since: (i) we no longer observed the increasing anisotropy at high concentrations of the mutant RBD, (ii) the level of the anisotropy plateau (which is related to the inertia of the RNA–protein complex, and hence to its mass) was lower than that observed with wt-RBD, which is consistent with one RBD only binding to the RNA, (iii) binding was weaker and less intimate, as judged by the less intense fluorescence enhancement (Figure 7B), and (iv) no oligomers were observed with T49V-RBD in gel-shift experiments (Figure 7C).
Furthermore, RNA binding induces in the RBD a structural change that is evidenced by the modified fluorescence spectrum of tryptophan residues W16 and W16′, in spite of the fact that both tryptophan residues are located far from the dsRNA in the structures that were previously determined for the RBD bound to model dsRNAs (32,48).
Relations with NS1 function
Even if interactions of NS1 with RNAs in vivo are probably more complex and dynamic than the binding we measured in vitro with aptamers, the patterns that we analysed likely represent biologically relevant targets of NS1. Firstly, the sequence-specific interaction with AGCAAAAG is in agreement with previously published observations showing that NS1 enhances translation of viral RNAs through interacting with both eIF4GI and the 5′ terminus of the viral mRNAs (58,59). Although both studies pointed to the extreme 5′-end of the 5′-UTR as a target of NS1 in enhancing the translation of viral mRNAs, our data strengthen these observations and show that NS1, by itself, specifically binds to the AGCAAAAG sequence in a manner that also depends on the structure of the RNA.
NS1 was also suggested to play a role in the nucleocytoplasmic export of viral mRNAs (60), and was shown to interact with both viral mRNAs and several factors of the mRNA export pathway (8,61,62). Our observations of the oligomerization of NS1 on its RNA ligands suggest a mechanism through which NS1 may at the same time favour the nucleocytoplasmic transport and regulate the splicing of viral mRNAs (10). Oligomerization of a viral protein on its specific RNA substrate has been observed for other viral RNA-binding proteins, notably for the Rev protein of Human Immunodeficiency virus (60,63). It was shown that, through its oligomerization on the Rev responsive element, Rev forms an organized ribonucleoprotein (RNP) that is suggested to function in the export of intron-containing viral RNAs (52). Whether oligomerization of NS1 on viral mRNAs could play a similar role in the nucleocytoplasmic export of viral mRNAs remains to be formally demonstrated, although published data (see above) seem to support this view. Introducing the T49V substitution that abolishes the oligomerization, either in transient expression of NS1 or in genetically engineered viruses, may help answer this point.
Specific binding to the AGCAAAAG motif may also play a role in the regulation of viral RNA synthesis, in which NS1 was shown to be involved (64–66), or in its interaction with the nucleoprotein (NP) and PA subunits of the polymerase complex (67,68). However, it remains to be determined whether the AGCAAAAG motif in the viral cRNAs and mRNAs is structurally competent to bind to the RBD (69).
The second virus-specific motif that we identified but did not explore, UGAUUGAAG, is also a highly conserved motif, located 15 nucleotides downstream of NS1 stop codon. Like several posttranscriptional modules that are present in the 3′-UTR of mRNAs, it may allow NS1 to regulate the fate of its own mRNA, and perhaps also of the other viral mRNAs that harbour this pattern or the related UAAUUGAAG (i.e. PA and PB2, see Table 1). This motif is much more prevalent in the positive-strand RNAs (mRNA and cRNA) than it is in the genomic RNAs (which are associated with the nucleoprotein NP). This agrees well with a specific interaction of NS1 with positive-strand viral RNAs which, like itself, are also absent from the virion. Because mRNAs encoding allele B-NS1 harbour a slightly different motif, this allele specificity could be one of the parameters controlling the reassortment: for instance PA or PB2 segments harbouring the UGAUUGAAG may be advantaged over those that do not, when NS1 is of allele A.
Beside virus-specific sequences, binding of NS1 to the double-stranded motifs 5′-GUAAC-3′/3′-CAUUG-5′ may also act in the regulation of splicing. Indeed, this motif closely resembles the canonical 5′-splice site, at the 5′ boundary of classical introns (70): the double-stranded structure 5′-GUAA-3′ / 3′-CAUU-5′ forms the core of the base-paired region that is established when the 5′-end of small nuclear RNA U1 anneals to pre-mRNAs and thereby selects the 5′-splice donor site (71,72). This double-stranded structure is bound by spliceosomal protein U1C, raising the possibility that NS1 may exert a general block on RNA splicing by competing with U1C. NS1 was indeed shown to associate with spliceosomes and inhibit pre-mRNA splicing (24). Although this inhibition was attributed to its binding to U6 snRNA, it may also rely on its competition with U1C. Regulation by NS1 of the splicing of its own mRNA might therefore involve two mechanisms, i.e. general block on splicing and specific binding of NS1 to the two virus-specific sequences. This may perhaps explain why different studies found that NS1 either down-regulated the splicing of its own mRNA (10), or had no effect on this splicing event (24,73). For instance, these contrasting effects could result from the presence or absence of the UGAUUGAAG motif in NS1 mRNA, downstream of its stop codon.
Finally, we notice that all of our NS1-specific aptamers have M fold predicted structures that are reminiscent of the pre-miRNA structures (51). We also found that AGCAAAAG is present in miRNA-511 and miRNA-548v in humans, whereas UGAUUGAAG is found in chicken miRNA-1434. Whether these can have any relation to influenza pathology in these species remains to be determined.
In conclusion, we have identified through SELEX a number of structural and sequential motifs that bind NS1 in vitro, and likely represent biologically relevant targets of NS1 activities. Although the AGCAAAAG motif corresponds to a previously known biological target of NS1 in vivo, the double-stranded motif (5′-GUAAC-3′/3′-CAUUG-5′), as well as other virus-specific patterns that we did not further investigate, was unsuspected. Our results indicate that NS1 binds RNAs in a manner that is far more specific than previously thought, with a strikingly high affinity. We reached these conclusions by studying model RNAs, and it will be necessary to explore, in the context of viral RNAs and of the viral cycle, the structural requirements and the biological significance of these interactions in vivo.
FUNDING
Direction Générale de l’Alimentation (Fonds de Recherche Influenza Aviaires, project 08) (in part); INRA and Région Centre (joint grant to S.B.). Funding for open access charge: Institut National de la Recherche Agronomique and Institut Carnot Santé Animale.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are indebted to William Dundon and Ilaria Capua for the gift of NS1 cDNAs. We thank Dominique Kerboeuf for her help with the spectrofluorimeter; Pradip Nandi, Yves Mély and Anny Slama-Schwok for their helpful advice in fluorescence anisotropy; and Nadia Naffakh for her critical reading of our manuscript. D.M. thanks Françoise Marc for her unfailing support.
REFERENCES
- 1.Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, et al. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science. 2012;337:199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palese P, Shaw M. Orthomyxoviridae: The viruses and their replication. In: Knipe D, Howley P, editors. Fields Virology. 5th edn. Philadelphia: Lippincott, Williams and Wilkins; 2007. [Google Scholar]
- 3.Wise HM, Foeglein A, Sun J, Dalton RM, Patel S, Howard W, Anderson EC, Barclay WS, Digard P. A complicated message: identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J. Virol. 2009;83:8021–8031. doi: 10.1128/JVI.00826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Obenauer JC, Denson J, Mehta PK, Su X, Mukatira S, Finkelstein DB, Xu X, Wang J, Ma J, Fan Y, et al. Large-scale sequence analysis of avian influenza isolates. Science. 2006;311:1576–1580. doi: 10.1126/science.1121586. [DOI] [PubMed] [Google Scholar]
- 5.Suarez DL, Perdue ML. Multiple alignment comparison of the non-structural genes of influenza A viruses. Virus Res. 1998;54:59–69. doi: 10.1016/s0168-1702(98)00011-2. [DOI] [PubMed] [Google Scholar]
- 6.Hale BG, Randall RE, Ortin J, Jackson D. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 2008;89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- 7.Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol. Cell. 1998;1:991–1000. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 8.Satterly N, Tsai PL, van Deursen J, Nussenzveig DR, Wang Y, Faria PA, Levay A, Levy DE, Fontoura BM. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc. Natl. Acad. Sci. USA. 2007;104:1853–1858. doi: 10.1073/pnas.0610977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shapira SD, Gat-Viks I, Shum BOV, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, et al. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garaigorta U, Ortin J. Mutation analysis of a recombinant NS replicon shows that influenza virus NS1 protein blocks the splicing and nucleo-cytoplasmic transport of its own viral mRNA. Nucleic Acids Res. 2007;35:4573–4582. doi: 10.1093/nar/gkm230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marion RM, Aragon T, Beloso A, Nieto A, Ortin J. The N-terminal half of the influenza virus NS1 protein is sufficient for nuclear retention of mRNA and enhancement of viral mRNA translation. Nucleic Acids Res. 1997;25:4271–4277. doi: 10.1093/nar/25.21.4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez-Sesma A, Marukian S, Ebersole BJ, Kaminski D, Park MS, Yuen T, Sealfon SC, Garcia-Sastre A, Moran TM. Influenza virus evades innate and adaptive immunity via the NS1 protein. J. Virol. 2006;80:6295–6304. doi: 10.1128/JVI.02381-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, Garcia-Sastre A. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egorov A, Brandt S, Sereinig S, Romanova J, Ferko B, Katinger D, Grassauer A, Alexandrova G, Katinger H, Muster T. Transfectant influenza A viruses with long deletions in the NS1 protein grow efficiently in Vero cells. J. Virol. 1998;72:6437–6441. doi: 10.1128/jvi.72.8.6437-6441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 16.Kochs G, Koerner I, Thiel L, Kothlow S, Kaspers B, Ruggli N, Summerfield A, Pavlovic J, Stech J, Staeheli P. Properties of H7N7 influenza A virus strain SC35M lacking interferon antagonist NS1 in mice and chickens. J. Gen. Virol. 2007;88:1403–1409. doi: 10.1099/vir.0.82764-0. [DOI] [PubMed] [Google Scholar]
- 17.Cauthen AN, Swayne DE, Sekellick MJ, Marcus PI, Suarez DL. Amelioration of influenza virus pathogenesis in chickens attributed to the enhanced interferon-inducing capacity of a virus with a truncated NS1 gene. J. Virol. 2007;81:1838–1847. doi: 10.1128/JVI.01667-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donelan NR, Basler CF, Garcia-Sastre A. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J. Virol. 2003;77:13257–13266. doi: 10.1128/JVI.77.24.13257-13266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiao P, Tian G, Li Y, Deng G, Jiang Y, Liu C, Liu W, Bu Z, Kawaoka Y, Chen H. A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice. J. Virol. 2008;82:1146–1154. doi: 10.1128/JVI.01698-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newby CM, Sabin L, Pekosz A. The RNA binding domain of influenza A virus NS1 protein affects secretion of tumor necrosis factor alpha, interleukin-6, and interferon in primary murine tracheal epithelial cells. J. Virol. 2007;81:9469–9480. doi: 10.1128/JVI.00989-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seo SH, Hoffmann E, Webster RG. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat. Med. 2002;8:950–954. doi: 10.1038/nm757. [DOI] [PubMed] [Google Scholar]
- 22.Hatada E, Saito S, Okishio N, Fukuda R. Binding of the influenza virus NS1 protein to model genome RNAs. J. Gen. Virol. 1997;78:1059–1063. doi: 10.1099/0022-1317-78-5-1059. [DOI] [PubMed] [Google Scholar]
- 23.Hatada E, Takizawa T, Fukuda R. Specific binding of influenza A virus NS1 protein to the virus minus-sense RNA in vitro. J. Gen. Virol. 1992;73:17–25. doi: 10.1099/0022-1317-73-1-17. [DOI] [PubMed] [Google Scholar]
- 24.Lu Y, Qian XY, Krug RM. The influenza virus NS1 protein: a novel inhibitor of pre-mRNA splicing. Genes Dev. 1994;8:1817–1828. doi: 10.1101/gad.8.15.1817. [DOI] [PubMed] [Google Scholar]
- 25.Qiu Y, Nemeroff M, Krug RM. The influenza virus NS1 protein binds to a specific region in human U6 snRNA and inhibits U6-U2 and U6-U4 snRNA interactions during splicing. RNA. 1995;1:304–316. [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W, Krug RM. U6atac snRNA, the highly divergent counterpart of U6 snRNA, is the specific target that mediates inhibition of AT-AC splicing by the influenza virus NS1 protein. RNA. 1998;4:55–64. [PMC free article] [PubMed] [Google Scholar]
- 27.Chien CY, Tejero R, Huang Y, Zimmerman DE, Rios CB, Krug RM, Montelione GT. A novel RNA-binding motif in influenza A virus non-structural protein 1. Nat. Struct. Biol. 1997;4:891–895. doi: 10.1038/nsb1197-891. [DOI] [PubMed] [Google Scholar]
- 28.Hatada E, Fukuda R. Binding of influenza A virus NS1 protein to dsRNA in vitro. J. Gen. Virol. 1992;73:3325–3329. doi: 10.1099/0022-1317-73-12-3325. [DOI] [PubMed] [Google Scholar]
- 29.Wang W, Riedel K, Lynch P, Chien CY, Montelione GT, Krug RM. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA. 1999;5:195–205. doi: 10.1017/s1355838299981621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chien CY, Xu Y, Xiao R, Aramini JM, Sahasrabudhe PV, Krug RM, Montelione GT. Biophysical characterization of the complex between double-stranded RNA and the N-terminal domain of the NS1 protein from influenza A virus: evidence for a novel RNA-binding mode. Biochemistry. 2004;43:1950–1962. doi: 10.1021/bi030176o. [DOI] [PubMed] [Google Scholar]
- 31.Someya T, Hosono K, Morimura K, Takaku H, Kawai G. Recognition of a bulged RNA by peptides derived from the influenza NS1 protein. J. Biochem. 2008;143:339–347. doi: 10.1093/jb/mvm225. [DOI] [PubMed] [Google Scholar]
- 32.Yin C, Khan JA, Swapna GV, Ertekin A, Krug RM, Tong L, Montelione GT. Conserved surface features form the double-stranded RNA binding site of non-structural protein 1 (NS1) from influenza A and B viruses. J. Biol. Chem. 2007;282:20584–20592. doi: 10.1074/jbc.M611619200. [DOI] [PubMed] [Google Scholar]
- 33.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 35.Arnold GS, Sasser AK, Stachler MD, Bartlett JS. Metabolic biotinylation provides a unique platform for the purification and targeting of multiple AAV vector serotypes. Mol. Ther. 2006;14:97–106. doi: 10.1016/j.ymthe.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 36.Rezaei H, Marc D, Choiset Y, Takahashi M, Hui Bon Hoa G, Haertle T, Grosclaude J, Debey P. High yield purification and physico-chemical properties of full-length recombinant allelic variants of sheep prion protein linked to scrapie susceptibility. Eur. J. Biochem. 2000;267:2833–2839. doi: 10.1046/j.1432-1033.2000.01347.x. [DOI] [PubMed] [Google Scholar]
- 37.Tarus B, Chevalier C, Richard CA, Delmas B, Di Primo C, Slama-Schwok A. Molecular dynamics studies of the nucleoprotein of influenza A virus: role of the protein flexibility in RNA binding. PLoS One. 2012;7:e30038. doi: 10.1371/journal.pone.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mercey R, Lantier I, Maurel MC, Grosclaude J, Lantier F, Marc D. Fast, reversible interaction of prion protein with RNA aptamers containing specific sequence patterns. Arch. Virol. 2006;151:2197–2214. doi: 10.1007/s00705-006-0790-3. [DOI] [PubMed] [Google Scholar]
- 39.Drolet DW, Jenison RD, Smith DE, Pratt D, Hicke BJ. A high throughput platform for systematic evolution of ligands by exponential enrichment (SELEX) Comb. Chem. High Throughput Screen. 1999;2:271–278. [PubMed] [Google Scholar]
- 40.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 42.Lakowicz JR. Principles of Fluorescence Spectroscopy. Springer, New York: 2006. 3rd ed. [Google Scholar]
- 43.Jameson DM, Ross JA. Fluorescence polarization/anisotropy in diagnostics and imaging. Chem. Rev. 2010;110:2685–2708. doi: 10.1021/cr900267p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Desselberger U, Racaniello VR, Zazra JJ, Palese P. The 3′ and 5′-terminal sequences of influenza A, B and C virus RNA segments are highly conserved and show partial inverted complementarity. Gene. 1980;8:315–328. doi: 10.1016/0378-1119(80)90007-4. [DOI] [PubMed] [Google Scholar]
- 45.Bornholdt ZA, Prasad BV. X-ray structure of NS1 from a highly pathogenic H5N1 influenza virus. Nature. 2008;456:985–988. doi: 10.1038/nature07444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hwang H, Kim H, Myong S. Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proc. Natl. Acad. Sci. USA. 2011;108:7414–7418. doi: 10.1073/pnas.1017672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng A, Wong SM, Yuan YA. Structural basis for dsRNA recognition by NS1 protein of influenza A virus. Cell Res. 2009;19:187–195. doi: 10.1038/cr.2008.288. [DOI] [PubMed] [Google Scholar]
- 49.Hoon S, Zhou B, Janda KD, Brenner S, Scolnick J. Aptamer selection by high-throughput sequencing and informatic analysis. Biotechniques. 2011;51:413–416. doi: 10.2144/000113786. [DOI] [PubMed] [Google Scholar]
- 50.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Starega-Roslan J, Koscianska E, Kozlowski P, Krzyzosiak WJ. The role of the precursor structure in the biogenesis of microRNA. Cell Mol. Life Sci. 2011;68:2859–2871. doi: 10.1007/s00018-011-0726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daugherty MD, D'Orso I, Frankel AD. A solution to limited genomic capacity: using adaptable binding surfaces to assemble the functional HIV Rev oligomer on RNA. Mol. Cell. 2008;31:824–834. doi: 10.1016/j.molcel.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M, Patel SS, Marcotrigiano J. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bayer TS, Booth LN, Knudsen SM, Ellington AD. Arginine-rich motifs present multiple interfaces for specific binding by RNA. RNA. 2005;11:1848–1857. doi: 10.1261/rna.2167605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu S, Ghalei H, Luhrmann R, Wahl MC. Structural basis for the dual U4 and U4atac snRNA-binding specificity of spliceosomal protein hPrp31. RNA. 2011;17:1655–1663. doi: 10.1261/rna.2690611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Okuda K, Shikanai T. A pentatricopeptide repeat protein acts as a site-specificity factor at multiple RNA editing sites with unrelated cis-acting elements in plastids. Nucleic Acids Res. 2012;40:5052–5064. doi: 10.1093/nar/gks164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de la Luna S, Fortes P, Beloso A, Ortin J. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs. J. Virol. 1995;69:2427–2433. doi: 10.1128/jvi.69.4.2427-2433.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park YW, Katze MG. Translational control by influenza virus. Identification of cis-acting sequences and trans-acting factors which may regulate selective viral mRNA translation. J. Biol. Chem. 1995;270:28433–28439. doi: 10.1074/jbc.270.47.28433. [DOI] [PubMed] [Google Scholar]
- 60.Schneider J, Wolff T. Nuclear functions of the influenza A and B viruses NS1 proteins: do they play a role in viral mRNA export? Vaccine. 2009;27:6312–6316. doi: 10.1016/j.vaccine.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 61.Bier K, York A, Fodor E. Cellular cap-binding proteins associate with influenza virus mRNAs. J. Gen. Virol. 2011;92:1627–1634. doi: 10.1099/vir.0.029231-0. [DOI] [PubMed] [Google Scholar]
- 62.Wang W, Cui ZQ, Han H, Zhang ZP, Wei HP, Zhou YF, Chen Z, Zhang XE. Imaging and characterizing influenza A virus mRNA transport in living cells. Nucleic Acids Res. 2008;36:4913–4928. doi: 10.1093/nar/gkn475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Daugherty MD, Liu B, Frankel AD. Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat. Struct. Mol. Biol. 2010;17:1337–1342. doi: 10.1038/nsmb.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Falcon AM, Marion RM, Zurcher T, Gomez P, Portela A, Nieto A, Ortin J. Defective RNA replication and late gene expression in temperature-sensitive influenza viruses expressing deleted forms of the NS1 protein. J. Virol. 2004;78:3880–3888. doi: 10.1128/JVI.78.8.3880-3888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Min JY, Li S, Sen GC, Krug RM. A site on the influenza A virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology. 2007;363:236–243. doi: 10.1016/j.virol.2007.01.038. [DOI] [PubMed] [Google Scholar]
- 66.Wang Z, Robb NC, Lenz E, Wolff T, Fodor E, Pleschka S. NS reassortment of an H7-type highly pathogenic avian influenza virus affects its propagation by altering the regulation of viral RNA production and antiviral host response. J. Virol. 2010;84:11323–11335. doi: 10.1128/JVI.01034-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuo RL, Krug RM. Influenza a virus polymerase is an integral component of the CPSF30-NS1A protein complex in infected cells. J. Virol. 2009;83:1611–1616. doi: 10.1128/JVI.01491-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robb NC, Chase G, Bier K, Vreede FT, Shaw PC, Naffakh N, Schwemmle M, Fodor E. The influenza A virus NS1 protein interacts with the nucleoprotein of viral ribonucleoprotein complexes. J. Virol. 2011;85:5228–5231. doi: 10.1128/JVI.02562-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Crow M, Deng T, Addley M, Brownlee GG. Mutational analysis of the influenza virus cRNA promoter and identification of nucleotides critical for replication. J. Virol. 2004;78:6263–6270. doi: 10.1128/JVI.78.12.6263-6270.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Burge CB, Tuschl T, Sharp PA. Splicing of precursors to mRNAs by the spliceosome. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 2nd edn. New York: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- 71.Muto Y, Pomeranz Krummel D, Oubridge C, Hernandez H, Robinson CV, Neuhaus D, Nagai K. The structure and biochemical properties of the human spliceosomal protein U1C. J. Mol. Biol. 2004;341:185–198. doi: 10.1016/j.jmb.2004.04.078. [DOI] [PubMed] [Google Scholar]
- 72.Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature. 2009;458:475–480. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Robb N, Jackson D, Vreede F, Fodor E. Splicing of influenza A virus NS1 mRNA is independent of the viral NS1 protein. J. Gen. Virol. 2010;91:2331–2340. doi: 10.1099/vir.0.022004-0. [DOI] [PubMed] [Google Scholar]