Abstract
MiR-210 is up-regulated in multiple cancer types but its function is disputable and further investigation is necessary. Using a bioinformatics approach, we identified the putative target genes of miR-210 in hypoxia-induced CNE cells from genome-wide scale. Two functional gene groups related to cell cycle and RNA processing were recognized as the major targets of miR-210. Here, we investigated the molecular mechanism and biological consequence of miR-210 in cell cycle regulation, particularly mitosis. Hypoxia-induced up-regulation of miR-210 was highly correlated with the down-regulation of a group of mitosis-related genes, including Plk1, Cdc25B, Cyclin F, Bub1B and Fam83D. MiR-210 suppressed the expression of these genes by directly targeting their 3′-UTRs. Over-expression of exogenous miR-210 disturbed mitotic progression and caused aberrant mitosis. Furthermore, miR-210 mimic with pharmacological doses reduced tumor formation in a mouse metastatic tumor model. Taken together, these results implicate that miR-210 disturbs mitosis through targeting multi-genes involved in mitotic progression, which may contribute to its inhibitory role on tumor formation.
INTRODUCTION
MicroRNAs (miRNAs) are a class of small non-coding RNA of 19–24 nucleotides in length, which play pivotal roles in various biological and pathologic processes, including cell proliferation, differentiation, apoptosis, metabolism, organ morphology, angiogenesis and cancer (1–8). There is mounting evidence that miRNA expression is distinct between normal and tumor tissues. In some instances, these differences are associated with the initiation and progression of cancer (9–13). By modulating oncogenic and tumor suppressor pathways, miRNAs can act as oncogenes or tumor suppressors, or oncomiRs or tumor-suppressors miRs, respectively (6,13–15).
Some miRNAs, such as miR-210, are up-regulated in multiple cancer types (16–19). However, they cannot simply be described as oncomiRs or tumor-suppressor miRs because of the wide array of targets involved in regulating different cellular functions. MiR-210 is a hypoxia-induced gene regulated by (hypoxia-inducible factor 1) HIF-1α and plays various roles in the cells (7,17,19–21). In a murine model of myocardial infarction, miR-210 inhibited apoptosis and improved angiogenesis and cardiac function through potential targets Efna3 and Ptp1b (22). MiR-210 also down-regulates mitochondrial function by repressing the expression of iron–sulfur cluster scaffold homolog and COX10 (cytochrome c oxidase assembly protein) (23). It has recently been reported that the expression of miR-210 is induced during erythroid differentiation in a time- and dosage-dependent manner. Consistent with increased expression of the fetal gamma-globin genes, miR-210 might be involved in erythropoiesis (24). The role of miR-210 in cancer is complex and disputable. Data from clinical cancer samples show that miR-210 expression is inversely correlated with disease-free states and overall survival rates (16,18–19). However, other studies have reported that a remarkably high frequency of miR-210 gene deletions was found in ovarian cancer patients (25). Over-expression of ectopic miR-210 could function as a tumor suppressor because it attenuates cancer cell proliferation through down-regulating E2F3, fibroblast growth factor receptor-like 1, Homeobox protein Hox-A1 (HOXA1), Homeobox protein Hox-A9 (HOXA9) and Max-binding protein (MNT) (17,26,27).
Through a combination of genome-wide scale analysis and bioinformatics approach, we identified two functional gene groups as the potential targets of miR-210. One is involved in cell cycle regulation and the other is related to RNA processing. Here, we investigated the molecular mechanism and biological consequences of miR-210 in cell cycle regulation. We found that hypoxia-induced up-regulation of miR-210 was highly correlated with the down-regulation of Plk1, Cdc25B, Cyclin F, Bub1B and Fam83D genes involved in mitotic regulation. MiR-210 suppressed the expression of these genes by directly targeting their 3′-untranslated regions (3′-UTRs). Over-expression of exogenous miR-210 also disturbed mitotic progression and caused aberrant mitosis. Moreover, miR-210 mimic with pharmacological doses reduced tumor formation in a mouse metastatic tumor model during the initial stage. These results indicate that the inhibitory role of miR-210 on tumor formation may be partially due to mitotic disturbances in cancer cells.
MATERIALS AND METHODS
Cell culture and hypoxia induction
HeLa or CNE cells (Kunming Cell Bank, China) were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum at 37°C with 5% carbon dioxide (CO2). Hypoxia was induced by treating cells with deferoxamine mesylate (DFOM, Sigma-Aldrich Co., MO, USA) at a final concentration of 130 μM.
mRNA expression profile
Total RNA was isolated using Trizol reagent (Invitrogen) and the samples were analysed by mRNA microarray (Capital Bio Corp., Beijing, China). Procedures were performed as described in detail on the website of CapitalBio (http://www.capitalbio.com) (28). For each test and control samples, two hybridizations were performed by using a reversal fluorescent strategy. Genes showing a greater than 2-fold induction or repression in both microarray were selected as differentially expressed genes.
MiRNAs, siRNAs and transfection
MiRNA mimics were designed according to the miRBase sequence database (http://microrna.sanger.ac.uk). miRNA mimics, miRNA inhibitors and small interfering RNA (siRNA) duplexes were synthesized and purified by Shanghai GenePharma Co. (Shanghai, China). The sequences of these inhibitors are the exact antisense copy of the mature miRNAs, and all the nucleotides in the inhibitors contain 2′-O-Methyl modification (2′-OMe) modifications at each base. SiRNA duplexes with random sequences were used as a negative control (NC). The sequence of these miRNA and siRNAs were included in the Supplementary Table S1. Cells were transfected with siRNA or miRNAs duplexes using Lipofectaime 2000 (Invitrogen Corp., Carlsbad, CA, USA) according to manufacturer’s instructions.
Lentiviral preparation and transfection
The recombinant lentivirus for miR-210 (Lv-miR210), which expresses mature human miR-210, and negative control (Lv-NC), which expresses a scrambled RNA, were purchased from GenePharma (Shanghai, China). Protocol of lentivirus infection is according to the GenePharma Recombinant Lentivirus Operation Manual (http://www.genepharma.com).
Immunocytochemistry
CNE cells transfected with miR-210 or NC were fixed in 4% paraformaldehyde in phosphate buffered saline (PBS) for 15 min, followed by permeabilization with 0.25% Triton X-100 in PBS for 10 min at room temperature. Fixed samples were blocked with 3% bovine serum albumin in PBS for 1 h at room temperature before incubation with primary and secondary antibodies. Primary antibodies used were α-tubulin and γ-tubulin (Sigma-Aldrich Co., MO, USA). Secondary antibodies used were Alexa Fluor 488 or 555 conjugates (Invitrogen, USA). Images were captured using an Olympus confocal microscope.
Cell synchronization and flow cytometry analysis
CNE cells transfected with miR-210, miR-9, Plk1-siRNA or NC were synchronized at mitosis by nocodazole (100 ng/ml) treatment for 12 h. The mitotic cells were released from nocodazole block by washing the cells three times with PBS. At 1 h after nocodazole release, the cells were fixed in 70% ethanol in PBS overnight. Cells were then counterstained with propidium iodide (PI) and analysed for DNA content by use of a BD InfluxTM flow cytometer.
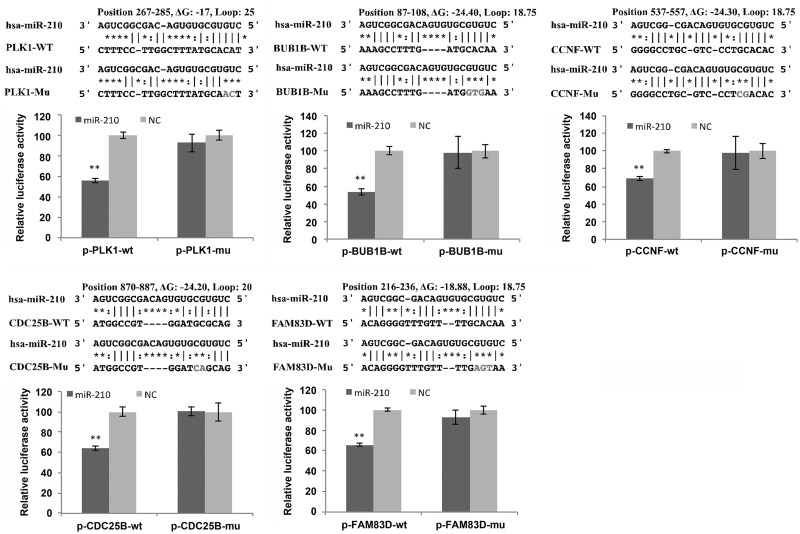
Luciferase activity assay
Reporter vectors were generated by inserting the 3′-UTRs of Plk1 (nt 267–285), Bub1B (nt 87–108), Cyclin F (nt 537–557), Cdc5B (nt 870–887), Fam83D (nt 216–236) and their corresponding mutated 3′-UTR fragments into [pRL-TK Vector(Promega Corporation) (pRL-TK)] plasmid (Promega, USA). Then, 3′-UTRs were amplified by PCR using the primers shown in Supplementary Table S1. Luciferase activities were assayed according to a previously published protocol (29). Briefly, CNE cells cultured in 24-well plate were transfected with 100 ng of plasmid and 20 nM of each miRNA using Lipofectamine 2000. Cells were collected at 30 h after transfection to perform Renilla luciferase activity assay.
Mice and tumor formation assays
Athymic nude mice aged 4 weeks were purchased from the Experimental Animal Center of Guang Zhou University of Chinese Medicine (Guang Zhou, China) and kept in pathogen-free animal facilities for 2 weeks at Tsinghua University Shenzhen Graduate School. The mice were divided into three groups for the treatment of miR-210, Plk1-siRNA or NC respectively, and each group had 8 mice. CNE cells were prepared for mouse tail vein injections. Briefly, the cells were suspended in PBS, and their viability assessed by trypan blue staining was typically >95%. Six-week-old mice were injected with approximately 5 × 105 CNE cells in the tail vein. Transferrin-polyethylenimine (TF-PEI)/modified-miRNA-mimics complex were delivered through tail vein injection every 3 days. After 6 injections, mice were euthanized and their lungs were removed and rinsed. The left lung lobes were collected to generate histological tablets to calculate the pulmonary metastases. The right lung lobes were fixed with formalin, embedded with paraffin for lung tissue sections, which were prepared at 5μm on a transverse plain. The sections were examined using H&E staining. The pulmonary metastases were counted and photographed. All experiments on animals were conducted according to the US Public Health Service’s Policy on Human Care and Use of Laboratory Animals.
RESULTS
Identification of miR-210 target genes in hypoxia-induced CNE cells
Results from mRNA microarray analysis revealed that 351 genes were down-regulated in hypoxia-induced CNE nasopharyngeal carcinoma cells. To clarify the potential targets of miR-210, we used FindTar, a prediction algorithm designed by our laboratory, to screen miR-210 targets from those down-regulated genes under hypoxia condition (30). Among them, 107 genes were predicted as the putative targets of miR-210 (Supplementary Table S2).
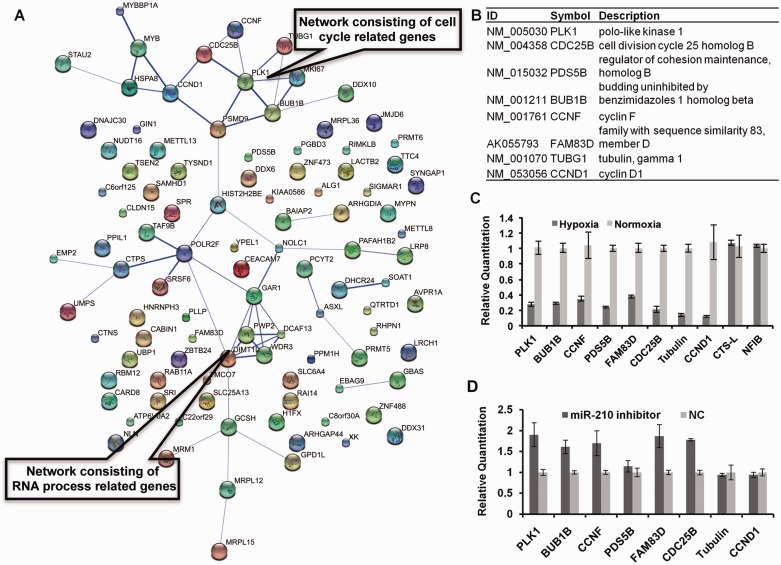
The putative targets of miR-210 were then uploaded into DAVID bioinformatics resources (http://david.abcc.ncifcrf.gov/) for functional annotation clustering and enrichment scoring. A higher enrichment score for a cluster indicated that the gene members within the cluster were involved in more important (enriched) terms in a given study (31). The results showed seven clusters with enrichment scores higher than 1.3, representing four biological functions, including structure molecules of organelle, RNA processing, cell cycle and a few methyltransferases (Supplementary Table S3). Search tool for the retrieval of interacting genes/proteins (http://string-db.org/) (STRING), a database of protein interactions, was then used to investigate functional associations among the candidate target genes of miR-210 and networks involved in the biological function of miR-210 (32). The 107 putative miR-210 genes were uploaded into STRING 9 (http://string-db.org/) and two areas where the genes cohered to form networks were identified. As shown in Figure 1A, one network consisted of cell cycle–related genes, and the other one related to RNA process. This result indicated that the role of miR-210 might be related to cell cycle regulation and RNA process.
Figure 1.
Identification of putative miR-210 target genes. (A) The functional association of miR-210 target genes. Totally 107 candidate target genes were uploaded into STRING 9 (http://string-db.org/), and two networks related to cell cycle and RNA process were identified. (B) Eight genes related to cell cycle were identified as putative miR-210 targets. (C) Hypoxia-induced down-regulation of PLK1, BUB1B, CCNF, PDS5B, FAM83D, CDC25B, CCND1 and TUBG1 mRNAs. CNE cells were treated with either Dimethyl Sulfoxide (DMSO) or DFOM at a final concentration of 130 µM. After 20 h of DFOM treatment, cells were harvested for qRT-PCR. Data are representative of three independent experiments. Cathepsin L (CTS-L) and Nuclear factor I/B (NFIB) were used as negative controls. (D) CNE cells were transfected with miR-210 inhibitor and treated with DFOM for 24 h. The down-regulation of PLK1, BUB1B, CCNF, FAM83D and CDC25B were reversed by miR-210 inhibitor. NC (small RNA with random sequence) was used as negative control.
In this investigation, we studied the function of miR-210 on cell cycle regulation. We analysed the functional cluster 4 from DAVID bioinformatics resources: 27 genes associated with the cell cycle were divided into 28 enriched functional annotations (Supplementary Table S3). After removing the overlapping genes, eight genes were identified from over 10 enriched functional annotations, including Plk1 [polo-like kinase 1 (PLK1)], Bub1B [budding uninhibited by benzimidazoles 1 homolog beta(yeast) or Mitotic checkpoint serine/threonine kinase BUB1 beta (BUB1B)], Cyclin F (CCNF), Pds5B [sister chromatid cohesion protein PDS5 homolog B or androgen induced inhibitor of proliferation (PDS5B)], Fam83D [family with sequence similarity 83, member D (FAM83D)], Cdc25B [cell division cycle 25 homolog B (S.pombe) (CDC25B)], γ-tubulin [tubulin, gamma 1 (TUBG1)] and Cyclin D1 (CCND1) (Figure 1B). All of the genes, except Cyclin D1, were related to mitosis. We examined their mRNA expression under normoxia or hypoxia condition. Results from quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) confirmed that hypoxic stress significantly suppressed the expression of these genes (Figure 1C). In contrast, inhibition of endogenous miR-210 with specific miRNA inhibitor could reverse the down-regulation of Plk1, Bub1B, Cyclin F, Pds5B and Cdc25B by hypoxic stress, suggesting that these genes might be miR-210 targets (Figure 1D).
MiR-210 inhibits tumor cell proliferation
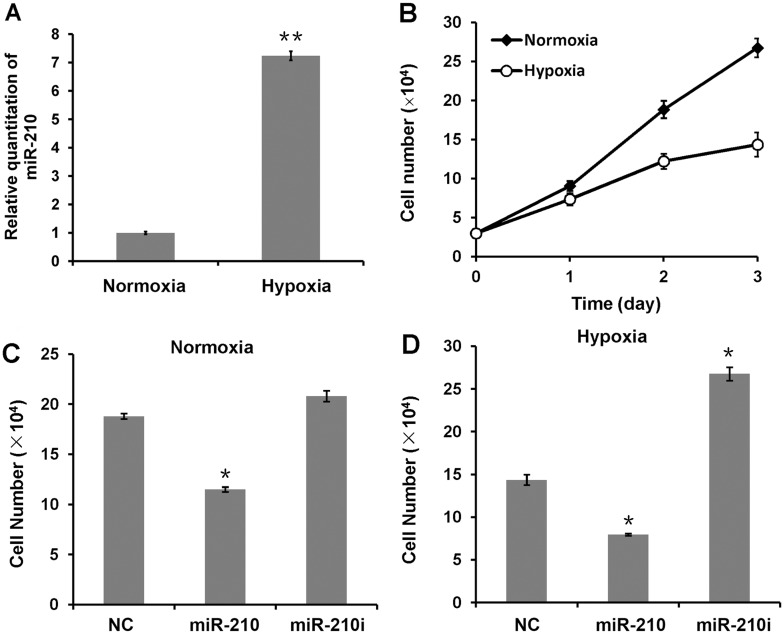
To investigate the correlation between miR-210 expression and cell proliferation, we examined the expression level of miR-210 under hypoxic stress. CNE cells were treated with the hypoxia mimetic agent DFOM for 24 h. The level of endogenous miR-210 was determined by qRT-PCR. As shown in Figure 2A, hypoxic stress induced a 7-fold increase of endogenous miR-210 expression. The cell proliferation assay also showed a retardation of cell growth in hypoxia-treated cells (Figure 2B). Then, we examined the effect of miR-210 over-expression on cell proliferation under normoxia or hypoxia condition. As shown in Figure 2C and D, over-expression of exogenous miR-210 significantly reduced the rate of cell proliferation. Conversely, inhibition of endogenous miR-210 restored cell proliferation rate, particularly in hypoxia-treated cells, when miR-210 is highly up-regulated (Figure 2D).
Figure 2.
MiR-210 inhibits tumor cell proliferation. (A) Hypoxia-induced up-regulation of miR-210 in CNE cells. Cells were treated with or without DFOM for 24 h. The samples were harvested for qRT-PCR to detect the expression of endogenous miR-210 under hypoxic stress or normoxia. (B) Hypoxic stress inhibits tumor cell proliferation. CNE cells were treated with or without DFOM for 3 days. The proliferation rate was determined by counting the cell density every 24 h. (C) Over-expression of miR-210 inhibits cell proliferation, and inhibition of endogenous miR-210 restores cell proliferation (D). CNE cells transfected with miR-210, miR-210 inhibitor or NC were treated with or without DFOM for 72 h, and the samples were harvested for proliferation assay. *P-value <0.05, two-tailed paired Student’s t-test.
MiR-210 down-regulates the expression of mitosis-related genes
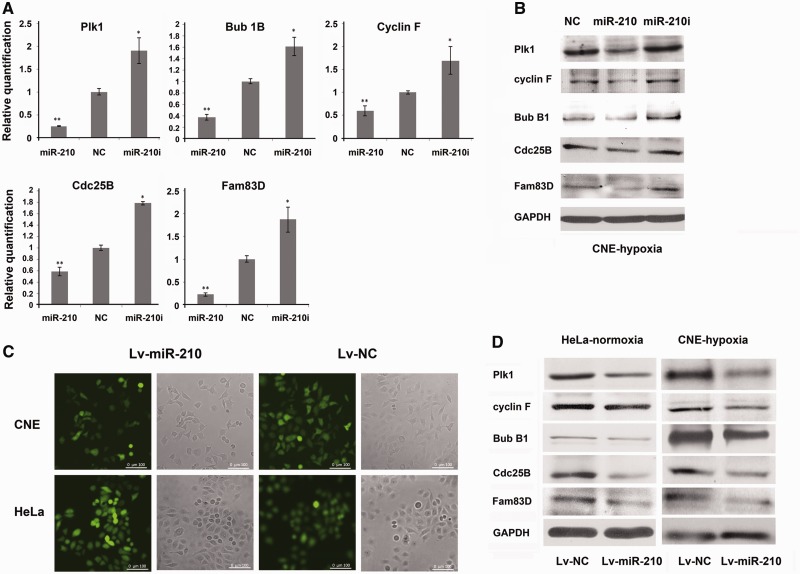
Bioinformatics analysis indicates that eight mitosis regulatory genes are putative targets of miR-210 in hypoxia-induced CNE cells. To validate, we over-expressed miR-210 in hypoxia-induced CNE cells and harvested cells for qRT-PCR and Western blotting. As shown in Figure 3A, miR-210 over-expression resulted in 40–80% reduction in transcript abundance for Plk1, Cdc25B, Bub1B, Cyclin F and Fam83D. Similarly, the protein expression of these genes was also down-regulated to various extents by exogenous miR-210. Among them, the protein level of Plk1, a key protein involved in both mitotic entry and spindle checkpoint, decreased almost 80% compared with control (Figure 3B). There was little, if any, reduction of Cyclin D1, Pds5B and γ-tubulin from both qRT-PCR and Western blotting (Data not shown).
Figure 3.
MiR-210 down-regulates the expression of mitosis-related genes. CNE cells transfected with miR-210, miR-210 inhibitor or NC were treated with DFOM for 24 h. Cells were harvested for qRT-PCR (A) or Western blotting (B) to detect the expression of PLK1, BUB1B, CCNF, FAM83D and CDC25B. (C) Lentivirus transfection of miR-210 or NC in CNE or HeLa cells; GFP was used as an indicator for transfection efficiency. (D) CNE or HeLa cells stably expressing miR-210 or NC were collected for Western blotting to detect the expression of PLK1, BUB1B, CCNF, FAM83D and CDC25B. *P-value <0.05, **P-value <0.01.
We also generated CNE and HeLa cells that stably expressed miR-210 and NC by lentiviral infection. For visualization, the GenePharma Supersilencing vector was designed to carry a green fluorescent protein (GFP) gene in its backbone (Figure 3C). We harvested the cells infected with either miR-210 or NC for Western blotting. It was shown that over-expression of miR-210 also down-regulated the protein expression of Plk1, Cyclin F, Bub1B, Cdc25B and Fam83D in both HeLa and CNE cells in normal culture condition (Figure 3D).
MiR-210 disturbs mitotic progression and causes defects of chromosome alignment and segregation
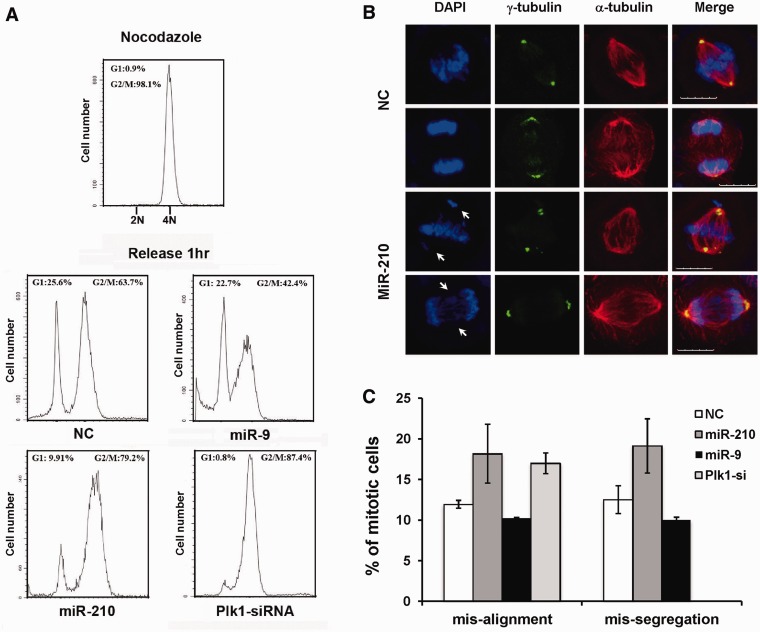
Having shown that miR-210 down-regulated the expression of some genes related to mitosis, we next investigated the function of miR-210 on mitotic progression. CNE cells transfected with NC, miR-210, Plk1-siRNA and miR-9 were synchronized at mitosis by nocodazole treatment, and the mitotic cells were released from nocodazole block for 1 h. The cells were fixed and stained with PI for flow cytometry analysis. As shown in Figure 4A, >25% of the NC-transfected cells exited mitosis and entered G1 phase at 1 h after nocodazole release. By contrast, there was a significant delay of mitotic exit in miR-210–transfected cells, as <10% of the cells entered G1. For comparison, we used miR-9 as a control miRNA because it had no predicted target genes related to mitosis. Similar to NC, 22.7% of G1 phase cells were observed in miR-9–transfected cells at 1 h after nocodazole release. Plk1, a major target of miR-210, plays an important role in mitotic spindle checkpoint. We found that depletion of Plk1 by siRNA almost completely blocked the mitotic exit and the majority of cells were arrested in mitosis after nocodazole release. These results indicate that miR-210 is involved in regulating mitotic progression.
Figure 4.
MiR-210 disturbs mitotic progression and causes aberrant mitosis. (A) CNE cells transfected with miR-210, miR-9, Plk1-siRNA or NC were synchronized at mitosis by nocodazole treatment. The mitotic cells were released from nocodazole block for 1 h. The cells were fixed and stained with PI for flow cytometry analysis. MiR-9, which has no predicted target genes related to mitosis, was used as a control miRNA. (B) CNE cells transfected with miR-210 or NC were fixed and subjected to co-immunofluorescence with α- and γ-tubulin antibodies, and the chromosomes were stained with DAPI. Scale bar: 10 µm. (C) CNE cells transfected with miR-210, miR-9, Plk1-siRNA or NC were partially synchronized by nocodazole treatment for 4 h, and then released for 1 h to quantify the number of aberrant mitosis. Data are representative of three independent experiments.
It is well known that some of the miR-210 targets, such as Plk1, Bub1B and cyclin F, are important regulators of mitotic spindle checkpoint. To investigate if miR-210 affected spindle checkpoint and chromosome segregation, we examined the mitotic spindle formation by co-immunofluorescence with α- and γ-tubulin antibodies. The morphology of the chromosomes was visualized by 4′,6-diamidino-2-phenylindole (DAPI) staining. As shown in Figure 4B, chromosome mis-alignment and mis-segregation were frequently observed in miR-210–transfected CNE cells. Aberrant centrosome amplification was also seen in a high frequency. The quantification of abnormal mitosis was shown in Figure 4C, and miR-210 induced a significant increase of chromosome alignment and segregation defects compared with NC and miR-9 controls. Silencing Plk1 resulted in metaphase arrest with a high percentage of chromosome mis-alignment. Therefore, miR-210 disturbs mitotic progression by inducing centrosome amplification, chromosome mis-alignment and mis-segregation, which might be due to its multi-target effect in mitosis.
MiR-210 regulates mitosis-related genes by targeting their 3′-UTRs
To examine if miR-210 regulates the expression of target genes via binding to the respective 3′-UTR regions, we designed pRLTK-luciferase reporter constructs, which contain the 3′-UTR regions of the target genes with putative miR-210 binding sites. Thus, miR-210 could bind to its target sites in the 3′-UTR and inhibit luciferase activity. As shown in Figure 5, miR-210 significantly suppressed luciferase activity when co-transfected with pRLTK-luciferase reporter constructs containing the 3′-UTRs of Plk1, Bub1B, Cyclin F, Cdc25B or Fam83D genes. To further confirm the direct interaction between miR-210 and its target genes, the binding sites in 3′-UTRs of the target genes were mutated. In comparison with wild-type reporter constructs, the mutations rescued the repressive effects of miR-210 on luciferase activity (Figure 5). These results demonstrated that miR-210 regulate the expression of multiple mitosis-related genes by directly binding to their 3′-UTR sites.
Figure 5.
MiR-210 directly targets the expression of Plk1, Bub1B, Cyclin F, CDC25B and Fam83D. Luciferase reporter vectors were generated by inserting the wild-type or mutated 3′UTRs of PLK1 (nt 267–285), BUB1B (nt 87–108), CCNF (nt 537–557), CDC25B (nt 870–887) or FAM83D (nt 216–236) into pRL-TK plasmid. The reporter vectors were then co-transfected into CNE cells with either miR-210 or NC. Cells were harvested for luciferase activity assay. Results shown are the mean±SD of triplicate determinations from three independent experiments. **P-value <0.01.
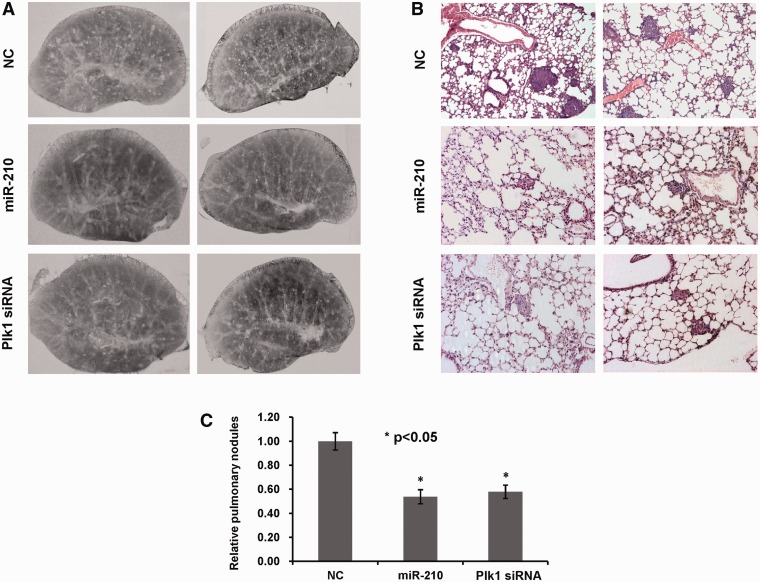
MiR-210 reduces tumor initiation in a mouse metastatic tumor model
To examine the effect of miR-210 on tumor initiation, we generated an experimental metastatic tumor model by intravenously injecting nude mice with CNE cells. From the next day of tumor cell injection, the mice were treated with miR-210, Plk1-siRNA or NC every 3 days. After 2 weeks, some white pulmonary nodules with 0.2–0.5 mm diameter were detected in histological tablets of mouse lung (Figure 6A). H&E staining from mouse lung tissue sections confirmed that these white pulmonary nodules were metastatic tumors (Figure 6B). We quantified the pulmonary nodules by microscopy and found that the number of pulmonary nodules in miR-210–treated mice was much less than that in NC groups. Plk1-siRNA showed a similar effect with miR-210 on tumor initiation (Figure 6C).
Figure 6.
MiR-210 reduces tumor initiation in a mouse metastatic tumor model. An experimental metastatic tumor model was generated though tail vein injection in nude mice with CNE cells. MiR-210 mimic or Plk1-siRNA was delivered intravenously. NC was used as negative control. After 6 times injection, mice were euthanized and their left lung lobes were collected to generate histological table (A) and the right lung lobes were removed for lung tissue sections. The sections were examined using H&E stain, and photographed (B). (C) The quantification of the white pulmonary nodules of metastatic tumors. *P-value <0.05. NC: small RNA with random sequence.
DISCUSSION
In patients with poor prognosis, the tumor growth rate is usually higher and the tumor size is also larger than that in patients with better prognosis (33). Oxygen homeostasis is frequently disrupted in tumor, and hypoxia appears to be strongly associated with tumor propagation and malignant progression. As a hypoxia-induced miRNA, miR-210 up-regulation is frequently observed in patients with poor prognosis (18,19). However, it does not mean that miR-210 is the cause of the poorer prognosis in cancer patients.
Multi-targets might be the most important biologic characteristic of miRNAs. Data from computational prediction and biological investigations of genome-wide scale suggest that one miRNA may have tens to hundreds target genes (34–36). Co-regulation of a group of functionally related genes to provoke detectable functional changes is a common mode of miRNA-mediated gene regulation (36). However, the identification of functionally related target gene groups of a miRNA is a challenge. Here, we combined the biologic approach of genome-wide scale and bioinformatics approach to identify the functionally related target genes of miR-210. We found that miR-210 target a group of mitosis-related genes. Through down-regulating several important mitotic regulators (Plk1, Cdc25B, Bub1B, cyclin F and Fam83D), miR-210 disturbs mitotic progression, exerts subtle effects on the key events of mitosis, and also inhibits tumor cells proliferation and tumor formation.
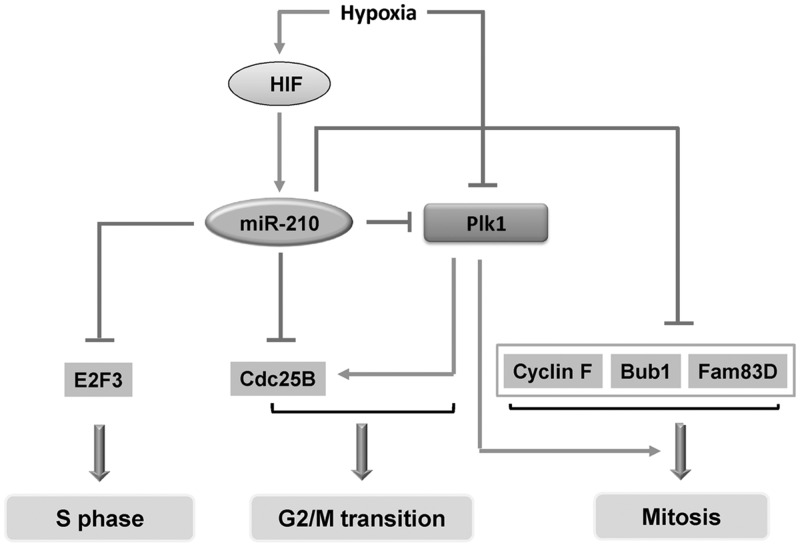
Numerous studies demonstrate that both Plk1 and Cdc25B control the onset of mitosis though a positive and negative feedback loop. The regulation of mitosis is complex. The Cdc25 phosphatase family plays an important role in G2-M transitions during unperturbed cell cycle (37,38). Among them, Cdc25B and Cdc25C are primarily required for entry into mitosis (39). An incipient activation of Cdk1/cyclin B is initiated by cdc25B during the G2/M transition, whereas the full activation of Cdk1/cyclin B is governed by Cdc25C at the onset of mitosis (39,40). The active Cdk1/cyclin B then phosphorylates Cdc25B and C, leading to an irreversible auto-amplification loop that drives cells into mitosis (41,42). On the other hand, Plk1 promotes the activation of Cdk1/cyclin B by activating Cdc25C (43). Plk1 also leads to a further increase in Cdk1 activity by additionally inhibiting Myt1 and inducing the degradation of Wee1 by phosphorylation (44–46). In addition to mitotic entry, Plk1 controls multiple steps during mitotic progression. Plk1 phosphoryaltes ninein-like protein and kizuna to promote the recruitment of γ-tubulin ring complexes and maintain the integrity of the centrosomes (45,47,48). Plk1 regulates sister chromatid resolution by promoting the removal of cohesins in prophase and facilitating their cleavage by separase (44,45). Plk1 is also essential for cytokinesis by activating the Rho GTPase, an activator of the actomyosin ring to stimulate the contraction of the contractile ring and promote cytokinesis (44,45). It has been demonstrated recently that over-expression of miR-210 induces growth suppression and G2/M arrest (49). Therefore, this investigation provides a potential mechanism that miR-210 may regulate G2/M transition by targeting Plk1 and Cdc25B (Figure 7).
Figure 7.
MiR-210 targets multiple genes involved in cell cycle regulation. MiR-210 controls G2/M transition by targeting Cdc25 B and Plk1. MiR-210 also govern mitotic progression through mediating the expression of Plk1, Bub1B, Cyclin F, Pds5B and Fam83D. In addition, miR-210 is involved in centrosome replication in S phase by targeting the expression of E2F3.
MiR-210 also suppresses the expression of Bub1B, Cyclin F and Fam83D. The checkpoint serine/threonine-protein kinase Bub1B localizes to the kinetochores and plays a role in the inhibition of the anaphase-promoting complex (50). Cyclin F physically associates with CP110, a protein essential for centrosome duplication. Knocking down cyclin F expression with siRNA induces multipolar spindles and asymmetric bipolar spindles with lagging chromosomes (51). Fam83D is also named as CHICA because it interacts with kinesin-like protein (KIF22)/chromokinesin Kid (KID) to the spindle microtubules and is required for proper chromosome congression and alignment during mitosis (52). In this study, we demonstrate that over-expression of miR-210 induces subtle effects on mitosis, including chromosome mis-alignment and mis-segregation, centrosome amplification. This effect could be due to the direct down-regulation of Bub1B, cyclin F and Fam83D. Recent study also reported that miR-210 can regulate centrosome duplication cycle by targeting the expression of E2F3 in renal carcinoma cells (49). Take together, the biological function of miR-210 on cell cycle progression, particularly mitosis, is determined by its multiple target effects. MiR-210 controls the onset of mitosis by regulating the expression of Cdc25B and Plk1, and it also governs the mitotic progression by targeting Plk1, Cyclin F, Bub1B and Fam83D (Figure 7).
Among the target genes of miR-210, Plk1 may play an integral role because of its multiple functions in mitosis and maintaining DNA integrity. Plk1 is frequently over-expressed in many tumor types and represents a target for anti-cancer therapy. Preclinical studies show that Plk1 siRNAs or small molecule inhibitors induce mitotic arrest and inhibit tumor growth (53). Here, we found that miR-210 reduced tumor formation in a mouse metastatic tumor model, and depletion of Plk1 by siRNA had a similar effect. One possibility might be that miR-210 exerts subtle effects on the key events of mitosis, which can destroy DNA integrity and eventually induce cancer cell death. On the other hand, miR-210–mediated down-regulation of other cell cycle regulatory genes, such as E2F3, may also play a role in repressing tumor cell proliferation (17,25,26).
Contradictory results, however, exist concerning the function of miR-210 as a pro- or anti-tumorigenesis gene. Analysing miR-210 expression in tumor samples indicates that miR-210 is over-expressed in most solid tumors (16,17,19,54); however, there is a high frequency of miR-210 gene copy deletion in ovarian cancer (25). As a robust hypoxia-inducible miRNA, miR-210 can either inhibit apoptosis (55,56), increase migration and invasion (57), or rather suppress tumor initiation (17) that depends on the tissue or cell models. In nude mice bearing nasopharyngeal carcinoma xenograft, we find that ectopic expression of miR-210 reduces tumor formation and growth during the initial stage, but not in the aggressive tumors. The possible explanation is that during the early stage of tumor formation, the level of endogenous miR-210 is relatively lower because the deprivation of oxygen is not significant. Thus, over-expression of high level of miR-210 could inhibit tumor cell proliferation though targeting multiple cell cycle regulatory genes. However, as a tumor grows, hypoxia induces a robust increase of endogenous miR-210, and tumor cells may gradually become tolerant to miR-210 and survive.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3.
FUNDING
China-Canada Joint Health Research (National Natural Science Foundation of China and Canadian Institutes of Health Research) [No. 30911120492 to Y.Z. and B.B.Y.]; Ministry of Health, China [2012ZX09102301-019]; International Cooperation Grant of Shenzhen [ZYA201007060079A]. Funding for open access charge: China-Canada Joint Health Research (National Natural Science Foundation of China and Canadian Institutes of Health Research) [No. 30911120492 to Y.Z. and B.B.Y.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The funding institutes had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 3.Shan SW, Lee DY, Deng Z, Shatseva T, Jeyapalan Z, Du WW, Zhang Y, Xuan JW, Yee SP, Siragam V, et al. MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat. Cell. Biol. 2009;11:1031–1038. doi: 10.1038/ncb1917. [DOI] [PubMed] [Google Scholar]
- 4.Kahai S, Lee SC, Lee DY, Yang J, Li M, Wang CH, Jiang Z, Zhang Y, Peng C, Yang BB. MicroRNA miR-378 regulates nephronectin expression modulating osteoblast differentiation by targeting GalNT-7. PLoS One. 2009;4:e7535. doi: 10.1371/journal.pone.0007535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hua Z, Lv Q, Ye W, Wong CK, Cai G, Gu D, Ji Y, Zhao C, Wang J, Yang BB, et al. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye G, Fu G, Cui S, Zhao S, Bernaudo S, Bai Y, Ding Y, Zhang Y, Yang BB, Peng C. MicroRNA 376c enhances ovarian cancer cell survival by targeting activin receptor-like kinase 7: implications for chemoresistance. J Cell Sci. 2011;124:359–368. doi: 10.1242/jcs.072223. [DOI] [PubMed] [Google Scholar]
- 8.Zhang JF, Fu WM, He ML, Xie WD, Lv Q, Wan G, Li G, Wang H, Lu G, Hu X, et al. MiRNA-20a promotes osteogenic differentiation of human mesenchymal stem cells by co-regulating BMP signaling. RNA Biol. 2011;8:829–838. doi: 10.4161/rna.8.5.16043. [DOI] [PubMed] [Google Scholar]
- 9.Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol. Med. 2006;12:580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 11.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sc.i USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 13.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu. Rev. Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 14.Garofalo M, Croce CM. microRNAs: master regulators as potential therapeutics in cancer. Annu. Rev. Pharmacol. Toxicol. 2011;51:25–43. doi: 10.1146/annurev-pharmtox-010510-100517. [DOI] [PubMed] [Google Scholar]
- 15.Medina PP, Slack FJ. microRNAs and cancer: an overview. Cell Cycle. 2008;7:2485–2492. doi: 10.4161/cc.7.16.6453. [DOI] [PubMed] [Google Scholar]
- 16.Greither T, Grochola LF, Udelnow A, Lautenschlager C, Wurl P, Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int. J. Cancer. 2010;126:73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 17.Huang X, Ding L, Bennewith KL, Tong RT, Welford SM, Ang KK, Story M, Le QT, Giaccia AJ. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol. Cell. 2009;35:856–867. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greither T, Wurl P, Grochola L, Bond G, Bache M, Kappler M, Lautenschlager C, Holzhausen HJ, Wach S, Eckert AW, et al. Expression of microRNA 210 associates with poor survival and age of tumor onset of soft-tissue sarcoma patients. Int. J. Cancer. 2012;130:1230–1235. doi: 10.1002/ijc.26109. [DOI] [PubMed] [Google Scholar]
- 19.Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, Harris AL, Gleadle JM, Ragoussis J. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin. Cancer Res. 2008;14:1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 20.Huang X, Le QT, Giaccia AJ. MiR-210–micromanager of the hypoxia pathway. Trends Mol. Med. 2010;16:230–237. doi: 10.1016/j.molmed.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Bian S, Yang CS. Green tea polyphenol EGCG suppresses lung cancer cell growth through upregulating miR-210 expression caused by stabilizing HIF-1alpha. Carcinogenesis. 2011;32:1881–1889. doi: 10.1093/carcin/bgr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu S, Huang M, Li Z, Jia F, Ghosh Z, Lijkwan MA, Fasanaro P, Sun N, Wang X, Martelli F, et al. MicroRNA-210 as a novel therapy for treatment of ischemic heart disease. Circulation. 2010;122:S124–S131. doi: 10.1161/CIRCULATIONAHA.109.928424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan SY, Zhang YY, Hemann C, Mahoney CE, Zweier JL, Loscalzo J. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab. 2009;10:273–284. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bianchi N, Zuccato C, Lampronti I, Borgatti M, Gambari R. Expression of miR-210 during erythroid differentiation and induction of gamma-globin gene expression. BMB Rep. 2009;42:493–499. doi: 10.5483/bmbrep.2009.42.8.493. [DOI] [PubMed] [Google Scholar]
- 25.Giannakakis A, Sandaltzopoulos R, Greshock J, Liang S, Huang J, Hasegawa K, Li C, O'Brien-Jenkins A, Katsaros D, Weber BL, et al. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol. Ther. 2008;7:255–264. doi: 10.4161/cbt.7.2.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biswas S, Roy S, Banerjee J, Hussain SR, Khanna S, Meenakshisundaram G, Kuppusamy P, Friedman A, Sen CK. Hypoxia inducible microRNA 210 attenuates keratinocyte proliferation and impairs closure in a murine model of ischemic wounds. Proc. Natl. Acad. Sci. USA. 2010;107:6976–6981. doi: 10.1073/pnas.1001653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsuchiya S, Fujiwara T, Sato F, Shimada Y, Tanaka E, Sakai Y, Shimizu K, Tsujimoto G. MicroRNA-210 regulates cancer cell proliferation through targeting fibroblast growth factor receptor-like 1 (FGFRL1) J. Biol. Chem. 2011;286:420–428. doi: 10.1074/jbc.M110.170852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo Y, Guo H, Zhang L, Xie H, Zhao X, Wang F, Li Z, Wang Y, Ma S, Tao J, et al. Genomic analysis of anti-hepatitis B virus (HBV) activity by small interfering RNA and lamivudine in stable HBV-producing cells. J. Virol. 2005;79:14392–14403. doi: 10.1128/JVI.79.22.14392-14403.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He J, Zhang JF, Yi C, Lv Q, Xie WD, Li JN, Wan G, Cui K, Kung HF, Yang J, et al. miRNA-mediated functional changes through co-regulating function related genes. PLoS One. 2010;5:e13558. doi: 10.1371/journal.pone.0013558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye W, Lv Q, Wong CK, Hu S, Fu C, Hua Z, Cai G, Li G, Yang BB, Zhang Y. The effect of central loops in miRNA:MRE duplexes on the efficiency of miRNA-mediated gene regulation. PLoS One. 2008;3:e1719. doi: 10.1371/journal.pone.0001719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 32.Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28:3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothe F, Ignatiadis M, Chaboteaux C, Haibe-Kains B, Kheddoumi N, Majjaj S, Badran B, Fayyad-Kazan H, Desmedt C, Harris AL, et al. Global microRNA expression profiling identifies MiR-210 associated with tumor proliferation, invasion and poor clinical outcome in breast cancer. PLoS One. 2011;6:e20980. doi: 10.1371/journal.pone.0020980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 35.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 36.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boutros R, Lobjois V, Ducommun B. CDC25 phosphatases in cancer cells: key players? Good targets? Nat. Rev. Cancer. 2007;7:495–507. doi: 10.1038/nrc2169. [DOI] [PubMed] [Google Scholar]
- 38.Lavecchia A, Di Giovanni C, Novellino E. CDC25A and B dual-specificity phosphatase inhibitors: potential agents for cancer therapy. Curr. Med. Chem. 2009;16:1831–1849. doi: 10.2174/092986709788186084. [DOI] [PubMed] [Google Scholar]
- 39.Gabrielli BG, De Souza CP, Tonks ID, Clark JM, Hayward NK, Ellem KA. Cytoplasmic accumulation of cdc25B phosphatase in mitosis triggers centrosomal microtubule nucleation in HeLa cells. J. Cell Sci. 1996;109(Pt 5):1081–1093. doi: 10.1242/jcs.109.5.1081. [DOI] [PubMed] [Google Scholar]
- 40.Lindqvist A, Kallstrom H, Lundgren A, Barsoum E, Rosenthal CK. Cdc25B cooperates with Cdc25A to induce mitosis but has a unique role in activating cyclin B1-Cdk1 at the centrosome. J. Cell Biol. 2005;171:35–45. doi: 10.1083/jcb.200503066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boutros R, Dozier C, Ducommun B. The when and wheres of CDC25 phosphatases. Curr. Opin. Cell Biol. 2006;18:185–191. doi: 10.1016/j.ceb.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 42.Gabrielli BG, Clark JM, McCormack AK, Ellem KA. Hyperphosphorylation of the N-terminal domain of Cdc25 regulates activity toward cyclin B1/Cdc2 but not cyclin A/Cdk2. J. Biol. Chem. 1997;272:28607–28614. doi: 10.1074/jbc.272.45.28607. [DOI] [PubMed] [Google Scholar]
- 43.Donaldson MM, Tavares AA, Hagan IM, Nigg EA, Glover DM. The mitotic roles of Polo-like kinase. J. Cell Sci. 2001;114:2357–2358. doi: 10.1242/jcs.114.13.2357. [DOI] [PubMed] [Google Scholar]
- 44.Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nat. Rev. Mol. Cell Biol. 2009;10:265–275. doi: 10.1038/nrm2653. [DOI] [PubMed] [Google Scholar]
- 45.Lens SM, Voest EE, Medema RH. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat. Rev. Cancer. 2010;10:825–841. doi: 10.1038/nrc2964. [DOI] [PubMed] [Google Scholar]
- 46.Lindqvist A, Rodriguez-Bravo V, Medema RH. The decision to enter mitosis: feedback and redundancy in the mitotic entry network. J. Cell Biol. 2009;185:193–202. doi: 10.1083/jcb.200812045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Casenghi M, Meraldi P, Weinhart U, Duncan PI, Korner R, Nigg EA. Polo-like kinase 1 regulates Nlp, a centrosome protein involved in microtubule nucleation. Dev. Cell. 2003;5:113–125. doi: 10.1016/s1534-5807(03)00193-x. [DOI] [PubMed] [Google Scholar]
- 48.Oshimori N, Ohsugi M, Yamamoto T. The Plk1 target Kizuna stabilizes mitotic centrosomes to ensure spindle bipolarity. Nat. Cell Biol. 2006;8:1095–1101. doi: 10.1038/ncb1474. [DOI] [PubMed] [Google Scholar]
- 49.Nakada C, Tsukamoto Y, Matsuura K, Nguyen TL, Hijiya N, Uchida T, Sato F, Mimata H, Seto M, Moriyama M. Overexpression of miR-210, a downstream target of HIF1alpha, causes centrosome amplification in renal carcinoma cells. J. Pathol. 2011;224:280–288. doi: 10.1002/path.2860. [DOI] [PubMed] [Google Scholar]
- 50.Davenport JW, Fernandes ER, Harris LD, Neale GA, Goorha R. The mouse mitotic checkpoint gene bub1b, a novel bub1 family member, is expressed in a cell cycle-dependent manner. Genomics. 1999;55:113–117. doi: 10.1006/geno.1998.5629. [DOI] [PubMed] [Google Scholar]
- 51.D'Angiolella V, Donato V, Vijayakumar S, Saraf A, Florens L, Washburn MP, Dynlacht B, Pagano M. SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010;466:138–142. doi: 10.1038/nature09140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Santamaria A, Nagel S, Sillje HH, Nigg EA. The spindle protein CHICA mediates localization of the chromokinesin Kid to the mitotic spindle. Curr. Biol. 2008;18:723–729. doi: 10.1016/j.cub.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 53.Degenhardt Y, Lampkin T. Targeting Polo-like kinase in cancer therapy. Clin. Cancer Res. 2010;16:384–389. doi: 10.1158/1078-0432.CCR-09-1380. [DOI] [PubMed] [Google Scholar]
- 54.Foekens JA, Sieuwerts AM, Smid M, Look MP, de Weerd V, Boersma AW, Klijn JG, Wiemer EA, Martens JW. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc. Natl Acad. Sci. USA. 2008;105:13021–13026. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, et al. A microRNA signature of hypoxia. Mol. Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fasanaro P, D'Alessandra Y, Di Stefano V, Melchionna R, Romani S, Pompilio G, Capogrossi MC, Martelli F. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J. Biol. Chem. 2008;283:15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ying Q, Liang L, Guo W, Zha R, Tian Q, Huang S, Yao J, Ding J, Bao M, Ge C, et al. Hypoxia-inducible microRNA-210 augments the metastatic potential of tumor cells by targeting vacuole membrane protein 1 in hepatocellular carcinoma. Hepatology. 2011;54:2064–2075. doi: 10.1002/hep.24614. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.