Abstract
Human DNA polymerase (pol) λ functions in base excision repair and non-homologous end joining. We have previously shown that DNA pol λ is involved in accurate bypass of the two frequent oxidative lesions, 7,8-dihydro-8-oxoguanine and 1,2-dihydro-2-oxoadenine during the S phase. However, nothing is known so far about the relationship of DNA pol λ with the S phase DNA damage response checkpoint. Here, we show that a knockdown of DNA pol λ, but not of its close homologue DNA pol β, results in replication fork stress and activates the S phase checkpoint, slowing S phase progression in different human cancer cell lines. We furthermore show that DNA pol λ protects cells from oxidative DNA damage and also functions in rescuing stalled replication forks. Its absence becomes lethal for a cell when a functional checkpoint is missing, suggesting a DNA synthesis deficiency. Our results provide the first evidence, to our knowledge, that DNA pol λ is required for cell cycle progression and is functionally connected to the S phase DNA damage response machinery in cancer cells.
INTRODUCTION
The genetic information stored into DNA is constantly subjected to potentially harmful alterations due to the action of genotoxic agents. It has been estimated that mammalian genomes undergo ∼100 000 modifications per day. The major source of DNA damage is reactive oxygen species (ROS), which are constantly produced during normal cell metabolism. When ROS react with DNA, they produce oxidized bases, some of which are highly mutagenic lesions. The most frequently generated lesions (103–104 per cell/per day) are 7,8-dihydro-8-oxoguanine (8-oxo-G) and 1,2-dihydro-2-oxoadenine (2-OH-A) (1). The presence of an oxidized 8-oxo-G base in the replicating strand is generally believed not to block replication, but it can easily mis-direct nucleotide incorporation by the replicative DNA polymerases (pols) α, δ and ε, so that mis-incorporation of A opposite 8-oxo-G occurs with a high frequency (2). On the other hand, if the 2-OH-A persists on the replicating strand during the S phase, it constitutes a strong block for replicative DNA pols (3). As a consequence of blocked fork progression, abnormal single-stranded (ss) DNA is generated by the uncoupling between DNA unwinding and DNA synthesis. This activates the ATR kinase, which phosphorylates its downstream effector kinase Chk1 at Ser 345, that, in turn, prevents the origin activation and delays S phase progression (4). Prolonged stalling of a replication fork is dangerous, since it can generate aberrant DNA structures, such as ss and double-stranded (ds) breaks. However, DNA strand breaks can be also generated as a consequence of impaired base excision repair (BER) of oxidized DNA bases. The BER involves the removal of the damaged base by a specific glycosylase, generating an abasic site, which is then converted into an ss DNA break through the action of the APE1 endonuclease. Delayed processing of this ss DNA break by the BER machinery might cause its persistence, which can result in DNA replication block.
In human cells, the potentially deleterious consequences of the accumulation of oxidized bases are counteracted by specialized repair pathways. When the replisome is blocked, for example upon encountering a 2-OH-A lesion, checkpoint activation drives the recruitment of specialized DNA pols, that are able to bypass the lesion and allow fork restart, in a process called translesion synthesis (TLS) (5). On the other hand, the A:8-oxo-G mismatches frequently generated during S phase are repaired by a specialized BER pathway, initiated by the MutYH glycosylase (6). Finally, broken DNA ends, eventually generated by replication fork stalling or delayed BER, are repaired by homologous recombination (HR) (only during S phase) and by non-homologous end joining (NHEJ). All three pathways TLS, BER and NHEJ, require the essential contribution of a specialized polymerase such as DNA pol λ (7). Our previous results showed that DNA pol λ is the most proficient, among human DNA pols, in performing error-free bypass of 2-OH-A and 8-oxo-G lesions. This feature is essential both for TLS at the fork and for correct repair of the A:8-oxo-G mismatches (8–10). Finally, a convincing set of data exists, suggesting to a role of DNA pol λ in NHEJ-dependent repair of ds DNA breaks (DSBs) (11).
Given the central role of DNA pol λ in the pathways responsible to counteract DNA damages during replication, it is reasonable to hypothesize that it is tightly regulated and functionally connected to the S phase checkpoint machinery. We have recently shown that the functional and physical interaction of DNA pol λ with MutYH during 8-oxo-G repair is regulated by a finely tuned balance of phosphorylation and ubiquitination, ensuring stabilization of DNA pol λ in late S phase and its recruitment to chromatin into active 8-oxo-G repair complexes, while DNA pol λ molecules that not engaged in active repair, are targeted to proteasomal degradation (12). In addition, other data suggested that human MutYH interacts with ATR and functions as a mediator of Chk1 phosphorylation in response to DNA damage, thus providing a link between oxidative DNA damage response and S phase checkpoint activation (13). However, an evidence for direct correlation between perturbation of DNA pol λ levels and the checkpoint response is still missing.
Here, we show that a knockdown of DNA pol λ causes replication fork stress and activates the ATR/Chk1-dependent S phase checkpoint, slowing S phase progression in different human cancer cell lines. Silencing of the major BER enzyme, DNA pol β, which is a close homologue of DNA pol λ, results in no induction of S phase checkpoint activation. We conclude that DNA pol λ protects cells from oxidative DNA damage and can also function in rescuing stalled replication forks. Its absence results in cell lethality in the absence of a functional Chk1. These results provide the first evidence, to our knowledge, that DNA pol λ is specifically required for cell cycle progression and is functionally connected to the S phase DNA damage response machinery in cancer cells.
MATERIALS AND METHODS
Cell lines and treatments
HeLaS3 (ovarian carcinoma) and U2OS (osteosarcoma) cell lines were grown at 37°C and 5% CO2 in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum. PEO1 (BRCA2-deficient) and PEO4 (BRCA2-proficient) ovarian cancer cell lines were cultured at 37°C and 5% CO2 in DMEM containing 15% fetal bovine serum. Stable-inducible U2OS Pol λ knockdown cell line, U2KDL, was grown at 37°C and 5% CO2 in DMEM supplemented with 10% tetracycline free-fetal bovine serum (Lonza) and 300 µg/ml G418 (Gibco). KBrO3, UCN01, hydroxyurea (HU) and etoposide were purchased from Sigma Aldrich. Cells growing on 10 cm dishes (∼6 × 105 cells per dish) were incubated for 2 h at 37°C in the absence or presence of 20 mM or 40 mM KBrO3. Similar amounts of cells were incubated for 24 h at 37°C in the presence of 10 mM HU alone or in combination with 100 nM UCN01, cells were then harvested either at the end of treatment or following 24 h of recovery. The DNA damage positive control with etoposide was obtained by treating cells for 3 h at 37°C in the presence of 100 µM etoposide.
Reverse transcriptase–polymerase chain reaction and reverse transcriptase–real-time polymerase chain reaction
RNA extraction was performed using SV Total RNA Isolation System (Promega) and total RNA was quantified using Nanodrop (Thermo Scientific). To ensure a complete reverse transcription of total RNA, the Transcriptor First Strand cDNA Synthesis kit (Roche), which mixed random priming and oligo-dT together, was used following the manufacturer’s protocol. Amplification conditions for each cycle were as follows: 10 min at 25°C; 30 min at 55°C and 5 min at 85°C. POLL and POLB gene expression profile was checked by dual real-time reverse transcriptase–polymerase chain reaction (RT–PCR) assay using hydrolysis probes. The β-actin (ACTB) and β-glucuronidase genes (GUSB) were used as the reference genes for POL and POLB, respectively. All probes and primers purchased from Roche were as follows: POLL probe (Cod. 04685059001); Pol λ 5′-end primer 5′-CATCAAAAGTACTTGCAAAGATTCC-3′; Pol λ 3′-end primer 5′-GGGAGCTCAGCCACTCTTC-3′; Pol β probe (Cod. 04689089001); Pol β 5′-end primer 5′-CTCGAGTTAGTGGCATTGGTC-3′; Pol β 3′-end primer 5′-TTTTAATTCCTTCATCTACAAACTTCC-3′; ACTB probe and primers (Cod. 05046165001) and GUSB probe and primers (Cod. 05190525001). The amplification reactions were performed using a LightCycler 480 instrument (Roche) and the following run programs: Pol λ pre-incubation (10 min at 95°C); 50 cycles (10 s at 95°C; 30 s at 57°C; 1 s at 72°C); cooling (30 s at 40°C). Pol β pre-incubation (10 min at 95°C); 50 cycles (10 s at 95°C; 30 s at 61°C; 1 s at 72°C); cooling (30 s at 40°C). Semi-quantitative RT (qRT)–PCR was performed using the same Pol λ primers as in real-time PCR and the DDX3 gene as a reference. DDX3 5′-end primer 5′-GGGGGATCCTTTGAGAAATACGATGACATTCCA GTT-3′; 3′-end primer 5′-CCCAAGCTTTTTGTCTGATTCTTCCACCCAAACTAC-3′. The amplification programs were as follows: Pol λ pre-incubation (10 min at 95°C); 30 cycles (1 min at 95°C; 1 min at 55°C; 1 min at 72°C); final extension (10 min at 72°C); DDX3 pre-incubation (10 min at 95°C); 25 cycles (1 min at 95°C; 1 min 30 s at 64°C; 1 min at 72°C) and final extension (10 min at 72°C). RT–PCR amplicons were separated and analysed using 2% agarose gel.
Protein extracts and immunoblotting
Cells were harvested, washed twice with 1 × phosphate buffered saline (PBS) and then lysed in cracking buffer. The obtained cell extracts were boiled for 5 min at 95°C and then sonicated. Whole cell extracts were separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis and analyzed by western blotting using specific antibodies. Immunoreactive bands were visualized using enhanced chemiluminescence (Pierce) and ChemiDoc XRS (BioRad). Rabbit polyclonal antibody raised against Pol λ was purchased from Bethyl Laboratories (Montgomery, TX, USA) and used at a dilution of 1:500. Anti-Pol β (ab2856) mouse polyclonal and anti-PARP1 (ab6079) rabbit polyclonal antibodies were purchased from Abcam and diluted 1:1000. Anti-Phospho-Chk1(S345), anti-Phospho-Chk2(Thr68) and anti-Phospho-ATR(Ser428) rabbit polyclonal antibodies were purchased by Cell Signalling (Danvers, MA, USA) and used both at a dilution of 1:1000. Anti-Chk1(G-4) and anti-Actin (2Q1055) mouse monoclonal antibodies were from Santa Cruz Biotech (Santa Cruz, CA, USA) and employed in the studies at a dilution of 1:1000. Anti-ChK2-clone 7 and anti-Phospho-Histone H2A.X (Ser139)-clone JBW301 mouse monoclonal antibodies were purchased from Upstate and used at a dilution of 1:500 and 1:5000, respectively. Anti-ATR rabbit polyclonal antibody was purchased from Calbiochem (used at the 1:5000 dilution). Anti-α-tubulin (clone DM 1 A) mouse monoclonal antibody was purchased from Sigma Aldrich (used at the 1:50 000 dilution).
Pol λ and Pol β knockdown using siRNA
All siRNA sequences targeting Pol λ purchased from the Ambion collection of Selective Validated siRNA were as follows: siRNA 26197 (si97) 5′-GUGACUUCCUGGAACGUAUtt-3′ and siRNA 26198 (si98) 5′-CAAAAGUACUUGCAAAGAUtt-3′. The siRNA targeting Pol β was purchased from Qiagen SI02663605: 5′-TACGAGTTCATCCATCAATTT-3′. Cells were transfected with 40 pmol of siRNA using the HiPerFect lipid transfection reagent (Qiagen). On Day 1, ∼2 × 105 cells were plated on 6-well dishes in 2.4 ml of antibiotic-free medium and incubated for 24 h in a siRNA–lipid complex (40 pmol siRNA + 12 μl of HiPerFect in 120 μl of DMEM medium). On Day 2, cells were washed with DMEM medium, and the transfection procedure was repeated. Cells were harvested every day with the last time point at 72 h post-transfection and Pol λ expression was analysed at both mRNA and protein level by semi-qRT–PCR and western blotting, respectively.
Pol λ overexpression
To induce overexpression of Pol λ in U2OS cells, the pcDNA3-polλ-Myc construct (14) and the corresponding empty vector were used. On Day 1, ∼1 × 106 U2OS cells were seeded on 10 cm dish. On Day 2, cells were treated with 10 µl Lipofectamine 2000 (Life technologies) in the absence or presence of 2 µg of a plasmid diluted in 500 µl of FBS-free medium. Following 20 min of incubation at room temperature, the mixture was added drop wise to a dish. The day after, cells were harvested and used for further analysis.
U2KDL-knockdown cell line
To increase the efficiency of a knockdown, the pSingletTS plasmid (Clontech) was used as it encodes both the specific shRNA sequences and the Tet-repressor, called tTS, within the same vector. The TRE/U6 promoter regulates the expression of a shRNA sequence while the tTS, which suppresses the production of shRNA by binding to the TRE/U6 sequence, is regulated by a CMV promoter. The sequence of siRNA 26198 was used to design the shRNA insert. The oligonucleotides contained a 5′-XhOI restriction site overhang on one DNA strand and a 5′-HindIII restriction site overhang on the other strand. These restriction site overhangs provided a direct binding of the annealed oligonucleotides to the XhOI/HidIII-digested pSingle tTS plasmid, ensuring the correct orientation. The loop sequence 5′-TTCAAGAGA-3′ jointed sense and antisense sequences together and the RNA Pol III terminator sequence, 6 poly(T) tract, at the 3′-end of the top strand encoded the transcriptional stop of the shRNA. The annealing of shRNA oligonucleotides required a mixture of 50 µM of ds-oligos assuming 100% theoretical annealing. The annealing reaction was carried out through incubating the solution at 95°C for 30 s and then allowing it to cool down gradually. The double digest reaction was performed overnight at 37°C in 1× Buffer 2 (NEB) supplemented with 100 µg/ml bovine serum albumin, 10 U of XhOI and HindIII (NEB) and 1 µg of undigested vector. DNA was extracted using a Qiagen kit. The ligation reaction (1× T4 buffer, 1.3 mM adenosine triphosphate, 50 ng of digested vector, 33.3 nM of shRNA oligo and 200 U of T4 ligase) was incubated overnight at 16°C. The day after, to increase the ligation efficiency, another addition of 100 U of T4 ligase followed by further incubation at room temperature for 1 h was performed. A high copy plasmid XL10 blue strain was further transformed using 2 µl of ligation mix, then the clones of interest were selected on LB agar supplemented with 50 µg/ml ampicillin. Colony PCR was set up using insert-specific forward primers (see below) and the following amplification program: pre-incubation (5 min at 95°C); 30 cycles (30 s at 95°C, 30 s at 50°C, 1 min at 72°C) and final extension (20 min at 72°C). The DNA of the selected positive clones was extracted using Mini-prep Qiagen kit and the correctness of shRNA sequences was validated by sequencing.
shPOLL-For: 5′-TCGAGGCAAAAGTACTTGCAAAGATTTCAAGAGAATCTTTGCAAGTACTTTTGTTTTTTA-3′
shPOLL-Rev: 5′-AGCTTAAAAAACAAAAGTACTTGCAAAGATTCTCTTGAAATCTTTGCAAGTACTTTTGCC-3′
Primer insert-specific POLL: 5′-GTACTTGCAAAGATTTCAAGAGAATCTTTGC-3′
pSingle tTS rev: 5′-GCTCATTAATGCAGACCCATAATACCC-3′
U2OS were grown on 10 cm dishes to 80% confluency and then the cells were transfected with Lipofectamine transfection reagent following the manufacturer’s protocol in the presence of 2 µg of purified plasmid. Following 24 h after transfection, cells were split to the optimal plating density of 3 × 105 cells/dish and incubated for further 24 h before the start of the selection process. The optimized selective concentration of G418 was 700 µg/ml. Following ∼2 weeks of the selection process, a few clones were picked using a cloning cylinder for further analysis. The obtained clones were grown in medium containing 300 µg/ml G418. An optimal minimal concentration of doxycycline that resulted in induction of shRNA expression was titrated at 1 µg/ml.
Cytotoxicity assay
The CellTiter 96 Aqueous One Solution cell proliferation assay was used as a colorimetric method for determination of the toxicity induced during various treatments. Briefly, cells were seeded on a 96-well plate at a density of 1000–10 000 cells/well ∼24 h before treatment. To evaluate the treatment effects, the medium was replaced with 100 µl of fresh complete medium and 20 µl of the cell titer reagent, cells were then incubated for at least 3 h at 37°C and the absorbance at 492 nm was recorded.
Cell cycle evaluation by flow cytometry
Cells were harvested and fixed in 2 ml of cold 70% ethanol at 4°C for 10 min, then stored in a total volume of 5 ml of 70% ethanol till flow cytometry analysis. Fixed cell pellets were re-suspended in 500 µl of PBS1X and stained with 10 µl of propidium iodide 20 µg/ml, hence analysed by FlowMax software on Partec PAS flow cytometer.
Comet assay
Cells were treated with 10 mM HU for 24 h and immediately after the end of the treatment they were harvested. Aliquots of 20 µl of 200 000 cell/ml suspension were mixed with 180 µl of low-melting agarose (1% in PBS) and added onto Trevigen slides. The lysis was performed by an alkaline lysis buffer (2.5 M NaCl, 10 mM Tris base, 100 mM ethylenediaminetetraacetic acid (EDTA), 0.2 mM NaOH, 1% of TritonX-100 and 1% Sarkosyl) at 4°C for 1 h and 30 min, followed by the electrophoresis into an alkaline electrophoresis solution (pH > 13) (200 mM NaOH and 1 mM EDTA) at constant 0.3 A for 30 min. Images of randomly selected SyBr-green stained cells were caught with 20× magnification and analysed by CometScore program (TriTek Corp.). Tail DNA (%) (tail intensity/total intensity of the comet) and tail length (in arbitrary units) were chosen for the presentation of the results.
Immunofluorescence
Cells grown on glass coverslips were fixed with cold methanol for 20 min. Primary antibody was anti-phospho-histone H2AX (Ser139) monoclonal antibody (clone 20E3, Cell Signalling) and secondary antibody was anti-rabbit Alexa Fluor 555 (Life Technologies). Nuclei were stained by ProLong Gold solution (Life Technologies).
RESULTS
Transient knockdown of DNA polymerase λ in HeLa cells induces S phase delay
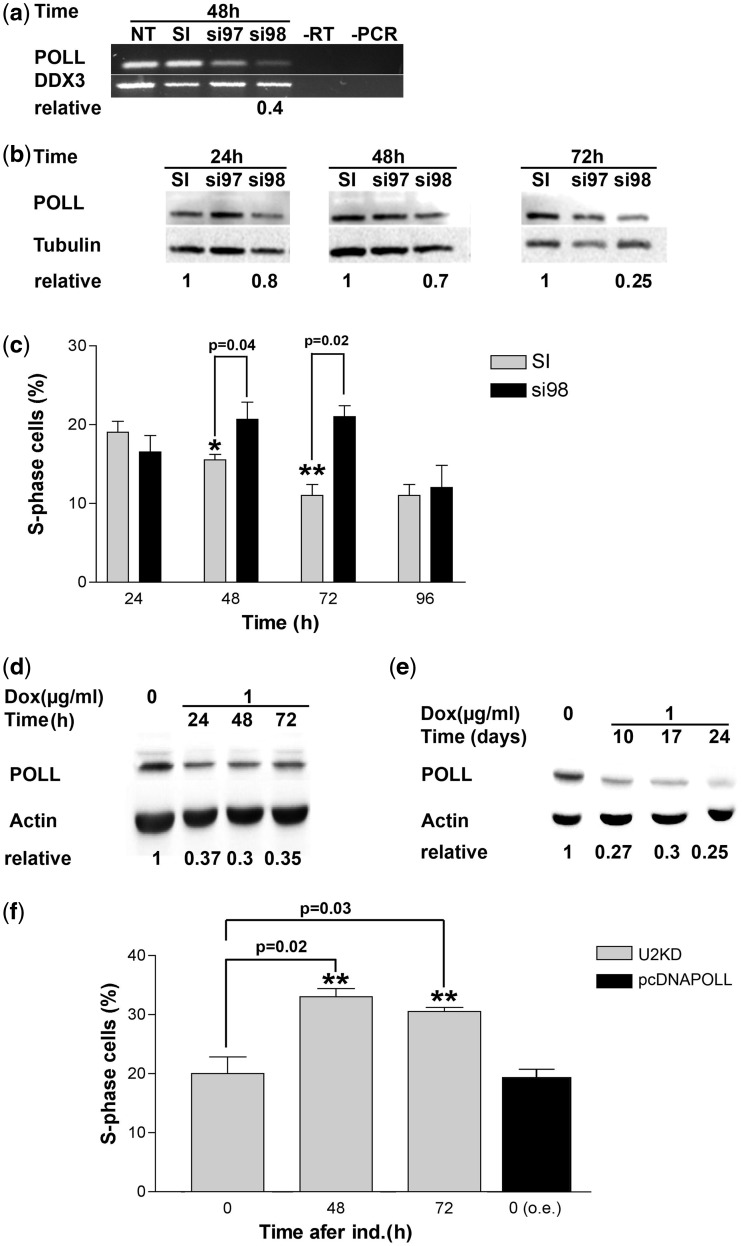
DNA pol λ expression was silenced in the HeLa S3 cell line. Two different siRNA sequences from the Ambion collection of Selective Validated siRNA were used to induce the knockdown, while a scrambled siRNA served as a negative control. Cells were harvested at 24, 48 and 72 h post-transfection and DNA pol λ levels were analysed at both mRNA and protein level by semi-qRT–PCR and western blotting, respectively. As shown in Figure 1, the most effective siRNA was si98, resulting in 75% down-regulation of DNA pol λ. We observed, however, a slight temporal difference between the mRNA and protein levels. In fact, while mRNA levels showed the maximal decrease already at 48 h (Figure 1a), protein levels reached maximal reduction at 72 h post-transfection (Figure 1b).
Figure 1.
DNA pol λ knockdown induces cell cycle delay. (a) RT–PCR measuring the mRNA levels for the POLL gene in HeLa cells after 48 h from transfection with siRNA oligonucleotides. NT, not transfected; SI, scrambled control; si97, si98, POLL-specific siRNAs. RT, PCR, negative controls. DDX3 (GenBank acc. no. NM_001193417) was used as a reference housekeeping gene. The normalized ratio of the intensity of POLL-specific products in SI versus si98 is indicated on bottom of the panel. (b) Western blot analysis of protein levels of DNA pol λ in HeLa cells after 24, 48 and 72 h from transfection with siRNA oligonucleotides. SI, scrambled control; si97, si98, POLL-specific siRNAs. Tubulin was used as the loading control. The normalized ratios of the intensity of DNA pol λ specific signal in SI versus si98 are indicated on bottom of the panel. (c) Percentage of S phase HeLa cells at different times after transfection with the SI scrambled control or the anti-POLL-specific si98 siRNA as determined by flow cytometry. Each measurement was performed in triplicate. Error bars are SD. P-values were calculated by two-tailed Student’s t-test. (d) Western blot analysis of protein levels of DNA pol λ in U2KDL cells after 24, 48 and 72 h of doxycycline induction. Actin was used as loading control. The normalized ratios of the intensity of DNA pol λ-specific signal in control (t0) versus silenced cells are indicated on bottom of the panel. (e) As in panel d, but at longer time points of induction. (f) Grey bars: percentage of S phase U2KDL cells at different times after doxycycline induction as determined by flow cytometry. Black bar, percentage of S phase U2OS cells ectopically overexpressing DNA pol λ at 48 h. Each measurement was performed in triplicate. Error bars are SD. P-values were calculated by two-tailed Student’s t-test.
To assess the effects of DNA Pol λ down-regulation on cell cycle progression, cells were treated with si98 or the scrambled control and further analysed by flow cytometry to assess the effects of DNA pol λ down-regulation on cell cycle progression. As shown in Figure 1c, knockdown of DNA pol λ levels induced progressive accumulation of cells in S phase, which was maximal at 72 h post-transfection, corresponding to the lowest DNA pol λ protein levels, and then decreased at 96 h, possibly due to reduced silencing efficiency.
DNA polymerase λ is required for normal cell cycle progression in a stable knockdown U2OS cell line
Next, we established a stable clone of U2OS osteosarcoma cells with a chromosomally integrated cassette expressing a specific shRNA targeting DNA pol λ, under the control of a doxycycline inducible promoter (see ‘Materials and Methods’ section). We called this stable-inducible DNA pol λ knockdown cell line as U2KDL. The si98 sequence, the efficiency of which in knocking down DNA pol λ expression in U2OS, was validated in transient transfection (Figure 2d), was used to design the shRNA insert. As shown in Figure 1d, upon induction with doxycycline, DNA pol λ protein levels were reduced by 60% already after 24 h. Stable knockdown in the presence of doxycycline could be detected during 24 days of continuous induction (Figure 1e). Control U2OS parental cells exposed to doxycycline for an equivalent period of time showed no S phase delay (data not shown). Flow cytometric analysis revealed a significant accumulation of cells in S phase in the U2KDL cell line (Figure 1f and Supplementary Figure S1a), confirming the results obtained using the transient siRNA approach in HeLaS3 cells and suggesting that S phase delay is not restricted to a certain cell type. Growth curves of U2KDL in the absence or presence of doxycycline showed slower growth kinetics when DNA pol λ was silenced, in agreement with the S phase delay observed by flow cytometry (Supplementary Figure S1b and c). Additionally, cell viability was not affected in both these cell lines (data not shown). These results revealed a correlation between diminished levels of DNA pol λ and cell cycle progression. In order to test whether a similar effect could also be observed in the presence of an increased DNA pol λ expression level, the parental cell line U2OS was transfected either with the pcDNA3-polλ-Myc construct or with the empty vector pcDNA3, the former overexpressing DNA pol λ (pcDNAPOLL). Overexpression of DNA pol λ did not affect cell cycle progression (Figure 1f), clearly indicating that the S phase delay was specifically induced by a reduction in DNA pol λ levels.
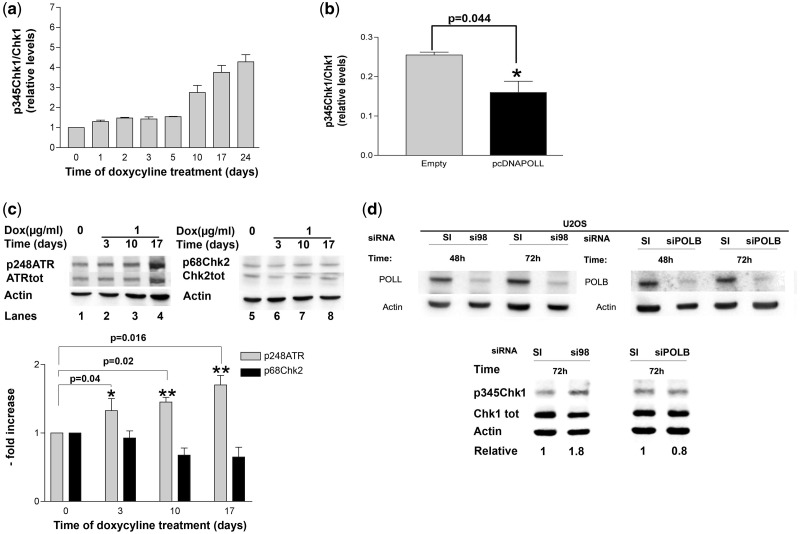
Figure 2.
DNA pol λ knockdown induces checkpoint activation. (a) Relative levels of p345Chk1 with respect to total Chk1 protein in UK2DL cells after various times of doxycycline induction. p345Chk1 and total Chk1 western blot signal intensities were normalized to actin controls before calculating their ratios. Each measurement was performed in triplicate. Error bars are SD. (b) Relative levels of p345Chk1 with respect to total Chk1 protein in U2OS parental cells (grey bar) or ectopically overexpressing DNA pol λ (black bar) at 48 h. p345Chk1 and total Chk1 western blot signal intensities were normalized to actin controls before calculating their ratios. Each measurement was performed in triplicate. Error bars are SD. P-values were calculated by two-tailed Student’s t-test. (c) Top panel: western blot analysis of protein levels of p248ATR and total ATR (lanes 1–4) or p68Chk2 and total Chk2 (lanes 5–8) in U2KDL cells after various times of doxycycline induction. Actin was used as loading control. Bottom panel: relative levels of p248ATR (grey bars) or p68Chk2 (black bars) with respect to total ATR and Chk2, respectively, in U2KDL cells after various times of doxycycline induction. Western blot signal intensities were normalized to actin controls before calculating their ratios. Each measurement was performed in triplicate. Error bars are SD. P-values were calculated by two-tailed Student’s t-test. (d) Top panel: western blot analysis of protein levels of DNA pol λ (left) and β (right) in U2OS cells after 48 h and 72 h from transfection with anti-POLL (si98) or anti-POLB (siPOLB) siRNA oligonucleotide or scrambled controls (SI). Bottom Panel: western blot analysis of protein levels of p345Chk1 and total Chk1 in U2OS cells after 72 h from transfection with anti-POLL (si98) or anti-POLB (siPOLB) siRNA oligonucleotide or scrambled controls (SI). Actin was used as loading control. Normalized ratios of the intensity of p345Chk1 and total Chk1 in control versus silenced cells were used to calculate the relative values indicated on bottom of the panel.
Silencing of DNA polymerase λ, but not DNA polymerase β, induces checkpoint activation
To investigate the mechanism underlying the observed delay in S phase in DNA pol λ knockdown cells, we analysed the phosphorylation status of the checkpoint kinase Chk1 in uninduced versus induced U2KDL cells. Chk1 is phosphorylated on Ser 345 by the kinase ATR, in response to DNA replication stress. Western blot analysis with specific antibodies against the Ser 345 phosphorylated form of Chk1 (p345Chk1) showed a basal level of p345Chk1 in U2OS cells, likely reflecting the steady-state level of checkpoint activation as a consequence endogenous DNA damage occurring during unperturbed replication. However, the levels of p345Chk1 significantly increased upon knockdown of DNA pol λ expression and the p345Chk1 form progressively accumulated as the repression of DNA pol λ was prolonged over time, reaching >4-fold increase after 24 days of continuous doxycycline treatment (Figure 2a). Control U2OS parental cells exposed to doxycycline for an equivalent time period showed no increase in p345Chk1 (Supplementary Figure 1d). No increase in p345Chk1 was observed in pcDNAPOLL cells overexpressing DNA pol λ, (Figure 2b) clearly indicating that checkpoint activation was specifically due to DNA pol λ down-regulation. On the opposite, a small, but statistically significant, decrease in p345Chk1 levels were observed in DNA pol λ overexpressing cells (Figure 2b). S phase checkpoint activation is initiated by phosphorylation of the kinase ATR on Ser 248, which, in turn, phosphorylates Chk1. The enrichment in the phosphorylated form of ATR (p248ATR), with respect to the total ATR present in the U2KDL cells, was next checked by western blotting with specific antibodies. As shown in Figure 2c, p248ATR levels increased after induction of DNA pol λ knockdown. While the ATR-Chk1 axis is specific for S phase DNA damage, another checkpoint pathway operates in response to DNA damage, which is dependent upon phosphorylation of the kinase Chk2 at Thr 68 by ATM. Under these conditions, no increase in p68Chk2 was observed upon induction of DNA pol λ knockdown (Figure 2c), indicating that down-regulation of DNA pol λ specifically triggered the activation of the ATR/Chk1 S phase checkpoint axis. DNA pol β is a close homologue of DNA pol λ, which also belongs to DNA pol family X, that plays a major role in BER. In order to assess whether down-regulation of DNA pol β was also capable of inducing checkpoint activation, we silenced its expression by transiently transfecting the U2OS parental cell line with a specific siRNA. As shown in Figure 2d, down-regulation of DNA pol β did not result in increased Chk1 phosphorylation, indicating that checkpoint activation was specifically triggered by reduced levels of DNA pol λ only.
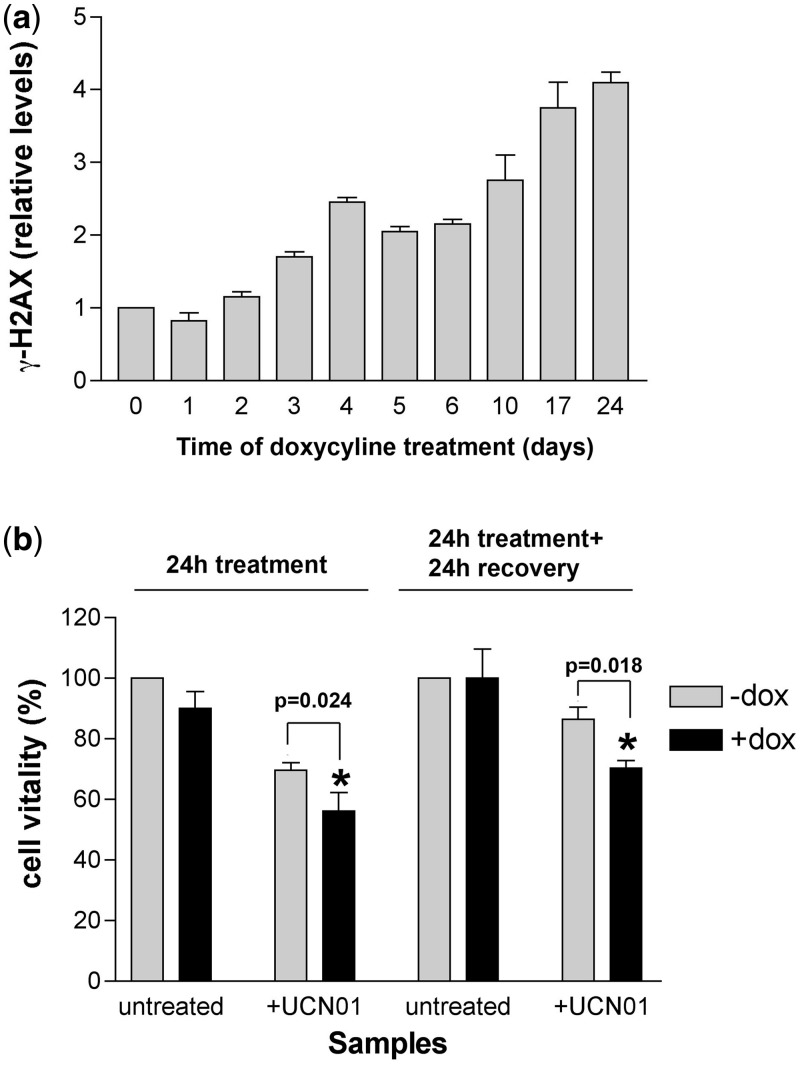
Silencing of DNA polymerase λ induces replicative stress and reduces cell viability when Chk1 is inhibited
The persistent activation of the S phase checkpoint observed in the induced U2KDL cells suggests that a reduced expression of DNA pol λ results in DNA replication stress. Accumulation of the H2AX histone variant phosphorylated at Ser139 (γ-H2AX) has long been recognized as an early marker of replication stress. Thus, we aimed to check whether γ-H2AX levels increased upon knockdown of DNA pol λ. As shown in Figure 3a, increased levels of γ-H2AX were detected in U2KDL cells upon doxycycline treatment. The γ-H2AX accumulation could be detected up to 24 days of continuous DNA pol λ knockdown, with a kinetic similar to that of p345Chk1 (compare Figure 3a with Figure 2a). An increase in the number of nuclear γ-H2AX foci in doxycycline-treated U2KDL cells was also detected by immunofluorescence (Supplementary Figure S1e). To understand whether persistent Chk1 phosphorylation was required to protect cells from the consequences of the replication fork stress generated in the absence of DNA pol λ, U2KDL cells were incubated in the absence or presence of the specific Chk1 inhibitor UCN01, for 24 h, followed by 24 h of recovery. To induce a DNA Pol λ knockdown both during treatment and in the recovery phase, a parallel experiment was performed in the continuous presence of doxycycline. It should be noted that UCN01 does not prevent Chk1 phosphorylation by ATR, but inhibits its kinase activity, thus blocking downstream signalling. As shown in Figure 3b, treatment with UCN01 together with DNA pol λ knockdown, resulted in a decrease in cell viability, both 24 h post-treatment and following the 24 h of recovery step, with respect to U2KDL cells expressing DNA pol λ. Thus, a functional checkpoint is important for cell survival in the presence of reduced levels of DNA pol λ.
Figure 3.
DNA pol λ knockdown induces replication stress and reduces cell viability when Chk1 is inhibited. (a) Levels of γ-H2AX in U2KDL cells after various times of doxycycline induction. γ-H2AX western blot signal intensities were normalized to the actin controls. Each measurement was performed in triplicate. Error bars are SD. (b) Effects of UCN01 on cell viability (expressed as % of viable cells) in U2KDL cells in the absence (grey bars) or in the presence (black bars) of doxycycline induction. Viability was measured at the end of 24 h treatment and after additional 24 h of recovery. Each measurement was performed in triplicate. Error bars are SD. P-values were calculated by two-tailed Student’s t-test.
These data clearly suggested a functional correlation among DNA pol λ knockdown, DNA replication stress and checkpoint activation, even in the absence of exogenous DNA damaging treatments.
Expression of DNA polymerase λ is induced by DNA damage
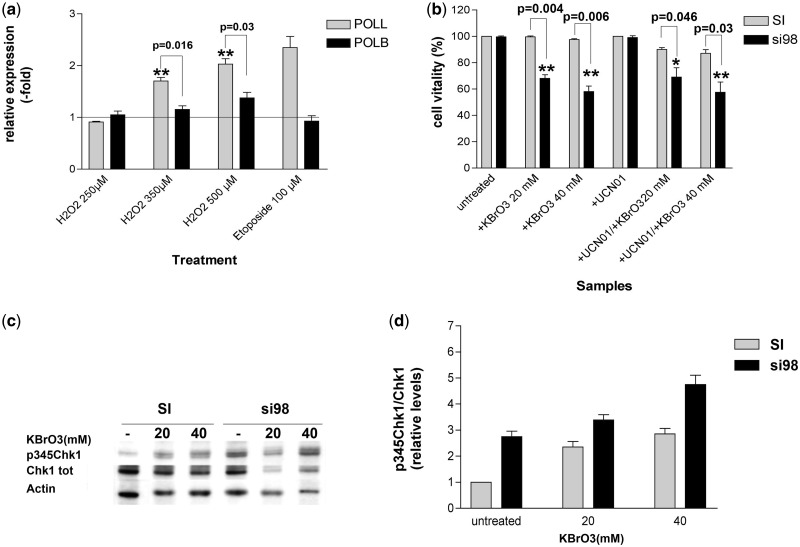
The results presented so far, indicated an involvement of DNA pol λ in the DNA damage response pathway during DNA replication. Since DNA pol λ has been shown to be involved in TLS and NHEJ DNA repair pathways, we aimed to evaluate whether the expression levels of DNA pol λ were changed upon induction using different DNA damaging agents. Since some of the functions of DNA pol λ can be also fulfiled by DNA pol β, we also tested the expression levels of the latter after DNA damaging treatments. To evaluate the response to oxidative stress, HeLaS3 cells were treated with increasing doses (250, 350 and 500 µM) of hydrogen peroxide (H2O2) for 4 h, whereas the induction of DSBs was performed using a single dose (100 µM) of etoposide for 3 h. Expression levels of DNA pols λ and β were measured following 24 h of recovery, by real-time qPCR and plotted as fold induction with respect to the expression levels of control untreated cells. As shown in Figure 4a, both oxidative stress and DSBs caused ∼2-fold increase in the levels of DNA pol λ. The levels of DNA pol β were not affected by etoposide, in agreement with the notion that this enzyme is not involved in DSBs repair, and only slightly increased by high doses of H2O2. Thus, it appears that only DNA pol λ levels are significantly regulated in response to different damaging treatments, further reinforcing its role in the cellular DNA damage response pathways.
Figure 4.
DNA pol λ protects cells from oxidative stress independently from checkpoint activation. (a) Expression levels (fold induction relative to untreated cells) of POLL (grey bars) and POLB (black bars) genes as determined by qRT–PCR in HeLa cells exposed to different genotoxic agents. Each measurement was performed in quadruplicate. Error bars are SD. P-values were calculated by two-tailed Student’s t-test. (b) Cell viability (expressed as % of viable cells) in HeLa cells exposed to KBrO3, UCN01 or both, after transfection with scrambled (SI, grey bars) or POLL-specific (si98, black bars) siRNA. Each measurement was performed in triplicate. Error bars are SD. P-values were calculated by two-tailed Student’s t-test. (c) Western blot analysis of protein levels of p345Chk1 and total Chk1 in HeLa cells treated with increasing doses of KBrO3 after transfection with scrambled (SI) or POLL-specific (si98) siRNA. Actin was used as a loading control. (d) Relative levels of p345Chk1 with respect to total Chk1 protein in HeLa cells treated with increasing doses of KBrO3 after transfection with scrambled (SI, grey bars) or POLL-specific (si98, black bars) siRNA. p345Chk1 and total Chk1 western blot signal intensities were normalized to actin controls before calculating their ratios. Each measurement was performed in triplicate. Error bars are SD.
DNA polymerase λ protects cells from oxidative stress independently of checkpoint activation
The observed induction of DNA pol λ expression following oxidative DNA damage prompted us to further investigate whether reduced levels of DNA pol λ increased cell sensitivity to oxidation. HeLaS3 cells, transfected with either the anti-DNA pol λ si98 or with the scrambled control (SI) oligonucleotides, were treated with increasing doses of the oxidation agent KBrO3 alone, or in combination with the Chk1 inhibitor UCN01 and cell vitality was assessed. Efficient silencing was checked by western blotting analysis using anti-DNA pol λ antibodies (Supplementary Figure S2a). As shown in Figure 4b, silencing of DNA pol λ increased cell sensitivity towards oxidative DNA damage, but simultaneous inhibition of Chk1 did not further increase cell death. KBrO3 treatment alone induced an accumulation of p345Chk1 (Figure 4c and d) indicating that oxidative DNA damage can trigger checkpoint activation. As expected, silencing of DNA pol λ alone resulted in 2.8-fold increase in the p345Chk1 levels, when compared with control cells. Combination of KBrO3 and DNA pol λ knockdown led to an additive accumulation of p345Chk1. Taken together, these data, in agreement with earlier studies, indicate that DNA pol λ can protect cells from oxidative DNA damage; however, inhibiting the downstream signalling of Chk1 did not result in increased mortality in the absence of DNA pol λ. While this might be partially due to intrinsic differences between U2OS and HeLa cells, it nonetheless suggests the existence of an alternative pathway for oxidative DNA damage tolerance, which can compensate for DNA pol λ absence and is independent from Chk1.
DNA polymerase λ protects cells during replication fork restart and its depletion is synthetically lethal with a defective checkpoint
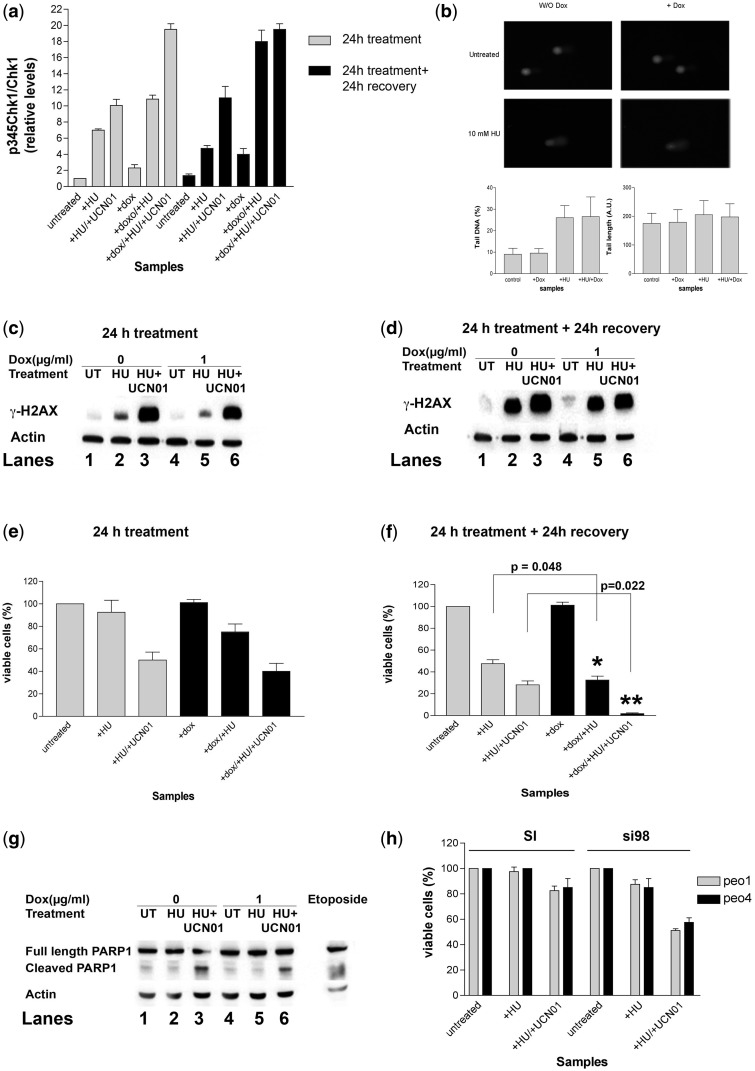
Next, we evaluated the effects of DNA pol λ knockdown under conditions of DNA replication stress, either in the presence or absence of a functional Chk1 kinase. To this aim, to cause replication fork stalling, U2KDL cells were treated with 10 mM HU for 24 h, in the absence or presence of doxycycline (to induce DNA pol λ silencing) as well as the Chk1 inhibitor UCN01. HU and UCN01 were then removed by changing the medium and cells were allowed to recover for 24 h, in the absence or presence of doxycycline. Thus, through this experiment, we aimed to evaluate the contribution of DNA pol λ under four different conditions: (i) during HU treatment; (ii) in the following recovery; (iii) during HU treatment together with Chk1 inhibition and (iv) in the recovery from the combined HU/UCN01 treatment. For each condition, we measured the three parameters as follows: (i) p345Chk1 levels, as an indicator of checkpoint activation; (ii) γ-H2AX levels as an indicator of replication stress and (iii) cell viability.
As shown in Figure 5a, HU treatment resulted in an increase of p345Chk1 levels, which also remained high in the following 24 h of recovery, consistent with the notion that checkpoint activation is necessary in order to restart fork progression. Inhibition of Chk1 during HU treatment caused a further increase in p345Chk1 levels, both at the end of treatment and in the following recovery stage. We also checked whether the HU treatment was inducing DNA damage by measuring the extent of DNA breaks by comet assay. As shown in Figure 5b, HU treatment significantly increased the number of DNA breaks with respect to untreated cells. On the other hand, DNA pol λ knockdown alone did not induce a significant accumulation of DNA breaks. Accordingly, combination of doxycycline with HU did not increase the amount of DNA breaks with respect to HU treatment alone.
Figure 5.
DNA pol λ protects cells during replication fork restart and its absence is synthetically lethal with a defective checkpoint. (a) Relative levels of p345Chk1 with respect to total Chk1 protein in U2KDL cells either untreated, or exposed to HU alone or in combination with UCN01, in the absence or presence of doxycycline induction. Samples were taken at the end of 24 h treatment (grey bars) and after additional 24 h of recovery (black bars). p345Chk1 and total Chk1 western blot signal intensities were normalized to actin controls before calculating their ratios. Each measurement was performed in triplicate. Error bars are SD. (b) Comet assay for the detection of DNA breaks in U2KDL cells either untreated or treated with doxycycline alone, HU alone or their combination. Samples were taken at the end of 24 h treatment. (c) Western blot analysis of protein levels of γ-H2AX in U2KDL cells either untreated, or exposed for 24 h to HU alone or in combination with UCN01, in the absence (lanes 1–3) or in the presence (lanes 4–6) of doxycycline induction. Actin was used as loading control. (d) As in panel b, but after 24 h of recovery. (e) Cell viability (expressed as % of viable cells) in U2KDL cells either untreated or exposed for 24 h to HU alone or in combination with UCN01, in the absence (grey bars) or presence (black bars) of doxycycline induction. Each measurement was performed in triplicate. Error bars are SD. (f) As in panel d, but after 24 h of recovery. Each measurement was performed in triplicate. Error bars are SD. P-values were calculated by two-tailed Student’s t-test. (g) Western blot analysis of protein levels of cleaved PARP1 and full-length PARP1 in U2KDL cells either untreated, or exposed to HU alone or in combination with UCN01, in the absence (lanes 1–3) or presence (lanes 4–6) of doxycycline induction. Samples were taken after 24 h of recovery. Actin was used as a loading control. (h) Cell viability (expressed as % of viable cells) in PEO1 (grey bars) or PEO4 (black bars) cells either untreated or exposed for 24 h to HU alone or in combination with UCN01, after transfection with scrambled (SI) or POLL-specific (si98) siRNA. Each measurement was in triplicate. Error bars are SD.
Following the 24 h of HU treatment, only a moderate accumulation of γ-H2AX was measured (Figure 5c, lane 2), which was increased dramatically in the presence of UCN01 (Figure 5c, lane 3). Interestingly, the levels of γ-H2AX significantly increased in the 24 h stage of recovery from the treatment with HU alone (Figure 5d, lane 2), indicating the presence of significant stress due to replication fork restart. Again, addition of the Chk1 inhibitor caused a further increase in γ-H2AX in the recovery phase as well (Figure 5d, lane 3). Overall, these data suggested that a functional checkpoint is important to limit the replication stress during HU treatment and in the recovery phase.
DNA pol λ knockdown caused an increased accumulation of the phosphorylated form of Chk1, both during the 24 h treatment with HU or HU/UCN01 and in the following 24 h of recovery, with respect to U2KDL cells treated in the absence of doxycycline (Figure 5a). Control parental U2OS cells treated with doxycycline showed no Chk1 activation during 6 days of continuous treatment (Supplementary Figure 2b), confirming that checkpoint activation was not due to antibiotic treatment. The experiments shown in Figure 3a clearly indicated that DNA pol λ silencing induced γ-H2AX accumulation after 72 h of induction. Thus, the contribution of DNA pol λ knockdown to the γ-H2AX increase observed after 24 h of HU (Figure 5c) and in the following 24 h of recovery (Figure 5d) was expected to be minimal. Indeed, no significant differences were noted in γ-H2AX levels in the different conditions tested, between doxycycline-induced and -uninduced cells (Figures 5c and d, compare lanes 4–6 with lanes 1–3), further confirming that DNA replication stress under these conditions was mainly due to HU treatment.
HU treatment alone did not cause a significant reduction in cell viability, while its combination with UCN01 induced ∼50% cell death (Figure 5e). Depletion of DNA pol λ under these conditions did not increase cell mortality (Figure 5e). When cell viability was measured following 24 h of recovery (Figure 5f), HU treatment alone induced ∼50% of cell mortality, while its combination with DNA pol λ silencing resulted in higher (70%) cell death that was proven to be statistically significant. Cells recovering from the combined HU/UCN01 treatment showed ∼70% mortality, consistently with the high level of the accumulated replicative stress (Figure 5c and d). Interestingly, DNA pol λ depletion during the recovery from the HU/UCN01 combination was lethal, causing almost complete (>98%) cell death. Analysis of PARP-1 cleavage by caspase 3 activity confirmed that, in the presence of the Chk1 inhibitor UCN01, cell death during recovery was due to apoptosis (Figure 5g).
These results show that DNA pol λ plays an important role in protecting cells from the lethal consequences of replicative stress during the recovery of stalled DNA replication forks and that this function becomes essential in the presence of a defective checkpoint.
The effects of DNA polymerase λ silencing and Chk1 inhibition during HU treatment are independent from HR
One of the consequences of stalled replication forks is the formation of DSBs, which are repaired by HR and NHEJ in mammalian cells, the latter requiring DNA pol λ. Thus, if DSBs persistence was the main cause of cell death in the absence of DNA pol λ and Chk1, one might speculate that cells defective in HR, and thus relying on NHEJ only for DSBs repair, should show a higher sensitivity to DNA pol λ knockdown following HU treatment. To test this hypothesis, ovarian cancer cells PEO1, deficient in the BRCA2 gene essential for HR, were incubated for 24 h in the absence or presence of HU and in the presence of either the specific anti-DNA pol λ si98 oligo or its scrambled control. Effective DNA pol λ silencing in the si98-treated cells was verified by western blotting (Supplementary Figure S2c). As shown in Figure 5h, PEO1 cells were highly resistant to checkpoint inhibition, showing moderate (20%) mortality in the presence of HU and UCN01. However, silencing of DNA pol λ under the same conditions significantly reduced cell viability (50% mortality) following 24 h of HU treatment. When a similar experiment was performed with the control PEO4 cell line, which is HR proficient, a similar decrease in cell viability (40%) was observed after HU treatment in the presence of the Chk1 inhibitor UCN01 (Figure 5h). Both PEO1 and PEO4 cell lines showed a similar increase in p345Chk1 in response to HU/UCN01 treatment (Supplementary Figure S2d), indicating that the checkpoint response was functional. Taken together, these results suggest that DNA pol λ protects cells from damages generated by replication fork stalling independently from the HR pathway.
DISCUSSION
In this work, we present evidence that DNA pol λ is functionally connected to the S phase checkpoint. Down-regulation of its expression resulted in the activation of the ATR-Chk1 branch of the cell cycle checkpoint, an S phase delay and DNA replication stress, as indicated by increased levels of γH2AX. Interestingly, no accumulation of DNA breaks or activation of Chk2 was observed, suggesting that accumulation of DSBs, which is a late consequence of perturbed replication fork progression, was not a major effect of DNA pol λ silencing. Thus, lack of DNA pol λ in S phase seems to induce a stress independently from its role in DSBs repair. This hypothesis is further supported by the observation that following the cell exposure to HU treatment, which was used to induce replication fork arrest, depletion of DNA pol λ in the presence of Chk1 inhibition caused significant lethality either in the presence or in the absence of a functional HR. This suggests that DNA pol λ becomes essential to counteract the consequences of replication fork stalling in the presence of a defective checkpoint, irrespectively from HR, indicating that DSBs accumulation was not a major effect of DNA pol λ inhibition. When the replication block is released, the S phase checkpoint allows forks to properly resume their functions (15). In fact, checkpoint mutants are unable to resume replication after removal of a HU block. Consistently, we showed that recovery from HU treatment in the absence of a functional Chk1 led to significant cell mortality. Down-regulation of DNA pol λ alone during the recovery also reduced cell viability, even in the presence of normal HR and checkpoint functions and resulted synthetically lethal when combined with Chk1 inhibition. These results reveal a previously undetected role of DNA pol λ in contributing to replication fork stability. It has to be mentioned that all the phenotypes reported here were observed in the presence of low DNA pol λ levels, since knockdown of the gene in our experimental setup was never complete. This ability of cells to sense variations in DNA pol λ levels during S phase and to react accordingly is consistent with the complex regulation of DNA pol λ recently described, which involves a finely tuned balance between phosphorylation by Cdk2/cyclinA in late S and G2 phases of the cell cycle, promoting its stability, and ubiquitination, targeting this enzyme for proteasomal degradation (12). Interestingly, the phenotype observed for DNA pol λ knockdown cells was distinct from the one observed following down-regulation of other specialized DNA pols. In fact, silencing of DNA pol η was found to inhibit the ATM/Chk2 pathway and to confer resistance to p53-dependent apoptosis upon UV irradiation (16). It has been shown that DNA pol ζ is required for cell proliferation of normal human cells (17) and that silencing of Rev3, the catalytic subunit of DNA pol ζ, activated the ATM/Chk2 pathway and caused G1 cell cycle arrest and senescence (18). Thus, it appears that DNA pol λ has a role in DNA replication, which is clearly distinct from other TLS and repair DNA pols.
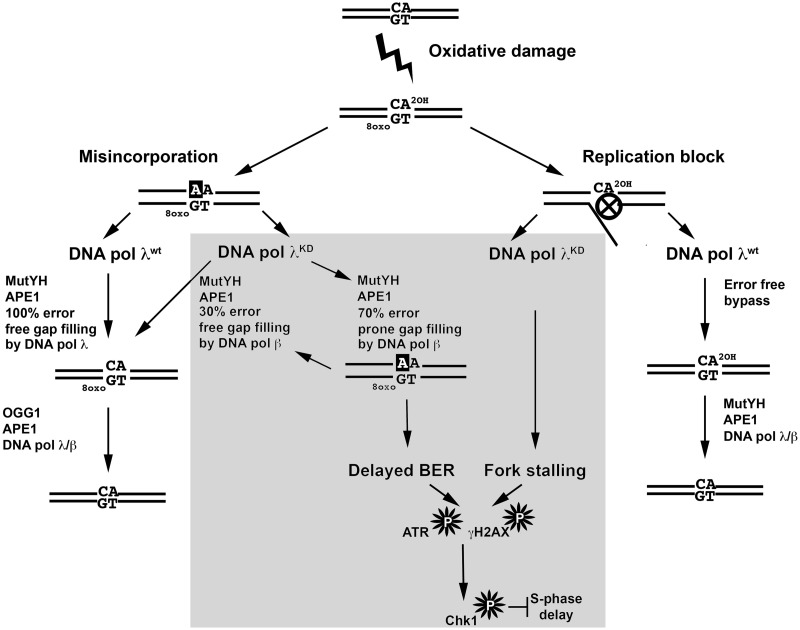
One possible mechanism through which DNA pol λ knockdown induced replicative stress could be a defective response to oxidative damage. Approximately 10 000 different oxidative DNA lesions are spontaneously generated per genome/per day. These, among many others, include the blocking 2-OH-A damage and the mutagenic 8-oxo-G lesion (1). Recently, we have shown that DNA pol λ is involved in TLS bypass of 2-OH-A and in the MutYH-initiated specialized BER sub-pathway acting during S phase to remove the A:8-oxo-G mismatches generated during DNA replication (19). In our hypothetical model, in the presence of DNA pol λ, 2-OH-A lesions can be bypassed by DNA pol λ-dependent TLS (Figure 6, right part). In addition, MutYH-initiated BER of A:8-oxo-G mismatches generated during replication proceeds with high efficiency, leading to removal of 8-oxo-G (Figure 6, left part). However, in the absence of DNA pol λ (Figure 6, middle panel) 2-OH-A can lead to replication fork pausing, in addition BER of A:8-oxo-G is performed by DNA pol β with low efficiency, resulting in delayed repair and accumulation of 1-nt gapped ss break intermediates. These effects together can trigger ATR–Chk1 activation, resulting in S phase delay. Consistently with a major role of DNA pol λ in oxidative DNA damage response, we found that the down-regulation of DNA pol λ sensitized cells to exogenous oxidative stress, which is also in line with previous findings (20). The S phase checkpoint was activated in response to oxidative DNA damage, but Chk1 inhibition combined with DNA pol λ silencing did not result in increased cell death. It is likely that DNA pol β functions as a backup for DNA pol λ in the S phase specific BER of oxidative DNA damage, without being affected by Chk1 inhibition. Accordingly, we show that silencing of DNA pol β, when DNA pol λ is expressed, does not lead to Chk1 phosphorylation. This suggests a functional hierarchy between these two DNA pols, where DNA pol λ plays a main role in S phase and is connected to the ATR/Chk1 pathway, while DNA pol β, the main BER DNA pol, is acting throughout the cell cycle and can eventually compensate for DNA pol λ absence under normal physiological conditions.
Figure 6.
A model for the interaction of DNA pol λ with the S phase checkpoint. For details see the ‘Discussion’ section.
Given the multiple roles of DNA pol λ in promoting genomic stability, as also unveiled in this study, it seems surprising that it is not essential for cell viability. In fact, DNA pol λ ablation does not affect viability and fertility of knockout (KO) mice (21,22) and mouse embryonic fibroblasts derived from DNA pol λ KO mice were more sensitive to oxidative stress, but responding normally to DNA alkylating agents or ionizing radiation (23,24). Our results provide the first insight, to our knowledge, into the mechanism which allows survival of cells with low DNA pol λ levels. We show that cells silenced for DNA pol λ for >20 generations do replicate, but they also constitutively activate the S phase checkpoint. This phenomenon could be viewed as a sort of adaptation mechanism, triggered by the higher levels of replicative stress caused by reduced levels of DNA pol λ with respect to normal cells, as visualized by constant γ-H2AX accumulation. Accordingly, inhibition of Chk1 reduced viability of DNA pol λ silenced cells. Constitutive activation of checkpoint as an adaptive response to defects in DNA replication proteins has been described so far. The 46BR.1G1 cell line, for example, has a genetic defect resulting in constitutively low levels of DNA ligase I, an enzyme involved in DNA replication and repair. These cells are viable and exhibit only a very minor cell cycle delay, in spite of a constant accumulation of ss DNA breaks and DSBs at each replicative cycle, and their survival is dependent upon constant activation of the DSBs-specific response mediated by ATM/Chk2 (25).
The absolute dependence of cells deficient in DNA pol λ expression from the ATR/Chk1 pathway renders them less able to tolerate DNA damage or replication stresses, and we showed that DNA pol λ knockdown is synthetically lethal when Chk1 is inhibited under perturbed DNA replication conditions in different model cancer cell lines. These results can have potential implications for anticancer chemotherapy. In fact, DNA pol λ has been found overexpressed in many cancer cell types (26). This observation can be, at least partially, explained by our data pointing to a role of DNA pol λ in protecting proliferating cells from DNA replication stress. In addition, the observation that the lack of DNA pol λ is well tolerated in normal cells, but can become synthetically lethal for cells with defects in the checkpoint pathways, paves the way to the exploitation of this enzyme as a novel therapeutic target (27).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1 and 2.
FUNDING
The Italian Cancer Research Association AIRC [IG-12084 to G.M.]; Swiss National Science Foundation [31003A_133100/1 to U.W. and U.H.]; University of Zurich (Switzerland) (to U.H., B.v.L. and N.G.). Fellowship from the PhD School in Biomolecular Sciences and Biotechnology—Institute for Advanced Studies IUSS, Pavia (Italy) (to E.Z.). Funding for open access charge: AIRC.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Simone Sabbioneda, IGM-CNR, Pavia (Italy), for critical reading of the article and Dr Carla Rohrer Bley for the support.
REFERENCES
- 1.Collins AR. Oxidative DNA damage, antioxidants, and cancer. Bioessays. 1999;21:238–246. doi: 10.1002/(SICI)1521-1878(199903)21:3<238::AID-BIES8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 2.Amoroso A, Crespan E, Wimmer U, Hubscher U, Maga G. DNA polymerases and oxidative damage: friends or foes? Curr. Mol. Pharmacol. 2008;1:162–170. doi: 10.2174/1874467210801020162. [DOI] [PubMed] [Google Scholar]
- 3.Barone F, McCulloch SD, Macpherson P, Maga G, Yamada M, Nohmi T, Minoprio A, Mazzei F, Kunkel TA, Karran P, et al. Replication of 2-hydroxyadenine-containing DNA and recognition by human MutSalpha. DNA Repair. 2007;6:355–366. doi: 10.1016/j.dnarep.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelly TJ, Brown GW. Regulation of chromosome replication. Annu. Rev. Biochem. 2000;69:829–880. doi: 10.1146/annurev.biochem.69.1.829. [DOI] [PubMed] [Google Scholar]
- 5.Hubscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu. Rev. Biochem. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- 6.Markkanen E, Hubscher U, van Loon B. Regulation of oxidative DNA damage repair: the adenine:8-oxo-guanine problem. Cell Cycle. 2012;11:1070–1075. doi: 10.4161/cc.11.6.19448. [DOI] [PubMed] [Google Scholar]
- 7.Hubscher U, Maga G. DNA replication and repair bypass machines. Curr. Opin. Chem. Biol. 2011;15:627–635. doi: 10.1016/j.cbpa.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 8.Crespan E, Hubscher U, Maga G. Error-free bypass of 2-hydroxyadenine by human DNA polymerase lambda with Proliferating Cell Nuclear Antigen and Replication Protein A in different sequence contexts. Nucleic Acids Res. 2007;35:5173–5181. doi: 10.1093/nar/gkm568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maga G, Villani G, Crespan E, Wimmer U, Ferrari E, Bertocci B, Hübscher U. 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature. 2007;447:606–608. doi: 10.1038/nature05843. [DOI] [PubMed] [Google Scholar]
- 10.Maga G, Crespan E, Wimmer U, van Loon B, Amoroso A, Mondello C, Belgiovine C, Ferrari E, Locatelli G, Villani G, et al. Replication protein A and proliferating cell nuclear antigen coordinate DNA polymerase selection in 8-oxo-guanine repair. Proc. Natl Acad. Sci. USA. 2008;105:20689–20694. doi: 10.1073/pnas.0811241106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol. Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 12.Markkanen E, van Loon B, Ferrari E, Parsons JL, Dianov GL, Hübscher U. Regulation of oxidative DNA damage repair by DNA polymerase lambda and MutYH by cross-talk of phosphorylation and ubiquitination. Proc. Natl Acad. Sci. USA. 2012;109:437–442. doi: 10.1073/pnas.1110449109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahm SH, Park JH, Ko SI, Lee YR, Chung IS, Chung JH, Kang LW, Han YS. Knock-down of human MutY homolog (hMYH) decreases phosphorylation of checkpoint kinase 1 (Chk1) induced by hydroxyurea and UV treatment. BMB Rep. 2011;44:352–357. doi: 10.5483/BMBRep.2011.44.5.352. [DOI] [PubMed] [Google Scholar]
- 14.Wimmer U, Ferrari E, Hunziker P, Hubscher U. Control of DNA polymerase lambda stability by phosphorylation and ubiquitination during the cell cycle. EMBO Rep. 2008;9:1027–1033. doi: 10.1038/embor.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Branzei D, Foiani M. The checkpoint response to replication stress. DNA Repair. 2009;8:1038–1046. doi: 10.1016/j.dnarep.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 16.Liu G, Chen X. DNA polymerase eta, the product of the xeroderma pigmentosum variant gene and a target of p53, modulates the DNA damage checkpoint and p53 activation. Mol. Cell. Biol. 2006;26:1398–1413. doi: 10.1128/MCB.26.4.1398-1413.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lange SS, Wittschieben JP, Wood RD. DNA polymerase ζ, is required for cell proliferation of normal human cells. Nucleic Acids Res. 2012;40:4473–4482. doi: 10.1093/nar/gks054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knobel PA, Kotov IN, Felley-Bosco E, Stahel RA, Marti TM. Inhibition of REV3 expression induces persistent DNA damage and growth arrest in cancer cells. Neoplasia. 2011;13:961–970. doi: 10.1593/neo.11828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Loon B, Hubscher U. An 8-oxo-guanine repair pathway coordinated by MUTYH glycosylase and DNA polymerase lambda. Proc. Natl Acad. Sci. USA. 2009;106:18201–18206. doi: 10.1073/pnas.0907280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braithwaite EK, Kedar PS, Lan L, Polosina YY, Asagoshi K, Poltoratsky VP, Horton JK, Miller H, Teebor GW, Yasui A, et al. DNA polymerase lambda protects mouse fibroblasts against oxidative DNA damage and is recruited to sites of DNA damage/repair. J. Biol. Chem. 2005;280:31641–31647. doi: 10.1074/jbc.C500256200. [DOI] [PubMed] [Google Scholar]
- 21.Bertocci B, De Smet A, Weill JC, Reynaud CA. Nonoverlapping functions of DNA polymerases mu, lambda, and terminal deoxynucleotidyltransferase during immunoglobulin V(D)J recombination in vivo. Immunity. 2006;25:31–41. doi: 10.1016/j.immuni.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 22.Bertocci B, De Smet A, Flatter E, Dahan A, Bories JC, Landreau C, Weill JC, Reynaud CA. Cutting edge: DNA polymerases mu and lambda are dispensable for Ig gene hypermutation. J. Immunol. 2002;168:3702–3706. doi: 10.4049/jimmunol.168.8.3702. [DOI] [PubMed] [Google Scholar]
- 23.Braithwaite EK, Kedar PS, Stumpo DJ, Bertocci B, Freedman JH, Samson LD, Wilson SH. DNA polymerases beta and lambda mediate overlapping and independent roles in base excision repair in mouse embryonic fibroblasts. PLoS One. 2010;5:e12229. doi: 10.1371/journal.pone.0012229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vermeulen C, Bertocci B, Begg AC, Vens C. Ionizing radiation sensitivity of DNA polymerase lambda-deficient cells. Radiat. Res. 2007;168:683–688. doi: 10.1667/RR1057R.1. [DOI] [PubMed] [Google Scholar]
- 25.Soza S, Leva V, Vago R, Ferrari G, Mazzini G, Biamonti G, Montecucco A. DNA ligase I deficiency leads to replication-dependent DNA damage and impacts cell morphology without blocking cell cycle progression. Mol. Cell. Biol. 2009;29:2032–2041. doi: 10.1128/MCB.01730-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albertella MR, Lau A, O'Connor MJ. The overexpression of specialized DNA polymerases in cancer. DNA Repair. 2005;4:583–593. doi: 10.1016/j.dnarep.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 27.Maga G, Hubscher U. Repair and translesion DNA polymerases as anticancer drug targets. Anticancer Agents Med. Chem. 2008;8:431–447. doi: 10.2174/187152008784220348. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.