Abstract
The large terminase subunit is a central component of the genome packaging motor from tailed bacteriophages and herpes viruses. This two-domain enzyme has an N-terminal ATPase activity that fuels DNA translocation during packaging and a C-terminal nuclease activity required for initiation and termination of the packaging cycle. Here, we report that bacteriophage SPP1 large terminase (gp2) is a metal-dependent nuclease whose stability and activity are strongly and preferentially enhanced by Mn2+ ions. Mutation of conserved residues that coordinate Mn2+ ions in the nuclease catalytic site affect the metal-induced gp2 stabilization and impair both gp2-specific cleavage at the packaging initiation site pac and unspecific nuclease activity. Several of these mutations block also DNA encapsidation without affecting ATP hydrolysis or gp2 C-terminus binding to the procapsid portal vertex. The data are consistent with a mechanism in which the nuclease domain bound to the portal switches between nuclease activity and a coordinated action with the ATPase domain for DNA translocation. This switch of activities of the nuclease domain is critical to achieve the viral chromosome packaging cycle.
INTRODUCTION
Nucleases hydrolyze the phosphodiester linkages present in nucleic acids. These enzymes are implicated in numerous DNA transactions such as DNA replication, repair and recombination. Many nucleases possess other associated catalytic properties such as DNA polymerase, ligase, helicase or kinase activities that combine together to achieve specific biological functions. One of the diverse biological processes where nucleases play a fundamental role is viral maturation. In most tailed double-stranded DNA (dsDNA) bacteriophages and herpes viruses, DNA is replicated as concatemers that are the substrate for encapsidation into preformed procapsids (1–3). Specific recognition and cleavage of the viral concatemeric, DNA during the packaging process is mediated by the terminase, a hetero-oligomeric complex composed in most phages of small and large subunits. Besides cleavage of the concatemeric DNA substrate, the terminase assembles at the procapsid portal vertex and possesses an ATPase activity necessary to fuel DNA translocation into the procapsid during the packaging process (4). The reaction is normally terminated by endonucleolytic cleavage of the concatemer and initiation of a subsequent packaging cycle at another procapsid. The process yields up to 12 or more encapsidation cycles per concatemer (5,6) revealing the high processivity of the viral packaging motor. The terminase large subunit is a multifunctional enzyme whose nuclease activity is required for an initial, most frequently sequence-specific DNA cleavage, followed by a switch to ATPase activity that powers dsDNA translocation. A switch back to nuclease activity in response to the level of DNA filling of the capsid achieves the dsDNA termination cut ending the packaging cycle. This cleavage is either sequence-specific or sequence-independent (headful packaging mechanism) depending on the viral system (7).
Structural information is available for the complete large terminase from phage T4 (8) and for nuclease domains from phages RB49 (8), SPP1 (9), P22 (10) and also nuclease domain from the human cytomegalovirus (HCMV-1) (11). The N-terminal ATPase domain is related to monomeric helicases, while the carboxyl terminal nuclease domain exhibits an RNase H fold. Structural conservation suggests a common, yet unknown mechanism, for coordination of the critical switches between nuclease and ATPase activities during the viral genome packaging cycle of tailed phages and herpes viruses.
The Bacillus subtilis bacteriophage SPP1 is a well-established model system for viruses that package their dsDNA by a headful packaging mechanism. The SPP1 terminase small subunit gp1 provides specificity for binding to the pac sequence in the DNA concatemer (12,13) whereas the large subunit, gp2, displays nuclease and ATPase activities (14–16). Gp2, which is a 422-amino acid long polypeptide, is a monomer in solution consisting of an N-terminal ATPase domain and a C-terminal nuclease domain (14). Both ATPase and nuclease activities are modulated by gp1 and the portal protein gp6 (15,17). The gp6 portal, at the procapsid, provides the interface for assembly of the packaging motor through a direct interaction with gp2 (18). Motor pumping of dsDNA to the procapsid interior through the gp6 central channel (19) relies on a complex cross-talk between gp1, gp2 and gp6 and conformational changes in the portal region surrounding the central DNA conduit channel (17,20). Genetic evidence shows that gp6 is also central for the sensor mechanism that measures the level of DNA capsid filling to trigger the switch of gp2 activity to exert the sequence-independent endonucleolytic cleavage that terminates the packaging cycle (21,22).
We report that gp2 nuclease activity and stability are modulated by divalent cations being preferentially enhanced by Mn2+ ions. We show that amino acid substitutions in the nuclease catalytic site impair both gp2-specific cleavage at pac and its unspecific nuclease activity but neither affect binding to the capsid portal vertex nor its ATPase activity. Significantly, several of these mutations block linear DNA packaging in vitro revealing that a correctly structured gp2 nuclease center is critical for the mechanism of DNA translocation into viral capsids.
MATERIALS AND METHODS
Enzymes and reagents
All chemicals used in this study were of p.a. grade. Sybr Gold was from Molecular Probes. Protein molecular weight markers and DNA restriction enzymes were from Biolabs. Proteinase K was from Roche. Lysozyme, DNase and Dextran were from Sigma. Adenosine-5′-triphosphate was purchased from Roche. [γ-32P]ATP was from Perkin Elmer.
Bacterial strains, bacteriophages and plasmids
SPP1 suppressor-sensitive mutants (sus70, sus19 and sus70sus115) were described previously (12,21,22). Bacillus subtilis HA101B (sup-3) and YB886 (sup°) were the permissive and non-permissive strains used for SPP1 amplification (12). The Escherichia coli strains used for production of the terminase subunits gp2 and gp1 were BL21(DE3) and BL21(DE3) (pLysS), respectively (23). Plasmids pCB191 (24), pBT115 (12,16) and pBT163 (12) have been described. Expression was controlled by the LacI repressor coded by plasmid pREP4 (QIAGEN) in E. coli. The E. coli XL-1 Blue strain (Stratagene) was used for gp2 site-directed mutagenesis. The gene 2 alleles coding for gp2 single and double mutants were engineered by site-directed mutagenesis using the QuickChange Site-Directed Mutagenesis Kit (Stratagene). The templates for mutagenesis were plasmid pCB191 for expression in E. coli and pBT163 for expression in B. subtilis. Residues 1–198 of gp2 were cloned into pET28a using the NdeI/HindIII restriction sites [N-terminal domain (NTD) with a C-terminal His tag]. The C-terminal gp2 construct (residues 232–422) was described previously (9). Complete sequencing of gene 2 alleles was carried out (GATC, Konstanz). Phage production with the different gp2 mutants was analyzed by in vivo complementation assays in the B. subtilis strain YB886 carrying the pBT163 constructs with different gene 2 alleles.
DNA manipulations
Plasmid DNA was purified with a plasmid purification kit from Qiagen. Mature SPP1 and T5 DNAs were extracted from CsCl-purified phage particles by phenol–chloroform extraction followed by precipitation with isopropanol. DNA concentration was determined using molar extinction coefficient of 6500 M/cm at 260 nm.
Protein purification
Gp1 and gp2 were purified as described previously (16). Protein concentration was determined by the Bradford method (25), using bovine serum albumin as a standard. Gp1 and gp2 concentrations were expressed as moles of protein decamers (13) and monomers (14), respectively.
Production and purification of SPP1 procapsids
Wild-type (wt) SPP1 procapsids or procapsids lacking the portal protein, gp6, were produced by infection of the B. subtilis YB886 strain with SPP1 sus70 and sus70sus115 suppressor-sensitive phages, respectively, and purified as described (17). The purity of the procapsid preparations was analyzed by SDS–PAGE and the presence or absence of gp6 was checked by western blotting using anti-gp6 antibody. Blots were developed using the ECL detection system (Amersham Pharmacia Biotech). The quality of the procapsid preparations was also checked by electron microscopy of negatively stained samples.
Fluorescence-based thermal shift assay
Thermal stability assays were performed with 1 µM gp2 in 20 mM Tris–HCl pH 7.5, 300 mM NaCl and the SYPRO Orange dye as a fluorescent probe (diluted ∼800-fold from a 5000-fold stock solution, Invitrogen). Fluorescence measures the binding of the SYPRO orange probe to exposed protein hydrophobic regions. This dye becomes highly fluorescent when bound to protein hydrophobic sites, which become accessible upon thermal unfolding (26). Reaction mixtures were made in duplicate in a 96-well fast PCR plate at a final volume of 10 µl. The temperature gradient was carried out in the range of 15–95°C at 1.5°C/min with a StepOnePlus real-time PCR system (Applied Biosystems). Fluorescence was recorded as a function of temperature in real time (excitation with a blue LED source and emission filtered through a 5-carboxy-X-rhodamine emission filter). The melting temperature (Tm) was calculated with StepOne software v2.2 as the maximum of the derivative of the resulting SYPRO Orange fluorescence curves. Data from duplicates were very reproducible (error in the Tms ranging from 0.0°C to 0.02°C).Then the mean from the duplicates was considered as the value obtained for each condition in each experiment.
In vitro nuclease assay
The in vitro nuclease activity assay was performed as described previously (16). Unless indicated elsewhere DNA was incubated with gp2 proteins (1 µM) in 50 mM Tris–HCl pH 7.5, 50 mM NaCl with or without metal ions at 37°C for 1 h. The reactions were stopped by addition of EDTA and the samples deproteineized by incubation with proteinase K and SDS for 45 min at 37°C. Analytical gel electrophoresis was carried out in 0.8% (w/v) agarose/Tris–borate EDTA horizontal slab gels.
In vivo pac cleavage assays
The assay was adapted from Chai et al. (12). Exponentially growing (OD600 = 0.8) B. subtilis YB886 strains carrying plasmid pBT163 (co-expressing gene 1 and different gene 2 alleles) were centrifuged and the pellet was resuspended in 1/10 of the initial culture volume in lysis solution (100 mM Tris–HCl pH 7.5, 10 mM EDTA, 400 µg/ml RNase, 1 mg/ml lysozyme). After 1-h incubation at room temperature, SDS was added to a final concentration of 0.5% and cell lysates incubated 10 min, followed by addition of pronase and further incubation for 30 min at 37°C. Total DNA was extracted with phenol/chloroform and dialyzed extensively against 10 mM Tris–HCl pH 8.0, 10 mM EDTA. DNA was then EcoRI-cleaved and the digests were electrophoresed in 1% agarose. DNA was transferred to a nylon membrane and analyzed by the Southern technique essentially as previously described (27). A DNA fragment from plasmid pBT115 containing the pac cleavage site was used as probe (see Figure 6A).
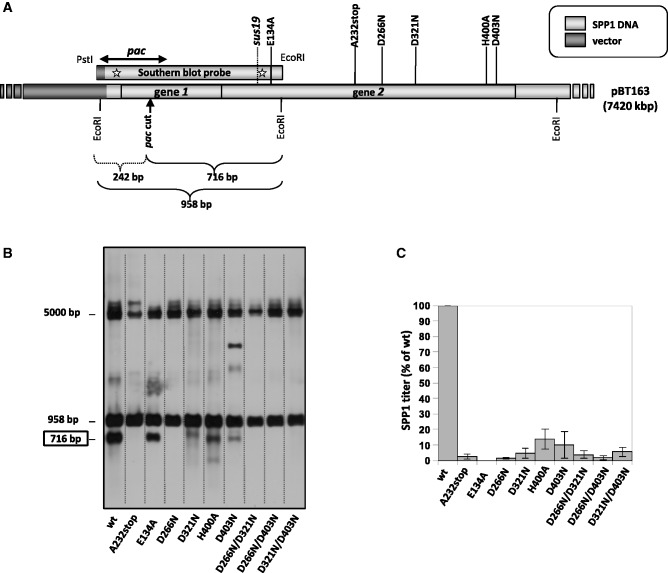
Figure 6.
Gp2 nuclease mutations inhibit pac cleavage in vivo. (A) Schematic representation of the pBT163 plasmid (7.4 kb) (12) which contains the 2.6-kb BanI-HindIII fragment from SPP1 DNA, comprising genes 1 and 2. The vertical arrow denotes the SPP1 pac cleavage site, situated within gene 1. EcoRI sites as well as position of gp2 mutations inserted by site-directed mutagenesis in pBT163 are indicated. The hybridization region of the Southern-blot probe used in the in vivo pac cleavage assay was from pBT115 carrying gene 1 with the pac cleavage site and part of gene 2) is shown. SPP1 DNA is depicted in light grey. (B) Southern blot of EcoRI-digested DNA isolated from B. subtilis strain YB886 carrying pBT163 coding gene 1 and different gene 2 alleles. The blot was hybridized with the DNA fragment probe schematized in panel (A). The band with an electrophoretic mobility close to the 5-kb marker is attributed to intact 7.4-kb supercoiled plasmid that was observed to exhibit this migration behavior. (C) Effect of gp2 mutations on phage yield. The effect of gp2 mutations on phage production was estimated in complementation assays by titration of SPP1sus19 phages [the position of the sus19 mutation in gene 2 is shown in panel (A)] in strains bearing plasmids coding for gene 2 mutant alleles and estimated as percentage of the titer (pfu, plaque-forming units/ml) obtained with the strain coding for wt gp2. The phage yield for the wt gp2 (considered as 100%) was 2.0 × 1011 pfu/ml. Values are the average of three independent experiments.
Pull-down assays
Affinity pull-down assays were carried out using purified hexa-histidine tagged-gp2 and Co2+-coated magnetic beads (Invitrogen, Norway) as described (18). Purified gp2 constructs and procapsids were incubated for 1 h at 30°C in 50 mM Tris–HCl pH 7.8, 50 mM NaCl and 10 mM MgCl2. After 10-min incubation with the beads, unbound material was removed, beads were washed, and bead-bound material was released by directly adding SDS–PAGE loading buffer and boiling the sample at 100°C for 5–10 min. Proteins present in the beads were analyzed by SDS–PAGE and western blotting using appropriate rabbit polyclonal antisera and the ECL-Kit (Amersham).
ATPase activity measurement
The ATPase activity was determined as described (17). Briefly, the ATPase standard reactions were incubated in 50 mM Tris–HCl, pH 7.8, 50 mM NaCl, 2 mM DTT and 10 mM MgCl2, in a final volume of 20 µl. The reactions were initiated by addition of [γ-32P]ATP after a pre-incubation of 5 min at 37°C and allowed to proceed for 15 min more at the same temperature. The ATPase-specific activity was determined by calculating the amount of Pi* produced per minute and expressed in terms of µmol/min/mg gp2.
DNA packaging reaction in vitro
DNA packaging reactions were performed using the SPP1 in vitro DNA packaging system as described (16). Briefly, unless stated otherwise, a standard 20 µl reaction mixture contained 1 mM ATP, 1 nM linear plasmid DNA (pBT115), 10 nM procapsids and terminase proteins (1 µM of gp2 and 1 µM of gp1) in 50 mM Tris–HCl, pH 7.8, 50 mM NaCl, 10 mM MgCl2, 10% dextran. Reactions were incubated for 1 h at 30°C. DNase was then added at 20 µg/ml and incubation continued for 10 min at 30°C. Reactions were stopped by addition of 50 mM EDTA and deproteinized. DNA was resolved by gel electrophoresis in 0.8% agarose gels and stained with Sybr Gold according to the manufacturer’s instructions.
RESULTS
Metal ion and substrate specificity of gp2 nuclease
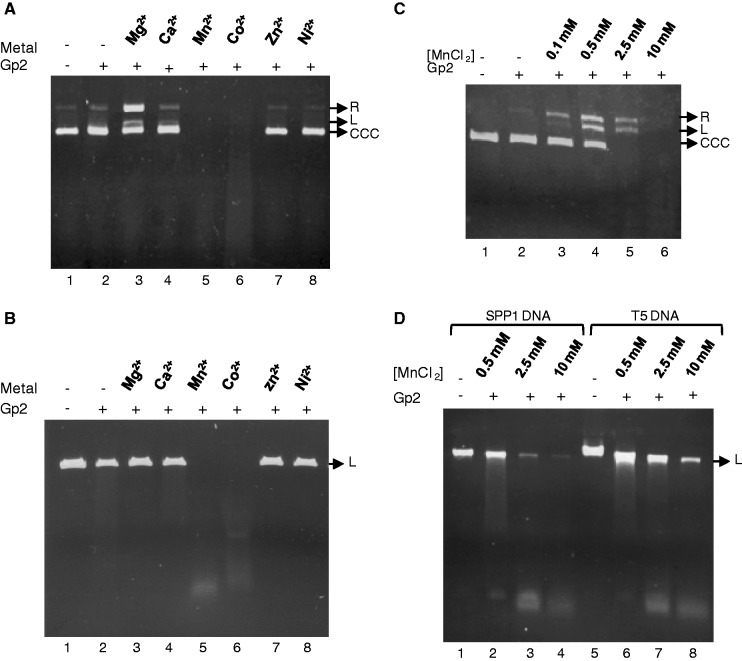
Gp2 has sequence-independent endonucleolytic activity in the presence of the divalent cation Mg2+, converting supercoiled circular plasmid DNA to nicked open circular DNA and to linear DNA, but is not active in the presence of Ca2+, Zn2+ or Ni2 (Figure 1A, lanes 3, 4, 7 and 8) (14–16). The gp2 nuclease activity is strongly increased by Mn2+ or Co2+ relatively to the effect observed with Mg2+, leading to complete DNA degradation in similar experimental conditions (Figure 1A, compare lanes 3, 5 and 6). Gp2 also displays a strong nuclease activity on linear dsDNA in the presence of Mn2+ and Co2+, degrading DNA to a smear of DNA fragments (Figure 1B, lanes 5 and 6). None of the other metal ions tested, including Mg2+, led to detectable cleavage of linear DNA (Figure 1B). Experiments performed with a circular DNA substrate and diverse Mn2+ concentrations ranging from 100 µM to 10 mM showed that as the Mn2+ dose increases, gp2 converts supercoiled circular plasmid DNA to nicked open circular DNA, subsequently to linear DNA and finally to completely degraded DNA (Figure 1C). In the presence of Mn2+, gp2 also digests long linear dsDNA substrates such as phage SPP1 (45 kb) and T5 (125 kb) DNAs (Figure 1D) in a dose-dependent manner.
Figure 1.
Metal ion specificity of gp2 nuclease. (A) Effect of divalent cations on gp2 circular dsDNA nuclease activity. Supercoiled pBT115 DNA was incubated with purified gp2 alone or with gp2 in the presence of 10 mM Mg2+, Ca2+, Mn2+, Co2+, Zn2+ or Ni2+. After 1 h at 37°C, the reactions were stopped and DNA analyzed as described under ‘Materials and Methods’ section. R, relaxed circular; L, linear; CCC, covalently closed circular DNA. (B) Effect of divalent cations on gp2 linear dsDNA nuclease activity. Linear plasmid pBT115 (PstI-cleaved) was incubated with gp2 with or without different divalent cations and analyzed as above. (C) Dose-dependent effect of Mn2+ on gp2 nuclease activity. Supercoiled pBT115 DNA was incubated with gp2 in the presence of increasing concentrations of Mn2+. (D) Nuclease activity of gp2 with long linear DNA substrates in the presence of different Mn2+ concentrations. SPP1 (45 kb) or T5 (125 kb) phage DNAs were incubated with gp2 in the presence of increasing concentrations of Mn2+.
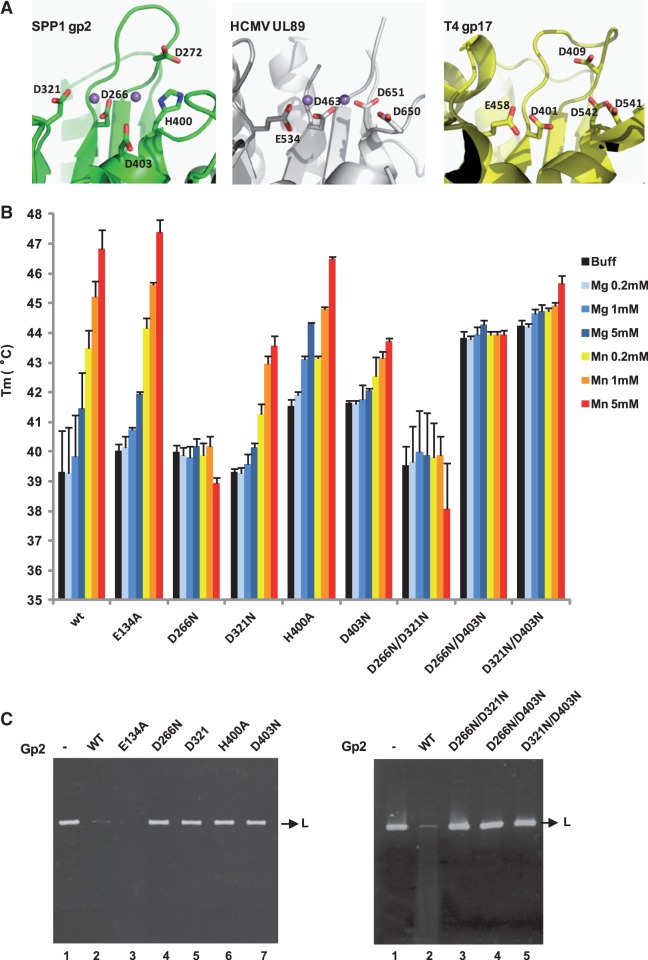
Figure 5.
Thermal stability assays and in vitro nuclease activity of gp2 mutant proteins. (A) Conservation of the terminase nuclease catalytic site of phage SPP1 (gp2; PDB accession code 2WC9), of HCMV (cytomegalovirus) (UL89; PDB 3N4Q) and of phage T4 (gp17; PDB 3CPE). The side chains of residues involved in metal coordination/catalysis are shown as sticks. The Mn2+ ions (magenta spheres) in SPP1 gp2 and in HCMV UL89 structures are 4 and 3.6 Å far apart, respectively. The structures were aligned with DALI and the figure prepared with PyMol. (B) Thermal stability of wild-type (wt) and gp2 mutant proteins analyzed as in Figure 4 in the absence of divalent metal ion (black bars) or in the presence of different concentrations (200 µM, 1 or 5 mM) of Mg2+ (light to dark blue bars) or Mn2+ (orange to red bars). The Tm values (°C) shown are the mean ± SD of results from at least three independent experiments. All proteins showed a single-transition denaturation curve. (C) In vitro nuclease activity of gp2 mutants. The nuclease activity of the gp2 single (left panel) and double (right panel) mutants was checked in an in vitro nuclease assay, as described under ‘Materials and Methods’ section, using a linear plasmid DNA substrate in the presence of 2.5 mM Mn2+.
The gp2-isolated carboxyl-domain has nuclease activity in the presence of Mn2+
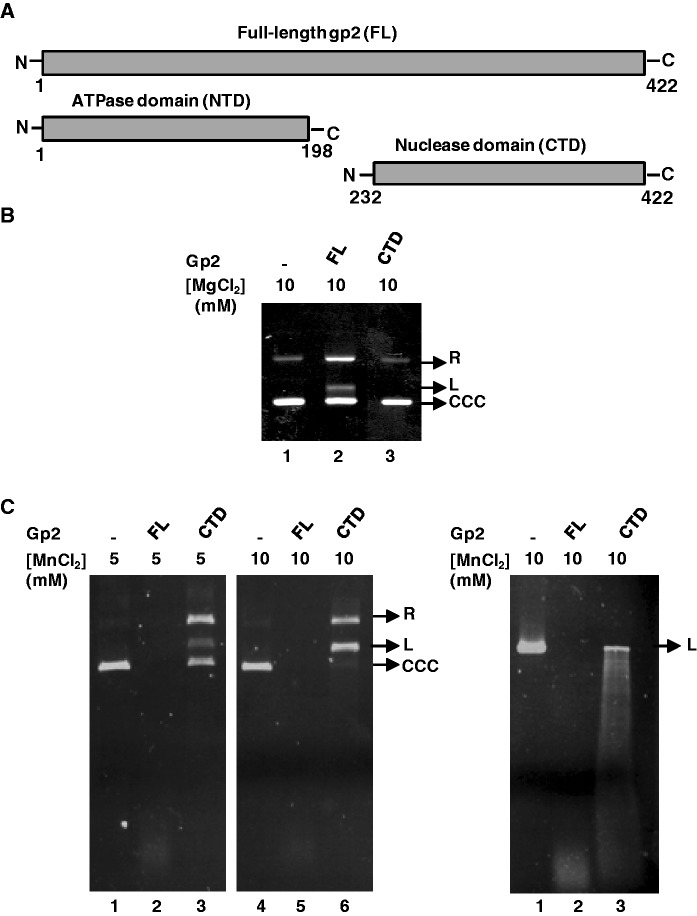
The gp2 nuclease center resides in the protein C-terminal domain (CTD) (9). To check whether the gp2 CTD alone [residues 232–422, as defined in Smits et al. (9); Figure 2A and Supplementary Figure S1] could cleave DNA, its nuclease activity was compared with that of the full-length (FL) protein. The isolated gp2 CTD showed no detectable nucleolytic activity in the presence of Mg2+ as metal ion (Figure 2B compare lanes 2 and 3). Interestingly, in the presence of Mn2+, the isolated gp2 CTD gained the capacity to cleave and degrade different DNA substrates (Figure 2C, circular DNA, left panel and linear DNA, right panel), although less efficiently and at a higher Mn2+ concentration, than found for the FL protein. The results confirm that the gp2 CTD harbors the enzyme nuclease activity and strengthens the gp2 Mn2+ preference to support nucleolytic cleavage of circular and linear dsDNA. They also reveal that optimal gp2 nuclease action requires the FL gp2 protein, as reported for other phage terminases (10,28).
Figure 2.
The isolated gp2 CTD has in vitro nuclease activity in the presence of Mn2+. (A) Representation of full-length (FL), N-terminal (NTD) and C-terminal (CTD) gp2 domains. (B) Nuclease assay of gp2 FL or CTD in the presence of Mg2+. Covalently closed circular pBT115 DNA (CCC) was incubated with FL or CTD gp2 (1 µM) for 1 h at 37°C in the presence of 10 mM Mg2+. Reactions were stopped and DNA analyzed as described under ‘Materials and Methods’ section. (C) Nuclease assay of the gp2 FL or CTD in the presence of Mn2+. CCC (left panel) or L (right panel) pBT115 DNA was incubated with FL or CTD gp2 in the presence of the indicated Mn2+ concentrations.
Binding of the gp2 nuclease domain to portal-containing procapsids
Besides its nuclease and ATPase activities, gp2 also plays a crucial role during assembly of the DNA packaging motor by directly docking at the procapsid portal vertex (18). Previous biochemical and genetics experiments in phages T3, T4 and λ suggested that residues of the CTD of the large terminase subunits are involved in the interaction with the portal (29–32). To address the question of whether the gp2 CTD mediates portal binding during motor assembly, we carried out affinity pull-down assays with the isolated gp2 CTD and NTD and purified SPP1 procapsids. The gp2 CTD is able to pull-down procapsids, although less efficiently than the FL protein (Figure 3, lanes 2 and 3). The specific interaction between procapsids and the gp2 CTD is mediated by the portal protein, as shown by parallel experiments with procapsids lacking the portal protein (Figure 3, compare lanes 3 and 7). A very faint signal corresponding to procapsid pull-down with the gp2 NTD was also detected. However, this binding to procapsids is unspecific as no difference was found between procapsids with or without the portal protein (Figure 3, lanes 4 and 8). The CTD of gp2 thus mediates anchoring of the packaging motor at the portal vertex in addition to its nuclease function.
Figure 3.
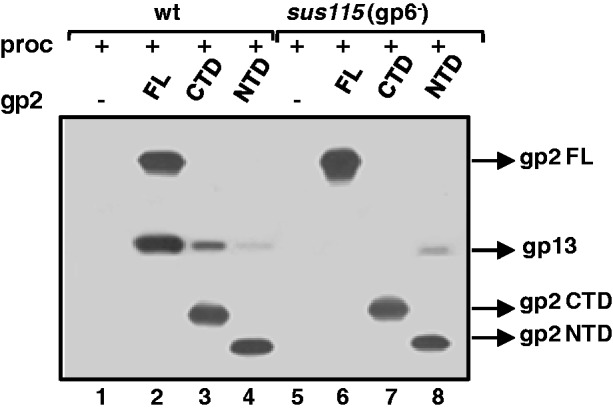
Interaction of the gp2 CTD with the procapsid portal vertex. Specific binding of FL, NTD or CTD gp2 to the procapsid (proc) was checked by affinity pull-down assays with purified wild-type (wt) or portal-less (sus115, gp6−) procapsids, as detailed in ‘Materials and Methods’ section. Proteins present in the beads fraction were resolved by SDS–PAGE followed by western blotting. Blots were incubated sequentially with anti-SPP1 (which recognizes the major head protein gp13) and anti-gp2 antibodies.
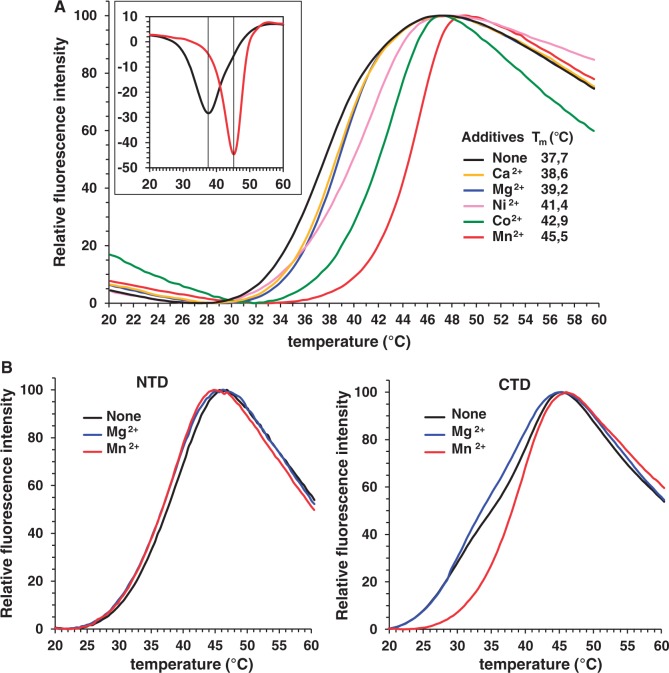
Divalent metal ions stabilize the nuclease domain and the gp2 overall structure
To investigate whether the effect of metal ions on gp2 nuclease activity results of an exclusive requirement for catalysis or if their binding correlates also with a stabilization effect on the gp2 structure we monitored the protein thermal stability by a fluorescence-based thermal shift assay (26). This method measures fluorescence emission upon binding of a probe to hydrophobic regions that become exposed during protein thermal denaturation. Gp2 melting curves are shown in Figure 4A. An increase in thermal stability was found upon addition of 1 mM Mn2+ leading to a Tm (apparent melting temperature) rise of ∼8°C, followed by Co2+, Ni2+ and, to a less extent, Ca2+ and Mg2+. Dose-dependent experiments with metal concentrations ranging from 320 nM to 5 mM confirmed these results (Supplementary Figure S2). We then compared the stability of the isolated gp2 NTD and CTD, in the absence or in the presence of Mg2+ or Mn2+, relatively to FL gp2 in the same experimental conditions. No significant difference was detected between the Tm of the metal-free NTD and FL gp2 (Supplementary Figure S3A). The metal-free gp2 CTD shows reproducibly a more complex denaturation behavior (black curve in Figure 4B, right panel) leading to double minima for the derivative of the fluorescence signal curve, which hampers an accurate determination of the Tm (Supplementary Figure S3B). Nevertheless, its denaturation occurs at ∼39°C, as the NTD and FL gp2. In contrast to the FL gp2, no increase in Tm of the NTD and CTD was detected following Mg2+ or Mn2+ addition (Figure 4 and Supplementary Figure S3). However, addition of Mn2+ at 200 µM led reproducibly to a unimodal denaturation curve of the gp2 CTD in contrast to the behavior found in absence of metal ion or at the same Mg2+ concentration (Figure 4B and Supplementary Figure S3).
Figure 4.
Effect of metals on gp2 thermal stability. (A) Assay of gp2 thermal stability in the presence of different divalent metal ions. The gp2 stability was analyzed by a fluorescence-based thermal shift assay as described in ‘Materials and Methods’ section, in the absence (black curve) or in the presence of 1 mM Mg2+, Mn2+, Ni2+, Co2+ or Ca2+ (blue, red, pink, green and yellow curves, respectively). The fluorescence signal is a function of protein unfolding due to the temperature increase. The melting temperature (Tm) is indicated for each experimental condition. The inset shows the opposite of the first derivatives of the fluorescence signal from the denaturation curves of gp2 without metal ions (black) or in the presence of Mn2+, (red), calculated with StepOne software v2.2. Identical melting curves were obtained in three independent experiments (B) Thermostability curves of NTD or CTD gp2 without metal ions (black curve) or in the presence of 200 µM Mg2+ (blue) or Mn2+ (red curves). Identical melting curves were obtained in three independent experiments.
Mutations in the nuclease active site have distinct effects on metal-induced gp2 stabilization
Soaking gp2 nuclease domain crystals with Mn2+ revealed two peaks in the anomalous difference maps, corresponding to bound metal ions (9). One ion is coordinated by D266, H400 and D403 [metal ion A (33), while the second ion is coordinated by D266 and D321 (metal ion B)] (9) (Figure 5A and Supplementary Figure S4). These residues are highly conserved in gp2-related proteins and a similar organization for coordination of metal ions is found in all four terminase nuclease domains whose structure is available (Figure 5A). They create an electronegative environment for two metal ion-dependent catalysis of endonucleolytic cleavage in a strand of dsDNA that is accommodated in the active site cleft (9). The finding that two Mn2+ are coordinated at the gp2 catalytic center led us to investigate if these two bound ions play a structural role responsible for the gp2 metal-induced enhanced stability. The thermal stability of gp2 carrying single and double mutations in residues D266, D321, H400 and D403 was analyzed with or without metal ions. Gp2 carrying a mutation to alanine in the conserved E134 of the Walker B motif in the N-terminal ATPase domain was characterized as a control. None of the mutations affected gp2 stable production (data not shown). Likewise, none of the mutations reduced protein thermal stability in the absence of divalent metal ions (Figure 5B, black bars). Two of the gp2 double mutants, D266N/D403N and D321N/D403N, were actually more stable than the wt protein in these experimental conditions (∼4°C increase in Tm), and a slight increase of Tm was observed for the H400A and D403N mutants. Addition of Mg2+ (light to dark blue bars) or Mn2+ (orange to red bars) caused distinct effects on the thermal stability of the different gp2 proteins. Wild-type gp2 and the N-terminal E134A mutant exhibited a similar behavior, i.e. a modest increase in Tm caused by Mg2+ and a marked Mn2+-induced Tm raise for both proteins. Addition of Mn2+ also increased the thermal stability of the D321N, H400A and D403N gp2 mutant proteins, although the effect was less pronounced than in wt and in the E134A gp2. In contrast, neither Mg2+ nor Mn2+ stabilized the D266N and D266N/D321N mutants. Instead, Mn2+, at the higher concentration used in these experiments (5 mM, dark orange bar) seems to render the two proteins slightly less stable. This small decrease in stability caused by high Mn2+ associated with D266N mutation was abolished when combined with D403N substitution. The latter mutation, isolated or combined with other mutations of aspartic acids in the active site, leads to a higher stability of gp2 in absence of metal ions but renders also gp2 stability poorly responsive to this type of ions (Figure 5B).
Gp2 nuclease mutations impair non-specific DNA cleavage
Gp2 proteins carrying amino acid substitutions D266, D321, H400 and D403 in the nuclease active site lost the capacity to digest linear dsDNA, contrarily to wt gp2, demonstrating that these residues are critical for gp2 non-specific nuclease activity (Figure 5C, left panel, lanes 4–7). The same holds true for the three double mutants D266N/D321N, D266N/D403N and D321N/D403N (Figure 5C, right panel, lanes 3–5). The fact that all the substitutions affected the gp2 nuclease shows in addition that the cleavage seen in the in vitro assays with the wt nuclease is inherently associated with gp2 and not with other eventual nuclease contaminants. In contrast, the N-terminal mutation E134A has no effect on nuclease activity, as gp2 E134A mutant protein could cleave DNA as efficiently as the wt (Figure 5C, left panel, lanes 2 and 3).
Gp2 nuclease mutations drastically reduce in vivo pac cleavage
Initiation of SPP1 DNA packaging in vivo occurs by specific recognition and cleavage of the pac site at the DNA concatemer. Cleavage at pac requires the presence of the small and large terminase subunits, gp1 and gp2 (12,34). To evaluate the role of the gp2-mutated residues in protein-specific nuclease activity, we set up a pac cleavage assay in vivo, using a plasmid system [pBT163, ∼7.4 kb; (12)] co-expressing genes 1 and 2. As schematized in Figure 6A, the SPP1 pac cleavage site is located within gene 1. Cleavage at pac divides a 958-bp EcoRI DNA fragment of pBT163 into the 242- and 716-bp DNA fragments. Total DNA from exponentially grown B. subtilis strains carrying the pBT163 constructs co-encoding gp1 and the wt or mutant gp2 proteins was isolated, digested with EcoRI and separated in agarose gels. Southern blots were then hybridized with the probe indicated in Figure 6A. As shown in Figure 6B, a fragment of ∼700 bp is detected in the presence of wt gp2, in agreement with the predicted fragment size following pac cleaved and EcoRI digestion of the pBT163 plasmid (Figure 6A). As a negative control, the ∼700-bp fragment was not present with a truncated gp2 protein (A232stop) lacking the gp2 CTD. As reported previously for pac cleavage during SPP1 infections (12), we failed to detect the small 242-bp fragment that could be expected following cleavage at pac (Figure 6A). This asymmetry may reflect the formation after or during pac cleavage of a complex between phage DNA and the terminase proteins that protected packageable DNA against nucleolytic attack from cellular exonucleases (12). The nuclease(s) that could be responsible for the observed degradation of the 242-bp fragment remain(s) unknown. The 958-bp fragment detected in the Southern blot corresponds to the pBT163 EcoRI DNA fragment with uncut pac whereas upper bands arise from uncut and partially digested plasmid DNA. The presence of the 958-bp fragment shows that the plasmid assay system reproduces faithfully also the control mechanism of pac cleavage observed during SPP1 infection that is not a frequent event, occurring only once ‘per’ SPP1 packaging series in a phage DNA concatemer (6,34).
No detectable cleavage at pac occurred with the single mutant protein D266N and the three double mutants D266N/D321N, D266N/D403N and D321N/D403N. In case of mutations D321N, H400A and D403N we observed a strong reduction in the generation of the ∼700-kb fragment when compared to gp2 wt, revealing a residual nuclease activity. None of the mutations affected gp2 production in the B. subtilis cells (data not shown). The deficiency of in vivo pac cleavage of gp2 nuclease mutants in the pBT163 system (Figure 6B) correlated with the inability of the plasmid-borne gp2 mutants to complement SPP1 sus19, a mutant defective for gp2 production (Figure 6C). The N-terminal gp2 E134A mutation, which affects neither the unspecific gp2 nuclease (Figure 5C) nor specific cleavage at pac (Figure 6B), strongly impairs phage production (Figure 6C). This can be explained by the lack of ATPase activity of this mutant gp2 (see below).
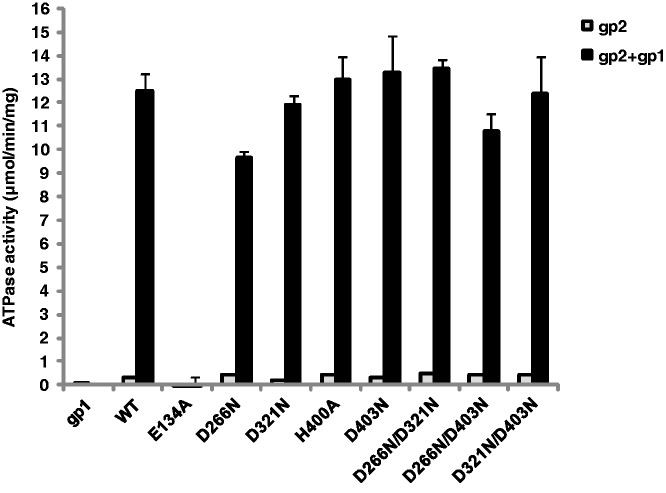
Nuclease mutants retain gp1-stimulated gp2 ATPase activity and gp2 portal binding
To control that mutations in the gp2 nuclease active center do not have a general impact on gp2 structure we assayed their ATPase activity, a requirement for gp2 function in DNA packaging. All nuclease mutants exhibited a low-basal ATPase activity similar to wt gp2 (Figure 7, light bars) and retained the gp1-stimulated ATPase activity (dark bars). No significant differences were observed when Mn2+ is used in place of Mg2+ as the metal ion (data not shown). On the contrary, the E134A mutation completely abrogated ATPase activity, as expected from its localization in the gp2 ATPase center. All mutant gp2 proteins exhibited normal binding to procapsids in a portal-mediated manner (Supplementary Figure S5, top and bottom panels).
Figure 7.
Effect of gp2 nuclease mutations on the terminase ATPase activity. The ATPase activity of wt and gp2 mutant proteins (1 µM) was determined in the absence (light bars) and in the presence (dark bars) of gp1 (1 µM) as described in ‘Materials and Methods’ section. The graphic shows the mean and SD of the results obtained for three independent experiments.
Gp2 nuclease mutants show distinct DNA packaging phenotypes
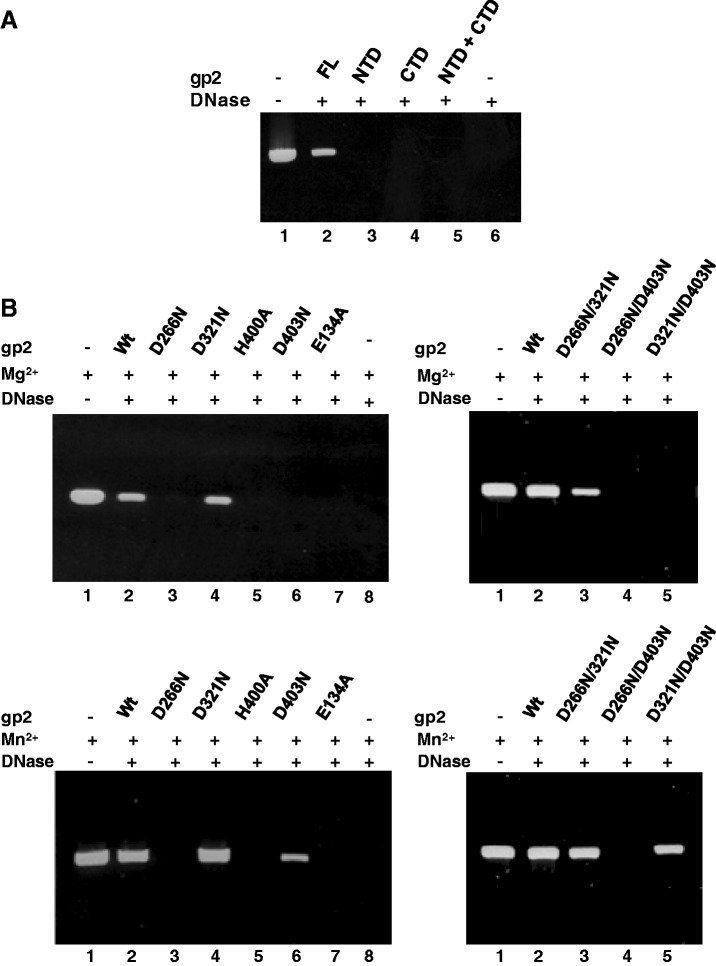
The gp2 nuclease activity is required in vivo for packaging initiation at the pac sequence and to terminate the packaging cycle by a non-specific endonucleoytic cleavage. After both cleavage events, this activity shall be switched off to allow for a DNA translocation step into the viral procapsid driven by the ATPase activity of the gp2 NTD. We hypothesized that mutations in the nuclease center could impact on the DNA translocation mechanism because the substrate DNA might remain in the vicinity of the gp2 nuclease center cleft during its pumping to the capsid interior. To address this question, we took advantage of the in vitro defined SPP1 DNA packaging system that, contrarily to the in vivo situation, neither requires cleavage at pac nor headful cut if a linear DNA molecule shorter than the capsid packaging capacity is used as substrate (16). The system only requirement for packaging is a free end on a linear DNA molecule. Here DNA encapsidation was monitored by a DNAse protection assay (16). Purified gp2 NTD or CTD, either alone or in combination, exhibited no DNA packaging activity, showing that SPP1 DNA packaging requires a FL gp2 (Figure 8A, lanes 3–5 versus lane 2), as also found for the large terminase of bacteriophage T4 (28). As could be expected from the ATPase assays (Figure 7), DNA packaging was fully abrogated when packaging reactions were carried out with the gp2 ATPase mutant E134A (Figure 8B, left panels, lane 7). Interestingly, diverse packaging phenotypes were registered with gp2 proteins carrying the nuclease mutations in packaging reactions carried out in the presence of Mg2+ or Mn2+. D321N and D266N/D321N substitutions do not significantly affect DNA packaging (Figure 8B). In contrast, mutations D266N, H400A (Figure 8B, left panels) and D266N/D403N (right panels) block DNA packaging in the presence of Mg2+ (top panels) or of Mn2+ (bottom panels). Interestingly, mutation D321N suppresses D266N yielding a double mutant competent for DNA packaging. Finally, mutant proteins D403N and D321N/D403N cannot package DNA in the presence of Mg2+ (left and right top panels in Figure 8B) but both mutants gain the capacity to package DNA in the presence of Mn2+ (lower left and right panels). Structural organization of the terminase nuclease active center and metal coordination thus plays an essential role in viral DNA encapsidation.
Figure 8.
DNA packaging in vitro of gp2 mutant forms. (A) DNA packaging in vitro was analyzed by a DNase protection assay as detailed under ‘Materials and Methods’ section in packaging reactions with the indicated purified gp2 constructs. Lane 1, control DNA; lane 6, control DNA treated with DNase. (B) Packaging efficiency of gp2 proteins carrying ATPase or nuclease mutations in packaging reactions performed in the presence of Mg2+ (upper panels) or Mn2+ (lower panels).
DISCUSSION
The two-domain terminase large subunit of tailed phages and herpesviruses is a dual activity enzyme of the viral DNA packaging motor. Its ability to switch between endonuclease and ATP-driven translocase activities is essential to achieve viral DNA packaging that occurs processively along a dsDNA concatemer substrate. The present study addressed the role of the nuclease catalytic center of SPP1 gp2 throughout the complete encapsidation cycle. Mutagenesis and functional analyses show that metal coordination at the nuclease center impacts on gp2 stability (Figures 4 and 5B) and, in addition to the expected function in DNA cleavage, it also plays a role in DNA encapsidation (Figure 8). The gp2 nuclease domain is thus the coordination center of the viral DNA packaging motor, a function that fits well to the finding of its direct binding to the capsid portal system through which DNA is pumped (Figure 3).
The gp2 nuclease activity shows a strong preference for Mn2+ and Co2+ over Mg2+ while the enzyme is inactive in the presence of Ca2+, Zn2+ or Ni2+ (Figure 1). Mn2+ has also a substantial and preferential stabilization effect on FL gp2 structure (Figure 4A). Mutagenesis of aspartate residues that coordinate metal ions for catalysis in the nuclease active center impaired or reduced the Mn2+-enhancement of gp2 stability (Figure 5). Binding of the metal at the catalytic center thus plays a structural role. This role depends on the inter-domain cross-talk with the ATPase domain of gp2 since strong stabilization by Mn2+ is observed only in the context of FL gp2 (Figure 4 and Supplementary Figure S3A). A similar dependence is found for the gp2 nuclease activity. The isolated gp2 CTD cleaves DNA in the presence of Mn2+ but not of Mg2+ (Figure 2). Mn2+-dependent DNA cleavage with the purified CTD is, however, much less efficient than with the FL protein, in agreement with previous data (9). The reduced Mn2+-enhancement of gp2 CTD stability relatively to FL gp2 may have an effect on the nuclease activity of the CTD domain which is weaker than in FL protein. However, high Mn2+ concentrations enhance the CTD nuclease activity without a major effect in the protein thermal stability. Altogether the results with the gp2 CTD suggest that although the ATPase NTD has no detectable nuclease activity, its presence is required for optimal gp2 conformation and/or catalysis, as proposed for other phage terminases (10,28). A strong preference for Mn2+ as co-factor in highly efficient nucleic acid cleavage appears to be a widespread feature of viral enzymes displaying DNA or RNA nuclease activities such as the large terminase of the herpes virus HCMV UL-89 (11), the influenza virus RNA-dependent polymerase (35,36) or the influenza-like endonuclease domains of Bunyaviridae and Arenaviridae RNA polymerases (37,38). A divalent metal cation requirement (Mg2+ or Mn2+) was also reported for the nuclease activity of phages λ (39), T5 (40) and T4 terminases (28).
Despite poor sequence conservation, the core of the gp2 nuclease domain has a fold similar to the nuclease domains of RNase H family proteins and binds two Mn2+ ions in its active site (9), suggesting a common two metal-ion-dependent catalysis mechanism to hydrolyze the phosphate ester bond of the nucleic acid chain (33). Conserved acidic residues are found in the catalytic centers of these nucleases (40). They coordinate a pair of divalent metal ions, often Mg2+, spaced approximately by 4 Å that are proposed to activate the attacking nucleophile and stabilize the transition state during the nucleic acid cleavage reaction (33,41–43). Three of these acidic residues are positioned at equivalent positions in previously determined structures of RNase H family proteins and correspond to gp2 D266, D321 and D403 (9). In addition, the active site histidine H400 is also conserved in SPP1 closely related terminases (9) or is replaced by aspartate in the structures of the T4, HCMV and P22 terminases (8,10,11). H400 could be important in conferring the Mn2+ preference found in our cleavage and protein stability assays because Mn2+ ions can be favorably coordinated by both acidic residues and histidines, whereas ligation by Mg2+ by histidines is less common (44,45). This is due to the higher affinity of Mn2+ to nitrogen relatively to Mg2+, which prefers oxygen (44). Mutation of the gp2 conserved residues (D266N, D321N, D403N and H400A) impaired the enzyme nuclease activity (Figures 5C and 6). All mutant gp2 proteins retained ATPase activity, gp1-stimulated ATPase and binding to portal-containing procapsids, showing that the mutations do not affect the gp2 overall folding. The most drastic effect was observed for the D266N substitution that completely abolishes gp2 cleavage at pac (Figure 6). This observation is consistent with the critical topology of D266 in the gp2 catalytic site (Figure 5A and Supplementary Figure S4). It is possible that defective metal-binding due to the other point mutations is poorly compensated by the presence of the remaining acidic residues in the active site, explaining some residual pac cleavage activity for the D321N, H400A and D403N gp2 proteins. In conformity with this possibility, double mutations of the active site aspartic acids totally abolished nuclease activity (Figure 6).
The precise location of Mn2+ ions found in the active site of the gp2 nuclease domain (Figure 5A) (9) might undergo some change during catalysis because full metal occupation in the correct geometry at the nuclease catalytic site is only expected upon substrate binding (33). However, structural conservation and our functional analyses provide a plausible organization for the gp2 nuclease catalytic site (Figure 5A). Its architecture is compatible with a two-metal ion catalytic pathway (33) in which metal B is coordinated by D266 and D321 and metal A by D266, H400 and D403 (Supplementary Figure S4). D266 is the aspartate residue strictly conserved in nucleases of the RNase H family that coordinates the two metal ions required for the phosphoryl transfer reaction leading to cleavage of the scissile phosphate of a nucleic acid or nucleotide (33). Its substitution in gp2 fully inactivates nuclease activity and Mn2+-enhancement of gp2 stability (Figure 5). This effect, which likely results of the inability of gp2 D266N to bind metal ions, is also found when combined with D321N. In contrast, mutation D403N and its combination with substitution of aspartates D266N and D321N lead to a high thermal stability independent of metal ions that is attributed to relieve of local destabilization of the structure due to repulsive forces between D403 and neighbor carboxylate residues (D266 and D396). This destabilization effect might be one of the roles of D403 to support metal coordination and catalysis. Coordination of metal ions to the gp2 catalytic center thus has the expected role in catalysis but plays also a specific structural role to stabilize the gp2 structure. The concentration of Mg2+ in the cytoplasm of exponentially growing B. subtilis was reported to be ∼50 mM, although a large fraction is complexed with cellular components primarily rRNA, while Mn2+ is found in micromolar amounts (46). A similar situation was described for eukaryotic cells (47). The precise role of both ions on terminase activity in the cell is thus difficult to assess. It is important to note, however, that Mn2+ increases gp2 stability at concentrations as low as 8 µM (Supplementary Figure S3) and that the endonuclease is active at 100 µM Mn2+ in our in vitro plasmid nicking/linearization assays (Figure 1C). The millimolar Mn2+ concentrations that induce a strong gp2 nuclease activity leading to complete degradation of linear DNA are not found in the cell. On the contrary, the terminase has an endonuclease activity that is a well regulated and not frequent event during the encapsidation cycle which is compatible with the gp2 properties found at physiological, micromolar, concentrations of Mn2+. Interestingly, the affinity of the endonuclease domain of the influenza virus polymerase PA subunit for Mn2+ ions (Kds for the two sites of 0.3 and 6.5 µM) was found to be 500- to 600-fold higher than for Mg2+ ions (35). A role for both ions in viral nucleases activity strict regulation and structure stabilization is thus plausible.
Our study showed that the gp2 nuclease domain plays essential roles at different steps of viral DNA encapsidation. The novel plasmid assay developed to measure in vivo the initiation event of SPP1 DNA packaging showed that the pac binding protein gp1, the terminase small subunit (13), and gp2 are necessary and sufficient to reproduce faithfully sequence-specific cleavage at pac (12) in absence of other phage products (Figure 6). Gp1 creates the structural context for gp2 cleavage at pac (13,48) and prevents non-specific nuclease attack to one of the DNA ends, the substrate for packaging. The SPP1 DNA end destined for packaging is protected from cellular exonucleases most likely by the gp1–gp2 complex where the nuclease activity is strictly shut off (15). The low frequency of pac cleavage observed (Figure 6) is consistent with the requirement that the sequence pac is used for cleavage only once ‘per’ packaging series along a substrate concatemer while the other pac sites remain intact (6,12,49). The gp1–gp2–pac nucleoprotein complex thus fully ensures the auto-regulated level of pac cleavage and directionality of the subsequent encapsidation event by selectively protecting one DNA end.
The gp2 nuclease CTD is then involved in specific docking to the procapsid portal vertex (Figure 3) as found in previous genetic and biochemical studies in phages T3, T4 and λ (29–32). As observed for the nuclease activity, binding of gp2 CTD to the portal is nevertheless less efficient relatively to FL gp2. It is possible that other gp2 residues, probably comprising the linker region between the NTD and CTD (8,50), are also involving in direct portal binding or are required to keep gp2 in an optimal conformation for the interaction with the portal. Docking of gp2 at the procapsid portal vertex leads to significant stimulation of the gp1:gp2 ATPase activity (17) consistent with switching on the gp2 translocase activity and suppression of any nuclease activity while DNA moves through the packaging motor. Pumping of the viral concatemeric DNA to the procapsid interior requires the hydrolysis of ATP that is catalyzed by the gp2 NTD, as observed for other viral terminases (28,51). Remarkably, some mutations in the conservative gp2 nuclease catalytic site residues (D266N, H400A and D266N/D403N) block DNA encapsidation. This was demonstrated in DNA packaging assays in vitro with a linear substrate, which does not involve a DNA cut [(16); Figure 8]. All the nuclease mutations have regular ATPase activity and binding to the procapsid portal vertex (Figure 7 and Supplementary Figure S5). In addition, the nature of the metal ion present affects the DNA packaging capacity of two gp2 nuclease mutant proteins D403N and D321N/D403N that are active in presence of Mn2+ but not of Mg2+. The role of the nuclease center in DNA encapsidation is not strictly dependent on binding of metal ions to the nuclease active site as shown by the observation that gp2 D266N/D321N supports DNA encapsidation. It rather results from the structural organization of the nuclease active site and probably of its harboring cleft to support DNA translocation that likely occurs in their vicinity.
After filling of the viral capsid with DNA, gp2 integrates signaling from the portal (21,52) to stop translocation and switching to endonucleolytic activity that cleaves the DNA substrate (the unspecific headful cut) to end the genome packaging cycle. A mutant in the SPP1 portal protein (gp6T319A) that packages small size DNA (22) leads to less efficient ATP hydrolysis by gp2 and slow packaging kinetics (17). Gp6 can thus render the gp2 DNA translocation machine less effective and cause a premature switch from gp2 translocase to headful nuclease that yields packaged viral chromosomes shorter than wt mature length. It is likely that this intimate cross-talk is mediated by interaction of the gp2 CTD with gp6. Our study together with available data show that the nuclease domain is the coordination center of the DNA packaging motor that integrates signals from the phage pac recognition terminase subunit, the ATPase domain, and the portal protein to switch between nuclease and translocase activities that are essential to achieve the viral genome encapsidation cycle in tailed phages and herpesviruses.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5.
FUNDING
Association Nationale pour la Recherche (ANR), PacVir [ANR-06-BLAN-0168] and DNA Gating [ANR-09-BLAN-0149-01] grants; Comunidad de Madrid [S2009MAT-1507 to J.C.A.]. Funding for open access charge: Centre National pour la Recherche Scientifique.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Pascale Boulanger for providing T5 phage and Drs Fred Antson (York University) and Marie Christine Vaney (Institut Pasteur) for discussions on the terminase structures.
REFERENCES
- 1.Casjens S, Hendrix R. Control mechanisms in dsDNA bacteriophage assembly. In: Calendar R, editor. The Bacteriophages. Vol. 1. New York: Plenum Press; 1988. pp. 15–91. [Google Scholar]
- 2.Black LW. DNA packaging in dsDNA bacteriophages. Annu. Rev. Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 3.Lo Piano A, Martínez-Jiménez M, Zecchi L, Ayora S. Recombination-dependent concatemeric viral DNA replication. Virus Res. 2011;160:1–14. doi: 10.1016/j.virusres.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Ann. Rev. Genet. 2008;42:19.1–19.35. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 5.Adams M, Hayden M, Casjens S. On the sequential packaging of bacteriophage P22 DNA. J. Virol. 1983;46:673–677. doi: 10.1128/jvi.46.2.673-677.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tavares P, Lurz R, Stiege A, Ruckert B, Trautner TA. Sequential headful packaging and fate of the cleaved DNA ends in bacteriophage SPP1. J. Mol. Biol. 1996;264:954–967. doi: 10.1006/jmbi.1996.0689. [DOI] [PubMed] [Google Scholar]
- 7.Tavares P, Zinn-Justin S, Orlova EV. Genome gating in tailed bacteriophage capsids. Adv. Exp. Med. Biol. 2012;726:585–600. doi: 10.1007/978-1-4614-0980-9_25. [DOI] [PubMed] [Google Scholar]
- 8.Sun S, Kondabagil K, Draper B, Alam TI, Bowman VD, Zhang Z, Hegde S, Fokine A, Rossmann MG, Rao VB. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135:1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 9.Smits C, Chechik M, Kovalevskiy OV, Shevtsov MB, Foster AW, Alonso JC, Antson AA. Structural basis for the nuclease activity of a bacteriophage large terminase. EMBO Reports. 2009;10:592–598. doi: 10.1038/embor.2009.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roy A, Cingolani G. Structure of P22 headful packaging nuclease. J. Biol. Chem. 2012;287:28196–28205. doi: 10.1074/jbc.M112.349894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nadal M, Mas PJ, Blanco AG, Arnan C, Solà M, Hart DJ, Coll M. Structure and inhibition of herpesvirus DNA packaging terminase nuclease domain. Proc. Natl Acad. Sci. USA. 2010;107:16078–16083. doi: 10.1073/pnas.1007144107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chai S, Bravo A, Luder G, Nedlin A, Trautner TA, Alonso JC. Molecular analysis of the Bacillus subtilis bacteriophage SPP1 region encompassing gene 1 to 6: the products of gene 1 and gene 2 are required for pac cleavage. J. Mol. Biol. 1992;124:87–102. doi: 10.1016/0022-2836(92)90578-8. [DOI] [PubMed] [Google Scholar]
- 13.Chai S, Lurz R, Alonso JC. The small subunit of the terminase enzyme of Bacillus subtilis bacteriophage SPP1 forms a specialized nucleoprotein complex with the packaging initiation region. J. Mol. Biol. 1995;252:386–398. doi: 10.1006/jmbi.1995.0505. [DOI] [PubMed] [Google Scholar]
- 14.Gual A, Camacho AC, Alonso JC. Functional analysis of the terminase large subunit, gp2, of Bacillus subtilis bacteriophage SPP1. J. Biol. Chem. 2000;275:35311–35319. doi: 10.1074/jbc.M004309200. [DOI] [PubMed] [Google Scholar]
- 15.Camacho AC, Gual A, Lurz R, Tavares P, Alonso JC. Bacillus subtilis bacteriophage SPP1 DNA packaging motor requires terminase and portal proteins. J. Biol. Chem. 2003;278:23251–23259. doi: 10.1074/jbc.M301805200. [DOI] [PubMed] [Google Scholar]
- 16.Oliveira L, Alonso JC, Tavares P. A defined in vitro system for DNA packaging by the bacteriophage SPP1: insights into the headful packaging mechanism. J. Mol. Biol. 2005;353:529–539. doi: 10.1016/j.jmb.2005.08.063. [DOI] [PubMed] [Google Scholar]
- 17.Oliveira L, Henriques AO, Tavares P. Modulation of the viral ATPase activity by the portal protein correlates with DNA packaging efficiency. J. Biol. Chem. 2006;281:21914–21923. doi: 10.1074/jbc.M603314200. [DOI] [PubMed] [Google Scholar]
- 18.Oliveira L, Cuervo A, Tavares P. Direct interaction of the bacteriophage SPP1 packaging ATPase with the portal protein. J. Biol. Chem. 2010;285:7366–7373. doi: 10.1074/jbc.M109.061010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lebedev AA, Krause MH, Isidro AL, Vagin A, Orlova EV, Turner J, Dodson EJ, Tavares P, Antson AA. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26:1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuervo A, Vaney MC, Antson A, Tavares P, Oliveira L. Structural rearrangements between portal protein subunits are essential for viral DNA translocation. J. Biol. Chem. 2007;282:18907–18913. doi: 10.1074/jbc.M701808200. [DOI] [PubMed] [Google Scholar]
- 21.Tavares P, Santos MA, Lurz R, Morelli G, de Lencastre H, Trautner TA. Identification of a gene in Bacillus subtilis bacteriophage SPP1 determining the amount of packaged DNA. J. Mol. Biol. 1992;225:81–92. doi: 10.1016/0022-2836(92)91027-m. [DOI] [PubMed] [Google Scholar]
- 22.Isidro A, Henriques AO, Tavares P. The portal protein plays essential roles at different steps of the SPP1 DNA packaging process. Virology. 2004;322:253–263. doi: 10.1016/j.virol.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 23.Studier FW. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol. 1991;219:37–44. doi: 10.1016/0022-2836(91)90855-z. [DOI] [PubMed] [Google Scholar]
- 24.Gual A, Alonso JC. Characterization of the small subunit of the terminase enzyme of the Bacillus subtilis bacteriophage SPP1. Virology. 1998;242:279–287. doi: 10.1006/viro.1997.9017. [DOI] [PubMed] [Google Scholar]
- 25.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 26.Ericsson UB, Hallberg BM, Detitta GT, Dekker N, Nordlund P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal. Biochem. 2006;357:289–298. doi: 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Maniatis T, Fritsch EF. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Ghosh-Kumar M, Alam TI, Draper B, Stack JD, Rao VB. Regulation by interdomain communication of a headful packaging nuclease from bacteriophage T4. Nucleic Acids Res. 2010;397:2742–2755. doi: 10.1093/nar/gkq1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morita M, Tasaka M, Fujisawa H. Structural and functional domains of the large subunit of the bacteriophage T3 DNA packaging enzyme: importance of the C-terminal region in prohead binding. J. Mol. Biol. 1995;245:635–644. doi: 10.1006/jmbi.1994.0052. [DOI] [PubMed] [Google Scholar]
- 30.Lin H, Rao VB, Black LW. Analysis of capsid portal protein and terminase functional domains: interaction sites required for DNA packaging in bacteriophage T4. J. Mol. Biol. 1999;289:249–260. doi: 10.1006/jmbi.1999.2781. [DOI] [PubMed] [Google Scholar]
- 31.Kanamaru S, Kondabagil K, Rossman MG, Rao V. The functional domains of bacteriophage T4 terminase. J. Biol. Chem. 2004;279:40795–40801. doi: 10.1074/jbc.M403647200. [DOI] [PubMed] [Google Scholar]
- 32.Yeo A, Feiss M. Specific interaction of terminase, the DNA packaging enzyme of bacteriophage λ, with the portal protein of the prohead. J. Mol. Biol. 1995;245:141–150. doi: 10.1006/jmbi.1994.0013. [DOI] [PubMed] [Google Scholar]
- 33.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 34.Bravo A, Alonso JC, Trautner TA. Functional analysis of the Bacillus subtilis bacteriophage SPP1 pac site. Nucleic Acids Res. 1990;18:2881–2886. doi: 10.1093/nar/18.10.2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crépin T, Dias A, Palencia A, Swale C, Cusack S, Ruigrok RW. Mutational and metal binding analysis of the endonuclease domain of the influenza virus polymerase PA subunit. J. Virol. 2010;84:9096–9104. doi: 10.1128/JVI.00995-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dias A, Bouvier D, Crépin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RW. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature. 2009;458:914–918. doi: 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- 37.Reguera J, Weber F, Cusack S. Bunyaviridae RNA polymerases (L-protein) have an N-terminal, influenza-like endonuclease domain, essential for viral cap-dependent transcription. PLoS Pathog. 2010;6:e1001101. doi: 10.1371/journal.ppat.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morin B, Coutard B, Lelke M, Ferron F, Kerber R, Jamal S, Frangeul A, Baronti C, Charrel R, de Lamballerie X, et al. The N-terminal domain of the arenavirus L protein is an RNA endonuclease essential in mRNA transcription. PLoS Pathog. 2010;69:e1001038. doi: 10.1371/journal.ppat.1001038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomka MA, Catalano CE. Physical and kinetic characterization of the DNA packaging enzyme from bacteriophage lambda. J. Biol. Chem. 1993;268:3056–3065. [PubMed] [Google Scholar]
- 40.Ponchon L, Boulanger P, Labesse G, Letellier L. The endonuclease domain of bacteriophage terminases belongs to the resolvase/integrase/ribonuclease H superfamily: a bioinformatics analysis validated by a functional study on bacteriophage T5. J. Biol. Chem. 2006;281:5829–5836. doi: 10.1074/jbc.M511817200. [DOI] [PubMed] [Google Scholar]
- 41.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl Acad. Sci. USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nowotny M, Yang W. Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J. 2006;259:1924–1933. doi: 10.1038/sj.emboj.7601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nowotny M, Gaidamakov SA, Ghirlando R, Cerritelli SM, Crouch RJ, Yang W. Structure of human RNase H1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription. Mol. Cell. 2007;28:264–276. doi: 10.1016/j.molcel.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 44.Bock CW, Kaufman, Katz A, Markham GD, Glusker JP. Manganese as a Replacement for Magnesium and Zinc, Functional Comparison of the Divalent Ions. J. Am. Chem. Soc. 1999;121:7360–7372. [Google Scholar]
- 45.Glusker J, Katz A, Bock CW. Metal ions in biological systems. Rigaku J. 1999;16:8–17. [Google Scholar]
- 46.Médicis ED, Paquette J, Gauthier JJ, Shapcott D. Magnesium and manganese content of halophilic bacteria. Appl. Environ. Microbiol. 1986;52:567–573. doi: 10.1128/aem.52.3.567-573.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao C, Lou Z, Guo Y, Ma M, Chen Y, Liang S, Zhang L, Chen S, Li X, Liu Y, et al. Nucleoside monophosphate complex structures of the endonuclease domain from the influenza virus polymerase PA subunit reveal the substrate binding site inside the catalytic center. J. Virol. 2009;83:9024–9030. doi: 10.1128/JVI.00911-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Büttner CR, Chechik M, Ortiz-Lombardía M, Smits C, Ebong IO, Chechik V, Jeschke G, Dykeman E, Benini S, Robinson CV, et al. Structural basis for DNA recognition and loading into a viral packaging motor. Proc. Natl Acad. Sci. USA. 2012;109:811–816. doi: 10.1073/pnas.1110270109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chai S, Szepan U, Alonso JC. Bacillus subtilis bacteriophage SPP1 terminase has a dual activity: it is required for the packaging initiation and represses its own synthesis. Gene. 1997;184:251–256. doi: 10.1016/s0378-1119(96)00609-9. [DOI] [PubMed] [Google Scholar]
- 50.Hegde S, Padilla-Sanchez V, Draper B, Rao VB. Portal-large terminase interactions of the bacteriophage T4 DNA packaging machine implicate a molecular lever mechanism for coupling ATPase to DNA translocation. J. Virol. 2012;86:4046–4057. doi: 10.1128/JVI.07197-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hang JQ, Tack BF, Feiss M. ATPase center of bacteriophage lambda terminase involved in post-cleavage stages of DNA packaging: identification of ATP-interactive amino acids. J. Mol. Biol. 2000;302:777–795. doi: 10.1006/jmbi.2000.4086. [DOI] [PubMed] [Google Scholar]
- 52.Orlova EV, Dube P, Beckmann E, Zemlin F, Lurz R, Trautner TA, Tavares P, van Heel M. Structure of the 13-fold symmetric portal protein of bacteriophage SPP1. Nat. Struct. Biol. 1999;6:842–846. doi: 10.1038/12303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.