Abstract
In every round of translation elongation, EF-G catalyzes translocation, the movement of tRNAs (and paired codons) to their adjacent binding sites in the ribosome. Previous kinetic studies have shown that the rate of tRNA–mRNA movement is limited by a conformational change in the ribosome termed ‘unlocking’. Although structural studies offer some clues as to what unlocking might entail, the molecular basis of this conformational change remains an open question. In this study, the contribution of intersubunit bridges to the energy barrier of translocation was systematically investigated. Unlike those targeting B2a and B3, mutations that disrupt bridges B1a, B4, B7a and B8 increased the maximal rate of both forward (EF-G dependent) and reverse (spontaneous) translocation. As bridge B1a is predicted to constrain 30S head movement and B4, B7a and B8 are predicted to constrain intersubunit rotation, these data provide evidence that formation of the unlocked (transition) state involves both 30S head movement and intersubunit rotation.
INTRODUCTION
The elongation phase of protein synthesis can be divided into three basic steps: binding of aminoacyl-tRNA to the ribosomal A site (decoding), transfer of the nascent peptide chain from the P-site tRNA to the A-site aminoacyl-tRNA (peptide bond formation) and movement of the tRNAs (with paired codons) to their adjacent sites in the ribosome (translocation). It was shown decades ago that poly-U-programmed ribosomes can synthesize polyphenylalanine, albeit slowly, in the absence of elongation factors EF-Tu and EF-G (1,2). This important finding suggests that the ribosome is fundamentally responsible for each step of elongation, and the factors act primarily to speedup the process.
Translocation involves the large-scale movement of the newly formed peptidyl-tRNA and deacyl-tRNA from the A and P sites to the P and E sites, respectively. A number of studies suggest that translocation occurs in a stepwise manner, with the acceptor ends of the tRNAs moving first with respect to the 50S subunit to occupy hybrid A/P and P/E sites, followed by movement of their anticodons (along with paired mRNA) with respect to the 30S subunit (3–15). Single-molecule Förster resonance energy transfer (FRET) (smFRET) studies indicate that, in the absence of EF-G, movement of the tRNAs within the 50S subunit is rapid and reversible, with fluctuations between classical (A/A and P/P) and hybrid (A/P and P/E) configurations occurring at rates of ∼3 s− 1 at room temperature (11). Movement of the codon–anticodon helices within the 30S subunit of the ribosome can also be observed in the absence of EF-G, in either the forward or the reverse direction, although the rate in this case is much slower (0.0002–0.002 s− 1) (16–18). Thus, this latter part of translocation represents the main free-energy barrier for the reaction, which EF-G helps to breach.
Wintermeyer and coworkers have studied EF-G–catalyzed translocation extensively, monitoring a number of observables including EF-G binding, GTP hydrolysis, Pi release, tRNA movement and mRNA movement (19–22). Their findings have led to the following kinetic model, which has garnered further support from other laboratories (15,23,24). Binding of EF-G•GTP to the ribosome in its pretranslocation (PRE) state results in rapid GTP hydrolysis (250 s− 1), followed by a slower step (35 s− 1), which limits the rate of both codon–anticodon movement and Pi release. This slow step is attributed to a conformational change in the ribosome termed ‘unlocking’. Mutations of EF-G slow codon–anticodon movement and Pi release by the same degree (21), consistent with common step limiting both events. Although both rate limited by unlocking, codon–anticodon movement and Pi release are independent of each other and probably occur in a random order. Evidence that these events are independent comes from the observation that several antibiotics inhibit codon–anticodon movement but not Pi release, whereas mutations in L7/12 inhibit Pi release but not codon–anticodon movement (19,21,22). Next, the ribosomal rearrangement must be reversed, which ‘relocks’ the tRNAs into their new sites. It is thought that during or just before this ‘relocking’ step, the extended domain 4 of EF-G moves into the 30S A site (25), thereby biasing tRNA movement in the forward direction (26). Finally, EF-G•GDP dissociates from the posttranslocation (POST) state ribosome.
An unfortunate source of confusion in the field is another distinct use of the term ‘unlocking’. Frank and coworkers have defined ‘unlocked’ ribosomes as those carrying a deacylated tRNA in the P site (14). Such ribosomes can readily adopt a conformation in which tRNA occupies the P/E site, the 30S subunit is rotated with respect the 50S subunit and the L1 stalk is positioned inward toward the 50S E site. By contrast, the presence of a peptidyl group restricts tRNA to the P/P site and ‘locks’ the ribosome in an unrotated conformation. That the acylation state of P-site tRNA (or more precisely, its ability to bind the P/E site) strongly influences the conformational dynamics of the ribosome has since been confirmed through smFRET studies (27–29). Although these studies speak to how peptidyl transfer alters the thermodynamic landscape of the ribosome, the term ‘unlocking’ as defined by Frank is synonymous to ‘peptidyl transfer’, at least in the context of the elongation cycle. In this article, we will use the term ‘unlocking’ as defined by Wintermeyer to describe the rate-limiting step of translocation (21), which clearly comes after peptidyl transfer.
The unlocking step of translocation is believed to entail a conformational change of the ribosome, but the nature of this unlocking rearrangement remains poorly understood. Potential clues come from cryo-EM and X-ray crystallographic studies, which have revealed a number of distinct ribosome conformations and hence allowed motions to be inferred (3,7,30–33). One motion is an overall rotation of the 30S subunit with respect to the 50S subunit in the counterclockwise (CCW) direction (solvent-side perspective). Another is an orthogonal swiveling of the 30S head domain about the neck helix in the direction of tRNA movement. Although these ribosomal motions can occur independently, both are coupled to movement of tRNA from the P/P to P/E site, suggesting their importance in at least the initial part of translocation (i.e. tRNA movement within the 50S subunit). A third ribosomal rearrangement, clearly pertinent to translocation within the 30S subunit, has been inferred from structural studies. Cate and coworkers pointed out that codon–anticodon movement minimally requires the opening of a ‘gate’ formed by nucleotides of the 30S head (G1338–U1341) and platform (A790) to allow passage of the tRNA from the 30S P to E site (30,31). The correlated ribosomal rearrangements that accompany the P/P-to-P/E transition (i.e. CCW intersubunit rotation and head swiveling; often referred to collectively as ‘ratcheting’) open the gate to some degree but are insufficient to allow codon–anticodon movement. Indeed, the ratcheted ribosome with hybrid-bound tRNAs corresponds to a substate of the PRE complex that is well represented and rapidly formed in the absence of EF-G (7), indicating that unlocking involves something more or something else.
The unlocked state corresponds to the high-energy transition state for translocation, which exists only transiently. Its inherent instability makes it refractory to conventional structural methods such as cryo-EM and X-ray crystallography; hence, other approaches will be necessary to gain insight into its structure. In this study, we analyzed the effects of six intersubunit bridge mutations on translocation. We found that four of these mutations increase the maximal rate of both forward (EF-G-catalyzed) and reverse (spontaneous) translocation and thus reduce the energy barrier for tRNA–mRNA movement. The corresponding bridges (B1a, B4, B7a and B8) are those predicted to constrain 30S head movement and intersubunit rotation to the largest degree, providing compelling evidence that these motions are part of unlocking.
MATERIALS AND METHODS
Mutant ribosomes
Mutations described in Table 1 were introduced into plasmid p278MS2 (34), using the QuikChangeTM (Stratagene) method. Plasmid p278MS2 and its derivatives carrying nonlethal mutations were moved into the E scherichia coli Δ7 prrn strain SQZ10, as described (35). Mutant 70S ribosomes were purified from each Δ7 prrn strain as follows. Cells were grown to mid-logarithmic phase (OD550 = 0.3–0.5) in 1 l Luria Broth at 37°C, chilled on ice for 30 min and harvested by centrifugation. The cells were resuspended in Buffer A (20 mM Tris–HCl, pH 7.5, 15 mM MgCl2, 100 mM NH4Cl, 0.5 mM EDTA, 6 mM βME) and lysed by passage through a French Press. The cell lysate was clarified by two sequential 15 min centrifugation runs at 18 000g, layered onto 10 ml sucrose cushions (1.1 M) in Buffer B (20 mM Tris-HCl pH 7.5, 15 mM MgCl2, 500 mM NH4Cl, 0.5 mM EDTA, 6 mM βME) and spun at 100 000g for 21 h in a Beckman Ti 50.2 rotor. The crude ribosome pellets were dissolved, diluted into Buffer B and repelleted by centrifugation at 100 000g for 3 h in a Beckman Ti 50.2 rotor. The resulting pellets were dissolved in Buffer C (20 mM Tris–HCl, pH 7.5, 15 mM MgCl2, 100 mM NH4Cl, 6 mM βME) and loaded onto 34 ml 10–40% sucrose gradients in Buffer C. Ribosomes were separated from subunits by centrifugation at 50 000g for 14 h in a Beckman SW32 rotor. The 70S fractions were collected, and the ribosomes were pelleted by centrifugation at 100 000g for 17 h in a Beckman Ti 50.2 rotor. Finally, the ribosomes were dissolved in Buffer D (20 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 100 mM NH4Cl, 6 mM βME), flash frozen in small aliquots and stored at −70°C.
Table 1.
Mutations targeting intersubunit bridges
| Bridge a | Components (50S-30S) | Mutation name(s) | Description | Growth defectb |
|---|---|---|---|---|
| B1a | H38-S13 | ΔB1a, H38d22 (44) | Nucleotides 876–901 of 23S rRNA replaced by GAGA | None |
| B2a | H69-h44 | ΔB2a, ΔH69 (42) | Nucleotides 1906–1930 of 23S rRNA replaced by U | Lethal |
| B3 | H71-h44 | ΔB3 | Nucleotides 1948–1958 of 23S rRNA replaced by GCAA | Lethal |
| B4 | H34-S15 | ΔB4, H34d (44) | Nucleotides 709–710 and 721–722 of 23S rRNA deleted | Moderate |
| B6 | H62-h44 | ΔB6 | Nucleotides 1685–1703 of 23S rRNA replaced by GAGA | Lethal |
| B7a | H68-h23 | ΔB7a, H68d (44) | Nucleotides 1845–1895 of 23S rRNA replaced by GGAA | Moderate |
| B8 | L14/L19-h14 | ΔB8, h14Δ2 bp (43) | Nucleotides 340–341 and 348–349 of 16S rRNA deleted | Moderate |
For the lethal mutations, p278MS2 variants were transformed into strain DH10(pcI857), and the corresponding mutant 50S subunits were purified using affinity chromatography as described (34). Subunits were otherwise purified using sucrose gradients (35). Small subunits were heat activated in the presence of 20 mM Mg2+ at 42°C for 20 min and then mixed with an equal amount of 50S subunits and further incubated at 37°C for 15 min to reassociate 70S ribosomes.
Kinetic experiments
EF-G–catalyzed translocation was measured essentially as described (15). Message m625 (5′-AAGGAAAUAAAAAUGGUAUAU-3′) with a 2′-amino-pyrene modification at the 3′-terminal uridine was purchased from Thermo Scientific. Ribosomes (1.5 μM) were incubated with tRNAMet (1.5 μM; Chemical Block) and m625 (1.25 μM) in Buffer E (50 mM Tris–HCl, pH 7.6, 15 mM MgCl2, 100 mM NH4Cl, 6 mM βME) at 37°C for 20 min to fill the ribosomal P site. Ac-Val-tRNAVal (1.5 μM), prepared from purified tRNAVal (Chemical Block) as described (36), was then added and the reaction was incubated at 37°C for 10 min to form the PRE complex. EF-G•GTP was formed in Buffer F (50 mM Tris–HCl, pH 7.6, 5 mM MgCl2, 30 mM NH4Cl, 70 mM KCl, 6 mM βME) by incubating EF-G (various concentrations) and GTP (1 mM) at 37°C for 5 min. The rate of mRNA movement was determined after rapid mixing of the PRE complex with EF-G•GTP in an SX20 stopped-flow spectrometer (Applied Photophysics) as described (15,37). For experiments involving reassociated ribosomes, the Mg2+ concentration in Buffers E and F was raised to 20 mM to promote subunit association.
Rates of reverse translocation were determined by monitoring the movement of mRNA (using toeprinting) and peptidyl-tRNA (using puromycin-reactivity assays) essentially as described (18). For toeprinting experiments, message m292 [5′-(N)41AAAGGAAAUAAAAAUGGUAUACUUUAAAUCU(N)67-3′, 0.5 μM], containing a pre-annealed radiolabeled primer near its 3′-end, was incubated in polymix buffer [5 mM potassium phosphate (pH 7.3), 95 mM KCl, 5 mM Mg(OAc)2, 0.5 mM CaCl2, 5 mM NH4Cl, 8 mM putriscine, 1 mM spermidine, 1 mM DTT] (38) with ribosomes (0.7 μM) and Ac-Val-tRNAVal (1 μM) at 37°C for 20 min to fill the P site. Reverse translocation was initiated by adding tRNAfMet (Chemical Block, various concentrations) at t = 0, and 2 μl aliquots were removed at various time points for primer extension analysis. For puromycin-reactivity assays, ribosomes (0.7 μM) were incubated with m292 (0.5 μM) and Ac-[14C]-Val-tRNAVal (0.3 μM) in polymix buffer at 37°C for 20 min to fill the P site. Reverse translocation was initiated by adding tRNAfMet (3 μM) at t = 0. At each time point, two 30 μl aliquots were removed and incubated at 37°C for 10 s with or without 1 mM puromycin, and immediately extracted with 1 ml ethyl acetate for 1 min. After centrifugation at 13 000 rpm for 1 min, 800 μl of the organic phase was removed for liquid scintillation counting to determine the amount of Ac-[14C]-Val-puromycin formed. The data were plotted versus time and fit to a single exponential function to obtain apparent rates of reverse translocation.
RESULTS
Experimental rationale
Studies of EF-G-dependent translocation suggest that codon–anticodon movement is rate limited by a ribosomal rearrangement termed ‘unlocking’ (21). It is envisaged that, in the unlocked state, the tRNAs can freely fluctuate, via Brownian motion, between PRE and POST configurations (26). A reasonable assumption is that, in the absence of EF-G, the ribosome can adopt a similar unlocked conformation (albeit at a much lower rate), and this determines the rate of the spontaneous translocation in either the forward or the reverse direction (39). Given this assumption, we reasoned that ribosomal mutations that promote the unlocking rearrangement would increase the maximal rate of both forward (EF-G-dependent) and reverse (spontaneous) translocation. Conversely, mutations that fail to speed either the forward or the reverse reaction must act in a different way. For example, E-site mutation S7ΔR77-Y84 specifically destabilizes the POST state and thereby increases the maximal rate of reverse but not forward translocation (40).
Although the molecular basis of unlocking remains unclear, structural studies suggest that disruption and/or distortion of bridges between the subunits might be involved (5,31–33). To shed light on the mechanism of unlocking, we mutagenized bridges B1a, B2a, B3, B4, B6, B7a and B8 and screened for those mutations that accelerate tRNA–mRNA movement in both directions.
Bridge mutations
A series of mutations targeting most of the intersubunit bridges were made (Figure 1 and Table 1), some of which have been characterized previously to varying degrees (41–44). The mutations were constructed in plasmid p278MS2, which contains the rrnB operon with an aptamer tag in the 23S gene (34). Those plasmids with nonlethal mutations were introduced into an E. coli strain lacking all chromosomal copies of the rRNA operons (Δ7 prrn; obtained from S. Quan and C. Squires). This resulted in a set of strains, each expressing a homogeneous population of ribosomes, from which control and mutant 70S ribosomes were purified. Mutations ΔB1a, ΔB7a and ΔB8 showed no obvious defects in subunit association, based on sucrose gradient sedimentation profiles seen during the purifications. Mutation ΔB4, on the other hand, caused a moderate defect in subunit association, in agreement with an earlier report (44).
Figure 1.
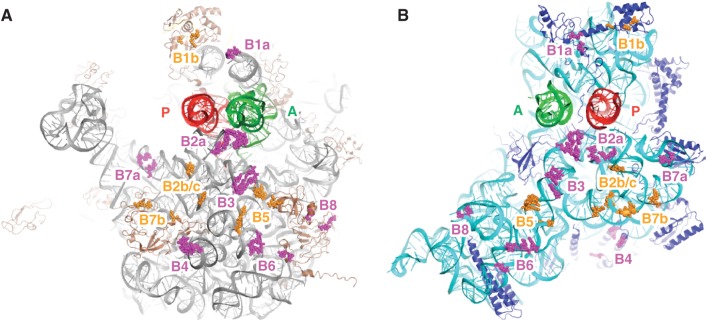
Locations of intersubunit bridges in the ribosome. Components of the bridges formed between the 50S (A) and 30S (B) subunits, viewed from the interface perspective. Bridges targeted in this study, magenta; other bridges, orange; 23S rRNA, gray; 5S rRNA, yellow; 50S proteins, brown; 16S rRNA, cyan; 30S proteins, dark blue; P-site tRNA, red; A-site tRNA, green. This image was generated in PyMOL using PDB entries 2WDG and 2WDI (58).
Mutations ΔB2a, ΔB3 and ΔB6 failed to support cell growth in strain Δ7 prrn. For these lethal mutations, plasmid-encoded rRNAs were expressed in E. coli strain DH10(pcI857), and the corresponding mutant 50S subunits were purified by affinity chromatography, using the aptamer tag in 23S rRNA (34). Primer extension analysis indicated >90% purity of the mutant subunits (data not shown). Affinity-purified 50S subunits were incubated with wild-type 30S subunits to form 70S ribosomes for the translocation studies described below. Mutations ΔB2a and ΔB3 conferred defects in subunit association, as expected, but the presence of tRNA and mRNA helped to compensate for these defects, allowing translocation to be analyzed. Large subunits carrying ΔB6, on the other hand, were unable to form 70S ribosomes even in the presence of tRNA and mRNA. This was evident because (i) addition of 50S(ΔB6) to 30S containing N-acetyl-Val-tRNAVal (AcVal-tRNAVal, an analog of peptidyl-tRNA) paired to GUA in the P site did not render the peptidyl group reactive to puromycin, and (ii) no evidence for EF-G–dependent translocation was observed in complexes formed with 50S(ΔB6) (data not shown). Consequently, ΔB6 was not further analyzed.
Effects of bridge mutations on reverse translocation
In certain mRNA contexts, the PRE state of the ribosome is thermodynamically favored over the POST state, allowing the rate of spontaneous reverse translocation to be easily measured (18). Reverse translocation was initiated by adding tRNAfMet to the E site of ribosomes containing P-site AcVal-tRNAVal, and movement of mRNA and tRNA was monitored using toeprinting and puromycin-reactivity assays, respectively. The fraction of ribosomes in the POST state, determined by either method, was plotted as a function of time, and the data were fit to a single-exponential equation to obtain apparent rates of reverse translocation (Figure 2A). Apparent rates were similar regardless of which method (toeprinting or puromycin reactivity) was employed. Experiments in which the concentration of E-tRNA was varied showed that the apparent rate (k app) begins to plateau at ∼1 µM (Figure 2B) in all cases tested. To compare the effects of the mutations, the apparent rate at a substantially higher concentration of E-tRNA (3 or 10 µM) was taken as an approximation of the maximal rate of reverse translocation (k rev) (Table 2). Our presumption that E-tRNA concentration was saturating under these conditions is supported by the fact that k app values obtained at 3 µM E-tRNA (puromycin-reactivity experiments) are comparable with those obtained at 10 µM E-tRNA (toeprinting experiments) in all cases.
Figure 2.
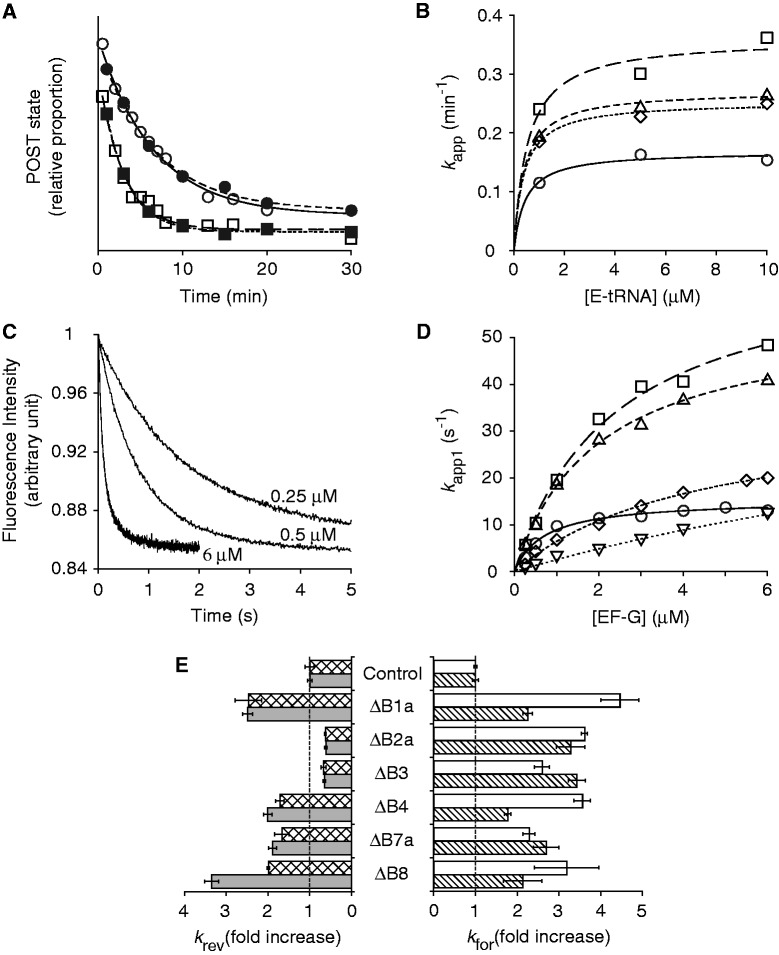
Effects of bridge mutations on translocation. (A) Examples of experiments in which rates of reverse translocation in control (circles) and mutant (ΔB1a; squares) ribosomes were measured, using toeprinting (open symbols) or puromycin-reactivity (closed symbols) assays. Data were fit to a single-exponential function to obtain apparent rates. (B) Apparent rates of reverse translocation (as measured by toeprinting) plotted against E-site tRNA concentration for the control (open circles) and mutant (ΔB1a, open squares; ΔB4, open triangles; ΔB7a, open rhombus) ribosomes. Curves represent hyperbolic fits to the data. (C) Examples of fluorescence traces obtained in measuring rates of forward translocation at different concentrations of EF-G (as indicated). Data were fit to a double-exponential function to obtain apparent rates for the fast (k app1) and slow (k app2) phases. (D) Plots of k app1 versus EF-G concentration for the control (open circles) and a number of mutant ribosomes (ΔB1a, open squares; ΔB4, open triangles; ΔB7a, open rhombus; ΔB8, open inverted triangles). Data were fit to the equation k app = k for•[EF-G]/(K 1/2 + [EF-G]) to yield the parameters shown in Table 2. (E) Summary of the effects of the bridge mutations on the maximal rate of spontaneous reverse translocation [k rev; toeprinting (checkered bars), puromycin-reactivity (gray bars)] and EF-G-catalyzed forward translocation (k for1, white bars; k for2, striped bars).
Table 2.
Rates of reverse translocation for control and mutant ribosomes
| Ribosomes | Mutation | k rev (min−1) (toeprinting)a | k rev (min−1) (puromycin reactivity)b |
|---|---|---|---|
| Purified 70S | Control | 0.16 ± 0.02 | 0.16 ± 0.01 |
| ΔB1a | 0.39 ± 0.05 | 0.39 ± 0.03 | |
| ΔB4 | 0.27 ± 0.02 | 0.31 ± 0.02 | |
| ΔB7a | 0.27 ± 0.03 | 0.30 ± 0.01 | |
| ΔB8 | 0.32 ± 0.03 | 0.53 ± 0.01 | |
| Reassociated 70S | Control | 0.39 ± 0.05 | 0.36 ± 0.01 |
| ΔB2a | 0.25 ± 0.01 | 0.22 ± 0.02 | |
| ΔB3 | 0.26 ± 0.03 | 0.24 ± 0.02 |
Data represent mean ± SEM from at least three independent experiments. aValues correspond to the apparent rate at 10 µM E-tRNA. bValues correspond to the apparent rate at 3 µM E-tRNA.
Control 70S ribosomes purified from E. coli Δ7 prrn gave k rev values of 0.16 min− 1, in line with earlier studies (18). Mutations ΔB1a, ΔB4, ΔB7a and ΔB8 each increased k rev significantly, by 2- to 3-fold (Table 2). When control ribosomes were reassociated from subunits (rather than isolated as intact 70S ribosomes), k rev was substantially higher (0.36–0.39 min− 1 versus 0.16 min− 1). This was not due to the affinity chromatography method used, because ribosomes made by reassociation of subunits prepared using conventional sucrose gradients gave the same k rev value (0.36 ± 0.04 min− 1). Reassociated ribosomes also exhibited a higher rate of forward translocation at saturating concentrations of EF-G (see below), suggesting that the unlocking rearrangement occurs more readily in these reassociated particles. Although the basis of this phenomenon remains unclear, it is probably due to a higher percentage of loose couples in the reassociated preparations (see Discussion). Importantly, this phenomenon does not hamper our analysis, because effects of mutations are always assessed with respect to the appropriate controls. Mutant ribosomes harboring ΔB2a or ΔB3 exhibited significantly lower k rev values than the reassociated control ribosomes (Table 2).
Effects of bridge mutations on forward translocation
To study the effect of bridge mutations on forward translocation, we measured the rate of EF-G–catalyzed codon–anticodon movement within the ribosome under single-turnover conditions, employing 3′-pyrene labeled mRNA as described previously (37). In this assay, the position of the pyrene fluorophore with respect to ribosome is such that translocation of the mRNA by three nucleotides is accompanied by a substantial decrease in fluorescence intensity, which can be monitored as a function of time in a stopped-flow machine. The observed decrease in fluorescence exhibits biphasic kinetics, with similar amplitudes for the fast and slow phases (15,40). No fluorescence change is seen in the presence of viomycin (37), an antibiotic known to block codon–anticodon movement without affecting EF-G binding, GTP hydrolysis or Pi release (20,21). Thus, the fast phase can be attributed to codon–anticodon movement. The slow phase may reflect a subsequent conformational change in the complex or a subpopulation of ribosomes in which codon–anticodon translocation occurs at a slower rate. Mutations of the 50S E site, which strongly inhibit movement of tRNA into the P/E site, do not alter the relative amplitudes of the two phases (15), suggesting that the biphasic kinetics are unrelated to equilibria between classical and hybrid tRNA-binding configurations of the PRE complex.
PRE complexes were formed by incubating control or mutant ribosomes with 3′ pyrene-labeled mRNA (m625) and tRNAMet to fill the P site and then adding AcVal-tRNAVal to bind the A site. Mixing the PRE complex with EF-G and GTP resulted in a biphasic decrease in fluorescence intensity (Figure 2C), with the fast phase accounting for 50–70% of the total amplitude, consistent with previous studies (15,40). Apparent rates were determined at various concentrations of EF-G to derive the kinetic parameters shown in Table 3 (Figure 2D). In general, maximal rates for the fast (k for1) and slow (k for2) phases were similarly influenced by the mutations (Table 3). For control 70S ribosomes purified from Δ7 prrn, k for1 was found to be 15 s− 1, with a corresponding K 1/2 value of 0.73 μM. Mutations ΔB1a, ΔB4 and ΔB7a increased k for1 by 5-, 4- and 2-fold and K 1/2 by 3-, 3- and 6-fold, respectively (Table 3). Mutation ΔB8 similarly increased k for1 (by 3-fold), although this was accompanied by a considerably larger effect on K 1/2 (25-fold). These four mutations also increase k rev (Table 2), suggesting that the corresponding bridges (B1a, B4, B7a and B8) normally restrain the unlocking rearrangement.
Table 3.
Kinetic parameters for EF-G-catalyzed translocation
| Ribosomes | Mutation | [Mg2+] (mM) | Fast phase |
Slow phase |
||
|---|---|---|---|---|---|---|
| k for1 (s−1) | K 1/2 (μM) | k for2 (s−1) | K 1/2 (μM) | |||
| Purified 70S | Control | 10 | 15 ± 1 | 0.73 ± 0.11 | 2.4 ± 0.2 | 0.22 ± 0.10 |
| ΔB1a | 10 | 68 ± 7 | 2.4 ± 0.5 | 5.4 ± 0.3 | 0.67 ± 0.12 | |
| ΔB4 | 10 | 55 ± 3 | 2.0 ± 0.2 | 4.3 ± 0.2 | 0.65 ± 0.11 | |
| ΔB7a | 10 | 35 ± 2 | 4.4 ± 0.5 | 6.5 ± 0.7 | 3.6 ± 0.8 | |
| ΔB8 | 10 | 49 ± 12 | 18 ± 5 | 5.2 ± 1.1 | 5.4 ± 2.0 | |
| Reassociated 70S | Control | 20 | 16 ± 1 | 0.53 ± 0.08 | 1.5 ± 0.1 | 0.15 ± 0.03 |
| ΔB2aa | 20 | 60 ± 2 | — | 5.0 ± 0.5 | — | |
| ΔB3a | 20 | 43 ± 3 | — | 5.2 ± 0.3 | — | |
| Controla | 10 | 35 ± 2 | — | 4.4 ± 0.5 | — | |
Reported values and their standard errors were derived from curve fits such as those shown in Figure 2D. aIn these cases, apparent rates at 6 μM EF-G were measured to approximate k for1 and k for2, and K 1/2 values were not determined.
Our initial attempts to characterize the lethal mutations ΔB2a and ΔB3 under the same conditions were unsuccessful. Amplitudes of the signal change were small, resulting in low signal-to-noise ratios and curves that could not be accurately evaluated. We suspected that the reduced amplitudes were due to defects in subunit association caused by the mutations. Indeed, when a higher concentration of Mg2+ was used (20 mM instead of 10 mM), the amplitudes were larger and the curves much better defined. At 6 µM EF-G, mutations ΔB2a and ΔB3 increased the rate of translocation significantly (Table 3). However, because these two mutations fail to stimulate reverse translocation (Table 2), they do not appear to act by promoting the unlocking rearrangement. They might destabilize the PRE state specifically and thereby accelerate only the forward reaction.
At 20 mM Mg2+, the concentration of EF-G was varied to obtain the kinetic parameters for the control reassociated ribosomes. The maximal rate (k for1 = 16 s− 1) was the same as that seen at 10 mM Mg2+ for the control ribosomes isolated as 70S couples (Table 3). Since high concentrations of Mg2+ are known to inhibit translocation (45), we compared the two preparations of control ribosomes under identical conditions. At 10 mM Mg2+, the apparent rate of translocation at 6 µM EF-G was >2-fold higher in the reassociated ribosomes than in those isolated as 70S couples (Table 3). As mentioned earlier, the fact that the reassociated ribosomes exhibit elevated rates of both forward and reverse translocation suggests that a conformational or compositional change in these ribosomes promotes the unlocking rearrangement.
DISCUSSION
A number of ribosomal motions have been inferred from structural studies and implicated in the mechanism of translocation. However, functional evidence to distinguish which movements are involved in the rate-limiting step of translocation (termed ‘unlocking’) has been lacking. Here, we analyzed six bridge mutations and found that four of them accelerate codon–anticodon movement in both directions. The corresponding bridges (B1a, B4, B7a and B8) are predicted to constrain 30S head movement and intersubunit rotation, suggesting that these motions are part of unlocking.
Evidence that unlocking involves swiveling (and/or tilting) of the 30S head
Two sites of contact, bridges B1a and B1b, link the head domain of the 30S subunit to the 50S subunit (Figure 1). B1a is formed by S13 and helix H38 of the 23S rRNA, which is also known as the A-site finger because it additionally contacts the elbow region of A-site tRNA (A-tRNA). Mutation ΔB1a truncates H38 by 10 base pairs, effectively removing contacts to both the 30S head and the A-tRNA. Suzuki and coworkers originally constructed this mutation and found that it stimulates poly-Phe synthesis in reactions containing EF-G at subsaturating concentrations (44). Subsequent single-molecule studies by the Blanchard laboratory showed that ΔB1a increases the maximal rate of EF-G–dependent translocation by about 2-fold and perturbs the conformational equilibria of the PRE complex, destabilizing tRNA in the classical configuration (A/A, P/P) compared with hybrid configurations (A/A, P/E and A/P, P/E) (41). In this study, we confirm that ΔB1a accelerates EF-G–dependent translocation and additionally show that it accelerates reverse translocation, providing evidence that the mutation promotes formation of the unlocked state. As swiveling of the head domain requires disruption of B1a, our data are consistent with the hypothesis that unlocking involves head swiveling (30). Earlier evidence for this hypothesis came from the finding that spectinomycin, which binds the neck helix of the 30S subunit, stabilizes an unswiveled conformation of the ribosome and inhibits both forward and reverse translocation (46). One cautionary note here is that these biochemical data do not rule out the possibility that a motion of the head other than swiveling is involved in unlocking. For example, tilting of the head away from the 50S would open the gate between the 30S P and E site, and this motion would also be promoted by ΔB1a and inhibited by spectinomycin. Indeed, unlocking may involve both swiveling and tilting. Consistent with this idea, a recent cryo-EM reconstruction of the ribosome containing tmRNA-SmpB and EF-G reveals a putative translocation intermediate in which the 30S head is highly rotated (by 19°) and tilted (by 12°) (47).
Evidence that unlocking involves rotation of the 30S body
Bridge B3 lies very near the center of intersubunit rotation (5,8,14,32,33). In principle, the degree to which 30S-50S contacts can constrain intersubunit rotation depends on their distance from B3. In this study, we targeted six bridges formed by the body/platform (B2a, B3, B4, B6, B7a and B8), by removing rRNA elements that contribute to each. Although the effects of ΔB6 on translocation could not be assessed due to a major defect in subunit association, effects of the other five mutations were determined. Mutations ΔB4, ΔB7a and ΔB8 accelerated both forward and reverse translocation, whereas ΔB2a and ΔB3 slow reverse translocation to some degree. The fact that mutations away from the center of intersubunit rotation generally reduce the energy barrier for codon–anticodon movement while those at or near the center do not provides compelling evidence that the mechanism of unlocking involves intersubunit rotation. Previously, Horan and Noller showed that ribosomes engineered with a disulfide bond between S6 and L2 (near B7b) are highly defective in EF-G–dependent translocation, and they concluded that intersubunit rotation is critical for the mechanism (48). Our findings support their conclusion and further suggest that intersubunit rotation plays a role in the rate-limiting unlocking step of translocation.
How mutations of the central bridges (ΔB2a and ΔB3) stimulate only the forward reaction remains unclear. One hypothesis is that these mutations destabilize the PRE state specifically, thereby reducing the energy barrier of translocation in a unidirectional manner. In line with this hypothesis, we saw a slight decrease in the extent of reverse translocation in these mutant ribosomes, when puromycin reactivity was used as the readout. The amount of puromycin-reactive complex observed at the start of the reaction was reduced to a final level of 2.1 ± 0.2% in the control case, compared to 4.8 ± 1.0% and 5.4 ± 1.2% in the ΔB2a and ΔB3 cases, respectively. These data are consistent with a ∼2-fold rightward shift in the PRE-to-POST equilibrium, conferred by each mutation. A change in the reaction endpoint was not apparent, though, when the reactions were measured by toeprinting, possibly due to increased background. Certainly, further experiments will be necessary to rigorously test this hypothesis.
Mutation ΔB8 decreases the apparent affinity of EF-G for the PRE complex
Mutation ΔB8 had an exceptionally large effect on the concentration dependence of EF-G–catalyzed translocation, increasing K 1/2 by 25-fold. We previously analyzed the effects of this same mutation on the initial phase of decoding, as ribosomes harboring ΔB8 are highly error prone (43). We found that ΔB8 accelerates EF-Tu–dependent GTP hydrolysis, particularly in the near-cognate case, but impacts K 1/2 for the ternary complex only modestly (cognate case; 3-fold increase) or not at all (near-cognate case). How might the differential effects of ΔB8 on K 1/2 for these two reactions be explained? One possibility is that ΔB8 more severely compromises the interaction of EF-G with the ribosome. Bridge B8 involves contacts between the terminal portion of h14 of 16S rRNA and proteins L14 and L19 of the large subunit. Mutation ΔB8 (previously termed h14Δ2) is a two-basepair truncation of h14 predicted to disrupt these contacts. Although no structure of EF-G bound to the PRE-state ribosome is available, a high-resolution structure of EF-G bound to the POST-state ribosome reveals a small area of contact between L14 (Q90, E92) and domain 3 of EF-G (R468) (25). One can imagine that ΔB8 alters the position of L14 and thereby reduces the affinity of EF-G for the PRE-state ribosome. Domain 3 of EF-Tu, which is structurally unrelated to domain 3 of EF-G, lies away from L14 in structures of the ternary complex–bound ribosome (49,50); hence, a ΔB8-induced change in L14 would not be expected to influence the affinity of the ternary complex in the same way. Another possible explanation for the distinct K 1/2 effects is that ΔB8 compromises an interaction formed by both factors, but one that is formed after GTP hydrolysis. Cryo-EM studies have provided evidence that the switch 1 motif of both factors can interact with the terminus of h14 (51,52). The role of this interaction remains unclear, although it is clearly unnecessary for GTPase activation and probably forms after GTP hydrolysis (43,50). In precluding this interaction, ΔB8 may influence a step subsequent to GTP hydrolysis, thereby impacting K 1/2 for EF-G–catalyzed translocation but not EF-Tu–dependent GTP hydrolysis.
A structural model for translocation
Spahn and coworkers have analyzed ribosome complexes containing EF-G, stabilized with either fusidic acid or GDPNP, using cryo-EM (53). Two distinct conformations of the ribosome were revealed. One (termed TIPRE) looked characteristically PRE-like, with a single tRNA occupying the P/E site, the 30S subunit rotated by 7° (with respect to 50S) and the head domain swiveled by 5° (with respect to the 30S body). The other (termed TIPOST) differed substantially—the intersubunit rotation was reduced to 4° while swiveling of the 30S head increased to 18°. This large degree of head swiveling in the absence of coupled intersubunit rotation shifted the anticodon of tRNA by 8-10 Å with respect to the platform domain, allowing the anticodon to interact simultaneously with the E-site components of the platform and the P-site components of the head. Positioned in this way, the tRNA was said to occupy an intrasubunit hybrid site, termed the pe/E site. It was suggested that this TIPOST conformation resembles the transition state for codon–anticodon movement. In line with this idea, this ribosomal conformation (highly swiveled head with little intersubunit rotation) is only detectable in complexes containing EF-G. In the absence of EF-G, adoption of this conformation is predicted to be highly unfavorable for either PRE- or POST-state ribosomes (7), as would be expected given the low rate of spontaneous translocation.
Our data can be easily rationalized in the context of the Spahn model (53). Assuming that the unlocked ribosome exhibits a highly swiveled 30S head with little intersubunit rotation, formation of the unlocked state from either the POST state or the hybrid PRE state requires changes in the degree of both head swiveling and intersubunit rotation (Figure 3). Unlocking the hybrid PRE state requires an increase in head swiveling and a decrease in intersubunit rotation, whereas unlocking the POST state requires an increase in both head swiveling and intersubunit rotation. Hence, mutations that remove constraints on either head swiveling (e.g. ΔB1a) or intersubunit rotation (e.g. ΔB4, ΔB7 a, ΔB8) should accelerate translocation in either direction, as we observe. It is important to point out that our data, while consistent with the Spahn model, lend no less support to other structural models in which formation of the unlocked state involves both intersubunit rotation and head movement. Also worth mentioning is that the model in Figure 3 is undoubtedly an oversimplified view, as it represents only one potential pathway between defined substates of PRE and POST. It is clear from cryo-EM and smFRET studies that PRE and (to a lesser extent) POST ribosomes are conformationally dynamic, so it seems most likely that the unlocked state(s) can be directly attained from multiple substates of PRE and POST.
Figure 3.
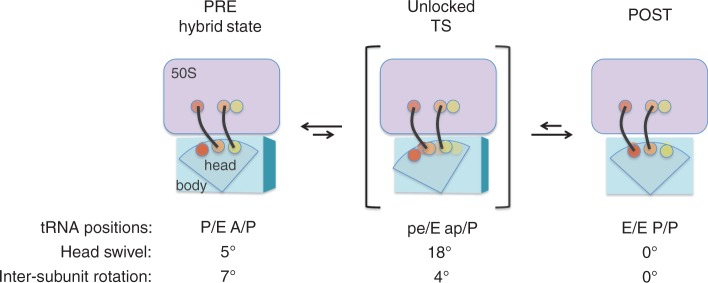
A structural model for translocation. Recent cryo-EM studies by Spahn and coworkers suggest that EF-G can stabilize a conformation of the ribosome in which the 30S head is highly swiveled (18°), while the 30S body/platform is only slightly rotated (4°) with respect to the 50S subunit (53). Such a conformation may resemble the unlocked (transition) state of translocation. 50S, lavender; 30S, blue; A site, yellow; P site, orange; E site, red; tRNAs, black.
Ribosomes reassociated from subunits exhibit a higher rate of unlocking
In the course of this study, we found that ribosomes made with purified subunits show higher k rev and k for1 values than ribosomes isolated as 70S couples, indicating that a conformational or compositional change in the reassociated ribosomes promotes unlocking. These data are consistent with our previous finding that rates of tRNA dissociation from the P site (k off) are also higher for reassociated ribosomes (54). Decades ago, Noll and coworkers characterized two biochemically distinct forms of the ribosome—termed tight and loose couples (55–57). Although both forms are active in vitro, the subunit association constant of tight couples is much higher than that of loose couples. In various functional assays, loose couples exhibit a shifted [Mg2+] dependence that reflects their association defect. Nearly all 70S ribosomes freshly prepared from cells in the presence of high Mg2+ (>10 mM) are tight couples. Exposure of these tight couples to low Mg2+ (1–2 mM) promotes a change in the 50S subunit that leads to formation of loose couples (55–57). Our reassociated ribosomes derive from subunits purified using 1 mM Mg2+ and hence are predicted to contain a substantial subpopulation of loose couples. We propose that the association defect characteristic of loose couples explains the larger k rev and k for1 values we observe in reassociated ribosomes. Consistent with this idea, loose couples bind P-tRNA less tightly and exhibit higher rates of factor-independent poly-Phe synthesis than tight couples (56). It is tempting to speculate that the conformational change(s) in the 50S subunit responsible for loose couple formation compromises one or more of bridges B1a, B4, B7a or B8 and thereby promotes unlocking.
FUNDING
National Institutes of Health (NIH) [GM072528 to K.F.] Funding for open access charge: NIH [GM072528].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank C. Squires and S. Quan for E. coli strain SQZ10, R. Green for plasmid p278MS2, D. Qin for technical help and S. McClory and R. Balakrishnan for providing some ΔB8 ribosomes and preliminary data.
REFERENCES
- 1.Gavrilova LP, Spirin AS. Stimulation of “non-enzymic” translocation in ribosomes by p-chloromercuribenzoate. FEBS Let. 1971;17:324–326. doi: 10.1016/0014-5793(71)80177-1. [DOI] [PubMed] [Google Scholar]
- 2.Pestka S. Studies on the formation of transfer ribonucleic acid-ribosome complexes. VI. Oligopeptide synthesis and translocation on ribosomes in the presence and absence of soluble transfer factors. J. Biol. Chem. 1969;244:1533–1539. [PubMed] [Google Scholar]
- 3.Agirrezabala X, Liao HY, Schreiner E, Fu J, Ortiz-Meoz RF, Schulten K, Green R, Frank J. Structural characterization of mRNA-tRNA translocation intermediates. Proc. Natl Acad. Sci. USA. 2012;109:6094–6099. doi: 10.1073/pnas.1201288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen C, Stevens B, Kaur J, Cabral D, Liu H, Wang Y, Zhang H, Rosenblum G, Smilansky Z, Goldman YE, et al. Single-molecule fluorescence measurements of ribosomal translocation dynamics. Mol. Cell. 2011;42:367–377. doi: 10.1016/j.molcel.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol. Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 7.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 8.Jin H, Kelley AC, Ramakrishnan V. Crystal structure of the hybrid state of ribosome in complex with the guanosine triphosphatase release factor 3. Proc. Natl Acad. Sci. USA. 2011;108:15798–15803. doi: 10.1073/pnas.1112185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGarry KG, Walker SE, Wang H, Fredrick K. Destabilization of the P site codon-anticodon helix results from movement of tRNA into the P/E hybrid state within the ribosome. Mol. Cell. 2005;20:613–622. doi: 10.1016/j.molcel.2005.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 11.Munro JB, Altman RB, O'Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol. Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Odom OW, Picking WD, Hardesty B. Movement of tRNA but not the nascent peptide during peptide bond formation on ribosomes. Biochemistry. 1990;29:10734–10744. doi: 10.1021/bi00500a004. [DOI] [PubMed] [Google Scholar]
- 13.Pan D, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol. Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 15.Walker SE, Shoji S, Pan D, Cooperman BS, Fredrick K. Role of hybrid tRNA-binding states in ribosomal translocation. Proc. Natl Acad. Sci. USA. 2008;105:9192–9197. doi: 10.1073/pnas.0710146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konevega AL, Fischer N, Semenkov YP, Stark H, Wintermeyer W, Rodnina MV. Spontaneous reverse movement of mRNA-bound tRNA through the ribosome. Nat. Struct. Mol. Biol. 2007;14:318–324. doi: 10.1038/nsmb1221. [DOI] [PubMed] [Google Scholar]
- 17.Semenkov YP, Shapkina TG, Kirillov SV. Puromycin reaction of the A-site bound peptidyl-tRNA. Biochimie. 1992;74:411–417. doi: 10.1016/0300-9084(92)90080-x. [DOI] [PubMed] [Google Scholar]
- 18.Shoji S, Walker SE, Fredrick K. Reverse translocation of tRNA in the ribosome. Mol. Cell. 2006;24:931–942. doi: 10.1016/j.molcel.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savelsbergh A, Mohr D, Kothe U, Wintermeyer W, Rodnina MV. Control of phosphate release from elongation factor G by ribosomal protein L7/12. EMBO J. 2005;24:4316–4323. doi: 10.1038/sj.emboj.7600884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 21.Savelsbergh A, Katunin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol. Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 22.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J. Mol. Biol. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 23.Pan D, Kirillov S, Zhang CM, Hou YM, Cooperman BS. Rapid ribosomal translocation depends on the conserved 18-55 base pair in P-site transfer RNA. Nat. Struct. Mol. Biol. 2006;13:354–359. doi: 10.1038/nsmb1074. [DOI] [PubMed] [Google Scholar]
- 24.Seo HS, Abedin S, Kamp D, Wilson DN, Nierhaus KH, Cooperman BS. EF-G-dependent GTPase on the ribosome. Conformational change and fusidic acid inhibition. Biochemistry. 2006;45:2504–2514. doi: 10.1021/bi0516677. [DOI] [PubMed] [Google Scholar]
- 25.Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodnina MV, Wintermeyer W. The ribosome as a molecular machine: the mechanism of tRNA-mRNA movement in translocation. Biochem. Soc. Trans. 2011;39:658–662. doi: 10.1042/BST0390658. [DOI] [PubMed] [Google Scholar]
- 27.Munro JB, Altman RB, Tung CS, Cate JH, Sanbonmatsu KY, Blanchard SC. Spontaneous formation of the unlocked state of the ribosome is a multistep process. Proc. Natl Acad. Sci. USA. 2010;107:709–714. doi: 10.1073/pnas.0908597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munro JB, Wasserman MR, Altman RB, Wang L, Blanchard SC. Correlated conformational events in EF-G and the ribosome regulate translocation. Nat. Struct. Mol. Biol. 2010;17:1470–1477. doi: 10.1038/nsmb.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol. Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 31.Zhang W, Dunkle JA, Cate JH. Structures of the ribosome in intermediate states of ratcheting. Science. 2009;325:1014–1017. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou J, Lancaster L, Trakhanov S, Noller HF. Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome. RNA. 2012;18:230–240. doi: 10.1261/rna.031187.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Youngman EM, Green R. Affinity purification of in vivo-assembled ribosomes for in vitro biochemical analysis. Methods. 2005;36:305–312. doi: 10.1016/j.ymeth.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 35.Qin D, Abdi NM, Fredrick K. Characterization of 16S rRNA mutations that decrease the fidelity of translation initiation. RNA. 2007;13:2348–2355. doi: 10.1261/rna.715307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker SE, Fredrick K. Preparation and evaluation of acylated tRNAs. Methods. 2008;44:81–86. doi: 10.1016/j.ymeth.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Studer SM, Feinberg JS, Joseph S. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J. Mol. Biol. 2003;327:369–381. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- 38.Ehrenberg M, Bilgin N, Kurland CG. Design and use of a fast and accurate in vitro translation system. In: Spedding G, editor. Ribosomes and Protein Synthesis—A Practical Approach. Oxford: IRL Press; 1990. pp. 101–129. [Google Scholar]
- 39.Shoji S, Walker SE, Fredrick K. Ribosomal translocation: one step closer to the molecular mechanism. ACS Chem. Biol. 2009;4:93–107. doi: 10.1021/cb8002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Devaraj A, Shoji S, Holbrook ED, Fredrick K. A role for the 30S subunit E site in maintenance of the translational reading frame. RNA. 2009;15:255–265. doi: 10.1261/rna.1320109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L, Altman RB, Blanchard SC. Insights into the molecular determinants of EF-G catalyzed translocation. RNA. 2011;17:2189–2200. doi: 10.1261/rna.029033.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ali IK, Lancaster L, Feinberg J, Joseph S, Noller HF. Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol. Cell. 2006;23:865–874. doi: 10.1016/j.molcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 43.McClory SP, Leisring JM, Qin D, Fredrick K. Missense suppressor mutations in 16S rRNA reveal the importance of helices h8 and h14 in aminoacyl-tRNA selection. RNA. 2010;16:1925–1934. doi: 10.1261/rna.2228510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Komoda T, Sato NS, Phelps SS, Namba N, Joseph S, Suzuki T. The A-site finger in 23 S rRNA acts as a functional attenuator for translocation. J. Biol. Chem. 2006;281:32303–32309. doi: 10.1074/jbc.M607058200. [DOI] [PubMed] [Google Scholar]
- 45.Spirin AS. Ribosomal translocation: Facts and models. Prog. Nucleic Acid Res. Mol. Biol. 1985;32:75–114. doi: 10.1016/s0079-6603(08)60346-3. [DOI] [PubMed] [Google Scholar]
- 46.Borovinskaya MA, Shoji S, Holton JM, Fredrick K, Cate JH. A steric block in translation caused by the antibiotic spectinomycin. ACS Chem. Biol. 2007;2:545–552. doi: 10.1021/cb700100n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramrath DJ, Yamamoto H, Rother K, Wittek D, Pech M, Mielke T, Loerke J, Scheerer P, Ivanov P, Teraoka Y, et al. The complex of tmRNA-SmpB and EF-G on translocating ribosomes. Nature. 2012;485:526–529. doi: 10.1038/nature11006. [DOI] [PubMed] [Google Scholar]
- 48.Horan LH, Noller HF. Intersubunit movement is required for ribosomal translocation. Proc. Natl Acad. Sci. USA. 2007;104:4881–4885. doi: 10.1073/pnas.0700762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FV, Weir JR, Ramakrishnan V. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330:835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Villa E, Sengupta J, Trabuco LG, LeBarron J, Baxter WT, Shaikh TR, Grassucci RA, Nissen P, Ehrenberg M, Schulten K, et al. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc. Natl Acad. Sci. USA. 2009;106:1063–1068. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Connell SR, Takemoto C, Wilson DN, Wang H, Murayama K, Terada T, Shirouzu M, Rost M, Schuler M, Giesebrecht J, et al. Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol. Cell. 2007;25:751–764. doi: 10.1016/j.molcel.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 53.Ratje AH, Loerke J, Mikolajka A, Brunner M, Hildebrand PW, Starosta AL, Donhofer A, Connell SR, Fucini P, Mielke T, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shoji S, Abdi NM, Bundschuh R, Fredrick K. Contribution of ribosomal residues to P-site tRNA binding. Nucleic Acids Res. 2009;37:4033–4042. doi: 10.1093/nar/gkp296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hapke B, Noll H. Structural dynamics of bacterial ribosomes. IV. Classification of ribosomes by subunit interaction. J. Mol. Biol. 1976;105:97–109. doi: 10.1016/0022-2836(76)90196-0. [DOI] [PubMed] [Google Scholar]
- 56.Noll H, Noll M, Hapke B, van Dieijen G. The mechanism of subunit interaction as a key to the understanding of ribosome function. In: Bautz EKF, Karlson P, Kersten H, editors. Regulation of Transcription and Translation in Eukaryotes. New York: Springer-Verlag; 1973. pp. 257–311. [Google Scholar]
- 57.Stahli C, Noll H. Structural dynamics of bacterial ribosomes. VI. Denaturation of active ribosomes by Mg2+ dependent conformational transitions. Mol. Gen. Genet. 1977;153:159–168. doi: 10.1007/BF00264731. [DOI] [PubMed] [Google Scholar]
- 58.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat. Struct. Mol. Biol. 2009;16:528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 60.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.