Abstract
Regulation of gene expression at the level of translation accounts for up to three orders of magnitude in its efficiency. We systematically compared the impact of several mRNA features on translation initiation at the first gene in an operon with those for the second gene. Experiments were done in a system with internal control based on dual cerulean and red (CER/RFP) fluorescent proteins. We demonstrated significant differences in the efficiency of Shine Dalgarno sequences acting at the leading gene and at the following genes in an operon. The majority of frequent intercistronic arrangements possess medium SD dependence, medium dependence on the preceding cistron translation and efficient stimulation by A/U-rich sequences. The second cistron starting immediately after preceding cistron stop codon displays unusually high dependence on the SD sequence.
INTRODUCTION
Translational control contributes as much as three orders of magnitude to the span of gene expression range (1). Since Shine and Dalgarno’s original discovery (2), a number of mRNA features critical for translation initiation in bacteria has been discovered. Among them are A/U-rich sequences most likely recognized by the ribosomal protein S1 (3), different initiation codons ranging from predominant AUG (4) to exceptional AUU (5,6) and secondary structure elements believed to inhibit or enhance translation depending on their position (1,7–9). In bacteria, an additional layer of complexity and potential for regulation is associated with gene organization into operons. Often, open reading frames overlap with the formation of particular stop and start codons arrangement (10). The efficiency of the following cistron initiation was studied on a limited set of examples (11–13). Systematic comparison of the translation initiation efficiency of leading and following cistrons is lacking.
The aim of the work presented here is a comprehensive and systematic comparison of various mRNA features contributing to the initiation of translation of a single gene and of a gene following another one in an operon in a single experimental system based on dual fluorescent proteins CER and RFP (14). For the following cistron initiation de novo and reinitiation are possible and their relative contribution to overall translation initiation could be distinguished. We found significant differences in the relative contribution of translation initiation region features to translation efficiency in single and polycistronic mRNA. Moreover, we demonstrated the exceptional SD-dependence of the second cistron translation if it follows the leading one without a gap or overlap.
MATERIALS AND METHODS
Strains and media
Escherichia coli strains BW25113 (15) were grown at 37°C in LB media, supplied with 100 µg/ml ampicillin if required. The JM109 E. coli strain was used for cloning procedures.
Plasmids
pRFPCER, the dual fluorescent protein reporter made in our laboratory (14) was used as the host vector for construction reporter plasmids. pRFPCER was digested with NdeI and SacII restriction enzymes, and the obtained linearized vector was directly ligated with pair of pre-annealed complementary oligonucleotides containing different translation initiation regions (Supplementary Table S1). Reporter constructs with bicistronic mRNA were made by PCR, while the region between stop codon of RFP and start codon of CER was replaced by PCR with specific oligonucleotides (Supplementary Table S2).
Dual fluorescent proteins reporter assay in a 96-well plate
Chemically competent cells made from BW25113 strain were aliquoted (50 μl) into a 96-well plate by a Janus (Perkin Elmer) automated workstation and 1 μl of appropriate plasmid (1 ng) was added to each well. Next, the plate was incubated 30 min at 4°C, and after heat-shock (2 min at 44°C), 200 μl of LB were added to each well. After 1 h incubation at 37°C, 20 μl of transformation solution were transferred into the 96-well plate with LB-agar media, supplied with 100 µg/ml ampicillin; this transfer was repeated three times, and the next day three 96-well agar plates for three independent inoculations were obtained. Inoculations were produced by the Janus automated workstation, and cells were grown overnight at 37°C in a 96-well 2 ml (Qiagen) plate with shaking (200 rpm); next cells were twice washed with 0.9% NaCl and the fluorescence of both proteins separately was measured by a Victor X5 2030 (Perkin Elmer) multifunctional reader using appropriate emission/excitation filters (430/486 nm for CER and 531/595 nm for RFP). Standard deviation was derived from at least three parallel independent measurements.
RNA purification
Overnight cell cultures were diluted 100 times and grown in a 96-well plate until all wells reached A590 = 0.4–0.6, then cells were centrifuged and RNA was purified by a SV Total RNA Isolation System (Promega).
cDNA synthesis and RT-PCR
cDNA libraries were constructed by means of a First strand cDNA synthesis kit (Fermentas) with random hexamer primer and RT-PCR reactions were carried out with primers for CER, RFP genes and with ‘RFP-CER’ primers in the case of bicistronic mRNA. Standard deviation was derived from at least three parallel independent reactions.
RESULTS
Dual fluorescent protein reporter for monitoring translation initiation
For this work we used a dual fluorescent protein reporter which was recently developed in our laboratory (14). Two fluorescent proteins, CER (16) and RFP (17), possess readily distinguishable spectral properties which allow their simultaneous measurement in the same bacterial culture. We inserted a set of translation initiation regions in front of CER, while preserving RFP as an internal control. The reporter system we use combines the advantage of internal control taken from dual luciferase reporter systems (18). The usage of fluorescent proteins is advantageous compared to a dual luciferase system, since it does not require any additional reagents, minimizes sample preparation procedures and allows to measure fluorescence directly in living bacterial cells. Both genes were transcribed from identical T5 phage promoters and their fluorescence was normalized using a control plasmid where both fluorescent proteins have similar 5′UTRs. This normalization allowed us to directly compare the efficiencies of translation. In an additional set of constructs we placed both genes under the control of a single T5 phage promoter as in polycistronic mRNA. RFP, as the first gene, was used as a reference; its translation initiation region was identical to that of the first set of constructs. CER followed RFP in the same mRNA in a number of arrangements, starting from 4 nt overlap to the 30 nt spacer between reading frames. When the 3′-end region of the RFP gene was changed in a number of bicistronic constructs, we used separate appropriate control constructs for normalization. Escherichia coli cells, transformed with the set of reporter plasmids, were grown in LB at 37°C to the stationary phase. All the strains reached essentially the same optical density at 590 nm.
Dependence of translation initiation efficiency on the start codon identity
To check the efficiency of the reporters, we applied it to systems which were already described in the literature (19,20). In the majority of genomes, AUG is the most frequent, or even predominant, start codon. In E. coli, other start codons such as GUG and UUG are quite frequent (Figure 1A). In rare, but significant cases, the AUU start codon is utilized, e.g. to begin infC gene coding for translation initiation factor 3 (IF3) (5). In our study, we substituted the AUG start codon with GUG, UUG and AUU codons in a ‘typical’ 4/7 mRNA containing a 4 nt long SD sequence and 7 nt distance between the center of SD and the start codon. The expression of CER driven by the most frequent AUG and GUG start codons was shown to be six to eight times higher than those of rare UUG and AUU codons (Figure 1B). Somewhat surprisingly initiation efficiency was marginally higher for less frequent GUG codon.
Figure 1.
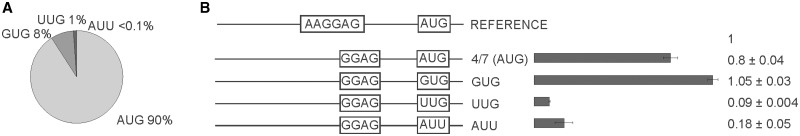
Influence of the start codon on expression efficiency. (A) Circular diagram of the start codon frequency distribution in the genome of E. coli MG1655 (20). (B) Schematic representation (left side of the panel) and translation efficiencies (right side of the panel) of the constructs. All constructs designations are indicated next to the schematic representations. Translation efficiencies of the CER reporter were normalized to the reference RFP construct (shown on top of the panel) and indicated as a diagram. Exact values are shown next to the corresponding bars. All constructs designations are indicated next to the schematic representations. Actual sequences could be found in Supplementary Table S1.
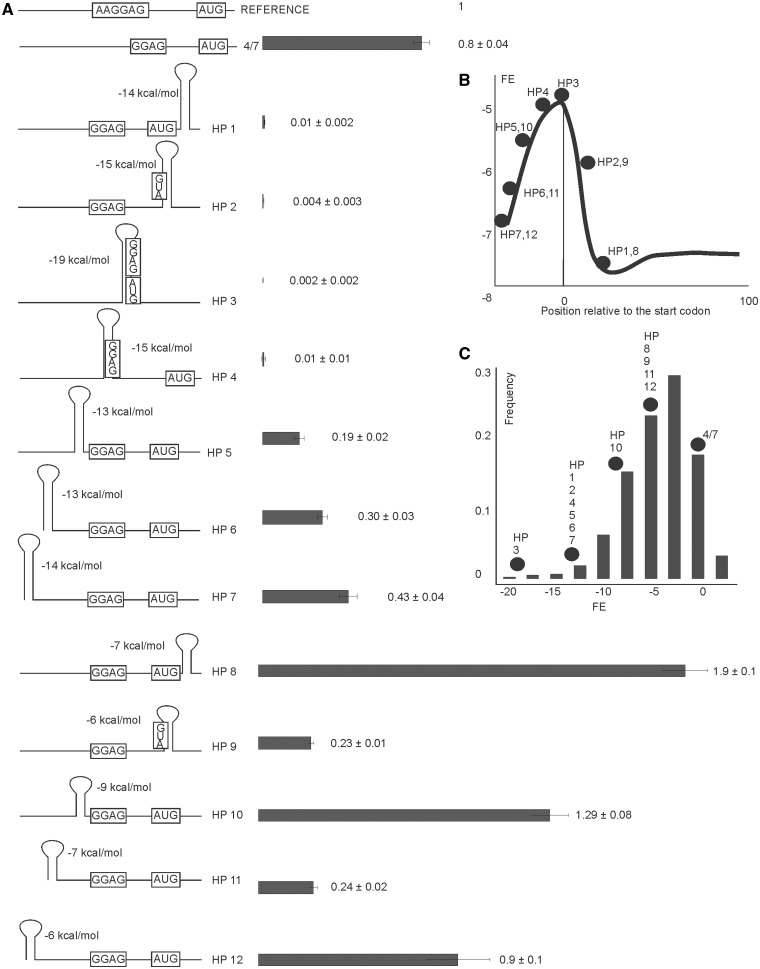
Dependence of translation initiation efficiency on position and strength of the secondary structure elements of the initiation region
Dependence of translation initiation on the mRNA secondary structure surrounding the initiation region was studied previously both experimentally and computationally. Genome-wide computational analysis suggested a low secondary structure content in the region surrounding the start codon in the majority of cellular mRNA, while the region immediately downstream of the start codon possesses on average a more stable secondary structure (Figure 2B) (7,8). Distribution of the mean folding energy (FE) of the secondary structure in the sequence window sliding along mRNA does not by itself say anything about translation efficiency. The most representative experimental study connecting FE with translation efficiency was based on the library of randomized GFP coding regions (1).
Figure 2.
Influence of strength and position of mRNA secondary structure elements in translation initiation region on expression efficiency. (A) Schematic representation (left side of the panel) and translation efficiencies (right side of the panel) of the constructs (similar to those on Figure 1B). All construct designations are indicated next to the schematic representations. Actual sequences could be found in Supplementary Table S1. (B) A plot of mean FE in kcal/mole of all E. coli mRNAs as a function of 40 nt sliding window position relative to the start codon (7). Dots represent the location of the secondary structure elements in the reporter constructs are listed on the panel (A) and marked. (C) Frequency distribution of translation initiation region folding energies in a complete set of E. coli mRNA (7). Dots represent the strength of the secondary structure elements in the reporter constructs are listed on the panel (A) and marked.
In this study, we studied the effects of high- and low-energy hairpin structures at specific positions of mRNA from the 5′-end to the beginning of a coding region (Figure 2A) on translation efficiency. Positions of secondary structure elements analysed here cover the region where the mean FE of the complete genomic mRNA set displays the most significant distortion from that of what is expected at random (Figure 2B) (7). We used two sets of hairpin structures differing in their FE (Figure 2A). The majority of natural mRNAs possesses either no secondary structure, like mRNA 4/7, used here for a comparison of weak secondary structures such as HP8, HP9, HP10, HP11 and HP12 mRNAs (Figure 2C). Highly structured mRNAs, like HP1, HP2, HP3, HP4, HP5, HP6 and HP7 could be found among the complete set of E. coli mRNA only scarcely (Figure 2C) (7). The reporter constructs used in our study represent a complete diversity of secondary structure elements’ strength and locations found in natural mRNA species. All secondary structures designed in this work were confirmed by a computational analysis of the translation initiation region (21), however, we could not totally exclude the possibility that some unexpected long-range secondary structure elements might form with the distant parts of the reporters mRNA.
We observed strong inhibition of translation by the stable hairpins located at any position in the region beginning with SD and ending in the coding region of mRNA (Figure 2A, HP1–4). Stable hairpins located upstream to SD gradually lose their influence on translation with increasing the distance between the hairpin and SD (Figure 2A, HP5–7). Weak hairpins affected translation in a somewhat more complex way. The hairpin covering the AUG codon inhibits translation (Figure 2A, HP9), whereas hairpins surrounding the translation initiation region from both the 5′ and 3′ sites stimulate expression (Figure 2A, HP8, HP10). The hairpin located far upstream of the translation initiation region had no effect on translation (Figure 2A, HP12).
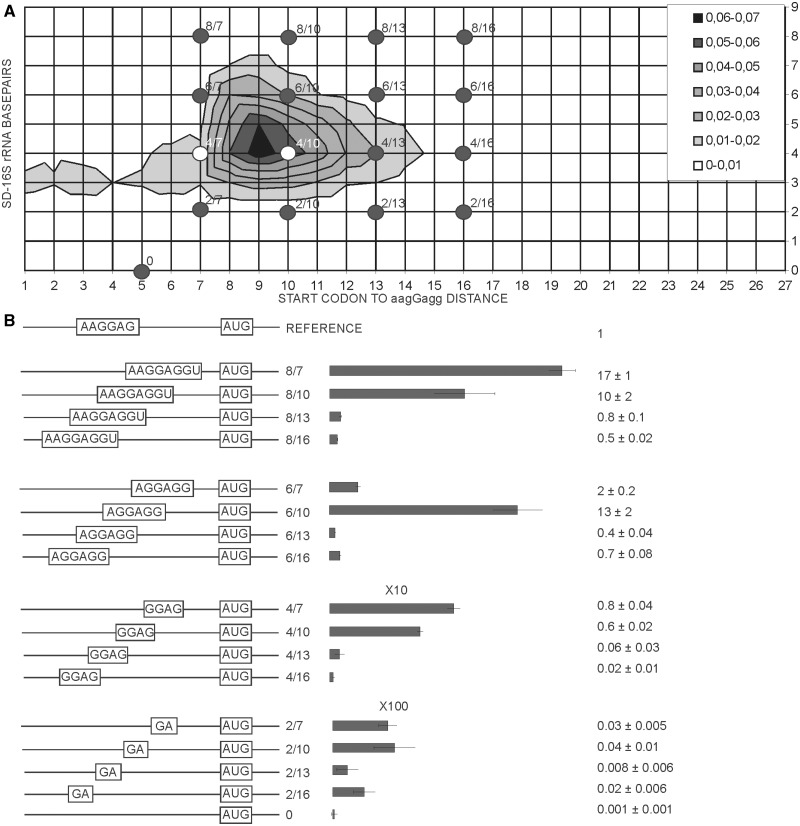
Dependence of translation initiation efficiency on the length of SD and the spacer between SD and start codon
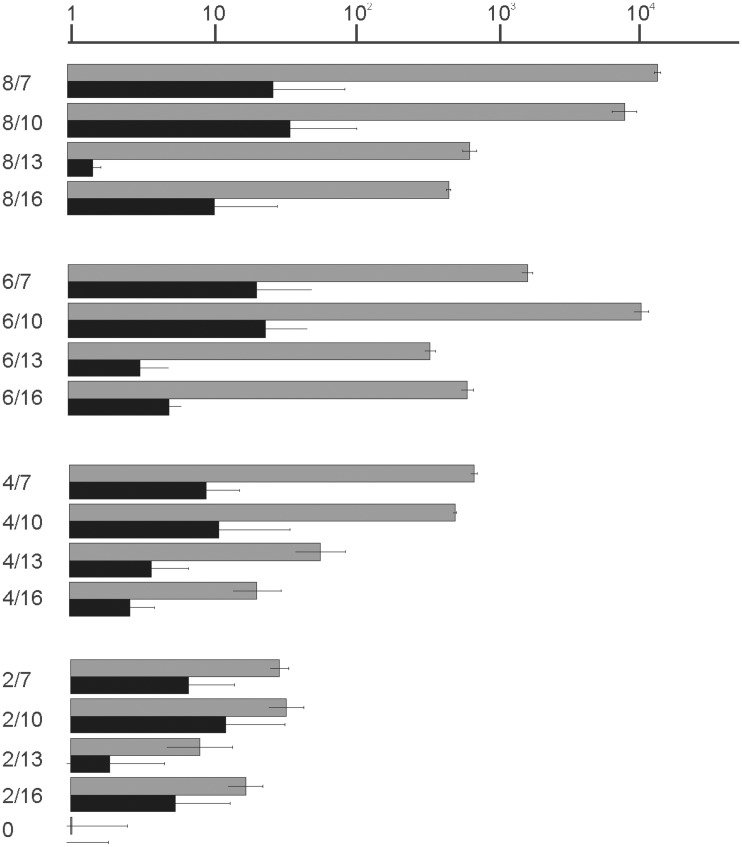
Two degrees of freedom had to be considered in analysis of SD-16S rRNA interaction, SD sequence length and length of the spacer between SD and the start codon. We used a distance between the first nucleotide of the start codon and median guanosine of aagGagg as the length of the spacer. To estimate the frequencies of each of such combinations we constructed the 2D plot using all annotated E. coli genes (Figure 3A). Only rare mRNAs have the SD center closer than 7 nt and farther than 15 nt to the start codon, whereas the complementarity region ranges from 2 to 8 (in a single case 9) nt (Figure 3A).
Figure 3.
Influence of SD sequence length and location on expression efficiency. (A) Distribution plot of SD length in basepairs (y-axis) and spacing from aagGagg to the first nucleotide of the start codon (x-axis). Frequency of occurrence of particular variant of translation initation region in the genome of E. coli MG1655 (22) is indicated by a color. The denser is the hue of gray the more frequent is the variant (see the key in the up right corner). Dots on the plot correspond to the constructs tested in this work. (B) Schematic representation (left side of the panel) and translation efficiencies (right side of the panel) of the constructs. All construct designations are indicated next to the schematic representations. Translation efficiencies of the CER reporter were normalized to the reference RFP construct (shown on top of the panel) and indicated as a diagram. Exact values are shown next to the corresponding bars. A scale was changed as indicated for the bottom two graphs for clarity of presentation. Actual sequences could be found in Supplementary Table S1.
An entire area representing a complete set of natural mRNA variations was evenly covered with a set of 16 model mRNAs (Figure 3A, black dots) numbered accordingly to SD length and distance to the start codon. The 17th mRNA, named ‘0’, contained no complementarity to the 16 S rRNA 3′region and was used as a control. The translation efficiency of mRNAs in the set ranged four orders of magnitude (Figure 3B). We found that depending on the spacer length, either the strongest (Figure 3B, 8/7 versus 6/7, 4/7 and 2/7; 8/13 versus 6/13, 4/13 and 2/13), or the moderately strong (Figure 3B, 6/10 versus 8/10, 4/10 and 2/10; 6/16 versus 8/16, 4/16 and 2/16), SD sequences gave better expression.
Dependence of the second cistron’s translation on mutual arrangement of the cistrons and the SD sequence
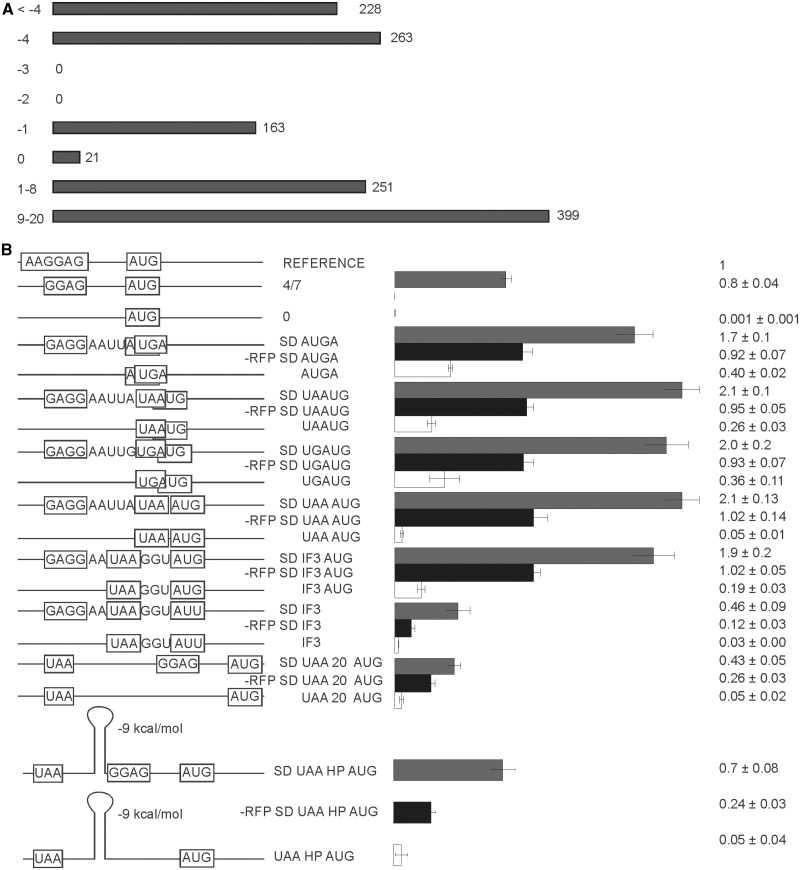
A large proportion of bacterial mRNAs are polycistronic, and often one following cistron overlaps a preceding one (Figure 4A) (10). Given the constraints on the stop and start codon sequences −4 and −1 overlaps are allowed at the expense of non-existing −3 and −2 nt overlaps. Reading frame overlaps by the −4 and −1 nt are quite frequent, while potentially possible juxtapositions of stop and start codons is selected against in real mRNAs (Figure 4A).
Figure 4.
Translation efficiency of second cistron (CER) in a set of bicistronic constructs. (A) Frequency distribution of intercistronic distances among the E. coli genes located on the same mRNA molecules. (B) Schematic representation (left side of the panel) and translation efficiencies (right side of the panel) of the constructs (similar to those on Figure 1B). All construct designations are indicated next to the schematic representations. Actual sequences could be found in Supplementary Tables S1 and S2. ‘SD’ indicates the presence of the Shine-Dalgarno sequence (gray bars). Construct names devoid of ‘SD’ indicate an absence of the Shine-Dalgarno sequence (white bars). ‘-RFP’ indicates premature termination of the first cistron precluding reinitiation (black bars).
Current models for translation initiation of the second cistron include either disassembly of post-termination complex and initiation de novo at the second cistron translation initiation region or migration of the post-termination ribosome or 30S subunit along mRNA in a search for the nearest initiation codon. It is possible that both possibilities could be realized at certain frequencies.
An important case of a 3 nt gap between cistrons corresponds to the infC gene coding for IF3. Another special feature of this gene is the unusual start codon AUU (5). To evaluate the translation efficiency of the second cistron in the context of various mutual arrangements of cistrons, we created a set of reporter constructs (Figure 4B). Eight constructs had the SD sequence while the other eight did not have any. An additional eight constructs had the SD sequence but lacked first gene translation, since RFP expression was inactivated by a premature stop codon at the place corresponding to the 14th amino acid. The complete set of eight reporters which lacked both the RFP expression and the SD sequence in front of CER was not tested in detail, since it had no expression of either fluorescent proteins at a detectable level.
Reading frame overlap by −4 nt can be realized in a single AUGA variant (Figure 4B, SD AUGA, AUGA), while a −1 overlap could be realized in two ways depending on a stop codon (Figure 4B, SD UAAUG, SD UGAUG, UAAUG and UGAUG). We included a rare case of a stop and start codon juxtaposition into the set (Figure 4B, SD UAAAUG, UAAAUG) and a special case, exactly matching the infC start site (Figure 4B, SD IF3). We also created the same reporter where the AUU start codon was replaced by AUG (Figure 4B, SD IF3 AUG). For both constructs we created control constructs without an SD sequence (Figure 4B, IF3 and IF3 AUG). To check the translation efficiency of the more distantly located second cistron, we separated RFP and CER genes along the 20 nt long single-stranded region with or without the SD sequence (Figure 4B, SD UAA 20 AUG and UAA 20 AUG). Reporter constructs containing a hairpin element between cistrons were tested as well (Figure 4B, SD UAA HP AUG, -RFP UAA HP AUG and UAA HP AUG).
All bicistronic constructs which have an SD sequence and an AUG start codon located between −4 and +3 nt relative to the stop codon of a preceding gene are expressed at approximately twice the efficiency of a comparable single cistron reporter (Figure 4B, compare 4/7 with SD AUGA, SD UAAUG, SD UGAUG, SD UAAAUG and SD IF3 AUG). Inhibition of the first cistron translation resulted in approximately a 2-fold drop in translation efficiency of the second cistron (Figure 4B, compare SD AUGA, SD UAAUG, SD UGAUG, SD UAAAUG and SD IF3 AUG with -RFP SD AUGA, -RFP SD UAAUG, -RFP SD UGAUG, -RFP SD UAAAUG and -RFP SD IF3 AUG), making it similar to that of independently translated mRNA (Figure 4B, compare 4/7 with -RFP SD AUGA, -RFP SD UAAUG, -RFP SD UGAUG, -RFP SD UAAAUG and -RFP SD IF3 AUG).
Increasing the distance to the second cistron up to 20 nt led to a significant decrease in translation efficiency in approximately one half of comparable single cistron reporters (Figure 4B, compare 4/7 with SD UAA 20 AUG). The dependence of the translation initiation of the second cistron on translation of the first one (Figure 4B, compare SD UAA 20 AUG with –RFP SD UAA 20 AUG) was less evident than the dependence for overlapped cistrons, but was still significant. Translation initiation efficiency on the natural start site of the infC gene (Figure 4B, SD IF3) was detected at a level twice as much as for a similar single cistron construct (Figure 2B, AUU) and four times lower than for the same construct with the AUG codon (Figure 4B, SD IF3 AUG).
The absence of an SD sequence upstream of the second cistron initiation site significantly reduced translation efficiency of the CER reporter. The translation efficiency of the CER gene started at the AUGA, UAAUG and UGAUG overlaps with the first RFP cistron (Figure 4B, AUGA, UAAUG, UGAUG) displayed three to eight times less efficient translation compared with the SD-containing constructs (Figure 4B, SD AUGA, SD UAAUG, SD UGAUG). While a single cistron reporter devoid of the SD sequence shows negligible translation (Figure 4B, 0) the translation efficiency of the second cistron remained significant, on the level of suboptimal single cistron constructs such as 4/10, 4/13 or 6/13 mRNAs, which are highly represented among natural mRNA species (Figure 1A). Since de novo translation initiation depend so highly on SD (Figure 4B, 0), the translation of the SD-less second cistron could only proceed via reinitiation. Reinitiation efficiency decays with an increase of the distance between the cistrons. At a 20 nt distance between genes efficiency of translation of the SD-less CER gene was 10 times lower than the corresponding SD-containing gene (Figure 4B, compare SD UAA 20 AUG with UAA 20 AUG).
Reinitiation of translation at a distance after the stop codon of a preceding cistron requires ribosome sliding along mRNA in the downstream direction (11). Secondary structure elements between cistrons might block the sliding (23). We introduced an RNA hairpin at an 8 nt distance from the stop codon, just before the SD of the second cistron (Figure 4, SD UAA HP AUG). Insertion of the hairpin did not inhibit translation of the second cistron. According to the control without the translation of the first cistron (Figure 4, -RFP SD UAA HP AUG), more than one half of the initiation events at the second cistron resulted from reinitiation. Thus, the hairpin between the cistrons could be efficiently melted by a sliding ribosome.
One notable exception from the general tendency was observed for the construct containing juxtaposed stop and start codons. The 42 times drop of translation efficiency accompanied the loss of the SD sequence in this case (Figure 4B, compare SD UAAAUG with UAAAUG), while overlap of gap between reading frames does not lead to such dependence of the second cistron translation on SD. It might be a mechanism to avoid translation reinitiation at a codon next to the stop codon of preceding gene. Earlier studies suggested that RRF could serve to prevent such reinitiation events (24).
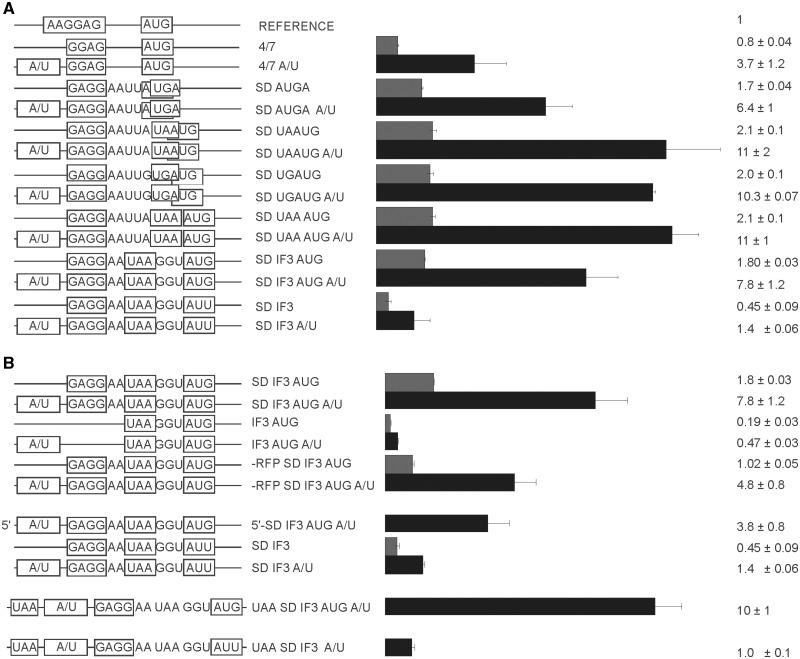
Dependence of translation initiation efficiency on an A/U-rich enhancer sequence for the first and second cistron
A/U-rich sequences contribute to translation efficiency presumably via enhanced interactions with the ribosomal protein S1 (19,25–27). We compared translation efficiencies of mono and bicistronic mRNAs in the presence and absence of an A/U-rich enhancer (Figure 5A). An A/U-rich enhancer derived from the highly expressed phoP gene increased expression of monocistronic mRNA almost five times (Figure 5A, compare 4/7 with 4/7 A/U), in agreement with many previously published observations (3,26). Similar enhancement of the translation efficiency was observed if A/U-rich element was inserted to the 3′-terminal part of the RFP gene in front of the second cistron in a set of reporters having different mutual cistron arrangements (Figure 5A). It should be noted, that insertion of an A/U-rich sequence into the C-terminal part of the RFP gene inhibited maturation of RFP fluorophore presumably due to misfolding of the betta barrel, so in these particular cases we had to normalize CER fluorescence by the cell density.
Figure 5.
Influence of the A/U-rich sequences upstream of the SD on efficiency of expression. (A) Schematic representation (left side of the panel) and translation efficiencies (right side of the panel) of the constructs (similar to those on Figure 1B). All constructs designations are indicated next to the schematic representations. ‘A/U’ indicates the presence of the A/U-rich sequence (black bars). Construct names devoid of ‘A/U’ indicate an absence of A/U-rich sequence (gray bars). (B) A more detailed examination of the stimulatory effect of A/U-rich enhancer dependence on the SD sequence, start codon identity, cistron location and previous cistron translation (if applicable). Schematic representation (left side of the panel) and translation efficiencies (right side of the panel) of the constructs (similar to those on Figure 1B). All construct designations are indicated next to the schematic representations. Actual sequences could be found in Supplementary Tables S1 and S2.
To pursue this further we created additional set of reporters based on IF3 and IF3 AUG constructs to see the interplay between an A/U-rich enhancer, the SD sequence, the start codon and the first cistron translation (Figure 5B). The stimulatory effect of A/U-rich sequences on the translation does not dependent on the SD sequence, the start codon identity and the previous cistron translation (Figure 5B). Additionally, we separated the translation initiation region of the second cistron from the first cistron (Figure 5B, UAA SD IF3 AUG A/U and SD IF3 AUG A/U). In these cases RFP translation ended by the UAA stop codon normally, when it was followed by A/U-rich sequence, the SD sequence and the spacer area identical to that of the SD IF3 AUG A/U construct, but located completely in the intercistronic area (Figure 5B). The sequences similar to these intercistronic area were also introduced on the 5′-UTR of the single cistron constructs (Figure 5B). For both single and bicistronic constructs we also can see an enhancement of the CER gene translation, indicating an efficient stimulation by an A/U-rich sequence. It seems that A/U-rich enhancer stimulate translation independently of the cistron arrangement, cistron order, SD sequence and start codon identity, however the extent of the translation stimulation was higher for efficiently translated mRNAs in agreement with previous findings (26).
Connection between translation and mRNA stability
Previous results suggested that mRNA stability is dependent on the level of its translation (28,29). To check the correlation of the translation level with the RNA stability we monitored RFP and CER mRNA levels when both mRNAs were produced from identical T5 promoters, but differed in translation efficiency (Figure 1B). As expected, the absence of translation (Figure 6A, 0) led to a decrease of the CER mRNA level while very effective translation (Figure 6A, 8/7, 6/7) led to mRNA level increase. The difference between the mRNA abundance was approximately one order of magnitude, whereas translation efficiency ranged four orders of magnitude. Such differences indicate that variability in the CER protein level could not be explained by variability in mRNA abundance and it is likely that variation in translation efficiency caused a difference in mRNA level.
Figure 6.
Comparison of translation efficiency and mRNA quantity for a set of model mRNAs. Shown are mRNA designations (left side) corresponding to those on Figure 1. The graph (right side) indicates expression efficiency in a logarithmic scale as determined by CER/RFP relative fluorescence (gray bars) and mRNA abundance in a logarithmic scale as determined by RT qPCP (black bars).
Additionally, we demonstrated bicistronic RFP-CER mRNA integrity. For all bicistronic constructs the ratio between CER, RFP and RFP-CER products was checked, and no significant differences were found—all differences lay within the margin of error.
DISCUSSION
Shine Dalgarno sequence influence on translation: ‘not that simple’
Since the original discovery in 1974 (2) the influence of SD on translation was studied many times in vivo (25,26,30) and in vitro (31). Experiments (31) carried out with the help of toe-printing method indicated that for the short SD the initiation efficiency is maximal for mRNA which would correspond to 5/8 mRNA in our classification. For long SD initiation efficiency is less sensitive to the spacer length. Most efficient mRNAs in toe-printing assay could be classified as 8/7—8/11 mRNA in our numbering system. Our results expanded upon previous studies since we sampled SD/spacer space evenly, covering almost all natural variations (Figure 3A, see dots corresponding to the reporter constructs). For large SD length, such as 8 nt, short spacers of 7–10 nt were shown to be preferential (Figure 3B, 8/7, 8/10). For the 6 nt SD sequence a short, 7 nt spacer is rather inhibitory, while a longer 10 nt spacer granted highly efficient translation (Figure 3B, compare 6/7 and 6/10) in agreement with previous results (25,26,31).
In the crystal structure of ribosomal complex with a synthetic mRNA containing 8 nt SD and oligoU (32,33) tRNA binds 14 nt downstream of SD. Shortening of the spacer to 9 nt (closest analog in our system is 8/10) led to the shift of the 3′-end region of the 16S rRNA toward the P-site and apparently to a more ‘tense’ conformation. Toe-printing assay of the the 30S complex with tRNALys and mRNA containing oligoA region downstream of 8 nt SD demonstrated that 6–7 nt spacer is optimal (31). Our results correspond better with those obtained by toe-printing (Figure 3B, 8/7).
As suggested by crystal structures (32,33), the spacer region is located in a channel where its conformation is restricted. It could be that composition of the spacer between SD and the start codon play a role in relative translation efficiency of mRNAs with long or short SD (34).
Another factor that should be considered is the efficiency of an SD-16S rRNA helix melting later in initiation or even further at the stage of elongation. This reason was mentioned as an argument for optimal rather than maximal SD length for efficient translation (25). In their study Milon et al. (35) found long SD, but not the short one delayed subunits association in the presence of IF1 and IF3 due to stabilization of IF3 interaction with the 30S subunit.
If mRNA with the long SD is readily engaged in initiation but rather slowly proceeds to elongation it could suppress translation initiation of other mRNAs since it engage a limiting component of translation initiation, such as free 30S subunits (36) or initiation factors. Accordingly, we observed a drop in the reference RFP gene expression relative to the optical density of the cells upon expression of the 8/7 CER construct which led to a high CER/RFP ratio. This might be the reason why such long SD sequences are rarely used in natural mRNAs. Only 18 E. coli mRNAs contain SD of 8 nt length and one, the antitoxin chpS mRNA, possesses SD of 9 nt length. The translation cost of such mRNAs would be higher for the cell. Another reason behind the fact that mRNAs with the small SD are prevalent in the cell (Figure 3A) is that majority of genes do not need exceptionally high expression.
It is obvious to expect that a difference in the mRNA sequence may change mRNA abundance even if it is transcribed from the same promoter (28). In this report we checked mRNA abundance and found it to correlate with translation efficiency. The abundance varied only one order of magnitude (for an extreme case of most inefficiently translated mRNA), while overall expression varied four orders of magnitude (Figure 6). Thus we can rely on the data obtained by the reporter constructs as indicating translation efficiency. Most likely translation efficiency is the cause of mRNA stability change.
The secondary structure of the translation initiation region: sometimes inhibiting, sometimes not
It is a matter of common opinion that the secondary structure could mask the translation initiation region and inhibit translation (1,37). The region surrounding the translation initiation site has in average reduced secondary structure as evidenced by an increase in the mean FE (Figure 2B). Distribution of the FE of the translation initiation site among natural mRNA species (Figure 2C) evidences in favor of no or very weak secondary structures of this region for the majority of mRNA species (7,37). Secondary structures more stable than −10 kcal/mol are present in only a marginal minority of mRNAs (7). In a recent study using a large random set of reporter constructs based on the GFP gene, a negative correlation was found between the strength of the initiation region secondary structure and translation efficiency (1). A study of kinetic parameters of translation initiation process revealed that mRNAs with stable secondary structure are slower in ribosome recruitment (38). An investigation of large genomic datasets revealed a complex distribution of mean FE along mRNA around the translation initiation site (Figure 2B). An initial increase in the mean FE (in weaker secondary structures) in the very start of translation was shown to be followed by a region where secondary structures are on average stronger (Figure 2B). The same region was shown to encode preferentially positively charged amino acids, presumed to slow down growing peptide passage through the peptide channel (8,39). Enrichment of this region by rare codons (with a lower codon adaptation index) added to the creation of the ‘ramp’ region believed to slow down translation for the sake of reduction in the following ribosome ‘traffic jams’ formation (7,8).
In our study, we systematically introduced RNA hairpins along the translation initiation region starting from the very 5′-end of mRNA up to the beginning of the coding region (Figure 2A). The location (Figure 2B) and FE (Figure 2C) of these hairpins covers a natural variety of secondary structure features of this region. The first conclusion from this part of the results was the expected one in that sequestration of SD and the start codon in the secondary structure resulted in a two to three orders of magnitude drop in translation efficiency (Figure 2A). Location of a small hairpin immediately downstream of the start codon did not have such an inhibitory effect in agreement with previous results (40) and even stimulated translation about the 2-fold (Figure 2A, HP8) in agreement of large scale computational analysis (7,8).
Hairpins located upstream of the SD sequence have only moderate inhibitory effects (Figure 2A, HP5–7) that decay steadily with an increase of the distance to SD from 2–14 nt up to the very 5′-end of mRNA (Figure 2A, HP7). In eukaryotic mRNA scanning of mRNA begins from the 5′-end, and it would be tempting to propose similar kind of process at least to contribute to translation initiation mechanism in prokaryotes as well. In bacteria, so far only leaderless mRNAs were shown to bind the ribosome by their 5′-end region (41–43). А hairpin located on the 5′-end of mRNA would occlude direct ribosome binding to this region but should not affect a direct interaction of the 30S subunit with a translation initiation site further downstream. Our results could not exclude some binding to the 5′-end followed by a scanning, but this mechanism can at least be bypassed.
Although the mechanism of scanning from the 5′-end in bacterial systems remains hypothetical and at least not essential, scanning for the next initiation site after termination of translation was described (11). Efficiency of translation reinitiation at the following cistron is known to decay with intercistonic length and the secondary structure (12,44). Here we introduced a weak hairpin between RFP and CER genes located at the same bicistronic mRNA (Figure 4B). A scanning ribosome would require to melt this hairpin to reach the initiation site of the CER gene or initiate translation of the second cistron de novo. As evidenced from the initiation efficiencies, the ribosome almost ignores this secondary structure, despite the fact that more than half of the initiation events of the second cistron could be attributed to the reinitiating ribosomes, while the rest of initiation events at the second cistron is contributed by de novo initiation.
Translation and operons: a balance between reinitiation and initiation de novo
To compare the efficiencies of reinitiation with de novo initiation we created a set of reporter constructs with and without SD sequences in front of the start codon of a second gene and devoid of the first cistron translation (Figure 4B). In the genome of E. coli, a substantial number of genes are located in operons (10). Intergenic distances between the genes located on the same transcripts are far from random (Figure 4A). Cases of an overlap between the stop and the start codons are particularly frequent (Figure 4A, −4 and −1). Due to the limitations applied by the stop and start codons identity, only −1 and −4 overlaps are allowed; the former one as UAAUG and UGAUG and the latter one as AUGA. Despite that no sequence restrictions apply for the juxtaposed stop and start codons, such cases are rare in the genome (Figure 4A, 0).
In our study we made the constructs with AUGA, UAAUG and UGAUG overlaps between RFP and CER reporter genes, juxtaposed stop and start UAAAUG and stop and start separated by 3 and 20 nt (Figure 4B). A special case of the infC gene started with the AUU codon (5) was modeled as the UAA GGU AUU junction (Figure 4B, SD IF3). The comparison of translation efficiency of bicistronic constructs with that of single cistron (Figure 4B 4/7) and with translation efficiency of the second CER cistron without translation of the first RFP gene makes it reasonable to suggest that for the set of AUGA, UAAUG, UGAUG, UAAAUG, UAA GGU AUG (IF3 AUG) and UAA GGU AUU (IF3) stop and start codon arrangement the total translation efficiency is a sum of the equal contribution of translation reinitiation and initiation de novo. Dependence on an SD sequence and independence on the preceding cistron translation is intermediate for the stop and start codons separated by the 20 nt spacer (Figure 4B, compare SD UAA 20 AUG with UAA 20 AUG and SD UAA 20 AUG with -RFP SD UAA 20 AUG). Lack of the first cistron translation led to a 40% drop in translation efficiency of CER. It is reasonable to suggest that such an arrangement of stop and start codons results in a 40/60 percent ratio of reinitiation to de novo initiation of the second cistron. For the juxtaposed stop and start codon arrangement UAAAUG efficiency of the second cistron translation depend substantially on the SD sequence (Figure 4B, compare SD UAAAUG with UAAAUG), much more than for other stop and start codons arrangements like AUGA, UAAUG, UGAUG and UAA GGU AUG (IF3 AUG). High dependence on the SD sequence makes the case of juxtaposed stop and start codons similar to the translation of single cistron mRNA. Notably, natural polycistronic mRNAs try to avoid UAAAUG sites (Figure 4A). It seems that reinitiation at such sites applies more stringent requirements on translation initiation signals.
The driving force of post-termination ribosome movement toward the 3′-end of mRNA is unknown. Is any type of scanning mechanism involved and does it consumes the energy of nucleoside −5′-triphosphate hydrolysis remains to be discovered. Here we demonstrated that a helix of −9 kcal/mol energy does not produce any obstacle for ribosome initiation downstream (Figure 4). The location of the start codon at the 20 nt distance downstream from the stop codon of the preceding cistron leads to a blend of ca. 60% de novo initiation and 40% reinitiation.
A/U-rich sequences enhance translation independently of other mRNA sequence elements
A/U-rich enhancers are located upstream of the SD sequences and increase translation efficiency by an order of magnitude (3,19,26). The influence of SD and enhancers is not additive but synergistic; most efficient SD sequences benefit predominantly from addition of the enhancer (26).
To check whether A/U-rich sequences affect the first and second cistron translation differently we inserted the A/U-rich enhancer sequence UAUUUUAAUAAUUAA from phoP mRNA upstream of the model mRNA 4/7 (Figure 5A, 4/7 A/U) and upstream of the initiation region of the second cistron in a number of our bicistronic constructs (Figure 5A). We found a 4.6-fold increase in translation efficiency by the A/U-rich enhancer in a single cistron construct (Figure 5A, compare 4/7 and 4/7 A/U). A putative A/U-rich enhancer inserted upstream of the second cistron stimulated translation in all mutual cistron arrangements (Figure 5A). Further analysis (Figure 5B) revealed that stimulation of translation by A/U-rich enhancer could not be eliminated by inactivation of the preceding cistron translation, lack of SD sequence and inefficient start codon. However, the extent of stimulation was higher for efficient translation initiation regions. For the case of the AUG start codon, the presence of SD and efficient preceding gene translation (Figure 5B, SD IF3 AUG) addition of A/U enhancer sequence increased translation by a factor of 4.3 (Figure 5B, SD IF3 AUG), from 1.8 to 7.8 relative units. Lack of SD sequence diminishes translation (Figure 5B, IF3 AUG), but addition of A/U enhancer (Figure 5B, IF3 AUG A/U) still contributed 2.5-fold to translation efficiency, from 0.19 to 0.47 relative units. Lack of preceding gene translation (Figure 5B, -RFP SD IF3 AUG) caused by a premature stop codon in RFP gene, or complete removal of preceding gene diminished translation of a reporter by one half, but addition of A/U sequence increased translation 3.7–4.7 times (Figure 5B, -RFP SD IF3 AUG A/U, 5′-SD IF3 AUG A/U). Inefficient AUU start codon decreased translation yield (Figure 5B, SD IF3), but A/U-rich enhancer activates translation 3.1 times (Figure 5B, SD IF3 A/U). Further testing (Figure 5B, UAA SD IF3 AUG A/U and UAA SD IF3 A/U) revealed that A/U-rich enhancer is still active even when RFP and CER genes were not overlapped, but are separated by a spacer.
CONCLUSIONS
We explored several mRNA features affecting translation initiation and reinitiation in a single experimental system, making possible a direct comparison of their contributions. We demonstrated that secondary structure elements are most inhibiting when sequestering the initiation codon and the SD sequence of the first cistron, while hairpins located further in the 5′ UTR of the single cistron or in the area between cistrons are rather irrelevant for translation. A/U-rich translation enhancers act independent from SD sequence and start codon identity, cistron location at a first or second place in an operon and independent on the second cistron arrangement relative to the first one. The efficiency of the following cistron translation moderately depend on SD and benefits highly from preceding cistron translation. However, the rarely found exact cistron juxtaposition depends much more on SD, which is indicative of more stringent requirements for translation initiation site in this case.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2.
FUNDING
Russian Foundation for Basic Research [10-04-01345-a, 11-04-01314-a, 11-04-01018-a, 11-04-12060-ofi and 11-04-91337-nnio-a]; Russian Ministry of Science [16.512.11.2108]; Federal Agency for Science and Innovations [02.740.11.0706]; Moscow University Development Program [PNR 5.13]. Funding for open access charge: Personal fund.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Authors are very thankful to Dr Alexey A. Bogdanov for valuable discussions and support and to Alexander Lebedeff for improvement of the language of the manuscript.
REFERENCES
- 1.Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl Acad. Sci. USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boni IV, Isaeva DM, Musychenko ML, Tzareva NV. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 1991;19:155–162. doi: 10.1093/nar/19.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dreyfus M. What constitutes the signal for the initiation of protein synthesis on Escherichia coli mRNAs? J. Mol. Biol. 1988;204:79–94. doi: 10.1016/0022-2836(88)90601-8. [DOI] [PubMed] [Google Scholar]
- 5.Butler JS, Springer M, Dondon J, Graffe M, Grunberg-Manago M. Escherichia coli protein synthesis initiation factor IF3 controls its own gene expression at the translational level in vivo. J. Mol. Biol. 1986;192:767–780. doi: 10.1016/0022-2836(86)90027-6. [DOI] [PubMed] [Google Scholar]
- 6.Binns N, Masters M. Expression of the Escherichia coli pcnB gene is translationally limited using an inefficient start codon: a second chromosomal example of translation initiated at AUU. Mol. Microbiol. 2002;44:1287–1298. doi: 10.1046/j.1365-2958.2002.02945.x. [DOI] [PubMed] [Google Scholar]
- 7.Tuller T, Waldman YY, Kupiec M, Ruppin E. Translation efficiency is determined by both codon bias and folding energy. Proc. Natl Acad. Sci. USA. 2010;107:3645–3650. doi: 10.1073/pnas.0909910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tuller T, Veksler-Lublinsky I, Gazit N, Kupiec M, Ruppin E, Ziv-Ukelson M. Composite effects of gene determinants on the translation speed and density of ribosomes. Genome Biol. 2011;12:R110. doi: 10.1186/gb-2011-12-11-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iserentant D, Fiers W. Secondary structure of mRNA and efficiency of translation initiation. Gene. 1980;9:1–12. doi: 10.1016/0378-1119(80)90163-8. [DOI] [PubMed] [Google Scholar]
- 10.Salgado H, Moreno-Hagelsieb G, Smith TF, Collado-Vides J. Operons in Escherichia coli: genomic analyses and predictions. Proc. Natl Acad. Sci. USA. 2000;97:6652–6657. doi: 10.1073/pnas.110147297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adhin MR, van Duin J. Scanning model for translational reinitiation in eubacteria. J. Mol. Biol. 1990;213:811–818. doi: 10.1016/S0022-2836(05)80265-7. [DOI] [PubMed] [Google Scholar]
- 12.Spanjaard RA, van Duin J. Translational reinitiation in the presence and absence of a Shine and Dalgarno sequence. Nucleic Acids Res. 1989;17:5501–5507. doi: 10.1093/nar/17.14.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andre A, Puca A, Sansone F, Brandi A, Antico G, Calogero RA. Reinitiation of protein synthesis in Escherichia coli can be induced by mRNA cis-elements unrelated to canonical translation initiation signals. FEBS Lett. 2000;468:73–78. doi: 10.1016/s0014-5793(00)01198-4. [DOI] [PubMed] [Google Scholar]
- 14.Osterman IA, Prokhorova IV, Sysoev VO, Boykova YV, Efremenkova OV, Svetlov MS, Kolb VA, Bogdanov AA, Sergiev PV, Dontsova OA. Attenuation-based dual-fluorescent-protein reporter for screening translation inhibitors. Antimicrob. Agents Chemother. 2012;56:1774–1783. doi: 10.1128/AAC.05395-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 17.Merzlyak EM, Goedhart J, Shcherbo D, Bulina ME, Shcheglov AS, Fradkov AF, Gaintzeva A, Lukyanov KA, Lukyanov S, Gadella TW, et al. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat. Methods. 2007;4:555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- 18.Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang JR, Deutscher MP. Analysis of the upstream region of the Escherichia coli rnd gene encoding RNase D. Evidence for translational regulation of a putative tRNA processing enzyme. J. Biol. Chem. 1989;264:18228–18233. [PubMed] [Google Scholar]
- 20.O'Donnell SM, Janssen GR. The initiation codon affects ribosome binding and translational efficiency in Escherichia coli of cI mRNA with or without the 5′ untranslated leader. J. Bacteriol. 2001;183:1277–1283. doi: 10.1128/JB.183.4.1277-1283.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 23.Jin H, Zhao Q, Gonzalez de Valdivia EI, Ardell DH, Stenstrom M, Isaksson LA. Influences on gene expression in vivo by a Shine-Dalgarno sequence. Mol. Microbiol. 2006;60:480–492. doi: 10.1111/j.1365-2958.2006.05110.x. [DOI] [PubMed] [Google Scholar]
- 24.Janosi L, Ricker R, Kaji A. Dual functions of ribosome recycling factor in protein biosynthesis: disassembling the termination complex and preventing translational errors. Biochimie. 1996;78:959–969. doi: 10.1016/s0300-9084(97)86718-1. [DOI] [PubMed] [Google Scholar]
- 25.Komarova AV, Tchufistova LS, Supina EV, Boni IV. Protein S1 counteracts the inhibitory effect of the extended Shine-Dalgarno sequence on translation. RNA. 2002;8:1137–1147. doi: 10.1017/s1355838202029990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vimberg V, Tats A, Remm M, Tenson T. Translation initiation region sequence preferences in Escherichia coli. BMC Mol. Biol. 2007;8:100. doi: 10.1186/1471-2199-8-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tzareva NV, Makhno VI, Boni IV. Ribosome-messenger recognition in the absence of the Shine-Dalgarno interactions. FEBS Lett. 1994;337:189–194. doi: 10.1016/0014-5793(94)80271-8. [DOI] [PubMed] [Google Scholar]
- 28.Komarova AV, Tchufistova LS, Dreyfus M, Boni IV. AU-rich sequences within 5′ untranslated leaders enhance translation and stabilize mRNA in Escherichia coli. J. Bacteriol. 2005;187:1344–1349. doi: 10.1128/JB.187.4.1344-1349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morse DE, Mosteller RD, Yanofsky C. Dynamics of synthesis, translation, and degradation of trp operon messenger RNA in E. coli. Cold Spring Harb. Symp. Quant. Biol. 1969;34:725–740. doi: 10.1101/sqb.1969.034.01.082. [DOI] [PubMed] [Google Scholar]
- 30.Lee K, Holland-Staley CA, Cunningham PR. Genetic analysis of the Shine-Dalgarno interaction: selection of alternative functional mRNA-rRNA combinations. RNA. 1996;2:1270–1285. [PMC free article] [PubMed] [Google Scholar]
- 31.Hartz D, McPheeters DS, Gold L. Influence of mRNA determinants on translation initiation in Escherichia coli. J. Mol. Biol. 1991;218:83–97. doi: 10.1016/0022-2836(91)90875-7. [DOI] [PubMed] [Google Scholar]
- 32.Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- 33.Jenner LB, Demeshkina N, Yusupova G, Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 2010;17:555–560. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- 34.de Boer HA, Comstock LJ, Hui A, Wong E, Vasser M. Portable Shine-Dalgarno regions; nucleotides between the Shine-Dalgarno sequence and the start codon affect the translation efficiency. Gene Amplif. Anal. 1983;3:103–116. [PubMed] [Google Scholar]
- 35.Milon P, Konevega AL, Gualerzi CO, Rodnina MV. Kinetic checkpoint at a late step in translation initiation. Mol. Cell. 2008;30:712–720. doi: 10.1016/j.molcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 36.Nomura M, Gourse R, Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu. Rev. Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- 37.Allert M, Cox JC, Hellinga HW. Multifactorial determinants of protein expression in prokaryotic open reading frames. J. Mol. Biol. 2010;402:905–918. doi: 10.1016/j.jmb.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Milon P, Maracci C, Filonava L, Gualerzi CO, Rodnina MV. Real-time assembly landscape of bacterial 30S translation initiation complex. Nat. Struct. Mol. Biol. 2012;19:609–615. doi: 10.1038/nsmb.2285. [DOI] [PubMed] [Google Scholar]
- 39.Lu J, Deutsch C. Electrostatics in the ribosomal tunnel modulate chain elongation rates. J. Mol. Biol. 2008;384:73–86. doi: 10.1016/j.jmb.2008.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balakin A, Skripkin E, Shatsky I, Bogdanov A. Transition of the mRNA sequence downstream from the initiation codon into a single-stranded conformation is strongly promoted by binding of the initiator tRNA. Biochim. Biophys. Acta. 1990;1050:119–123. doi: 10.1016/0167-4781(90)90151-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balakin AG, Skripkin EA, Shatsky IN, Bogdanov AA. Unusual ribosome binding properties of mRNA encoding bacteriophage lambda repressor. Nucleic Acids Res. 1992;20:563–571. doi: 10.1093/nar/20.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andreev DE, Terenin IM, Dunaevsky YE, Dmitriev SE, Shatsky IN. A leaderless mRNA can bind to mammalian 80S ribosomes and direct polypeptide synthesis in the absence of translation initiation factors. Mol. Cell. Biol. 2006;26:3164–3169. doi: 10.1128/MCB.26.8.3164-3169.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moll I, Grill S, Gualerzi CO, Blasi U. Leaderless mRNAs in bacteria: surprises in ribosomal recruitment and translational control. Mol. Microbiol. 2002;43:239–246. doi: 10.1046/j.1365-2958.2002.02739.x. [DOI] [PubMed] [Google Scholar]
- 44.Karamyshev AL, Karamysheva ZN, Yamami T, Ito K, Nakamura Y. Transient idling of posttermination ribosomes ready to reinitiate protein synthesis. Biochimie. 2004;86:933–938. doi: 10.1016/j.biochi.2004.08.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.