Abstract
The Escherichia coli lactose (lac) operon encodes the first genetic switch to be discovered, and lac remains a paradigm for studying negative and positive control of gene expression. Negative control is believed to involve competition of RNA polymerase and Lac repressor for overlapping binding sites. Contributions to the local Lac repressor concentration come from free repressor and repressor delivered to the operator from remote auxiliary operators by DNA looping. Long-standing questions persist concerning the actual role of DNA looping in the mechanism of promoter repression. Here, we use experiments in living bacteria to resolve four of these questions. We show that the distance dependence of repression enhancement is comparable for upstream and downstream auxiliary operators, confirming the hypothesis that repressor concentration increase is the principal mechanism of repression loops. We find that as few as four turns of DNA can be constrained in a stable loop by Lac repressor. We show that RNA polymerase is not trapped at repressed promoters. Finally, we show that constraining a promoter in a tight DNA loop is sufficient for repression even when promoter and operator do not overlap.
INTRODUCTION
It is difficult to overstate the historical significance of the Escherichia coli lactose (lac) operon as a paradigm for negative and positive control of gene expression (1). This short segment of the bacterial chromosome encodes a genetic switch that senses and responds to glucose and lactose concentrations to produce lactose digestion enzymes only when glucose is absent and lactose is present (2). Central to this function is the homotetrameric Lac repressor protein that binds DNA operator sequences in the lac operon (3). Lac repressor binding is weakened in the presence of allolactose or its analog, isopropyl β-D-1-thiogalactopyranoside (IPTG), relieving repression. In the absence of glucose, RNA polymerase binds cooperatively with catabolite activator protein at the lac promoter (positive control). In simplest terms, the mechanism of negative control involves Lac repressor binding to occlude access of RNA polymerase holoenzyme to the lac promoter (4).
Of particular significance to the present work is the fascinating observation that two remote auxiliary operators (Oaux) exist in the lac operon (5). It has been proposed and demonstrated (6–13) that bidentate repressor tetramers bound at auxiliary operators increase the effective repressor concentration at the regulatory operator (O) through DNA looping (Figure 1). This concept has been exploited as an approach to probe the physical properties of bacterial DNA in vitro (14) and within the bacterial nucleoid in vivo (12,15–17).
Figure 1.
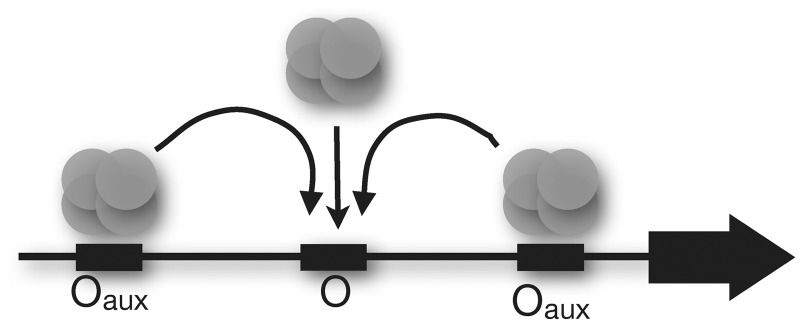
Hypothetical contributions to local bidentate repressor concentration at a bacterial operator (O) include contributions because of free repressor and contributions because of DNA-bound repressor at auxiliary operators (Oaux) through DNA looping.
The natural auxiliary operators of the lac operon have different affinities and occur such that one is just upstream and one is far downstream of the regulatory operator immediately proximal to the promoter. We have designed experiments in living bacteria to clarify the role of DNA looping in repression. We first tested the hypothesis that increasing the local concentration of repressor at the proximal operator is necessary and sufficient to explain promoter repression (13). This hypothesis implies that identical auxiliary operators positioned equivalent distances either upstream or downstream of the regulatory operator should provide comparable enhancement of repression by equally increasing the effective local repressor concentration at the regulatory operator. Artificial lac operators have been shown to enhance repression when upstream or downstream of the promoter (18), but the equivalence of upstream and downstream auxiliary operators has not been previously tested in systematic experiments. Second, we determined the smallest possible DNA loop involving Lac repressor in living bacteria. Third, we tested the early proposal that favorable repressor-polymerase contacts trap RNA polymerase at repressed promoters (19,20). Fourth, we tested in vivo a recent hypothesis that promoter DNA bending strain is intrinsically repressive (21).
MATERIALS AND METHODS
Bacterial strains
FW102 (the kind gift from F. Whipple) is a streptomycin resistant derivative of CSH142 [araD(gpt-lac)5] and is designated as wild-type in this study (22). Gene deletions and the presence of looping assay episomes were confirmed by diagnostic polymerase chain reaction (PCR) amplification after conjugation and selection (23).
DNA constructs
DNA looping constructs were based on plasmid pJ992, created by modifications of pFW11-null (22) as previously described (23). The relationship between lac operator sequence and repression in the absence of DNA looping were measured in preliminary experiments (Supplementary Figure S1). Sequences of new experimental and control promoters are shown in Supplementary Figure S2 with descriptions in Supplementary Table S1. The O2 operator normally present within the lacZ coding region was destroyed by site-directed mutagenesis (15). The experimental strong UV5 promoter does not contain a catabolite activator protein binding site. lacZ looping constructs were placed on the single copy F128 episome by homologous recombination between the constructed plasmids and bacterial episome followed by mating. F128 carries the lacI gene producing wild-type levels of repressor. Bacterial conjugation and selections were as previously described (23).
In vivo DNA looping assay and data fitting
Analysis of lac reporter gene expression was performed as described (23). Raw β-galactosidase reporter activity (E) is presented in Miller units. Normalized E′ values are then obtained by dividing E values by E obtained for a test construct where specific looping is not possible because only a single proximal O2 operator is present in the absence of an auxiliary operator. The repression ratio (‘RR’) is given by Einduced/Erepressed, where induction is obtained by addition of 2 mM IPTG. Best fits to the thermodynamic model of lac gene regulation produced estimates of seven parameters (Table 1) as described (23). The LacI Y282D mutation was created using QuikChange Lightning Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA). Upper and lower mutagenic primers were LJM-4110 5′-GACGATAC2GA2GACAGCTCATGTGACATC3GC2GT2A2C2AC2ATCA3CAG and LJM-4111 5′-CTGT3GATG2TG2T2A2CG2CG3ATGTCACATGAGCTGTCT2CG2TATCGTC, with the mutation underlined. LacI Y282D mutant looping constructs were placed on the single-copy F128 episome by homologous recombination. After mating and selection, correct strain recombinants were confirmed by PCR amplification to detect the inactivated internal lacZ O2 sequence with PCR primers LJM-1930 (5′-CGTCGT4ACA2CGTCG) and LJM-1931 (5′-CAT2GA3GT2A2TGA2TAGCAC). The LacI Y282D mutation was confirmed by PCR amplification followed by PCR product sequencing using primers LJM-4112 (5′-A2G2CGACTG2AGTGC2ATG) and LJM-4113 (5′-GA3C2TGTCGTGC2AGCTG). All data are included in Supplementary Table S2. For studies of T7 RNA polymerase promoters, promoter–reporter constructs were created based on plasmid pJ992, with modifications illustrated in Supplementary Figure S4, and were placed on the single-copy F128 episome. Cells were transformed with a plasmid expressing an arabinose inducible T7 RNA polymerase gene (a kind gift from Troy A. Lionberger) before lac reporter gene analysis. To determine β-galactosidase activity, bacterial cultures were grown in MOPS minimal buffered media (Teknova) supplemented with 0.8% of glycerol, 1.32 mM of dibasic potassium phosphate, 10 mM of NaHCO3, 0.2% of casamino acids and 12.5 µg/ml of thiamine. Cultures were grown either in the presence or absence of 2 mM IPTG and/or 0.02% arabinose. β-Galactosidase assays were performed as described (23). T7 RNA polymerase promoter–reporter constructs are illustrated schematically in Supplementary Figure S4, with data shown in Supplementary Table S3.
Table 1.
Parameters (95% confidence interval) fit to a thermodynamic model of Lac repressor looping
| Looping Construct | hr (bp/turn) |
Capp (×10−19 erg-cm) |
spoptimal (bp) |
KO2 |
Knull |
K°max |
K°NSL |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −IPTG | +IPTG | −IPTG | +IPTG | −IPTG | +IPTG | −IPTG | −IPTG | −IPTG | +IPTG | −IPTG | +IPTG | −IPTG | +IPTG | |
| Osym O2 upstreama | 11.4 ± 0.7 | 10.7 ± 0.7 | 0.76 ± 0.35 | 0.64 ± 0.26 | 78.3 ± 0.5 | 78.8 ± 0.6 | 2.45 | 0.13 | 68.6 ± 17.3 | 17.9 ± 4.6 | 10.4 ± 10.4 | 0 ± 2.6 | ||
| Osym alone upstreama | 11.4 ± 0 | 10.7 ± 1.5 | 0.52 ± 0 | 0.34 ± 0.04 | 74.3 ± 0 | 74.4 ± 1.3 | 0.17 | 0.16 | 4.6 ± 0 | 2.9 ± 0.5 | 0 ± 0 | 0 ± 0.2 | ||
| O2 Osym downstreamb | 11.3 ± 0.1 | 11.2 ± 0.2 | 2.46 ± 0.18 | 2.39 ± 0.25 | 68.4 ± 0 | 68.4 ± 0.1 | 3.80 | 1.42 | 343.8 ± 41.9 | 13.6 ± 2.7 | 6.6 ± 1.4 | 0.1 ± 0.3 | ||
| Osym alone downstreamb | 11.3 ± 0 | 11.2 ± 0.2 | 1.29 ± 0.04 | 0.89 ± 0.07 | 70.2 ± 0.2 | 70.5 ± 0.3 | 1.92 | 2.06 | 269.3 ± 9.5 | 17.3 ± 8.9 | 4.4 ± 0.1 | 0 ± 0.8 | ||
| O2 Osym mutantc | 11.1 ± 0.2 | 11.1 ± 0.1 | 1.82 ± 1.43 | 2.05 ± 0.85 | 68.7 ± 0.8 | 68.7 ± 0.2 | 0.04 | 0.09 | 176.2 ± 61.1 | 68.6 ± 11.8 | 0 ± 1.3 | 0 ± 0.3 | ||
| Osym alone mutantc | 11.2 ± 0.4 | 11.2 ± 0.4 | 0.90 ± 0.27 | 0.82 ± 0.10 | 60.2 ± 0.3 | 71.4 ± 0.5 | 5.40 | 5.71 | 15.9 ± 6.0 | 16.9 ± 3.3 | 0 ± 0.6 | 0 ± 0.1 | ||
Parameters not well determined by fitting are indicated in italics.
aFits based on data reported by Becker et al. (2005).
bFits based on current data set after eliminating data for spacings <41.5 bp.
cFits based on downstream data set with mutant LacI after eliminating data for spacings <41.5 bp.
Chromatin immunoprecipitation
Escherichia coli cultures were grown to log phase in 50 ml cultures of LB medium at 37°C in the presence or absence of 2 mM IPTG. Cross-linking of DNA and protein complexes was accomplished with the addition of 37% formaldehyde (Sigma) to make a final concentration of 1% in the presence of 10 mM sodium phosphate (pH 7.6). Cultures were maintained at room temperature with constant gentle swirling for 20 min. Reactions were quenched with cold 2 M glycine (200 mM of final concentration). Cells were harvested by centrifugation, washed three times with 4 ml of cold phosphate buffered saline and were resuspended in 1 ml of IP buffer [100 mM of Tris–HCl pH 8.0, 300 mM of NaCl, 2% of Triton X-100, 1 mM of PMSF and an additional protease inhibitor mix (Roche)]. Cells were lysed, and cellular DNA was sheared by sonication and was further analysed as described in the Supplementary Materials.
RESULTS AND DISCUSSION
Experimental design
The experimental design for this work is illustrated in Figure 2. We have previously studied a series of artificial lac UV5 promoters, where repression by a weak proximal operator (O2) is dramatically enhanced in a distance-dependent manner by a strong auxiliary operator (Osym) positioned upstream of the promoter (Figure 2A) (15–17,24,25). Repression loops formed by Lac repressor trap the promoter within a tightly bent segment, where O2 occupancy by repressor is assumed to occlude polymerase binding (dashed region in Figure 2B). We now create a series of promoter constructs illustrated in Figure 2C (see Supplementary Figure S2). In these constructs, the same promoter and proximal O2 elements are supplemented with the auxiliary Osym operator positioned ‘downstream’ such that looping again enhances repressor occupancy at O2 but does so without constraining the promoter within the loop (Figure 2D). Effects of Osym operators studied in the absence of DNA looping are shown in Supplementary Figure S3. The hypothesis to be tested is that promoter repression is equivalent for DNA loops of comparable size, regardless of whether the regulated promoter lies within the loop (Figure 2B) or adjacent to the loop (Figure 2D); ‘it is only the DNA loop-dependent increase in effective Lac repressor concentration at O2 that determines repression’. The particular alternative hypothesis of interest is that promoters entrapped within strained DNA loops (Figure 2B) are repressed more completely than when the promoter is not strained (Figure 2D).
Figure 2.
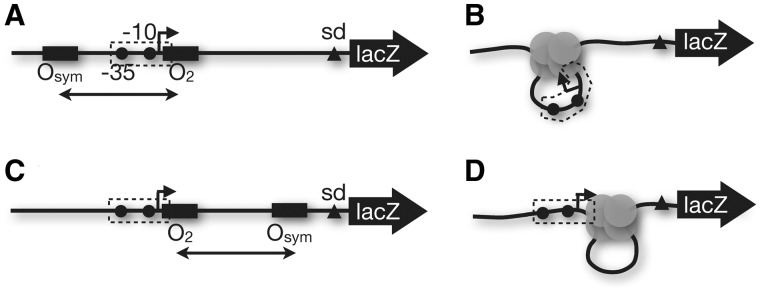
Promoter–reporter constructs to compare repression by upstream (A and B) versus downstream (C and D) DNA loops in the presence (A and C) or absence (B and D) of IPTG inducer. The dashed region indicates the RNA polymerase footprint, emphasizing that repressor binding at O2 occludes promoter access.
Upstream and downstream loops in control studies
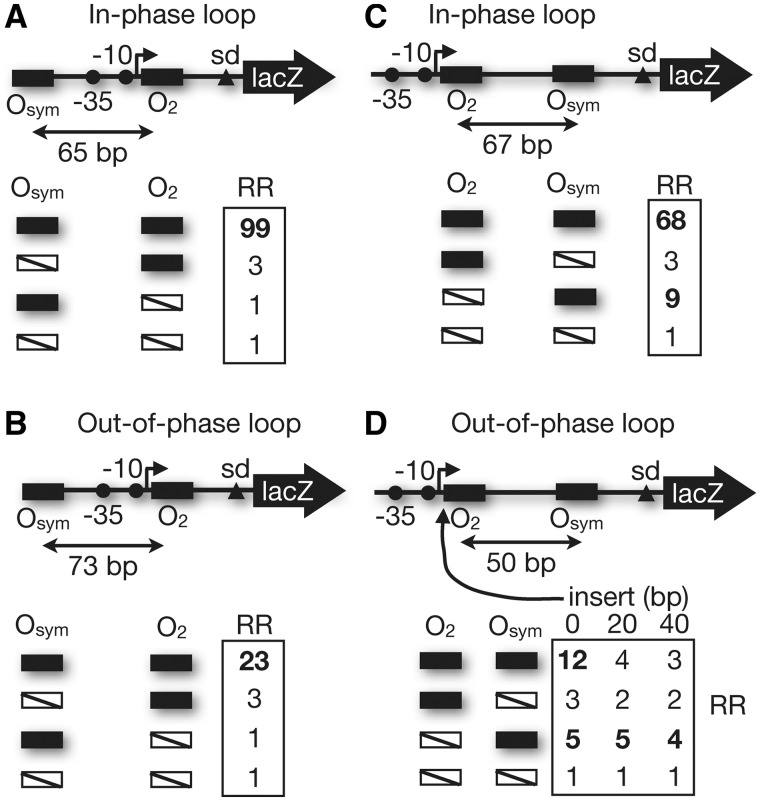
Experiments were undertaken by placing a series of spacing constructs (Supplementary Figure S2 and Table S2) in a single copy on the F′ episome of E. coli and measuring reporter gene (lacZ) expression in the absence or presence of IPTG inducer. Data were collected as a function of operator spacing and were fit to a thermodynamic model to estimate parameters describing the physical properties of the intervening DNA (12,15,23). We began with a set of control experiments to assess the effect of DNA loops formed upstream (Figure 3A and B) or downstream (Figure 3C and D) of the proximal O2 operator. The data in Figure 3 show how the promoter RR (induced reporter expression divided by repressed reporter expression) depends on the number and location of operators. Given that the apparent DNA helical repeat for the supercoiled lac promoter region in vivo is between 10.8 and 11 bp/turn (25), for an upstream Oaux, with O2 and Osym operators separated by 65 bp (center-to-center), the operators are on the same DNA face, yielding strong control and an RR value of 99 (Figure 3A). Repression by occupancy of the weak O2 operator without DNA looping drops by >30-fold, and no repression is detectable when O2 is disrupted (Figure 3A). Results are similar, but the strength of repression is reduced to ∼4-fold, for operators spaced by 73 bp (Figure 3B). Energetically unfavourable DNA twisting is required for closure of the repression loop in this case. We then tested the construct shown in Figure 3C, where the relationship of the lac UV5 promoter and proximal O2 operator is unchanged, but the auxiliary Osym operator is placed downstream. For an operator spacing (67 bp centre-to-centre) that places repressors on the same DNA face, repression is again seen to be strong (RR value 68) and dependent on the presence of both operators (Figure 3C). Interestingly, in the absence of DNA looping, the presence of Osym ∼50–90 bp downstream from the promoter yields RR values of 4–9 (Figure 3C and D). These results suggest that a tightly bound Lac repressor interferes weakly, but detectably, with RNA polymerase elongation (18,26). Loop-dependent repression remains detectable, but weaker, for an unfavorable downstream loop spacing (50 bp, Figure 3D). Importantly, loops involving an O2 operator positioned 20 or 40 bp downstream from the promoter were no more inhibitory than isolated repressor–operator complexes at these locations (Figure 3D). This result demonstrates that enhanced repression in experiments where Osym is downstream of the promoter depends on O2 adjacent to the promoter. This result also suggests that like upstream repression looping, downstream loop formation principally represses through inhibition of transcription initiation. This demonstrated similarity in mechanism allows direct comparison of quantitative data from upstream and downstream loops.
Figure 3.
RR behavior of the indicated promoter–reporter constructs. UV5 promoter elements (−35, −10), center-to-center spacing of weak and strong lac operators (O2 and Osym, respectively), Shine-Dalgarno (sd) and reporter (lacZ) are shown, as well as the length of any insert between the transcription start site (broken arrow) and proximal O2 operator. Boxed data show RR values for the indicated operator combinations, where filled black rectangles indicate intact operators and open slashed rectangles indicate disrupted operators. RR values in bold highlight comparisons. (A and B) Upstream loops with operators in phase (A) or out of phase (B). (C and D) Downstream loops with operators in phase (C) or out of phase (D).
Length-dependent upstream and downstream lac loop stabilities
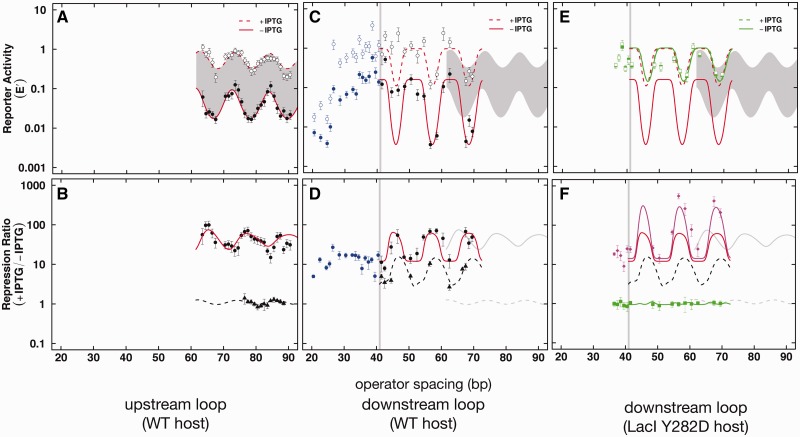
We have previously studied apparent DNA physical properties in living bacteria by monitoring promoter repression as a function of operator spacing for upstream loops (Figure 2A and B). We reported an oscillating pattern of repression as a function of operator spacing (Figure 4A and B) consistent with unexpectedly low DNA bending resistance, but strong DNA twist resistance, residual looping even in the presence of IPTG and a deduced DNA helical repeat parameter that differed between repressing and inducing conditions (15–17,24,25). New constructs with downstream auxiliary operators were studied, and the data are shown in Figure 4C and D with fitting to a thermodynamic model of DNA looping (Table 1). Figure 4C shows reporter activity normalized to a construct with only a proximal operator (E′) as a function of operator spacing (center-to-center). In Figure 4C, data for both repressing (filled black circles, solid red fit curve) and inducing (open black circles, dashed red fit curve) conditions are compared with data for loops involving upstream auxiliary operators (shaded region). The pattern is similar, although repressed E′ values tend to be lower for constructs with downstream auxiliary operators. We noted that oscillations in repressed and induced E′ values are somewhat irregular for operator spacings between 40 and 70 bp (Figure 4C), whereas oscillation of their RR (filled black circles and red line fit in Figure 4D) is regular and similar to the result for upstream loops (Figure 4D, solid grey curve). We interpret the irregular oscillation of downstream E′ values (Figure 4C) and regular oscillation of RR values (Figure 4D) as evidence of mRNA sequence-dependent gene expression for downstream auxiliary operators; sequence changes involving downstream auxiliary operators result in different 5′-untranslated mRNA leaders that may affect RNA stability and/or translation rate. Normalization removes such effects in the RR data (Figure 4D). Also different from results for upstream auxiliary operators in the absence of proximal operators (Figure 4B, filled triangles and dashed curve), constructs carrying a strong downstream auxiliary Osym operator in the absence of a proximal O2 operator show oscillating phase-dependent repression (Figure 4D, filled triangles, dashed black curve). This loop-independent repression is higher than for upstream auxiliary operators (Figure 4D, dashed grey line fit), again suggesting that an occupied Osym represents a barrier to transcription elongation by E. coli RNA polymerase. This oscillation could reflect Osym sequence effects or residual DNA looping anchored by weak recognition of the disabled proximal O2 operator or some other unknown cryptic operator sequence (10).
Figure 4.
Repression data and fits to thermodynamic model for loops with upstream (A and B) versus downstream (C–F) auxiliary Osym operators. Reporter activity is shown as E' (A, C and E) where the shaded envelope in (A) indicates behavior of upstream loop constructs under induced (open symbols, dashed fit curve) or repressed (filled symbols, solid fit curve) conditions with fits to thermodynamic model. Blue symbols for spacings closer than the vertical line at 41 bp (C–F) indicate constructs that do not show a canonical looping pattern. Panels (B, D and F) show RR data and fits to the thermodynamic model (see Supplementary Figure S3). Filled triangles and dashed fits indicate RR data for strains containing only an auxiliary Osym operator in the absence of a proximal operator. Grey fits in panels (D) and (F) indicate RR behaviour of upstream loops from (B). Green symbols and fits in panels (E) and (F) show data obtained in the absence of functional Lac repressor (LacI Y282D). Data and fit in magenta (panel F) show the modified RR for downstream loop constructs where the numerator reflects reporter expression in the absence of functional repressor (LacI Y282D) and the denominator reflects repressed reporter expression in the presence of wild-type Lac repressor.
To resolve this issue, we reproduced the study of downstream Osym effects in an E. coli strain with a totally disabled Lac repressor [LacI Y282D (27–29)]. The results are shown in Figure 4 (panels E and F). The E′ expression data (Figure 4, panel E, compare green to dashed red) show a comparable oscillation in the complete absence of Lac repressor relative to the original (dashed red) induced (+IPTG) E′ data. In panel F of Figure 4, these RRs data (green) do not oscillate, confirming the absence of active repressor. This result indicates that the residual E′ oscillation (green in Figure 4E) is because of some feature of the Osym operator sequence, as it rotates about the helix axis, ‘not’ residual weak looping because of weak repressor binding in the presence of IPTG. Given that the effect of the downstream Osym remains even in the complete absence of repressor, the origin of the oscillating pattern is mysterious and interesting. Oscillation with the helical repeat of DNA implies a face-of-the-helix effect. Possible explanations could include interactions between the promoter and a DNA structure at the strong inverted repeat at Osym or a strong hairpin structure in the nascent RNA transcript or even weak binding by some other protein at Osym. Thus, there is evidence for an oscillating effect of the palindromic operator sequence, per se, on gene expression.
The data in the complete absence of functional Lac repressor also provide the opportunity to express the RR as fully induced (no repressor) divided by fully repressed. These data are included in Figure 4F (magenta) and show the much larger dynamic control range when residual repression looping is eliminated, as previously suggested (30).
The effect of isolated auxiliary Osym was also measured in the absence of either a proximal operator or residual repressor (LacI Y282D mutant). This is shown in panels C and D of Supplementary Figure S3. Supplementary Figure S3C (light solid grey) shows weak oscillation previously observed for upstream Osym auxiliary operators, suggesting little position effect of upstream Osym, as reported (15). In Supplementary Figure S3C, the E′ (green) data in the absence of functional repressor show that isolated Osym operators downstream of the promoter cause an oscillating repression effect that is similar to the wild-type LacI induced by IPTG, indicating that there is an oscillating effect of the Osym operator sequence alone, that is magnified when Lac repressor binds to it. Supplementary Figure S3D (RR) shows that repressor binding to the isolated downstream Osym operator amplifies the effect of the operator (compare bLack dashed trace with green dashed trace).
It is important to note that loop-independent repression by a transcribed downstream Osym operator can produce RR values of 4–9 (Figure 4D, filled triangles), equivalent to cases where a proximal O2 operator and downstream Osym operator are out of phase. This repression in the absence of looping is because of collisions between repressor bound tightly at Osym and elongating RNA polymerase. This means that the 68-fold repression for the downstream loop observed in Figure 3C is only 7–14-fold higher than for the isolated Osym operator (compared with the 33-fold ratio related to isolated Osym for the upstream loop in Figure 3A). However, compared with O2 alone, the upstream RR increases by 33-fold and downstream by a similar 23-fold.
By fitting to a thermodynamic model of the Lac control loop (23), these data produce estimates for the seven parameters given in Table 1. Most striking is the difference in fit values for the apparent loop twist constant, Capp, whose estimates are >3-fold higher for downstream loops than for upstream loops. Because a common value for the DNA twist constant in vitro is ∼2.4 × 10−19 erg-cm, this suggests that it is the twist flexibility of the upstream loop that is anomalously high, even given the fact that the Lac repressor protein constitutes part of the loop. Because this parameter is dominated by the behavior of the UV5 promoter sequence in the upstream loops, it is possible that this result reflects a relatively high twist flexibility for the promoter sequence. In this regard, it has been previously shown by DNaseI accessibility (31) and cyclization kinetics (32) that TATA-like sequences display anomalous mechanical anisotropic flexibility that might explain our observations. Also noteworthy is the 5-fold higher fit value of K0max (the equilibrium constant for loops without twisting strain) for downstream loops. It is unclear why optimal downstream loops are more energetically favorable (as implied by this value). It is again possible that unique aspects of the Lac UV5 promoter could be reflected in this parameter.
Smallest detectable Lac repression loop in vivo
The design of Lac loop constructs with an auxiliary Osym operator downstream of O2 (Figure 2C and D) provided an opportunity to identify the smallest possible Lac repression loop. In previous work with upstream loops, we minimized the spacing of operators flanking an E. coli promoter until interference was detected (25). For operators flanking an E. coli promoter, the smallest centre-to-centre operator spacing for a repression loop was ∼60 bp (25). Examination of the data in Figure 4C and D shows that DNA looping fails between 40 and 45 bp (about four turns of the double helix, two of which are between the operators). For operator spacings <40 bp (center-to-center), there is no evidence of loop-dependent promoter repression (Figure 4C and D, blue symbols). This suggests that Lac repressor can constrain four turns of DNA into a stable loop. Interestingly, Hochschild and Ptashne (33) previously reported that loops driven by protein–protein interactions between cooperatively bound of phage λ repressor dimers could be detected for center-to-center spacings of as small as five DNA turns. Thus, the flexible tetrameric Lac repressor seems to be capable of stabilizing more highly strained DNA loops than phage λ repressor. Such tight DNA looping is likely to be facilitated by the high affinity of Lac repressor, its stable tetrameric structure and the flexibility of the Lac repressor tetramer, which has the potential to form an extended linear structure in bridging operators within a micro-loop (25,34–36).
Fate of RNA polymerase at Lac repression loops in vivo
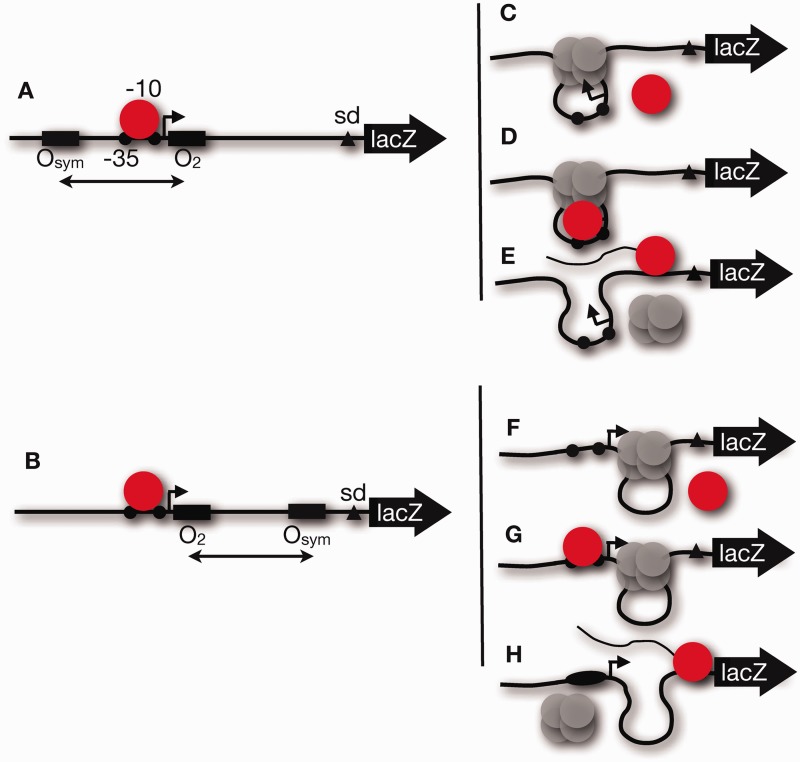
The data in Figures 3 and 4 and Table 1 support the hypothesis that promoter repression is principally by binding occlusion at O2, with enhanced local repressor concentration through DNA looping. Because aspects of this concept have previously been challenged (19–21), we considered other possible scenarios involving RNA polymerase (Figure 5). Under inducing conditions, RNA polymerase (red circle) initiates transcription by binding the Lac UV5 promoter flanked by unoccupied operators (Figure 5A) or when the unoccupied operators are both downstream (Figure 5B). Possible outcomes for RNA polymerase under repressing conditions are shown in Figure 5C–H. The conventional model is that RNA polymerase is occluded in both cases (Figure 5C and F), but RNA polymerase might be trapped at the promoter as suggested (19,20) (Figure 5D and G) or might engage the promoter and dispLace bound repressor (Figure 5E and H). We designed chromatin immunoprecipitation (ChIP) experiments to distinguish these models.
Figure 5.
Models for possible in vivo behavior of upstream (A) or downstream (B) looping constructs bearing a lac UV5 promoter (black dots indicate −35 and −10 elements) for RNA polymerase (red circle) where the proximal lac O2 operator impinges on the promoter. Possible outcomes under repressing conditions include polymerase exclusion (C and F), polymerase trapping (D and G) or polymerase read-through (E and H).
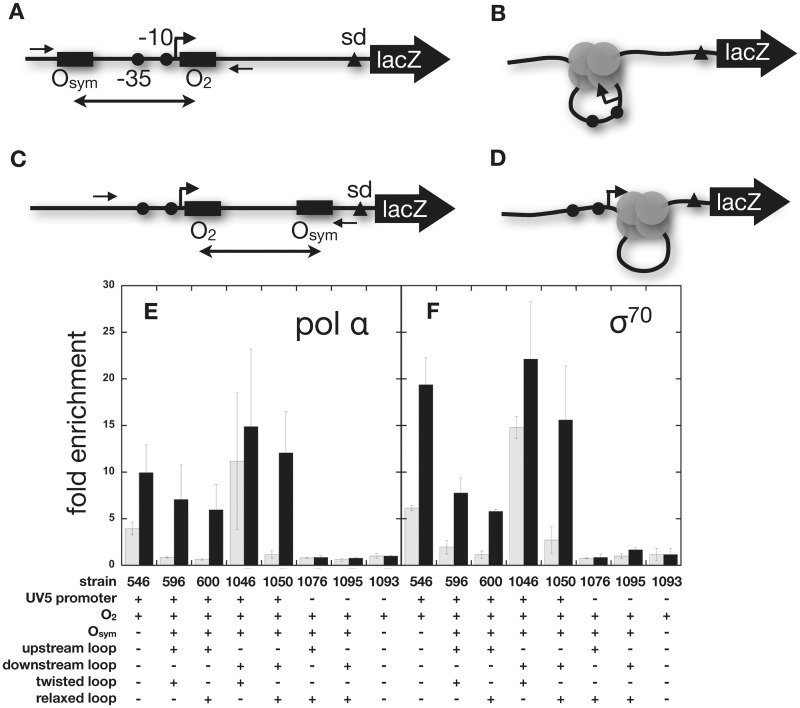
Antibodies against the α or σ70 subunits of E. coli RNA polymerase holoenzyme were used to immunoprecipitate promoter DNA fragments that had been cross-linked by formaldehyde in vivo. A combination of mechanical shearing and micrococcal nuclease treatment was used to reduce DNA fragment size to ∼200–400 bp before reversal of cross-linking and quantitative PCR with promoter-specific primers. Signals were normalized to total input DNA. Results are shown in Figure 6 and Supplementary Figure S5. Three constructs Lacking E. coli promoters (BL1076, BL1095 and BL1093) served as negative controls for RNA polymerase occupancy in the absence or presence of IPTG. The low background ChIP signal from BL1093 was set to 1.0 (Figure 6). Enrichment of α or σ70 subunits of E. coli RNA polymerase was then determined for five different constructs containing E. coli promoters under repressed or induced conditions. In all cases, in vivo promoter occupancy by RNA polymerase corresponded well to β-gaLactoside reporter expression. On induction, all constructs with E. coli promoters showed RNA polymerase occupancy 6–20-fold higher than background (Figure 6). For promoter constructs with a single weak O2 operator, leaky repression is only 2–3-fold (15,16), and this was confirmed for RNA polymerase occupancy (Figure 6, BL546). For constructs with upstream auxiliary operators without (BL600) or with (BL596) twisting strain in the repression loop, RNA polymerase occupancy (as measured by promoter cross-linking of α or σ70 subunits) was similar and at background levels before induction. Assuming that antibody recognition epitopes remain accessible in cross-linked repressed complexes, this result rules out RNA polymerase capture at repressed promoters in vivo. For repression loops formed with auxiliary operators downstream of the proximal O2 operator, results were similar (Figure 6). If the downstream loop was stable (BL1050; operators phased so the looped DNA is not twisted), repression was tight and RNA polymerase was excluded under repressed conditions (Figure 6). For downstream loops that are unstable because of dephased operators, and the requirement for DNA twisting strain (BL1046) repression is leaky and RNA polymerase holoenzyme occupancy is relatively high even under repressing conditions (Figure 6). Together these results favor the repression models shown in Figure 5C and F. RNA polymerase is prevented from binding to the Lac UV5 promoter to the extent that Lac repressor is bound at the proximal O2 operator. As expected, there is general correlation between IPTG-induced gene expression and RNA polymerase ChIP signal, and between RR and RNA polymerase ChIP signal ratio comparing induced with uninduced conditions (Supplementary Figure S5).
Figure 6.
RNA polymerase and σ70occupancy of lac promoters as detected by ChIP and quantitative PCR. Upstream loop constructs are studied under induced (A) or repressed (B) conditions. Downstream loop constructs are studied under induced (C) or repressed (D) conditions. Black dots indicate −35 and −10 elements. PCR primer sites are indicated by single-headed arrows. ChIP results under conditions of repression (grey bars) or induction (black bars) are shown for α subunit of RNA polymerase (E) or σ70 protein (F).
Repression without promoter/operator occlusion
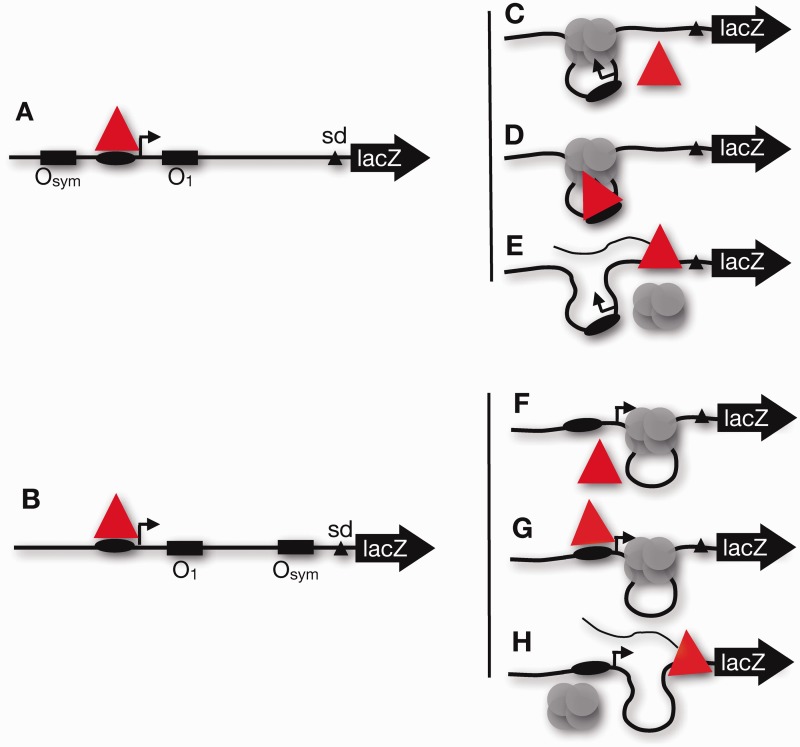
The results described earlier in the text confirm that when repressor and RNA polymerase compete for overlapping binding sites, repression is based on the effective local repressor concentration with contributions by free repressor and repressor looping with similar distance dependence from either upstream or downstream auxiliary operators. What if repressor and RNA polymerase do not compete for overlapping binding sites? Using the single subunit T7 RNA polymerase as a model, it has been shown in vitro that promoter bending strain, per se, can be repressive (21). To test this idea in vivo, we created the constructs shown in Figure 7A and B. When the auxiliary operator is upstream (Figure 7A), possible T7 RNA polymerase (red triangle) fates under repressing conditions are illustrated in Figure 7C–E. T7 RNA polymerase might be excluded (Figure 7C), captured (Figure 7D) or it might disrupt repressor and initiate transcription (Figure 7E). Similar scenarios might occur when the auxiliary operator is downstream of the proximal operator (Figure 7F–H).
Figure 7.
Models for possible in vivo behavior of upstream (A) or downstream (B) looping constructs bearing a T7 RNA polymerase (red triangle) promoter (oval) that does not overlap with lac O1 or Osym operators (rectangles). The promoter DNA is curved by looping in (A) but not (B). Possible outcomes under repressing conditions include polymerase exclusion (C and F), polymerase trapping (D and G) or polymerase read-through (E and H).
To test these ideas, three constructs were created for in vivo analysis using T7 RNA polymerase. In one construct (pJ1906; Figure 7A, Supplementary Figure S4), a T7 RNA polymerase promoter was pLaced between O1 and an upstream Osym operator, such that the promoter was approximately two DNA turns from either flanking operator. Lac repressor binding should not directly influence T7 RNA polymerase binding at this distance (37). The operator spacing (86.5 bp center-to-center) allows formation of a stable loop. In a second construct (pJ1940; Figure 7B, Supplementary Figure S4), T7 RNA polymerase promoter and O1 positions were preserved, but the auxiliary Osym operator was positioned downstream by 58.5 bp (center-to-center). As a control, an unlooped construct pLaced the T7 RNA polymerase promoter approximately two DNA turns upstream of an isolated O1 operator (pJ1938; Supplementary Figure S4).
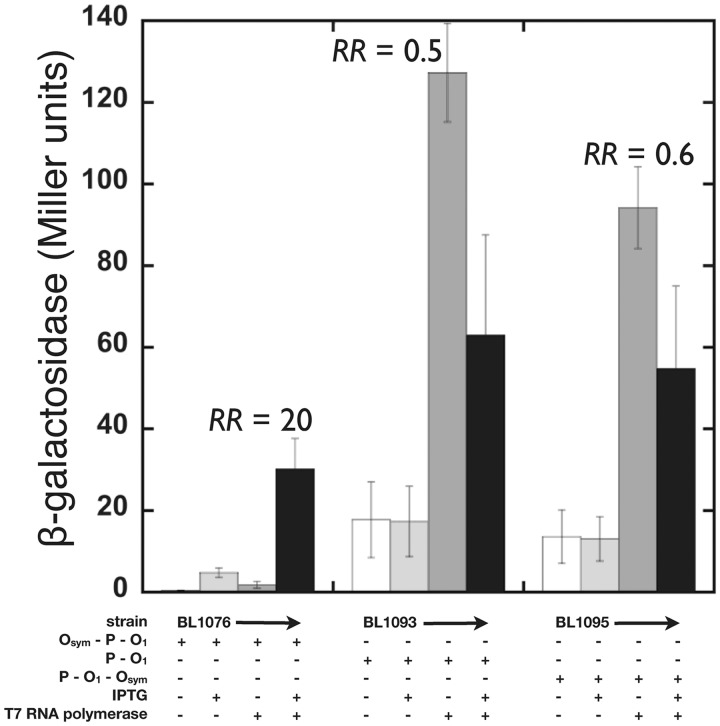
Constructs were pLaced on the E. coli F' episome for testing in the presence of a plasmid encoding arabinose-inducible T7 RNA polymerase. In vivo testing allowed determination of β-gaLactosidase expression and the RR. Data are shown in Figure 8 and Supplementary Table S3. In the absence of arabinose, T7 RNA polymerase is not induced, and the reporter signal is low (first two columns of each set). On T7 RNA polymerase induction, the results are striking. Even when the T7 RNA polymerase promoter is well separated from Lac operators, a RR of ∼20 is observed when the promoter is constrained within the repression loop (Figure 8; strain BL1076). In contrast, the same promoter gives RRs not statistically different from 1.0, when it is pLaced upstream from an isolated O1 operator (Figure 8; strain BL1093) or upstream from a stable DNA loop formed by Lac repressor binding to O1 and Osym operators (Figure 8; strain BL1095). Slightly reduced reporter gene expression on full induction of T7 transcription could indicate some favorable interaction between Lac repressor and T7 RNA polymerase or may reflect RNA destabilization because of uncoupling of transcription and translation (38).
Figure 8.
In vivo reporter gene expression from constructs bearing a T7 RNA polymerase promoter (P) two helical turns of DNA upstream of O1 (BL1093) and with an additional Osym either further upstream (BL1076) or further downstream (BL1095). Promoter/operator configurations are summarized later, and details are provided in Supplementary Figure S4. In each case, the RR corresponds to the ratio of the height of the final bar to that of the penultimate bar.
These results show that DNA looping can repress T7 transcription initiation, but not T7 transcription elongation, and that ‘repression of T7 transcription initiation can occur in vivo simply by promoter presentation in tightly bent DNA’. The models shown in Figure 7C and H are supported by these data. With respect to this mechanism, promoter bending deformation may impede T7 RNA polymerase binding, and/or it is possible that T7 RNA polymerase cannot initiate from the tightly bent promoter because the enzyme Lacks sufficient binding energy to untwist the constrained DNA within the loop. It remains to be determined whether promoter pLacement on the ‘inside’ of the looped DNA (as was the case here) is important for repression.
CONCLUSIONS AND FUTURE DIRECTIONS
Although perhaps the first and best-studied genetic switch in biology, several fundamental aspects of Lac promoter control remain incompletely understood. Using in vivo studies of simplified constructs that focus on negative control, this work has sought to clarify four basic principles of bacterial promoter repression by DNA looping. First, is the distance-dependence of DNA loop energetics similar for auxiliary operators positioned upstream or downstream from the promoter? Second, what is the smallest possible DNA loop involving Lac repressor? Third, is RNA polymerase trapped at repressed promoters? Fourth, when RNA polymerase and Lac repressor do not compete for overlapping binding sites, can a promoter be repressed in vivo simply by constraining it in a strained DNA loop?
Our results suggest that upstream and downstream auxiliary operators enhance the concentration of Lac repressor at a promoter-proximal operator with similar distance dependence. By studying DNA loops not containing promoters, we show that Lac repressor can constrain a stable loop with as few as four turns of double-helical DNA in vivo. ChIP results confirm that RNA polymerase exclusion, not RNA polymerase trapping, is the repression mechanism. Finally, using a T7 RNA polymerase promoter that does not overlap with Lac operators, we show in vivo that promoter pLacement within a strained DNA loop is sufficient to repress transcription initiation, confirming a previous proposal (21).
Future experiments are planned to extend these results in two ways. First, it is important to apply ChIP to study the issue of RNA polymerase exclusion versus trapping in the context of the wild-type Lac promoter region with an intact catabolite activator binding protein recognition sequence under conditions where both positive and negative control are operative. The spacing between the −10 promoter element and proximal operator is 6 bp in the wild-type Lac promoter versus 9 bp in our experimental constructs, and it will be important to determine whether this subtle difference affects RNA polymerase fate at the repressed promoter, especially given evidence of favorable repressor/RNA polymerase interaction (39). Second, we will study Lac promoter occupancy by architectural DNA binding proteins to explore the extent to which the apparent flexibility of the DNA in the Lac repression loop reflects the participation of DNA bending and kinking by these accessory proteins (15–17,24), a phenomenon that has previously been demonstrated in looping control at the E. coli gal operon (40–42).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5, Supplementary Figures 1–5 and Supplementary Methods.
FUNDING
Mayo Graduate School, the Mayo Foundation; National Institutes of Health [GM75965 to L.J.M.]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Molly Nelson-Holte for technical assistance.
REFERENCES
- 1.Müller-Hill B. The Lac Operon: A Short History of a Genetic Paradigm. Berlin, New York: Walter de Gruyter; 1996. [Google Scholar]
- 2.Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 3.Gilbert W, Muller-Hill B. Isolation of the Lac repressor. Proc. Natl. Acad. Sci. USA. 1966;56:1891–1898. doi: 10.1073/pnas.56.6.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmitz A, Galas DJ. The interaction of RNA polymerase and Lac repressor with the Lac control region. Nucleic Acids Res. 1979;6:111–137. doi: 10.1093/nar/6.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reznikoff WS, Winter RB, Hurley CK. The location of the repressor binding sites in the Lac operon. Proc. Natl Acad. Sci. USA. 1974;71:2314–2318. doi: 10.1073/pnas.71.6.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kramer H, Niemoller M, Amouyal M, Revet B, von Wilcken-Bergmann B, Müller-Hill B. Lac repressor forms loops with linear DNA carrying two suitably spaced Lac operators. EMBO J. 1987;6:1481–1491. doi: 10.1002/j.1460-2075.1987.tb02390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller J, Oehler S, Müller-Hill B. Repression of Lac promoter as a function of distance, phase and quality of an auxiliary Lac operator. J. Mol. Biol. 1996;257:21–29. doi: 10.1006/jmbi.1996.0143. [DOI] [PubMed] [Google Scholar]
- 8.Müller-Hill B. The function of auxiliary operators. Mol. Microbiol. 1998;29:13–18. doi: 10.1046/j.1365-2958.1998.00870.x. [DOI] [PubMed] [Google Scholar]
- 9.Oehler S, Eismann ER, Kramer H, Müller-Hill B. The three operators of the Lac operon cooperate in repression. EMBO J. 1990;9:973–979. doi: 10.1002/j.1460-2075.1990.tb08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Müller JJ, Barker AA, Oehler SS, Müller-Hill BB. Dimeric Lac repressors exhibit phase-dependent cooperativity. J. Mol. Biol. 1998;284:851–857. doi: 10.1006/jmbi.1998.2253. [DOI] [PubMed] [Google Scholar]
- 11.Oehler S, Müller-Hill B. High local concentration: a fundamental strategy of life. J. Mol. Biol. 2009;395:242–253. doi: 10.1016/j.jmb.2009.10.056. [DOI] [PubMed] [Google Scholar]
- 12.Bellomy G, Mossing M, Record M. Physical properties of DNA in vivo as probed by the length dependence of the Lac operator looping process. Biochemistry. 1988;27:3900–3906. doi: 10.1021/bi00411a002. [DOI] [PubMed] [Google Scholar]
- 13.Mossing MC, Record MT., Jr Upstream operators enhance repression of the Lac promoter. Science. 1986;233:889–892. doi: 10.1126/science.3090685. [DOI] [PubMed] [Google Scholar]
- 14.Han L, Garcia HG, Blumberg S, Towles KB, Beausang JF, Nelson PC, Phillips R. Concentration and length dependence of DNA looping in transcriptional regulation. PLoS One. 2009;4:e5621. doi: 10.1371/journal.pone.0005621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Becker NA, Kahn JD, Maher LJ., III Bacterial repression loops require enhanced DNA flexibility. J. Mol. Biol. 2005;349:716–730. doi: 10.1016/j.jmb.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 16.Becker NA, Kahn JD, Maher LJ., III Effects of nucleoid proteins on DNA repression loop formation in Escherichia coli. Nucleic Acids Res. 2007;35:3988–4000. doi: 10.1093/nar/gkm419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becker NA, Kahn JD, Maher LJ., III Eukaryotic HMGB proteins as repLacements for HU in E. coli repression loop formation. Nucleic Acids Res. 2008;36:4009–4021. doi: 10.1093/nar/gkn353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Besse M, von Wilcken-Bergmann B, Müller-Hill B. Synthetic Lac operator mediates repression through Lac repressor when introduced upstream and downstream from Lac promoter. EMBO J. 1986;5:1377–1381. doi: 10.1002/j.1460-2075.1986.tb04370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Straney SB, Crothers DM. Lac repressor is a transient gene-activating protein. Cell. 1987;51:699–707. doi: 10.1016/0092-8674(87)90093-6. [DOI] [PubMed] [Google Scholar]
- 20.Lee J, Goldfarb A. Lac repressor acts by modifying the initial transcribing complex so that it cannot leave the promoter. Cell. 1991;66:793–798. doi: 10.1016/0092-8674(91)90122-f. [DOI] [PubMed] [Google Scholar]
- 21.Lionberger TA, Meyhofer E. Bending the rules of transcriptional repression: tightly looped DNA directly represses T7 RNA polymerase. Biophys. J. 2010;99:1139–1148. doi: 10.1016/j.bpj.2010.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whipple FW. Genetic analysis of prokaryotic and eukaryotic DNA-binding proteins in Escherichia coli. Nucleic Acids Res. 1998;26:3700–3706. doi: 10.1093/nar/26.16.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters JP, Becker NA, Rueter EM, Bajzer Z, Kahn JD, Maher LJ. Quantitative methods for measuring DNA flexibility in vitro and in vivo. Meth. Enzymol. 2011;488:287–335. doi: 10.1016/B978-0-12-381268-1.00012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sebastian NT, Bystry EM, Becker NA, Maher LJ., III Enhancement of DNA flexibility in vitro and in vivo by HMGB box A proteins carrying box B residues. Biochemistry. 2009;48:2125–2134. doi: 10.1021/bi802269f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bond LM, Peters JP, Becker NA, Kahn JD, Maher LJ. Gene repression by minimal Lac loops in vivo. Nucleic Acids Res. 2010;38:8072–8082. doi: 10.1093/nar/gkq755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oehler S, Eismann ER, Kramer H, Müller-Hill B. The three operators of the Lac operon cooperate in repression. EMBO J. 1990;9:973–979. doi: 10.1002/j.1460-2075.1990.tb08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakerian AE, Matthews KS. Characterization of mutations in oligomerization domain of Lac repressor protein. J. Biol. Chem. 1991;266:22206–22214. [PubMed] [Google Scholar]
- 28.Chen J, Matthews KS. T41 mutation in Lac repressor is Tyr282—Asp. Gene. 1992;111:145–146. doi: 10.1016/0378-1119(92)90618-y. [DOI] [PubMed] [Google Scholar]
- 29.Schmitz A, Schmeissner U, Miller JH. Mutations affecting the quaternary structure of the Lac repressor. J. Biol. Chem. 1976;251:3359–3366. [PubMed] [Google Scholar]
- 30.Muller J, Oehler S, Müller-Hill B. Repression of Lac promoter as a function of distance, phase and quality of an auxiliary Lac operator. J. Mol. Biol. 1996;257:21–29. doi: 10.1006/jmbi.1996.0143. [DOI] [PubMed] [Google Scholar]
- 31.Brukner I, Sanchez R, Suck D, Pongor S. Sequence-dependent bending propensity of DNA as revealed by DNase I: parameters for trinucleotides. EMBO J. 1995;14:1812–1818. doi: 10.1002/j.1460-2075.1995.tb07169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis NA, Majee SS, Kahn JD. TATA box DNA deformation with and without the TATA box-binding protein. J. Mol. Biol. 1999;291:249–265. doi: 10.1006/jmbi.1999.2947. [DOI] [PubMed] [Google Scholar]
- 33.Hochschild A, Ptashne M. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell. 1986;44:681–687. doi: 10.1016/0092-8674(86)90833-0. [DOI] [PubMed] [Google Scholar]
- 34.Friedman AM, Fischmann TO, Steitz TA. Crystal structure of Lac repressor core tetramer and its implications for DNA looping. Science. 1995;268:1721–1727. doi: 10.1126/science.7792597. [DOI] [PubMed] [Google Scholar]
- 35.Haeusler AR, Goodson KA, Lillian TD, Wang X, Goyal SS, Perkins NC, Kahn JD. FRET studies of a landscape of Lac repressor-mediated DNA loops. Nucleic Acids Res. 2012;40:4432–4445. doi: 10.1093/nar/gks019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, McEwen AE, Crothers DM, Levene SD. Analysis of in vivo LacR-mediated gene repression based on the mechanics of DNA looping. PLoS One. 2006;1:e136. doi: 10.1371/journal.pone.0000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikeda R, Richardson CC. Interactions of the RNA polymerase of bacteriophage T7 with its promoter during binding and initiation of transcription. Proc. Natl Acad. Sci. USA. 1986;83:3614–3618. doi: 10.1073/pnas.83.11.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iost I, Dreyfus M. The stability of Escherichia coli LacZ mRNA depends upon the simultaneity of its synthesis and translation. EMBO J. 1995;14:3252–3261. doi: 10.1002/j.1460-2075.1995.tb07328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia HG, Sanchez A, Boedicker JQ, Osborne M, Gelles J, Kondev J, Phillips R. Operator sequence alters gene expression independently of transcription factor occupancy in bacteria. Cell Rep. 2012;2:150–161. doi: 10.1016/j.celrep.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aki T, Adhya S. Repressor induced site-specific binding of HU for transcriptional regulation. EMBO J. 1997;16:3666–3674. doi: 10.1093/emboj/16.12.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis DE, Adhya S. In vitro repression of the gal promoters by GalR and HU depends on the proper helical phasing of the two operators. J. Biol. Chem. 2002;277:2498–2504. doi: 10.1074/jbc.M108456200. [DOI] [PubMed] [Google Scholar]
- 42.Lia G, Bensimon D, Croquette V, Allemand JF, Dunlap D, Lewis DE, Adhya S, Finzi L. Supercoiling and denaturation in Gal repressor/heat unstable nucleoid protein (HU)-mediated DNA looping. Proc. Natl Acad. Sci. USA. 2003;100:11373–11377. doi: 10.1073/pnas.2034851100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.