Abstract
Transcription factors modulate expression primarily through specific recognition of cognate sequences resident in the promoter region of target genes. AraR (Bacillus subtilis) is a repressor of genes involved in l-arabinose metabolism. It binds to eight different operators present in five different promoters with distinct affinities through a DNA binding domain at the N-terminus. The structures of AraR-NTD in complex with two distinct operators (ORA1 and ORR3) reveal that two monomers bind to one recognition motif (T/ANG) each in the bipartite operators. The structures show that the two recognition motifs are spaced apart by six bases in cases of ORA1 and eight bases in case of ORR3. This increase in the spacing in the operators by two base pairs results in a drastic change in the position and orientation of the second monomer on DNA in the case of ORR3 when compared with ORA1. Because AraR binds to the two operators with distinct affinities to achieve different levels of repression, this observation suggests that the variation in the spacing between core recognition motifs could be a strategy used by this transcription modulator to differentially influence gene expression.
INTRODUCTION
The ability to perceive stimuli and respond appropriately is the hallmark of any living organism. In many instances, such responses are regulated at the level of transcription. A valuable and informative system to study transcription regulation involves modulators that are sensitive to the presence of small metabolites. AraR is one such transcription factor, which regulates carbon catabolism in Bacillus subtilis. In the absence of l-arabinose, AraR represses transcription of approximately 13 genes required for arabinose utilization. DNAase I footprinting and mutation experiments have shown that AraR binds to eight different operator sites within five different promoters present in the araABDLMNPQ-abfA operon plus araR, araE, abnA and xsa genes (1,2,3). It has been shown that binding of AraR to operator pairs ORA1–ORA2 and ORE1–ORE2 is cooperative and promotes DNA looping (1–3). The loop formation prevents transcription initiation and exerts tight repression. In contrast, binding to ORR3 shows no co-operativity and does not result in DNA distortion (3). Consequently, this mode of repression is less effective and allows basal transcription of araR gene.
In the presence of l-arabinose, repression of the genes regulated by AraR is abolished. The transcription of metabolic operon (araABDLMNPQ-abfA) and araE gene increases by 50-fold and that of araR gene by 4-fold (3–6). The consensus sequence recognized by AraR is palindromic 5′ATTTGTACGTACAAAT3′ and is 16 bp in length (1). Although there is high sequence similarity between the eight operators recognized by AraR, the repressor binds to each operator with distinct affinities ranging from 40 nM (ORR3) to 250 nM (ORA1) (1).
AraR comprises of two domains, the smaller N-terminus domain (NTD; 1–68 residues) and a larger C-terminus domain (CTD; 71–362). The CTD is the receptor for l-arabinose and is termed as the effector domain (8). The structure of this domain in complex with l-arabinose was determined recently and shows that the CTD mediates homodimerization of AraR (9). Random and site-directed mutagenesis experiments and in vivo effects of amino acid substitution have shown that the interaction of AraR with DNA is mediated by the NTD (7,8). We have determined crystal structures of AraR-NTD (ArRNTD) in complex with two different operators, ORA1 (5′-AAAATTGTTCGTACAAATATT-3′) and ORR3 (5′-AAATTTGTCCGTATACATTTT-3′) (Table 1 and Supplementary Figure S1). The structures show that each monomer binds to one core recognition motif present twice in the bipartite operators and also provide a basis for the observed difference in affinities for the two operators. Comparison of the two complexes shows that the spacing between the two core recognition motifs is a critical determinant of the relative orientation of the two monomers on DNA. The implications of this observation for repression by AraR and transcription regulation in general are discussed.
Table 1.
Data collection and Refinement statistics
| ArRNTDORR3 |
ArRNTDORA1, native | ArRNTDmORR3, native | ||
|---|---|---|---|---|
| SeMet | Native | |||
| Wavelength | 0.97864 Å | 1.0 Å | 1.0 Å | 1.0 Å |
| Space group | P43 | P43 | C2 | C2 |
| Cell constants (Å) | 46, 46, 171.9 | 46.2, 46.2, 171.2 | 138.8, 42.4, 67.4 | 137.8, 42.6, 67.4 |
| (°) | 90, 90, 90 | 90, 90, 90 | 90, 114.7, 90 | 90, 114.9, 90 |
| Resolution (Å) | 2.79 (2.9–2.79) Å)a | 2.3 (2.47–2.3) | 2.3 (2.42–2.3) | 1.97 (2.08–1.97) |
| Rsym or Rmergeb | 7.2 (38) | 5.0 (27.9) | 9.0 (37.8) | 7.3 (4.1) |
| I/δI | 19.7(6.0) | 14.1 (3.6) | 6.9 (2.4) | 9.9 (3.0) |
| Completeness (%) | 99.5(96.5) | 99.0 (100) | 97.6 (97.0) | 97.4 (96.1) |
| Redundancy | 10.7 (10.2) | 3.9 (3.8) | 3.0 (3.0) | 4.6 (4.6) |
| Refinement | ||||
| Resolution (Å) | 40–2.30 | 36.0–2.30 | 40–1.97 | |
| No. reflections | 15 670 | 15 886 | 24 593 | |
| Rworkd/Rfreec | 21.9/27.1 | 22.2/26.2 | 21.9/25.3 | |
| No. atoms | ||||
| Protein | 1281 | 1181 | 1181 | |
| DNA | 855 | 855 | 855 | |
| Water | 145 | 103 | 156 | |
| Average B-factors (Å2) | ||||
| Protein: A | 62.2 | 51.5 | 33.5 | |
| Protein: B | 63.8 | 46.2 | 38.4 | |
| DNA | 65.6 | 48.9 | 36.2 | |
| Water | 65.5 | 46.2 | 38 | |
| Rms deviations | ||||
| Bond lengths (Å) | 0.010 | 0.01 | 0.013 | |
| Bond angles (°) | 1.208 | 1.27 | 1.35 | |
aHighest resolution shell is shown in parentheses.
bRmerge = Σ|I – <I>|/ΣI, where I is the integrated intensity of a given reflection.
cRfree was calculated using 5% of data excluded from refinement.
dRwork = Σ||Fobs| − |Fcalc||/Σ|Fobs|.
MATERIALS AND METHODS
ArRNTD cloning, expression and purification
The gene segment corresponding to ArRNTD (residues 1–68) was amplified from genomic DNA (Bangalore Genie) and cloned using EcoR1 and Not1 sites, into a modified pET21b plasmid (Novagen). The gene segment was fused to an upstream six-Histidines-tag with a PreScission-cleavable linker (pDJN1). The fusion protein was expressed in C41DE3 cells induced with 0.8 mM isopropyl-D-thiogalactopyranoside (IPTG) at 18°C. Harvested cells were resuspended in lysis buffer (25 mM Tris pH 8.0, 500 mM NaCl and 5% glycerol) and lysed by sonication. Cell lysate was clarified by centrifugation and was loaded onto a HisTrap column (GE Healthcare). Protein was eluted using a step gradient to 100% elution buffer containing 1 M Imidazole. ArRNTD was further purified by gel filtration (Superdex 75, GE Healthcare). The purified protein was concentrated by centrifugal filtration (Argos Technologies) to 54 mg/ml and flash frozen and stored at −80°C. To prepare selenomethionine-labeled ArRNTD, the protein was expressed in B834 strain of Escherichia coli (methionine auxotroph), and the cells were grown using a Se-Met media kit (Molecular Dimensions). Purification and crystallization of the selenomethionyl-ArRNTD was identical to native protein, except buffers included 5 mM dithiothreitol.
Nucleic acid preparation
Four oligonucleotides corresponding to complementary sequences for the ORA1 and ORR3 (with a T/A overhang) were purchased from Keck Biotechnology Resource Laboratory (Yale University). These oligonucleotides were purified by ion-exchange chromatography (monoQ column) and then desalted (column) and lyophilized. Next, the oligonucleotides were dissolved in appropriate volumes of autoclaved and filtered (0.22 μm) water to achieve a final concentration of 4 mM. Equimolar amounts of complementary oligonucleotides were annealed by heating to 90°C for 5 min followed by cooling to 25°C, and the final concentration of the duplex DNA was 2 mM. Two oligonucleotides corresponding to the modified ORR3 (mORR3) operator were purchased from Sigma Genosys in a purified form, dissolved in sterile autoclaved water and then annealed to obtain a final concentration of 2 mM for duplex DNA.
Crystallization
Complex co-crystals were obtained using vapor diffusion by mixing the duplex DNA and ArRNTD (1–68) in the ratio 1.2:1, with the final concentration of DNA at 0.67 mM. The mixture was incubated on ice for 20 min. The complex crystallized in solutions of PEG 8 K (20%) buffered with 0.1 M sodium acetate (pH = 4.5) and containing 200 mM KCl. Crystals were cryoprotected by serial transfers using increasing concentrations of glycerol from 5% to 25% in steps of 5%. The crystals were flash frozen in liquid nitrogen. Crystallization condition for selenomethionine-labeled ArRNTD in complex with ORR3 was identical to that for the complex with native protein. Also, crystallization condition and the cryoprotection strategy for the ArRNTDORA1 and ArRNTDmORR3 complexes were identical to that for ArRNTDORR3.
Structure determination
The ArRNTDORR3 complex structure was solved by single-wavelength anomalous diffraction (SAD) method using crystals prepared from SeMet-labeled protein. SAD data were collected at peak wavelength (0.97864) of the X-ray absorption spectrum (Table 1) at the European Synchrotron Radiation facility (beamline BM14), Grenoble. In addition, data from native crystals of ArRNTDORR3, ArRNTDORA1 and ArRNTDmORR3 (PXIII, Swiss Light Source) were collected to a maximal resolution of 2.3, 2.3 and 1.97 Å, respectively (Table 1). The data were processed using HKL2000 (10). For the ArRNTDORR3 complex, using the anomalous signal from SeMet, one of the possible two Se sites in the asymmetric unit was located using ShelxD (11). An initial experimental electron density map could be computed using ShelxE using anomalous data up to 3.0 Å. Using this map, the polypeptide chain corresponding to ArRNTD could be built in Coot (12). Next, the DNA model was built using the make-na server (http://structure.usc.edu/make-na/server.html) and was docked into the electron density. The map was improved through iterative cycles of rigid body refinement in CNS (13). The model obtained was used for molecular replacement in MOLREP (14) against the native data set and gave a unique and unambiguous solution. This was followed by iterative cycles of refinement [CNS/PHENIX (15)] and model building (Coot) with constant monitoring of geometrical parameters. The final round of refinement was performed in REFMAC incorporating TLS restraints (16,17). For the final model, the Rfree is 27.1% and Rwork is 21.9% (Table 1).
The ArRNTDORA1 and ArRNTDmORR3 complex structures were solved by molecular replacement in PHASER using one molecule of ArRNTD and double-stranded DNA as the model from the ArRNTDORR3 and ArRNTDORA1 structures, respectively. The second molecule of ArRNTD was docked into the electron density, and map was improved through iterative cycles of rigid body refinement in CNS. Using Coot, the DNA sequence of ORR3 was modified to match that of ORA1 and mORR3 sequences. This was followed by cycles of model building (Coot) refinement (CNS and PHENIX) and water picking (PHENIX). Final rounds of refinement were performed in REFMAC incorporating TLS restraints. The Rfree and Rwork converged to final values of 26.2/22.2% and 25.3/21.9% for the ArRNTDORA1 and ArRNTDmORR3, respectively (Table 1).
Electron density maps of the protein–DNA interface for ArRNTDORA1 and ArRNTDORR3 complexes are shown in Supplementary Figure S1. For the final refined models of the three complexes, PROCHECK (18) revealed that 98% of the residues of the NTD are in the allowed regions of the Ramachandran plot. The structure was analyzed using the CONTACT (CCP4 suite), and area of the surface buried at the interface was calculated using CNS. The bend in the operator DNAs was calculated using CURVES (19) program, and shape complementarity index (Sc) was calculated using the SC program (CCP4 suite) (17,20).
Data deposition
Atomic coordinates and structure factors have been deposited in the PDB with accession codes 4EGY (ArRNTDORA1), 4EGZ (ArRNTDORR3) and 4H0E (ArRNTDmORR3).
RESULTS
Overall structure of ArRNTDORA1 and ArRNTDORR3
We have determined crystal structures of ArRNTD in complex with two different natural operators, ORA1 (5′-AAAATTGTTCGTACAAATATT-3′) and ORR3 (5′-AAATTTGTCCGTATACATTTT-3′) (Table 1 and Supplementary Figure S1). The DNA duplexes in the ArRNTDORA1 and ArRNTDORR3 complexes contain a T/A overhang and pack head to tail to form a pseudo continuous double helix. The duplexes used span from −5 to +15 with respect to the transcription start sites of the respective promoters. In both the complexes, there are two monomers of ArRNTD (monomers A and B) and one molecule of double-stranded DNA in the asymmetric unit (Figure 1). There is no interaction between the two monomers of ArRNTD in either of the complexes (Figure 1B and C). The structures of the two monomers bound to DNA are nearly identical. Monomer A aligns with monomer B with rmsds of 0.25 Å and 0.19 Å for the ArRNTDORA1 and ArRNTDORR3 complexes (68 Cα atoms), respectively. In addition, the structure of ArRNTD is identical in both ArRNTDORA1 and ArRNTDORR3 complexes. The superimposition of monomer A and monomer B of ORA1 onto the corresponding monomers of ORR3 yields rmsds of 0.37 Å and 0.43 Å (68 Cα atoms), respectively.
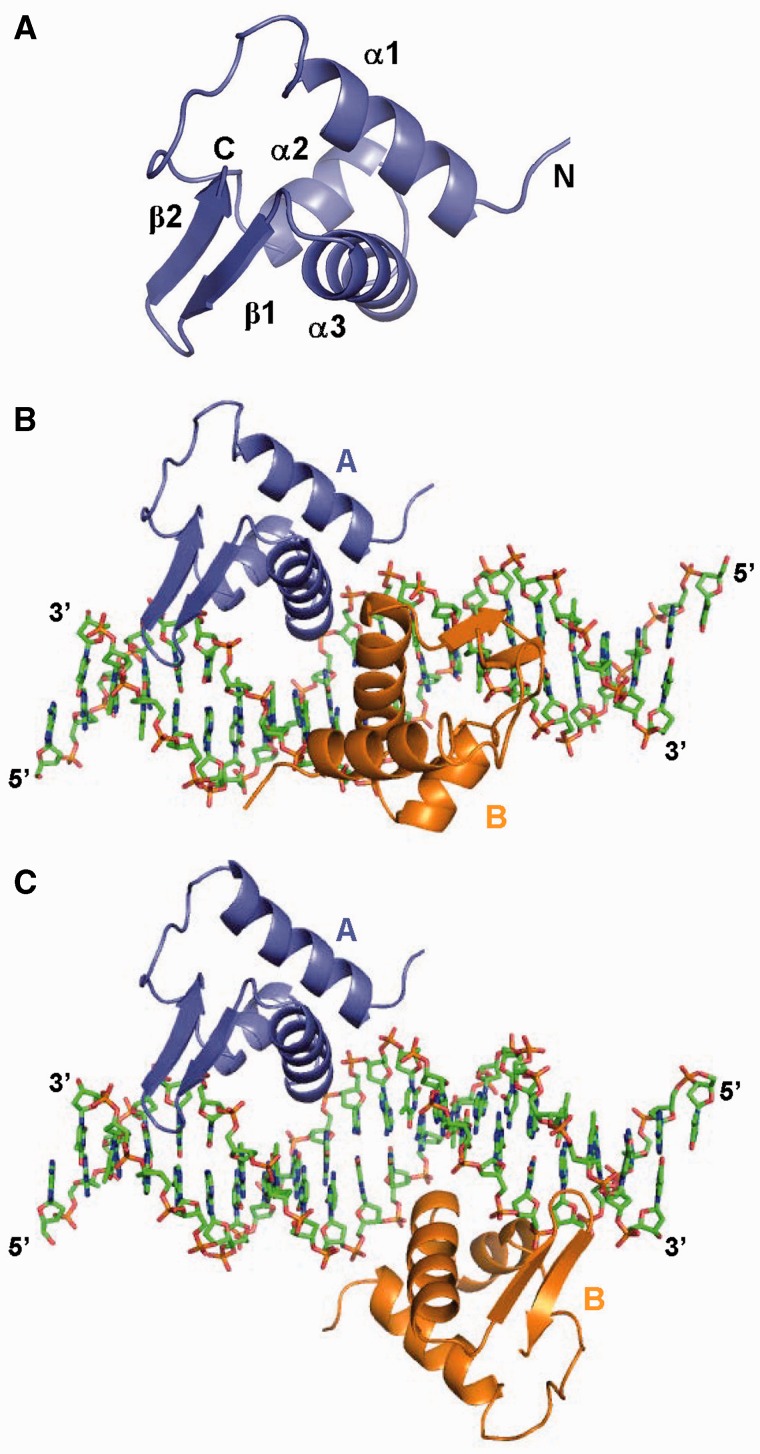
Figure 1.
Structure of ArRNTD-operator DNA complex. (A) Structure of a monomer of ArRNTD shown in ribbon representation with different secondary structural elements labeled. (B) Structure of ArRNTDORA1 complex. Protein is shown as ribbon representation and monomer A colored slate blue and monomer B colored orange. DNA is shown in stick representation and the 5′- and 3′-ends are labeled. (C) Structure of ArRNTDORR3. The figure is color coded as in (B).
ArRNTD monomers show the presence of the characteristic winged helix-turn-helix (HTH) motif (Figure 1A). Each monomer shows three consecutive alpha helices α1 (4–19), α2 (30–38) and α3 (41–54) followed by a β-sheet comprising of two strands β1 (56–61) and β2 (63–67). The HTH motif is formed by α2 and α3 and the latter forms the recognition helix and is oriented towards the major groove. The β-sheet represents the wing domain and residues in the loop (residues 60–63) in the β-sheet form a type II β-turn, which is inserted in the adjacent minor groove (Figure 1B and C).
ArRNTD: ORA1 interactions
The structure of the ArRNTDORA1 complex shows that the interactions of monomer A with DNA are similar to interactions of monomer B. Hence, a description of key interactions is restricted to monomer A (Figure 2A and C). Base-specific interactions are formed between residues of monomer A with nucleotides Ade3′ (Q61:A) and Gua5 (R41:A). R41:A is part of the recognition helix α3 and forms a bidentate hydrogen bond with the Hoogsteen edge of Gua5 (Figure 2A). The importance of this observed interaction is supported by the fact that the mutation Gua5→Thy in the ORA1 has an adverse effect on the affinity for AraR in vitro and also reduces repression by AraR in vivo(7). Q61:A is present in the β-turn of the wing motif that is inserted into the minor groove and interacts with Ade3′. This interaction is essential for function as the mutation Q61→A completely abolished repression activity in vivo and drastically decreased DNA-binding ability in vitro (7). Identical interactions are seen for corresponding residues of monomer B with G12’ (R41:B) and A14 (Q61:B) (Figure 2C).
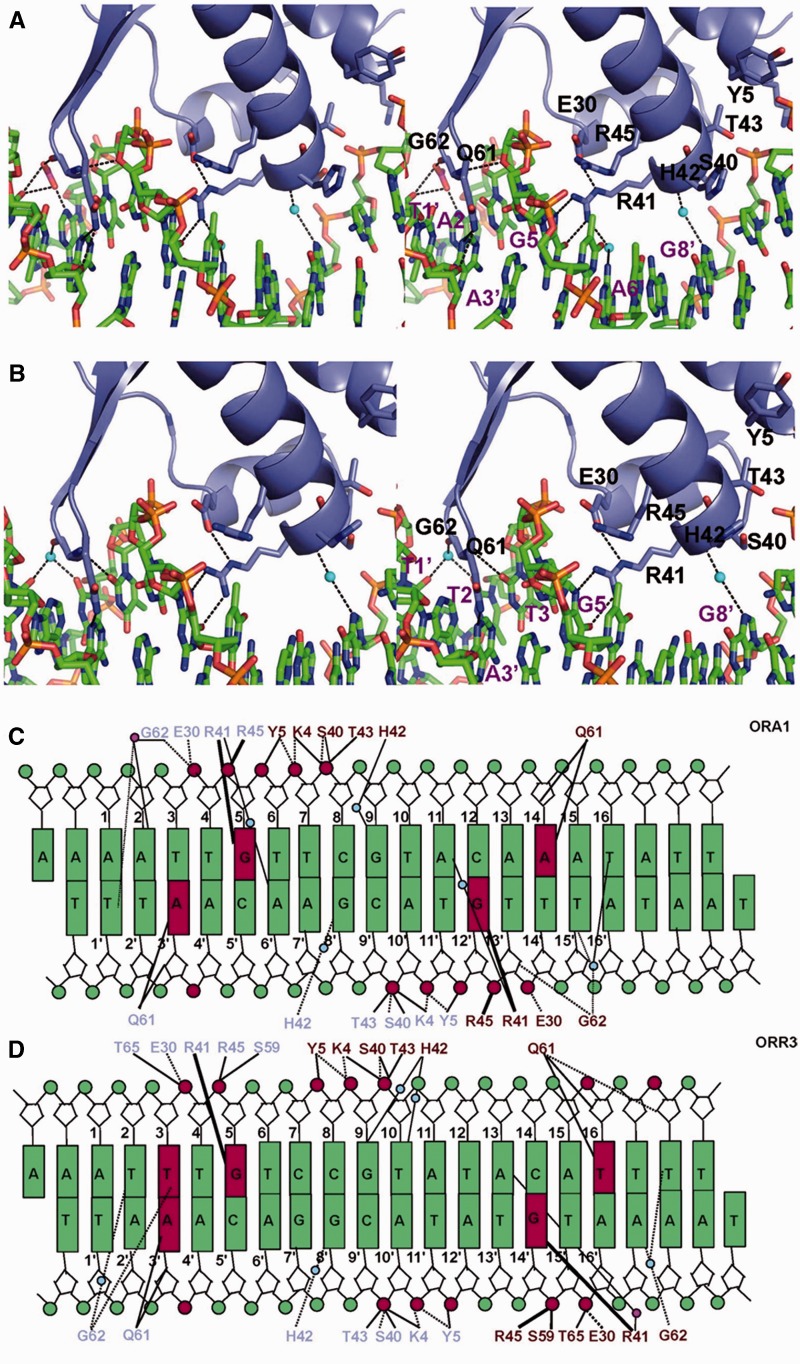
Figure 2.
Protein–DNA interactions. Stereoview of the direct and indirect base-specific interactions formed between AraR and DNA in (A) ArRNTDORA1 and (B) ArRNTDORR3 complexes. The interactions of monomer A are shown in the two complexes. Side chains of the interacting protein residues are displayed. Direct and water-mediated hydrogen bonds (<3.5Å) are shown with dashed lines, and the water molecules are shown as blue spheres. The acetate ion is displayed in stick representation. Schematic representation of the protein–DNA interactions is shown for the (C) ArRNTDORA1 and (D) ArRNTDORR3 complexes. The interacting protein residues from monomer A and monomer B are displayed in slate blue and dark red, respectively. The continuous lines represent the hydrogen bonds between protein side chains and DNA. The dash lines denote the hydrogen bonds with main chain of the protein, and the bold lines represent more than one hydrogen bond with the same residue. Blue and magenta circles represent the water molecules and acetate ions, respectively.
The key interaction that defines specificity of AraR towards cognate DNA is that of R41 with the guanine base. Surprisingly, the R41→A mutation does not have a drastic effect on repression in vivo. However, the mutation of the R45 (present in close proximity to R41) → L/A results in complete loss of regulatory activity (7,8). It is seen that both R41 and R45 interact with residue E30 (Figure 2A and B). The side chain of E30 forms a hydrogen bond with R41 and holds the Arg side chain in position to interact with DNA. In addition, E30 also forms a hydrogen bond with R45, which in turn contacts the DNA phosphate backbone at Gua5. E30 appears to be instrumental in orienting both R45 and R41 to interact with DNA. Consistent with the observed critical role of this residue, the mutation E30→A reduces repression in vivo 5-fold and affects affinity towards ORA1 in vitro by an order of magnitude (7). As a result of their close proximity, it is possible that the guanidinium group of R45 can swing in and substitute for that of R41 in the mutant R41→A and thus rescue function.
Additional base-specific interactions in the form of water-mediated hydrogen bonds are present for Ade6’ (R41:A), Gua8’ (H42:A), Gua9 (H42:B), Ade11 (R41:B), Thy15′ (G62:B) and Thy16 (G62:B) (Figure 2A and C). These water-mediated interactions are important for productive operator binding as the mutations Gua8’→Thy, Gua9→Thy/Ade/Cyt, Ade6’→Cyt, Ade11→Thy/Gua/Cyt and Thy16→Gua in ORA1 adversely affect DNA binding (tested in vitro) and transcriptional repression (tested in vivo) (7). Consistent with the observed interactions for H42, H42→A substitution in AraR led to a decrease in repression in vivo and affinity toward ORA1 in vitro (7). In addition to water-mediated interactions, an acetate ion forms bridging interactions between G62:A and the bases Ade2 and Thy1’ (Figure 2A and C). Also, the two monomers of ArRNTD show similar interactions and direct sequence recognition through base-specific interactions are largely restricted to two TNG recognition motifs (AAAATTGTTCGTACAAATATT) present within the ORA1 operator (Figure 2).
ArRNTD: ORR3 interactions
As in the case of ArRNTDORA1, monomer A of ArRNTDORR3 forms base-specific interactions with Ade3′ (Q61:A) and Gua5 (R41:A) (Figure 2B and D). However, in the case of monomer B, the equivalent interactions are with Thy16 (Q61:B) and Gua14’ (R41:B) (Figure 2D). In ArRNTDORA1, the interacting nucleotides for monomer B were Ade14 and Gua12’. Thus, there is an increase in the spacing between the two recognition motifs (TNG and ANG) within ORR3 by two base pairs (Figure 2C and D). In the case of monomer B of ORR3, the interaction of R41:B with the nucleotide corresponding to Gua12’, i.e. Gua14’ is conserved. In ORR3, at a position analogous to that of Ade3′, Thy16 is present in the other recognition site (Ade14 in ORA1) but Q61:B still forms a base-specific interaction with the thymine base (Figure 2D). Thus, Q61 exhibits plasticity as far as the identity of the interacting base (purine or pyrimidine) is concerned. An additional base-specific interaction is seen in ArRNTDORR3 with Thy3 (G62:A) (Figure 2B and D). An acetate ion forms bridging interactions between R41:B with Ade13 (Figure 2D). Also, base-specific recognition through water-mediated interactions are observed in case of Thy2(G62:A), Gua8’(H42:A), Gua9(H42:B), Thy10(H42:B), Ade17’(G62:B) and Thy18(G62:B) (Figure 2B and D). G62 is located in the minor groove and forms both direct and water-mediated base-specific interactions (Figure 2). This position cannot be occupied by any other residue than glycine as the presence of a side chain will lead to steric clashes with the DNA atoms.
In both ArRNTDORR3 and ArRNTDORA1 complexes, stabilizing hydrogen bonds are formed with the backbone atoms of DNA by residues K4, Y5 and T43 (Figure 2). The non-specific interactions formed by K4 and Y5 are important for function as the mutations K4→A and Y5→F affect repression by AraR up to 30-fold and 3.3-fold, respectively (7). Overall, the comparison of the two complexes shows that critical base-specific interactions formed by R41 and Q61 are present in all four engagements of ArRNTD with DNA (Figure 2C and D). Consequently, direct sequence recognition by AraR is largely restricted to the core T/ANG recognition motif present within the two half sites of each operator (Figures 1 and 3).
Figure 3.
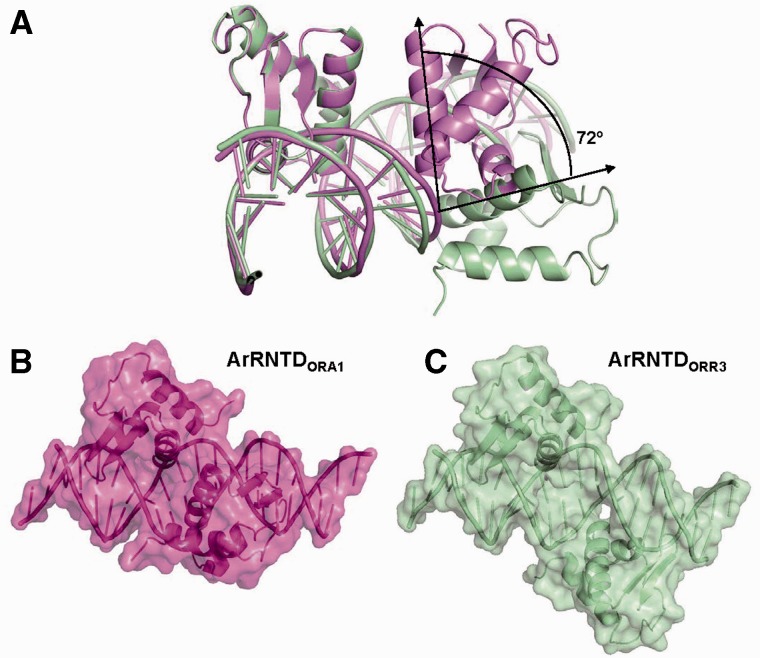
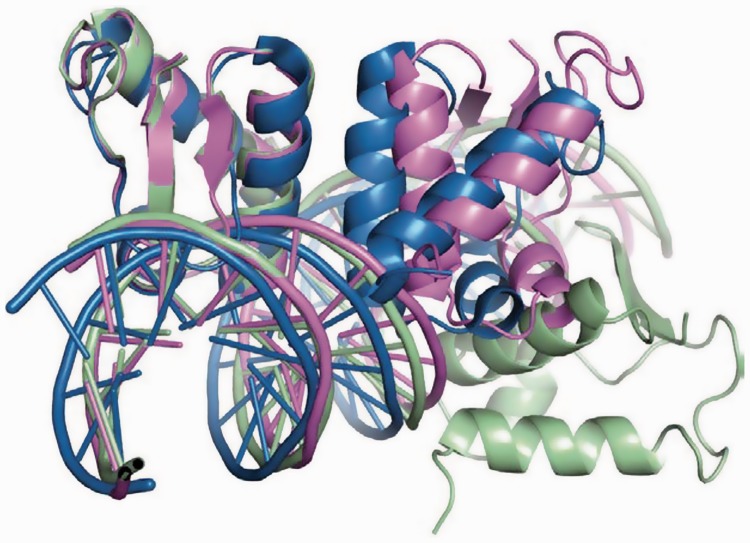
Relative orientation of monomers on DNA. (A) Structural alignment of the ArRNTD–DNA complexes. Monomers A from ArRNTDORA1 and ArRNTDORR3 complexes have been superimposed. The ArRNTDORA1 complex is shown in magenta, and the ArRNTDORR3 complex is shown in green color. The angle between the recognition helix of the B-monomer in the two complexes is shown. The surface of the ArRNTD monomers when bound to DNA is displayed for (B) ArRNTDORA1 and (C) ArRNTDORR3.
Comparison of the structures of ArRNTDORA1 and ArRNTDORR3 complexes
In both the complexes, the area of the interface between each ArRNTD monomer and DNA is approximately the same (∼1750 Å2). The protein and DNA surfaces in the two complexes exhibit similar shape complementarity (Sc values of 0.68) with that for the B monomer of ArRNTDORR3 being marginally higher (0.69). However, the spacing between the core recognition motifs is six bases in the case of ORA1 and eight bases in the case of ORR3. As a result, the relative orientation of the two monomers on DNA is significantly different in the two complexes (Figures 1B and C and 3A). Alignment of the two structures by superimposition of the A monomers shows that monomer B in ArRNTDORR3 spins along the DNA axis further away from monomer A, such that the angle between the α3 helix of the B monomers is 72° (Figure 3A). This corresponds well with the observed rotation of 36° per base for B-DNA.
An additional outcome of the increase in spacing is the difference in the extent to which the helical axis of DNA is bent on ArRNTD binding. The bend is 6.5° in case of ArRNTDORA1 but increases to 20° in the ArRNTDORR3 complex. Although the interactions with the core recognition motif are largely conserved, ArRNTD monomers form more direct hydrogen bonds with ORR3 than with ORA1. There are 34 hydrogen bonds formed between protein and DNA residues in ArRNTDORR3 when compared with 30 in ArRNTDORA1. The direct interactions unique to the ArRNTDORR3 complex are largely through residues present in the wing of monomer B. It appears that the increased spacing in ArRNTD ORR3 improves the grip of the two monomers on DNA resulting in the increased bend of the helical axis and greater number of polar interactions leading to higher affinity. Another important effect of the increase in spacing is the presentation of distinct surfaces in the two nucleoprotein complexes (Figure 3B and C). The Connolly surfaces of the two complexes shows non-uniform features due to the difference in the orientation of monomer B and the differences in the bend of the DNA helical axis.
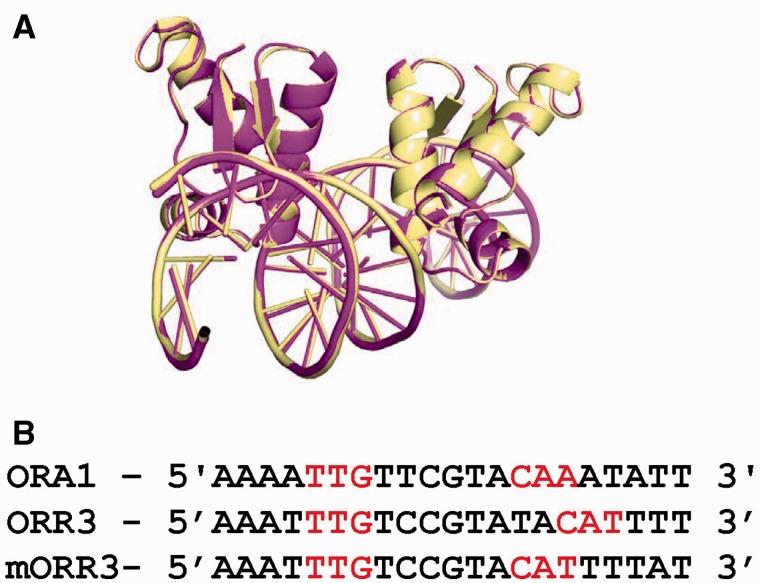
To verify whether the new position of the second monomer is a consequence of the increase in the spacing between the two core recognition motifs, we designed a variant of the ORR3 operator called mORR3. In this modified operator, two nucleotides Ade and Thy at positions 11 and 12 were removed between the core recognition motifs to give rise to a spacing that is equal to that in the case of ORA1 (six bases). The complex ArRNTDmORR3 crystallized in the space group C2 and X-ray diffraction data could be collected to a maximal resolution of 1.97 Å. The structure of the ArRNTDmORR3 complex (Table 1) was identical to that of the ArRNTDORA1 complex and the two structures superimpose with a rmsd of 0.181 Å for 136 Cα atoms (Figure 4). The position of monomer B is identical to that seen in case of the ArRNTDORA1 complex. As expected, the interactions between protein and DNA in the two structures are almost identical with the only difference arising due to the presence of Thy at position 14 in mORR3 when compared with Ade in the case of ORA1. Consequently, in the ArRNTDmORR3 complex, Q61 interacts with the O2 atom of thymine. Overall, this structure proves that the difference in relative orientation of monomer B between ArRNTDORA1 and ArRNTDORR3 is a direct consequence of the different spacing between the core recognition motifs in these two operators.
Figure 4.
Comparison of ArRNTDORA1 and ArRNTDmORR3. (A) Structural alignment of ArRNTDORA1 (magenta) and ArRNTDmORR3 (yellow) complexes. (B) Alignment of the DNA sequences used for crystallization of ArRNTDORA1 (ORA1), ArRNTDORR3 (ORR3) and ArRNTDmORR3 (mORR3). The recognition motif in each case is colored red.
Comparison with FadRFadB complex
The NTD of AraR (ArRNTD) belongs to the GntR family of transcription regulators (21). The GntR family is further subdivided into seven subfamilies (FadR, HutC, MocR, YtrA, DevA, PlmA and AraR), depending on the topologies of the effector binding C-terminal domain (22,23). The only member of the GntR family for which the crystal structure is available in complex with DNA is FadR (fatty acid degradation regulator) (24,25). The DNA-binding domain of FadR recognizes cognate operator FadB (CATCTGGTACGACCAGATC) through a winged helix-turn-helix motif in a manner similar to AraR. The rmsd for superimposition of the NTD of monomer A of FadR (residues 5–72) on monomer A in the ArRNTDORR3 complex is 1.9 Å, (68 Cα atoms). R45 in FadR forms a bidentate hydrogen bond with the Hoogsteen edge of Gua5 (similar to R41 in case of ArRNTD) (24,25). Thus, the recognition of Gua5 through a bidentate hydrogen bond with Arg present in α3 seems to be a conserved feature of this family. In the FadRFadB complex, direct base-specific protein–DNA interactions occur mainly through the TGG recognition motif present in each half site (CATCTGGTACGACCAGATC). The NTD of members of the GntR family is predicted to bind a signature sequence 5′-(N)yGT(N)xAC(N)y-3′ where the number x and y vary (22,23). On the basis of the base-specific interactions observed in structures of the ArRNTDORA1, ArRNTDORR3 and FadRFadB complexes, the signature sequence can be modified to 5′-(N)yTNG-(N)x-CNA/T-(N)y-3′.
Although there is similarity in the overall structure for each monomer of ArRNTD and FadR-NTD bound to DNA, the position of monomer B on DNA with respect to monomer A shows divergence in the two cases (Figure 5). The spacing between the two recognition motifs is 5 bp in the case of FadRFadB complex. Consequently, the two monomers are closer to each other than in case of ArRNTDORA1 and ArRNTDORR3 and α3 of the two monomers interact with each other to form a dimer interface. Overall, a comparison of the three complexes shows that the position and orientation of monomer B with respect to monomer A changes as the spacing between the core recognition motifs varies.
Figure 5.
Comparison with the FadRFadB complex. The structures were aligned through superimposition of monomer A from the ArRNTDORA1 (magenta), ArRNTDORR3 (green) and FadRFadB (blue) complexes.
DISCUSSION
The ArRNTDORA1 and ArRNTDORR3 structures show that the relative orientation of two monomers of a dimeric transcription factor on DNA is a function of the extent of spacing between corresponding recognition motifs in a bipartite operator. The importance of spacing between core binding sites on structure and function has been seen in case of two transcriptional regulators Pit-1 and Oct-1 (26,27). These regulators show the presence of two domains POUh and POUs connected by a flexible linker that are both able to bind to distinct sites on DNA. Pit-1 plays a critical role in ensuring cell-type-specific restriction of growth hormone expression (26). This is primarily achieved through distinct modes of interaction with two bipartite sites that show differences in the spacing between the two half sites. The POUh and POUs domains of Pit-1 are organized differently on binding to the GH-1 site (spacing: 4 bp) and Prl-P1 (spacing: 6 bp). This allows the protein to recruit repressor complex in case of GH-1 site and possibly an activator complex in case Prl-P1 (26). Similarly, for a related transcription factor Oct-1, the POUh and POUs domains exhibit distinct modes of organization on two different cognate sequences—termed MORE and PORE—that allow presentation of distinct protein surfaces probably for recruitment of unique downstream effectors (27). Transcription factors such as p53 and RFX-1 that are known to regulate transcription by binding to a number of different operators wherein the two half sites are separated by spacers of different lengths (28,29). In case of the activator p53, variable spacing in different cognate sequences has been shown to influence affinity of binding to cognate sites (30) and could result in different levels of activation for different genes.
Based on the observations made in case of Pit-1, Oct-1 and p53, it is possible that the two different binding modes of ArRNTD to ORA1 and ORR3 might be exploited to generate different levels of repression. Although there are no interactions between the NTD monomers bound to DNA, the full length protein is expected to bind to the symmetric cognate operators as a dimer formed through interactions between the effector binding domain (8,9). Mota and coworkers (1,3) have shown that binding of AraR to the ORA1–ORA2 and the ORE1–ORE2 pairs is cooperative and ultimately results in the formation of a DNA loop to achieve tight repression. The affinity of AraR for individual operators ORA1 (250 nM) and ORA2 (>250 nM) increases by approximately 8- and >5-fold when the two operators are present together (Kd of 34 nM and 47 nM for ORA1 and ORA2, respectively). Also, in the case of ORE1 and ORE2, the affinity for the individual operators increases by 3- and 2 fold with a decrease in Kd from 108 and 127 nM to 35 and 55 nM, respectively, when the two operators are present together. The authors suggested that co-operative binding and looping occur because the spacing between the ORA1 and ORA2 operators (42 bp) and that between ORE1 and ORE2 operators (43 bp) will place the bound AraR dimers in phase. It is expected that this would allow the formation of stabilizing contacts between the effector domains of AraR dimers bound at each operator. This interaction would lead to an increase in the affinity of binding at the two sites and also facilitate looping. In case of ORR3, the nearest AraR operator ORE1 is 81 bp upstream. Mota and coworkers (1,3) suggest that this places the dimers bound to ORE1 and ORR3 out of phase thereby preventing looping. However, studies probing repression by LacR show that an inter-operator spacing of 81 bp does not present a barrier to looping in vivo (31,32).
The structures presented here raise the possibility that when the two AraR monomers are closer to each other (as in case of ORA1 complex), they attain a dimer organization through their effector domains that presents a distinct molecular surface to facilitate contacts between AraR dimers bound to ORA1–ORA2 or ORE1–ORE2 and hence promote DNA looping. On the other hand, when the two AraR monomers are bound further apart on the operator (as in case of ORR3), the dimer organization of the effector domains is such that interaction with other dimers bound at proximal operators ORE1 is not possible. As a result, looping and the consequent enhanced repression will not happen, and this allows basal transcription of araR gene. In summary, the increased intra-operator spacing observed in case of ORR3 may help avoid adventitious stringent repression of the araR gene. It is vital to allow baseline expression of the araR gene as strong repression would disrupt the regulatory network that prevents expression of corresponding metabolic genes in the absence of arabinose.
A number of transcription factors are known to bind to DNA in a similar manner as AraR- one monomer/dimer binds each one of the two recognition motifs in a symmetric operator. The structures of ArRNTD in complex with ORA1 and ORR3 suggest that condensation in the form of oligomers on double helical DNA allows these transcription factors to amplify small changes in the linear DNA sequence into large changes in three dimensional structure. The difference in the structures probably leads to presentation of distinct surfaces for further oligomerization through space or the recruitment of different trans factors to differentially influence and regulate gene expression. The structures also show that the presentation of the linear sequence of DNA in a double helical form adds another layer to the information resident in DNA. This additional layer can be read and exploited by the same transcription factor to achieve functionally diverse outcomes.
ACCESSION NUMBERS
4EGY, 4EGZ and 4H0E.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure 1.
FUNDING
Department of Atomic Energy, Government of India; Department of Science & Technology [Sanction Order number: SR/FT/LS-122/200, for 3 years starting from 24 September 2009]; Ramanujan fellowship from the Department of Science and Technology, Government of India (to D.T.N.); Young Investigator Grant from Department of Science and Technology, Government of India (to D.J.); the BM14 project—a collaboration between Department of Biotechnology, Government of India, EMBL and ESRF (for data collection at the BM14 beamline of ESRF [Grenoble, France]). Funding for open access charge: Department of Atomic Energy, Government of India.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
D.T.N. thanks the X-ray diffraction facility located in the Molecular Biophysics Unit of the Indian Institute of Science [funded by Department of Biotechnology (DBT) and Department of Science and Technology (DST), Government of India] for facilitating screening and data collection. D.T.N. acknowledges the help rendered by Dr. Hassan Belrhali (ESRF) and Dr. Babu Manjashetty during data collection at the BM-14 beamline of ESRF and by Dr. Meitian Wang during data collection at the PXIII beamline of SLS. Figures were generated using PyMOL (Schroedinger Inc.). The authors thank Prof. Jayant Udgaonkar (NCBS) for critically reading the manuscript.
REFERENCES
- 1.Mota LJ, Tavares P, Sa-Nogueira I. Mode of action of AraR, the key regulator of L-arabinose metabolism in Bacillus subtilis. Mol. Microbiol. 1999;33:476–489. doi: 10.1046/j.1365-2958.1999.01484.x. [DOI] [PubMed] [Google Scholar]
- 2.Raposo MP, Inacio JM, Mota LJ, de Sa-Nogueira I. Transcriptional regulation of genes encoding arabinan-degrading enzymes in Bacillus subtilis. J. Bacteriol. 2004;186:1287–1296. doi: 10.1128/JB.186.5.1287-1296.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mota LJ, Sarmento LM, de Sa-Nogueira I. Control of the arabinose regulon in Bacillus subtilis by AraR in vivo: crucial roles of operators, cooperativity, and DNA looping. J. Bacteriol. 2001;183:4190–4201. doi: 10.1128/JB.183.14.4190-4201.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sa-Nogueira I, Mota LJ. Negative regulation of L-arabinose metabolism in Bacillus subtilis: characterization of the araR (araC) gene. J. Bacteriol. 1997;179:1598–1608. doi: 10.1128/jb.179.5.1598-1608.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sa-Nogueira I, Nogueira TV, Soares S, de Lencastre H. The Bacillus subtilis L-arabinose (ara) operon: nucleotide sequence, genetic organization and expression. Microbiology. 1997;143(Pt 3):957–969. doi: 10.1099/00221287-143-3-957. [DOI] [PubMed] [Google Scholar]
- 6.Sa-Nogueira I, Ramos SS. Cloning, functional analysis, and transcriptional regulation of the Bacillus subtilis araE gene involved in L-arabinose utilization. J. Bacteriol. 1997;179:7705–7711. doi: 10.1128/jb.179.24.7705-7711.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franco IS, Mota LJ, Soares CM, de Sa-Nogueira I. Probing key DNA contacts in AraR-mediated transcriptional repression of the Bacillus subtilis arabinose regulon. Nucleic Acids Res. 2007;35:4755–4766. doi: 10.1093/nar/gkm509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franco IS, Mota LJ, Soares CM, de Sa-Nogueira I. Functional domains of the Bacillus subtilis transcription factor AraR and identification of amino acids important for nucleoprotein complex assembly and effector binding. J. Bacteriol. 2006;188:3024–3036. doi: 10.1128/JB.188.8.3024-3036.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prochazkova K, Cermakova K, Pachl P, Sieglova I, Fabry M, Otwinowski Z, Rezacova P. Structure of the effector-binding domain of the arabinose repressor AraR from Bacillus subtilis. Acta Crystallogr. D Biol. Crystallogr. 2012;68:176–185. doi: 10.1107/S090744491105414X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Z. Otwinowski and Minor, M., (1997) Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology, 276: C.W. Carter, Jr. & R. M. Sweet, Eds., Academic Press (New York) 307–326. [DOI] [PubMed]
- 11.Sheldrick GM. A short history of SHELX. Acta Crystallogr. A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 12.Emsley P, Cowtan K. Coot: model building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 13.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 14.Vagin A, Teplyakov A. MOLREP and automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 15.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 17.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laskowski RA, MacArthur MW, Moss DS, Thornton J. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26 [Google Scholar]
- 19.Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: curves+ Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 21.Haydon DJ, Guest JR. A new family of bacterial regulatory proteins. FEMS Microbiol. Lett. 1991;63:291–295. doi: 10.1016/0378-1097(91)90101-f. [DOI] [PubMed] [Google Scholar]
- 22.Rigali S, Derouaux A, Giannotta F, Dusart J. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J. Biol. Chem. 2002;277:12507–12515. doi: 10.1074/jbc.M110968200. [DOI] [PubMed] [Google Scholar]
- 23.Hoskisson PA, Rigali S. Chapter 1: Variation in form and function the helix-turn-helix regulators of the GntR superfamily. Adv. Appl. Microbiol. 2009;69:1–22. doi: 10.1016/S0065-2164(09)69001-8. [DOI] [PubMed] [Google Scholar]
- 24.Xu Y, Heath RJ, Li Z, Rock CO, White SW. The FadR.DNA complex. Transcriptional control of fatty acid metabolism in Escherichia coli. J. Biol. Chem. 2001;276:17373–17379. doi: 10.1074/jbc.M100195200. [DOI] [PubMed] [Google Scholar]
- 25.van Aalten DM, DiRusso CC, Knudsen J. The structural basis of acyl coenzyme A-dependent regulation of the transcription factor FadR. EMBO J. 2001;20:2041–2050. doi: 10.1093/emboj/20.8.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scully KM, Jacobson EM, Jepsen K, Lunyak V, Viadiu H, Carriere C, Rose DW, Hooshmand F, Aggarwal AK, Rosenfeld MG. Allosteric effects of Pit-1 DNA sites on long-term repression in cell type specification. Science. 2000;290:1127–1131. doi: 10.1126/science.290.5494.1127. [DOI] [PubMed] [Google Scholar]
- 27.Remenyi A, Tomilin A, Pohl E, Lins K, Philippsen A, Reinbold R, Scholer HR, Wilmanns M. Differential dimer activities of the transcription factor Oct-1 by DNA-induced interface swapping. Mol. Cell. 2001;8:569–580. doi: 10.1016/s1097-2765(01)00336-7. [DOI] [PubMed] [Google Scholar]
- 28.Horvath MM, Wang X, Resnick MA, Bell DA. Divergent evolution of human p53 binding sites: cell cycle versus apoptosis. PLoS Genet. 2007;3:e127. doi: 10.1371/journal.pgen.0030127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gajiwala KS, Chen H, Cornille F, Roques BP, Reith W, Mach B, Burley SK. Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature. 2000;403:916–921. doi: 10.1038/35002634. [DOI] [PubMed] [Google Scholar]
- 30.Kitayner M, Rozenberg H, Kessler N, Rabinovich D, Shaulov L, Haran TE, Shakked Z. Structural basis of DNA recognition by p53 tetramers. Mol. Cell. 2006;22:741–753. doi: 10.1016/j.molcel.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 31.Muller J, Barker A, Oehler S, Muller-Hill B. Dimeric lac repressors exhibit phase-dependent co-operativity. J. Mol. Biol. 1998;284:851–857. doi: 10.1006/jmbi.1998.2253. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, McEwen AE, Crothers DM, Levene SD. Analysis of in-vivo LacR-mediated gene repression based on the mechanics of DNA looping. PLoS One. 2006;1:e136. doi: 10.1371/journal.pone.0000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.