Abstract
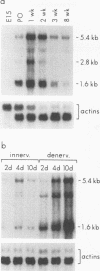
Synaptic nuclei of innervated muscle transcribe acetylcholine receptor (AChR) genes at a much higher level than extrasynaptic nuclei. To isolate candidate synaptic regulatory molecules responsible for the unique transcriptional potential of synaptic nuclei, we have taken a subtractive hybridization approach. Here, we report the cloning and characterization of a novel synapse-associated RNA, 7H4. 7H4 is expressed selectively in the endplate zone of skeletal muscle and is upregulated during early postnatal development and after denervation. Interestingly, the 7H4 gene has no introns, and yet two different-size RNAs with identical polyadenylated 3' ends are generated. Most intriguingly, the nucleotide sequence does not contain any significant open reading frames, suggesting that 7H4 may function as a noncoding RNA.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Askew D. S., Li J., Ihle J. N. Retroviral insertions in the murine His-1 locus activate the expression of a novel RNA that lacks an extensive open reading frame. Mol Cell Biol. 1994 Mar;14(3):1743–1751. doi: 10.1128/mcb.14.3.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei M. S., Zemel S., Tilghman S. M. Parental imprinting of the mouse H19 gene. Nature. 1991 May 9;351(6322):153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- Binder R., Hwang S. P., Ratnasabapathy R., Williams D. L. Degradation of apolipoprotein II mRNA occurs via endonucleolytic cleavage at 5'-AAU-3'/5'-UAA-3' elements in single-stranded loop domains of the 3'-noncoding region. J Biol Chem. 1989 Oct 5;264(28):16910–16918. [PubMed] [Google Scholar]
- Brannan C. I., Dees E. C., Ingram R. S., Tilghman S. M. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990 Jan;10(1):28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner H. R., Witzemann V., Sakmann B. Imprinting of acetylcholine receptor messenger RNA accumulation in mammalian neuromuscular synapses. Nature. 1990 Apr 5;344(6266):544–547. doi: 10.1038/344544a0. [DOI] [PubMed] [Google Scholar]
- Brockdorff N., Ashworth A., Kay G. F., Cooper P., Smith S., McCabe V. M., Norris D. P., Penny G. D., Patel D., Rastan S. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991 May 23;351(6324):329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- Brockdorff N., Ashworth A., Kay G. F., McCabe V. M., Norris D. P., Cooper P. J., Swift S., Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992 Oct 30;71(3):515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- Brown C. J., Hendrich B. D., Rupert J. L., Lafrenière R. G., Xing Y., Lawrence J., Willard H. F. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992 Oct 30;71(3):527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Brunkow M. E., Tilghman S. M. Ectopic expression of the H19 gene in mice causes prenatal lethality. Genes Dev. 1991 Jun;5(6):1092–1101. doi: 10.1101/gad.5.6.1092. [DOI] [PubMed] [Google Scholar]
- Buonanno A., Merlie J. P. Transcriptional regulation of nicotinic acetylcholine receptor genes during muscle development. J Biol Chem. 1986 Sep 5;261(25):11452–11455. [PubMed] [Google Scholar]
- Burden S. J. Synapse-specific gene expression. Trends Genet. 1993 Jan;9(1):12–16. doi: 10.1016/0168-9525(93)90066-Q. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Crowder C. M., Merlie J. P. DNase I-hypersensitive sites surround the mouse acetylcholine receptor delta-subunit gene. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8405–8409. doi: 10.1073/pnas.83.21.8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. L., Weintraub H., Lassar A. B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987 Dec 24;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- DeNardi C., Ausoni S., Moretti P., Gorza L., Velleca M., Buckingham M., Schiaffino S. Type 2X-myosin heavy chain is coded by a muscle fiber type-specific and developmentally regulated gene. J Cell Biol. 1993 Nov;123(4):823–835. doi: 10.1083/jcb.123.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis M. J., Ziskind-Conhaim L., Harris A. J. Development of neuromuscular junctions in rat embryos. Dev Biol. 1981 Jan 30;81(2):266–279. doi: 10.1016/0012-1606(81)90290-6. [DOI] [PubMed] [Google Scholar]
- Drapkin R., Merino A., Reinberg D. Regulation of RNA polymerase II transcription. Curr Opin Cell Biol. 1993 Jun;5(3):469–476. doi: 10.1016/0955-0674(93)90013-g. [DOI] [PubMed] [Google Scholar]
- Edmondson D. G., Olson E. N. Helix-loop-helix proteins as regulators of muscle-specific transcription. J Biol Chem. 1993 Jan 15;268(2):755–758. [PubMed] [Google Scholar]
- Edwards J. B., Delort J., Mallet J. Oligodeoxyribonucleotide ligation to single-stranded cDNAs: a new tool for cloning 5' ends of mRNAs and for constructing cDNA libraries by in vitro amplification. Nucleic Acids Res. 1991 Oct 11;19(19):5227–5232. doi: 10.1093/nar/19.19.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falls D. L., Rosen K. M., Corfas G., Lane W. S., Fischbach G. D. ARIA, a protein that stimulates acetylcholine receptor synthesis, is a member of the neu ligand family. Cell. 1993 Mar 12;72(5):801–815. doi: 10.1016/0092-8674(93)90407-h. [DOI] [PubMed] [Google Scholar]
- Fontaine B., Changeux J. P. Localization of nicotinic acetylcholine receptor alpha-subunit transcripts during myogenesis and motor endplate development in the chick. J Cell Biol. 1989 Mar;108(3):1025–1037. doi: 10.1083/jcb.108.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine B., Sassoon D., Buckingham M., Changeux J. P. Detection of the nicotinic acetylcholine receptor alpha-subunit mRNA by in situ hybridization at neuromuscular junctions of 15-day-old chick striated muscles. EMBO J. 1988 Mar;7(3):603–609. doi: 10.1002/j.1460-2075.1988.tb02853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frail D. E., Musil L. S., Buonanno A., Merlie J. P. Expression of RAPsyn (43K protein) and nicotinic acetylcholine receptor genes is not coordinately regulated in mouse muscle. Neuron. 1989 Jan;2(1):1077–1086. doi: 10.1016/0896-6273(89)90232-8. [DOI] [PubMed] [Google Scholar]
- Goldman D., Brenner H. R., Heinemann S. Acetylcholine receptor alpha-, beta-, gamma-, and delta-subunit mRNA levels are regulated by muscle activity. Neuron. 1988 Jun;1(4):329–333. doi: 10.1016/0896-6273(88)90081-5. [DOI] [PubMed] [Google Scholar]
- Goldman D., Carlson B. M., Staple J. Induction of adult-type nicotinic acetylcholine receptor gene expression in noninnervated regenerating muscle. Neuron. 1991 Oct;7(4):649–658. doi: 10.1016/0896-6273(91)90377-c. [DOI] [PubMed] [Google Scholar]
- Goldman D., Staple J. Spatial and temporal expression of acetylcholine receptor RNAs in innervated and denervated rat soleus muscle. Neuron. 1989 Aug;3(2):219–228. doi: 10.1016/0896-6273(89)90035-4. [DOI] [PubMed] [Google Scholar]
- Gundersen K., Sanes J. R., Merlie J. P. Neural regulation of muscle acetylcholine receptor epsilon- and alpha-subunit gene promoters in transgenic mice. J Cell Biol. 1993 Dec;123(6 Pt 1):1535–1544. doi: 10.1083/jcb.123.6.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall Z. W., Sanes J. R. Synaptic structure and development: the neuromuscular junction. Cell. 1993 Jan;72 (Suppl):99–121. doi: 10.1016/s0092-8674(05)80031-5. [DOI] [PubMed] [Google Scholar]
- Hao Y., Crenshaw T., Moulton T., Newcomb E., Tycko B. Tumour-suppressor activity of H19 RNA. Nature. 1993 Oct 21;365(6448):764–767. doi: 10.1038/365764a0. [DOI] [PubMed] [Google Scholar]
- Hunter D. D., Shah V., Merlie J. P., Sanes J. R. A laminin-like adhesive protein concentrated in the synaptic cleft of the neuromuscular junction. Nature. 1989 Mar 16;338(6212):229–234. doi: 10.1038/338229a0. [DOI] [PubMed] [Google Scholar]
- Jo S. A., Burden S. J. Synaptic basal lamina contains a signal for synapse-specific transcription. Development. 1992 Jul;115(3):673–680. doi: 10.1242/dev.115.3.673. [DOI] [PubMed] [Google Scholar]
- Kay G. F., Penny G. D., Patel D., Ashworth A., Brockdorff N., Rastan S. Expression of Xist during mouse development suggests a role in the initiation of X chromosome inactivation. Cell. 1993 Jan 29;72(2):171–182. doi: 10.1016/0092-8674(93)90658-d. [DOI] [PubMed] [Google Scholar]
- Klarsfeld A., Bessereau J. L., Salmon A. M., Triller A., Babinet C., Changeux J. P. An acetylcholine receptor alpha-subunit promoter conferring preferential synaptic expression in muscle of transgenic mice. EMBO J. 1991 Mar;10(3):625–632. doi: 10.1002/j.1460-2075.1991.tb07990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991 Nov;115(4):887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993 Dec 3;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Martinou J. C., Merlie J. P. Nerve-dependent modulation of acetylcholine receptor epsilon-subunit gene expression. J Neurosci. 1991 May;11(5):1291–1299. doi: 10.1523/JNEUROSCI.11-05-01291.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinsma D., Holthuizen P. E., Van den Brande J. L., Sussenbach J. S. Specific endonucleolytic cleavage of IGF-II mRNAs. Biochem Biophys Res Commun. 1991 Sep 30;179(3):1509–1516. doi: 10.1016/0006-291x(91)91743-v. [DOI] [PubMed] [Google Scholar]
- Meinsma D., Scheper W., Holthuizen P. E., Van den Brande J. L., Sussenbach J. S. Site-specific cleavage of IGF-II mRNAs requires sequence elements from two distinct regions of the IGF-II gene. Nucleic Acids Res. 1992 Oct 11;20(19):5003–5009. doi: 10.1093/nar/20.19.5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlie J. P., Isenberg K. E., Russell S. D., Sanes J. R. Denervation supersensitivity in skeletal muscle: analysis with a cloned cDNA probe. J Cell Biol. 1984 Jul;99(1 Pt 1):332–335. doi: 10.1083/jcb.99.1.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlie J. P., Sanes J. R. Concentration of acetylcholine receptor mRNA in synaptic regions of adult muscle fibres. Nature. 1985 Sep 5;317(6032):66–68. doi: 10.1038/317066a0. [DOI] [PubMed] [Google Scholar]
- Moss S. J., Darlison M. G., Beeson D. M., Barnard E. A. Development expression of the genes encoding the four subunits of the chicken muscle acetylcholine receptor. J Biol Chem. 1989 Dec 5;264(34):20199–20205. [PubMed] [Google Scholar]
- New H. V., Mudge A. W. Calcitonin gene-related peptide regulates muscle acetylcholine receptor synthesis. 1986 Oct 30-Nov 5Nature. 323(6091):809–811. doi: 10.1038/323809a0. [DOI] [PubMed] [Google Scholar]
- Nielsen F. C., Christiansen J. Endonucleolysis in the turnover of insulin-like growth factor II mRNA. J Biol Chem. 1992 Sep 25;267(27):19404–19411. [PubMed] [Google Scholar]
- Okada N. SINEs. Curr Opin Genet Dev. 1991 Dec;1(4):498–504. doi: 10.1016/s0959-437x(05)80198-4. [DOI] [PubMed] [Google Scholar]
- Pfeiffer S. E., Wechsler W. Biochemically differentiated neoplastic clone of Schwann cells. Proc Natl Acad Sci U S A. 1972 Oct;69(10):2885–2889. doi: 10.1073/pnas.69.10.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piette J., Huchet M., Houzelstein D., Changeux J. P. Compartmentalized expression of the alpha- and gamma-subunits of the acetylcholine receptor in recently fused myofibers. Dev Biol. 1993 May;157(1):205–213. doi: 10.1006/dbio.1993.1124. [DOI] [PubMed] [Google Scholar]
- Porteus M. H., Brice A. E., Bulfone A., Usdin T. B., Ciaranello R. D., Rubenstein J. L. Isolation and characterization of a library of cDNA clones that are preferentially expressed in the embryonic telencephalon. Brain Res Mol Brain Res. 1992 Jan;12(1-3):7–22. doi: 10.1016/0169-328x(92)90063-h. [DOI] [PubMed] [Google Scholar]
- Rastinejad F., Blau H. M. Genetic complementation reveals a novel regulatory role for 3' untranslated regions in growth and differentiation. Cell. 1993 Mar 26;72(6):903–917. doi: 10.1016/0092-8674(93)90579-f. [DOI] [PubMed] [Google Scholar]
- Rastinejad F., Conboy M. J., Rando T. A., Blau H. M. Tumor suppression by RNA from the 3' untranslated region of alpha-tropomyosin. Cell. 1993 Dec 17;75(6):1107–1117. doi: 10.1016/0092-8674(93)90320-p. [DOI] [PubMed] [Google Scholar]
- Rubenstein J. L., Brice A. E., Ciaranello R. D., Denney D., Porteus M. H., Usdin T. B. Subtractive hybridization system using single-stranded phagemids with directional inserts. Nucleic Acids Res. 1990 Aug 25;18(16):4833–4842. doi: 10.1093/nar/18.16.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A. B. Messenger RNA degradation in eukaryotes. Cell. 1993 Aug 13;74(3):413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- Sanes J. R., Johnson Y. R., Kotzbauer P. T., Mudd J., Hanley T., Martinou J. C., Merlie J. P. Selective expression of an acetylcholine receptor-lacZ transgene in synaptic nuclei of adult muscle fibers. Development. 1991 Dec;113(4):1181–1191. doi: 10.1242/dev.113.4.1181. [DOI] [PubMed] [Google Scholar]
- Sargent T. D. Isolation of differentially expressed genes. Methods Enzymol. 1987;152:423–432. doi: 10.1016/0076-6879(87)52049-3. [DOI] [PubMed] [Google Scholar]
- Schubert D., Harris A. J., Devine C. E., Heinemann S. Characterization of a unique muscle cell line. J Cell Biol. 1974 May;61(2):398–413. doi: 10.1083/jcb.61.2.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetze S. M., Role L. W. Developmental regulation of nicotinic acetylcholine receptors. Annu Rev Neurosci. 1987;10:403–457. doi: 10.1146/annurev.ne.10.030187.002155. [DOI] [PubMed] [Google Scholar]
- Shatkin A. J. Capping of eucaryotic mRNAs. Cell. 1976 Dec;9(4 Pt 2):645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- Simon A. M., Hoppe P., Burden S. J. Spatial restriction of AChR gene expression to subsynaptic nuclei. Development. 1992 Mar;114(3):545–553. doi: 10.1242/dev.114.3.545. [DOI] [PubMed] [Google Scholar]
- Spiegelman B. M., Frank M., Green H. Molecular cloning of mRNA from 3T3 adipocytes. Regulation of mRNA content for glycerophosphate dehydrogenase and other differentiation-dependent proteins during adipocyte development. J Biol Chem. 1983 Aug 25;258(16):10083–10089. [PubMed] [Google Scholar]
- Straus D., Ausubel F. M. Genomic subtraction for cloning DNA corresponding to deletion mutations. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1889–1893. doi: 10.1073/pnas.87.5.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun J. W., Schwarzbauer J. E., Hynes R. O. A single rat fibronectin gene generates three different mRNAs by alternative splicing of a complex exon. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5140–5144. doi: 10.1073/pnas.81.16.5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp W. C. Normal rat cell lines deficient in nuclear thymidine kinase. Virology. 1981 Aug;113(1):408–411. doi: 10.1016/0042-6822(81)90168-9. [DOI] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993 Dec 3;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Witzemann V., Barg B., Criado M., Stein E., Sakmann B. Developmental regulation of five subunit specific mRNAs encoding acetylcholine receptor subtypes in rat muscle. FEBS Lett. 1989 Jan 2;242(2):419–424. doi: 10.1016/0014-5793(89)80514-9. [DOI] [PubMed] [Google Scholar]
- Witzemann V., Barg B., Nishikawa Y., Sakmann B., Numa S. Differential regulation of muscle acetylcholine receptor gamma- and epsilon-subunit mRNAs. FEBS Lett. 1987 Oct 19;223(1):104–112. doi: 10.1016/0014-5793(87)80518-5. [DOI] [PubMed] [Google Scholar]
- Witzemann V., Brenner H. R., Sakmann B. Neural factors regulate AChR subunit mRNAs at rat neuromuscular synapses. J Cell Biol. 1991 Jul;114(1):125–141. doi: 10.1083/jcb.114.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W. I., Gitschier J., Lasky L. A., Lawn R. M. Base composition-independent hybridization in tetramethylammonium chloride: a method for oligonucleotide screening of highly complex gene libraries. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1585–1588. doi: 10.1073/pnas.82.6.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe D. Retention of differentiation potentialities during prolonged cultivation of myogenic cells. Proc Natl Acad Sci U S A. 1968 Oct;61(2):477–483. doi: 10.1073/pnas.61.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]