Abstract
In the yeast Saccharomyces cerevisiae, the aminoacyl-tRNA synthetases (aaRS) GluRS and MetRS form a complex with the auxiliary protein cofactor Arc1p. The latter binds the N-terminal domains of both synthetases increasing their affinity for the transfer-RNA (tRNA) substrates tRNAMet and tRNAGlu. Until now, structural information was available only on the enzymatic domains of the individual aaRSs but not on their complexes with associated cofactors. We have analysed the yeast Arc1p-complexes in solution by small-angle X-ray scattering (SAXS). The ternary complex of MetRS and GluRS with Arc1p, displays a peculiar extended star-like shape, implying possible flexibility of the complex. We reconstituted in vitro a pentameric complex and demonstrated by electrophoretic mobility shift assay that the complex is active and contains tRNAMet and tRNAGlu, in addition to the three protein partners. SAXS reveals that binding of the tRNAs leads to a dramatic compaction of the pentameric complex compared to the ternary one. A hybrid low-resolution model of the pentameric complex is constructed rationalizing the compaction effect by the interactions of negatively charged tRNA backbones with the positively charged tRNA-binding domains of the synthetases.
INTRODUCTION
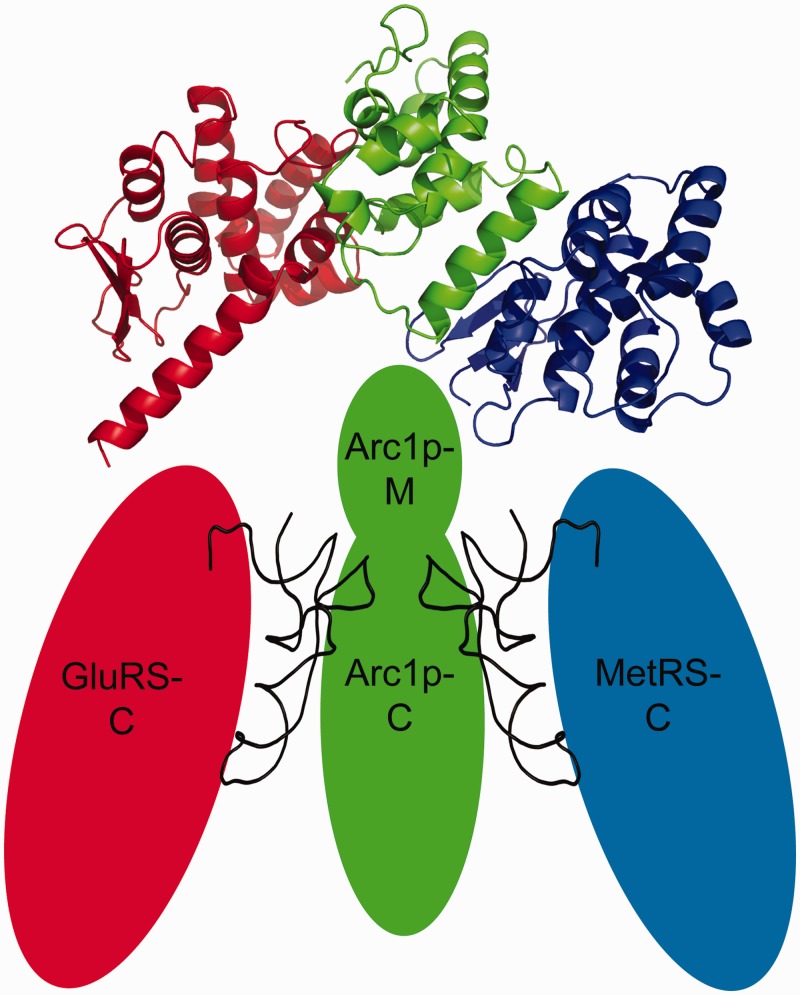
During protein translation aminoacyl-tRNA synthetases (aaRSs) catalyse the charging of a transfer-RNA (tRNA) with its cognate amino acid. The aaRSs ensure the accuracy of the aminoacylation of the appropriate tRNA via nucleotide determinants, which define the identity of the tRNA (1,2). Whereas prokaryotic aaRSs typically function as monomers or homo-dimers, eukaryotic aaRSs show heteromeric interactions (3–6). Eukaryotic aaRSs must be integrated into an efficient tRNA nuclear export- and shuttling machinery and are interconnected closely with the translation machinery, because of the compartmentalization and bigger size of the eukaryotic cell. This requires the presence of additional protein-tRNA-binding domains (TRBDs), in contrast to the prokaryotic aaRS, and a higher degree of complex formation (7). In some aaRS also a rearrangement of the tRNA binding and catalytic domain was found to be essential for the aminoacylation (8,9). In higher eukaryotes a multi-enzyme complex exists, built out of nine aaRSs, including MetRS and GluProRS, and three cofactors (10–12). In yeast there exists an evolutionary intermediate aminoacyl-tRNA synthetase complex formed by two class I synthetases, MetRS and GluRS and the aminoacyl cofactor Arc1p (10,12). While the biochemistry of yeast Arc1p-complex is well-characterized (13), there is no structure of the whole complex available until now. It has been shown by structural (6) and biochemical (13,14) studies that both synthetases (MetRS and GluRS) bind through their N-terminal domains to the N-terminal domain of Arc1p and that the synthetases bind to largely independent sites (Figure 1) (14,15), forming the MetRS*Arc1p*GluRS-complex (MAG). This interaction increases the affinity for the cognate tRNAs, due to the presence of a TRBD formed by the middle- and C-terminal domains of Arc1p (16–19). tRNAMet and tRNAGlu simultaneously bind to their cognate synthetase as well as to Arc1p. The synthetases bind the acceptor stem of their cognate tRNAs from the minor groove site via their C-terminal domains (1). For our studies we only used tRNAMet elongator and not tRNAMet initiator, because the literature indicates that the tRNAMet initiator poorly binds to the Arc1p-complex (18).
Figure 1.
Schematic picture of the Arc1p-complex. MetRS (in blue) and GluRS (in red) are binding through their N-termini to Arc1p (shown in green). The N-terminal domains of all three proteins are displayed as ribbon models of their X-ray crystallographic structures. Both tRNAs are shown in between their corresponding synthetase and Arc1p.
In the present work, quaternary structure of the ternary Arc1p-complex and the full pentameric assembly with the two tRNAs in solution is characterized using synchrotron small-angle X-ray scattering (SAXS). The low-resolution ab initio models are obtained and hybrid models are constructed using the high-resolution structural data and homology models for the individual components. A dramatic compaction of the complex is observed upon the tRNA binding and the effect is correlated with the electrostatic interactions within the complexes.
MATERIALS AND METHODS
Cloning of the proteins and tRNAs
The cofactor Arc1p and the two synthetases GluRS and MetRS were cloned from the plasmids pETHIS6/pET8c-Arc1p, pEMBLyex4-His8-GluRS and pEMBLyex4-His8-MetRS into a modified expression vector pETm (Novagen). The proteins were expressed and purified following the protocol described in an earlier publication (20).
Both tRNAs, tRNAMet and tRNAGlu, were PCR amplified from a pZErO2 vector by producing a 5′-overhang with an EcoRI and a 3′-overhang with a PstI recognition site. The tRNA genes and the expression vector pBSTNAV2 were cut with EcoRI and PstI and ligated. Expression was carried out in JM101 cells.
Sample preparation
All proteins were expressed in E scherichia coli and purified using Ni-beads. After cleavage with TEV protease, the protein was buffer exchanged and loaded again on a Ni-column, followed by gel filtration. Each purification step was checked by SDS-PAGE. The tRNAs were expressed in JM101 cells for 20 h at 37°C in 4 l TB medium (constitutive tRNA expression). The cell pellet was resuspended in a buffer containing 5 mM Tris pH 7.4 and 10 mM Mg(CH3COO)2. After a phenol extraction, as previously described elsewhere (21), the samples were concentrated to 1 ml and loaded on a Superdex 75 column (GE Healthcare), equilibrated in buffer (20 mM HEPES pH 7.2, 150 mM NaCl and 5 mM MgCl2). The peak was analysed by UREA-PAGE and the fractions of interest were pooled together and concentrated for further experiments. These fractions did not contain only the desired tRNAs (tRNAMet and tRNAGlu), but also other tRNA species in minor concentrations. These tRNAs will be named tRNAGlu or tRNAMet mixture in the following text.
In vitro reconstruction of the Arc1p-complex
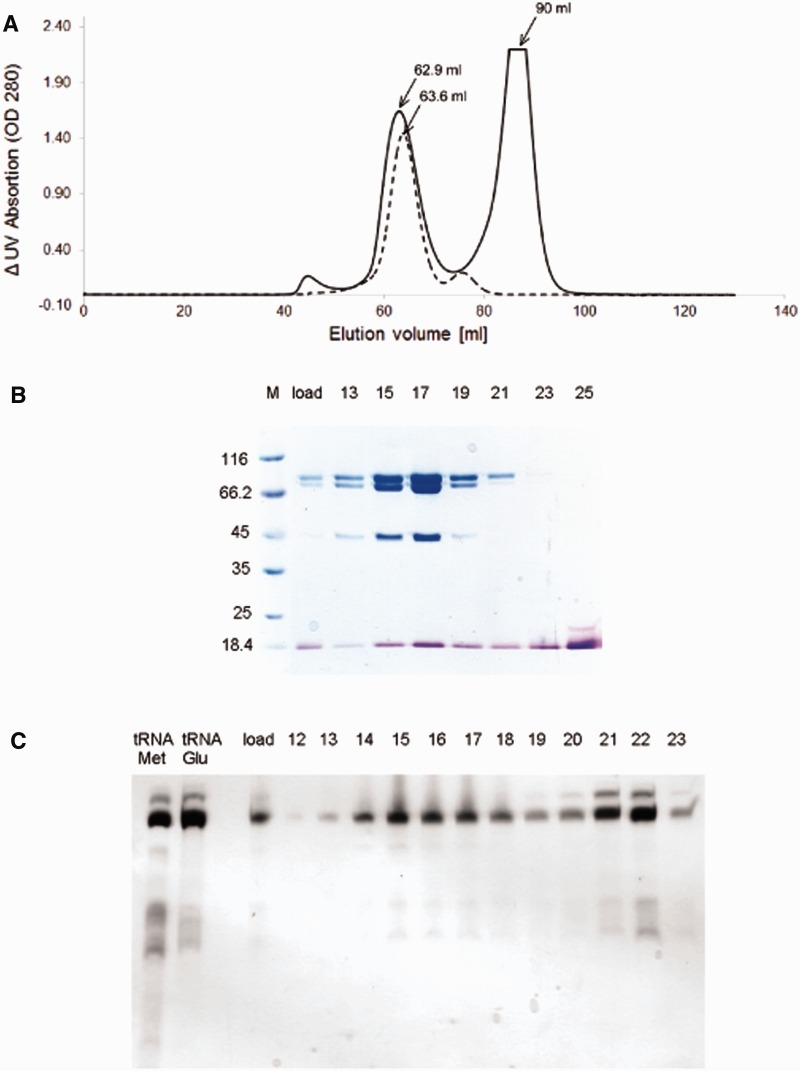
For building the protein complex we first made a complex of two proteins, MetRS and Arc1p. Arc1p was mixed in excess (1:1.5 molar ratio) with MetRS and then the sample was loaded onto a Superdex 200 column (GE Healthcare) followed by SDS-PAGE analysis. Then we concentrated the peak fractions. We mixed this complex with an excess of GluRS (1:1.5) and loaded it again onto a Superdex 200 (Figure 2A, dashed line). After the Superdex run we obtained a very clean trimeric complex (MAG=MetRS:Arc1p:GluRS). After SDS-PAGE we concentrated the peak fractions and the tRNAMet and tRNAGlu enriched tRNA mixtures were added in a 1:1.5:1.5 molar ratio (complex:tRNAMet:tRNAGlu). The mixture was purified by a Superdex 200 run, which resulted in two peaks (Figure 2A). The first peak of the continuous line is at the elution volume of 62.9 ml, which corresponds to the pentameric complex consisting of MetRS, Arc1p, GluRS and both tRNAs (MAG*tRNAs). The second peak of the continuous line occurs at 90 ml, which corresponds to the size of a tRNA.
Figure 2.
Sample preparation and quality control. (A) Chromatogram of the Superdex 200 run. The dashed line is showing the chromatogram of the MAG purification. The continuous line shows the result of the Superdex run of the pentameric complex. (B) SDS-PAGE. The gel shows the sample (load) containing all the three proteins, as well as both tRNAs, which was loaded on S200, followed by the peak fractions 13–25. Gel was stained first with Coomassie Blue (blue) to visualize proteins and then with Toluidinium Blue (violet) to show tRNAs. (C) UREA-PAGE after Superdex 200 run. The first lane shows tRNAMet mixture after purification, the second lane shows tRNAGlu mixture. The sample (load) consists of all three proteins and both tRNA mixtures. Fractions were collected and loaded on this gel (fractions 12–23). Fractions 13–19 contain tRNA enriched in tRNAMet and tRNAGlu.
As it is visible in the chromatogram (Figure 2A) there is a movement of the first peak from 63.8 ml elution volume (dashed line) to 62.9 ml elution volume (continuous line). The reason of this movement is the binding of the tRNAs to the trimeric complex.
Gel analysis of the protein:tRNA complex
After the last Superdex 200 run, we analysed the fractions of the two peaks by SDS-PAGE (Figure 2B) and UREA-PAGE (Figure 2C). The molecular weights of the proteins are as follows: GluRS 84.5 kDa, MetRS 85.5 kDa and Arc1p 43 kDa. On both gels we loaded the sample which we injected on the column (indicated on gel with: load) and the peak fractions, which we collected during the run. On the UREA-PAGE we loaded in addition samples of purified tRNAMet and tRNAGlu mixtures as controls. To visualize the proteins as well as the tRNAs on one gel, we stained the SDS-PAGE (Figure 2B) two times. The first staining step was with Coomassie Blue G250 solution, followed by washing with water. After this step the proteins were visible on the gel. The second staining was with a 0.1% Toluidinium Blue solution. We incubated the gel for 10 min in this solution and washed it afterwards with water. After this procedure the proteins are still blue stained and the tRNAs are visible as violet bands. With the extra Toluidinium Blue staining of the SDS-PAGE, there is no further need to run the samples on an extra UREA-PAGE. After comparing both gels, we took the fractions 13–19 of the first peak, which consist of all the three proteins as well as both tRNAs. The second peak (fractions 20–25) contains the excess tRNAs, which were added to the complex. Interestingly, the tRNA mixtures in the first peak, which are bound to the complex, are purer than the tRNA mixtures in the second, as one can see on the UREA-PAGE (Figure 2C). There are additional bands visible on the gel, what leads us to the conclusion, that it is a more pure tRNA, which is bound to the complex.
The sample was concentrated in a filter device up to 10 mg/ml. The sample was then shock frozen and stored at −80°C for the SAXS experiments. Dynamic light scattering (data not shown) was done prior to the SAXS measurements to control the sample homogeneity.
The SAXS experiments compared the trimeric complex (which consists of the two synthetases, MetRS and GluRS, and the cofactor Arc1p) with the pentameric complex formed by simultaneous binding of the two cognate tRNAs (tRNAMet and tRNAGlu) to the trimeric complex. In order to identify the individual components of the trimeric complex, samples of dimeric complexes (MetRS*Arc1p and GluRS*Arc1p) of subunits were also prepared and investigated under identical conditions.
Electrophoretic mobility shift assay
The tRNA mixtures enriched by tRNAGlu or by tRNAMet, were labelled with 32P using T4PNK, followed by UREA-PAGE gel purification. The corresponding tRNA bands were cut out of the gel and incubated overnight in buffer (0.6 M AcNH4, 1 mM EDTA, 0.1% SDS). After an Ethanol precipitation the tRNAs were taken up in water.
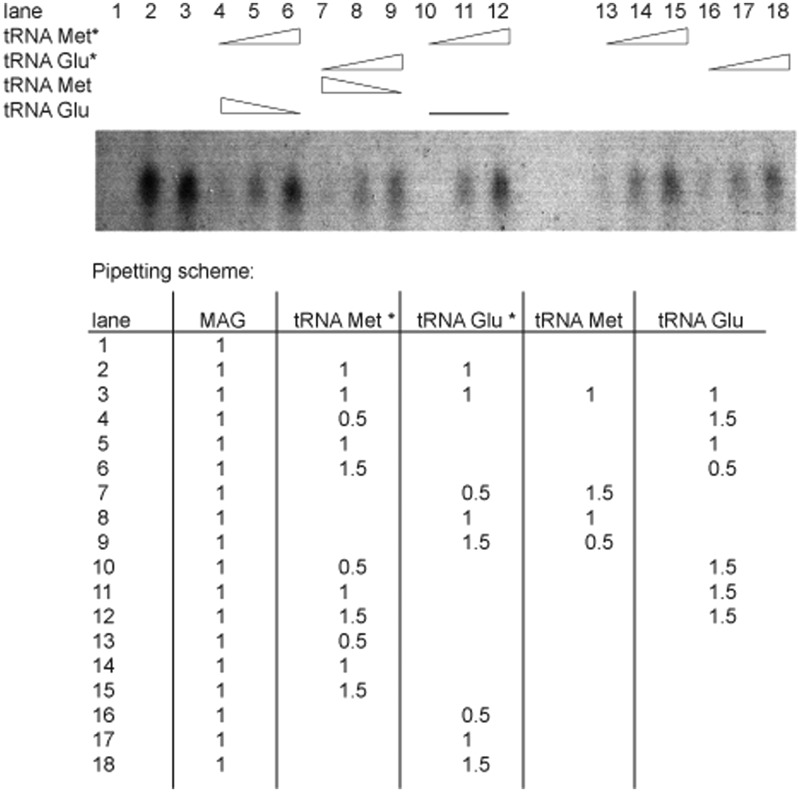
To show the binding of the tRNAs to the complex, different concentrations of tRNA mixtures were added to the complex and incubated for 30 min on ice. First unlabelled tRNAGlu mixture was given to the trimeric complex and labelled tRNAMet mixture was added. The concentration of tRNAGlu mixture was reduced while the concentration of tRNAMet mixture was increased. Then a competition assay was performed. Therefore, unlabelled tRNAGlu mixture was first given to the trimeric complex in a constant ratio and labelled tRNAMet mixture was then added in different concentration steps. The samples were analysed on a 6% native gel and the bands detected by autoradiography (22) (Figure 3). For more detailed information see Supplementary Experimental Procedures.
Figure 3.
EMSA of the pentameric complex. The binding of the tRNA mixtures, which are enriched in tRNAMet or tRNAGlu, to the trimeric complex can be followed up by autoradiography bands. 32P-labelled tRNA mixtures are indicated with a star (*), unlabelled tRNA mixtures without. The lanes are numbered from 1 to 18, corresponding to the pipetting scheme. The triangles indicate increasing or decreasing concentrations of tRNA mixtures. The line stands for a constant concentration of tRNAGlu. The pipetting scheme shows the amount of components added for each lane in µl. Always 1 µl of the trimeric complex (MAG) was taken for each reaction and different concentration of labelled or unlabelled tRNA mixtures were added. One tRNA is not influencing the binding efficiency of the other one (lanes 4–9 and 13–18) and both tRNAs can bind simultaneously to the complex (lanes 10–12). For more detailed information see Supplementary Experimental Procedure.
SAXS data collection
The SAXS data were recorded at the EMBL beamline X33 on the DORIS storage ring at DESY in Hamburg (23) using a robotic sample changer (24) at the X-ray wavelength of 1.5 Å. A MAR345 image plate detector (MAR Research GmbH) was used at a sample-detector distance of 2.7 m to cover a momentum transfer range s of 0.012–0.45 Å−1 (s = 4π sin (θ)/λ, where 2θ is the scattering angle). The scattering patterns of the trimeric and pentameric Arc1p-complex were measured for at least three solute concentrations ranging from 2 mg/ml to 10 mg/ml in the buffer containing 20 mM HEPES buffer pH 7.2, 150 mM NaCl, 5 mM MgCl2 and 1 mM DTT. The solute concentration was verified with a NanoDrop 1000 instrument (Thermo Scientific) immediately prior to the data collection. To check for effects of radiation damage, a second collection of 2-min exposure was taken and no systematic differences were noted between the images showing the exposure to X-rays had no significant effect on the data collected.
The data were analysed and extrapolated to infinite dilution following standard procedures using the programs from the ATSAS suite (25). The radius of gyration R g was evaluated using the Guinier approximation in PRIMUS (26) and from the entire scattering patterns using the indirect transform package GNOM (27), the latter program also provides the distance distribution function p(r). The molecular mass (MM) of the constructs were estimated from both I(0) estimates calibrated against BSA (66 kDa). The excluded volumes of the hydrated particles were calculated by using the Porod invariant (28). The ab initio low-resolution models were calculated using DAMMIN (29). Multiple models were created for each construct and averaged using DAMAVER (30). For the trimeric complex, three-phase models depicting each protein were obtained using MONSA (29,31) by simultaneous fitting of the SAXS data for the full-length construct along with the data for the individual proteins and the binary complexes of Metrs:Arc1p and Glurs:Arc1p. The bead models were processed to produce surface envelopes using SITUS (32).
The scattering from the tentative model of the pentameric complex was calculated by CRYSOL (33). The rigid body modelling of the complex was done using SASREF (34). Starting from the arrangement of the subunits as in the interactively constructed tentative complex, their positions and orientations were refined using a low-temperature-simulated annealing minimization and employing the appropriate molecular constraints to maintain the integrity of the protein-RNA-binding sites. The ab initio and rigid body models of all constructs were constructed by fitting the experimental data in the range of scattering vectors up to s = 0.25 Å−1, responsible for the quaternary structure.
Structures used for molecular modelling
In order to determine the interactions between the three proteins, as well as their behaviour in binding the two tRNAs, we fitted high-resolution structures of the proteins and tRNAs into the shapes of the complexes. However, since there are no 3D-structures available for the whole yeast proteins, MetRS, GluRS and Arc1p, we fit structural homologues of the middle and C-terminal domains into the SAXS-derived low-resolution shapes. For the interacting N-terminal domains of the three proteins we use the model previously proposed by us on the basis of X-ray studies of dimeric complexes (PDB-ID: 2HSM) (6). For the Arc1p1-121 domain (PDB-ID: 2HQTT), as well as for the GluRS1-180 (PDB-ID: 2HRAA) and MetRS1-159 (PDB-ID: 2HSNA) domains we used the structures previously determined (35). For searching for structural homologues for the C-termini of the proteins we used the SWISS MODEL WORKSPACE (36–40), which suggested for Arc1p residues 205-376 to use PDB-ID: 1E7ZA (P43 EMAP2 RNA binding domain from human (41)) as a structural homologue with a sequence identity of 45.3%. For GluRS residues 206-710: PDB-ID: 1EUQ (Glutaminyl-tRNA synthetase from E. coli (42)), which has a sequence identity to the yeast GluRS of 35.7% and for MetRS residues 196-704 it suggested the structural homologue MetRS from Aquifex aeolicus (PDB-ID: 2CSX (43)), which shows a sequence identity to S. cerevisiae MetRS of 23%. In case of tRNAMet and tRNAGlu we use the tRNAs, which are already included in the PDB files of the structural homologues. For tRNAGlu we take the tRNAGln from E. coli, which is bound to Glutaminyl-tRNA synthetase (PDB-ID: 1EUQ) and for tRNAMet we use the tRNAMet from Aquifex aeolicus (PDB-ID: 2CSX). Surface charge analysis was done using PyMOL.
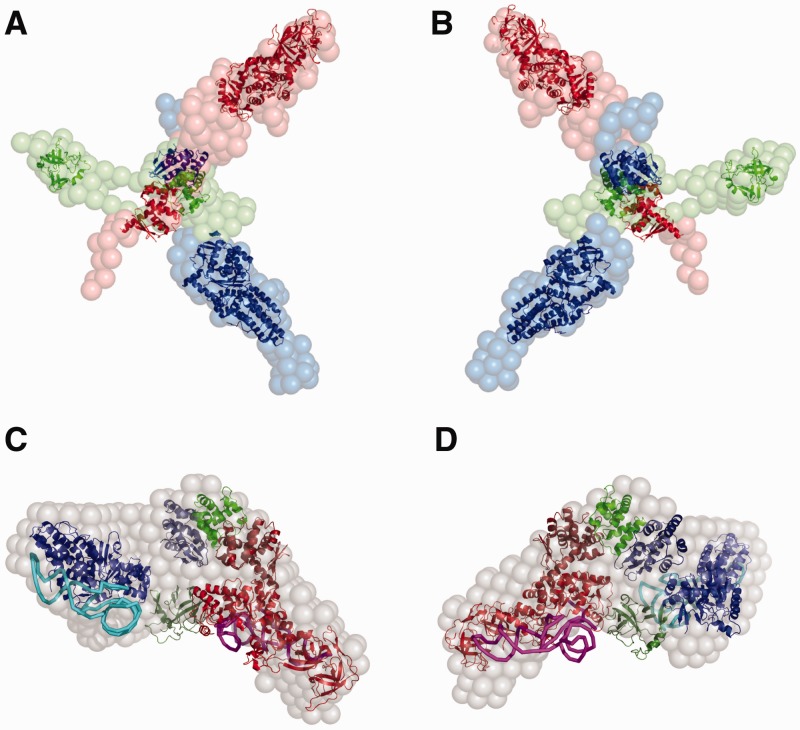
For the fitting of the structures we also took in account the missing linker regions. We analysed the secondary structure for the missing amino acids for GluRS 181-205 and MetRS 160-195 with the Network Protein Sequence Analysis server (44) (see Supplementary Data, Supplementary Figure S1A and B). The prediction shows for the missing amino acids of GluRS 181-200 a random coil region and for GluRS 201-205 an extended strand. The distance in our pentameric model (Figure 4C and D) between the N-terminal and C-terminal region of GluRS is 18 Å, so that the N-termini can easily be connected to the C-terminus via the 25 missing amino acids. For the residues 160–172 of MetRS a random coil region was predicted, for the residues 173–175 an extended strand and for the residues 176–195 again a random coil region. The distance between the N-terminus and C-terminus of MetRS in our model is 65 Å, which can be bridged with the predicted secondary structure of 35 missing amino acids.
Figure 4.
Pentameric and trimeric complex. The structures of the homologues and/or the known structures of the proteins and tRNAs are inserted into the ab intio model obtained by MONSA (29,31). The colours, which are used for the proteins are the same in the case of the pentamer and the trimer. GluRS is shown in red, Arc1p in green and MetRS in blue. Panels (A) and (B) show the trimeric complex from different sides. The surface of GluRS is shown in red spheres, MetRS in blue and Arc1p in green. Panels (C) and (D) display the pentameric complex in two different views. The ab intio model is illustrated in grey spheres, tRNAGlu is shown in magenta and tRNAMet in cyan.
RESULTS
Sample quality control
All proteins and tRNAs were purified by gel filtration and purity was checked by SDS-PAGE and UREA-PAGE. Also the trimeric and pentameric complexes were loaded on a Superdex 200 column to separate the complexes from unbound proteins or tRNAs. Only the peak fractions, which contain the desired complex were used for SAXS experiments. Figure 2B and C shows the peak fractions of the Superdex 200 run of the pentameric complex on SDS-PAGE and UREA-PAGE, respectively, where one can clearly see, that there is a preferential binding of tRNAMet and tRNAGlu to the complex (fractions 13–19). The excess of tRNAs elute later together with some impurities (fractions 20–25), which do not bind to the complex.
To check the expression level of the desired tRNA, a comparative expression of native tRNA in JM101 cells was carried out. Therefore an expression with and without the plasmid carrying the tRNA gene was done side by side and the expression level with the tRNA gene on the plasmid was 10-fold higher than without the tRNA gene (data not shown).
Radioactive electrophoretic mobility shift assay
The electrophoretic mobility shift assay (EMSA) was performed to prove that the yeast Arc1p-complex can bind tRNAMet and tRNAGlu simultaneously and independently from each other. In the assay, a titration for each tRNA mixture was done first and the two tRNA mixtures were subsequently competing with each other for binding to the complex. The titration indicated, that both tRNAs, which are enriched in the tRNA mixture, can bind to the complex and that the binding is concentration dependent. Moreover, the competition assay revealed that a high concentration of tRNAGlu did not influence the binding capacity of tRNAMet to the complex. Together with the sample quality control by SDS-PAGE and UREA-PAGE these data demonstrated that tRNAMet and tRNAGlu did bind to the complex and that both tRNAs were bound independently from each other (Figure 3).
As an additional check we performed a pyrrophosphate assay, to verify that the synthetase binds the tRNA and activates the amino acid to transfer it to the tRNA, which leads to a pyrrophosphate that can be measured by a spectrometer. If there is no tRNA, the activation of the amino acid does not take place and no pyrrophosphate can be detected. The same is the case, if there is tRNA but no amino acid in the reaction. These results (data not shown) provide an additional (albeit qualitative) evidence that the trimeric complex binds the tRNAs and can activate amino acids.
SAXS experiments and ab initio shape determination
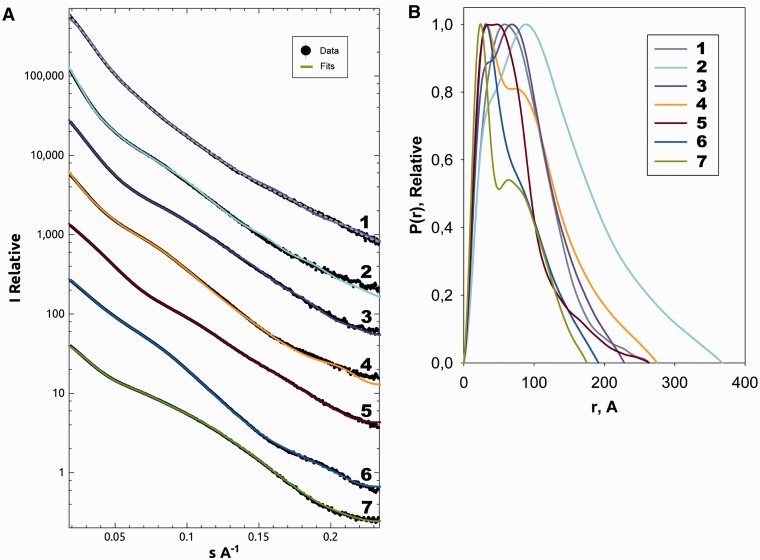
To comprehensively characterize the Arc1p-complex formation, SAXS patterns were recorded on several available constructs. We measured the three individual proteins, Arc1p, GluRS and MetRS, two binary complexes MetRS:Arc1p and GluRS:Arc1p, as well as the trimeric and pentameric complexes. All the experimental data were measured at multiple solute concentrations, normalized and processed following standard procedures (Figure 5A). All the measured constructs displayed the behaviour characteristic for monodisperse solutions (including linear Guinier plots presented in Supplementary Figure S2). The overall parameters of the constructs summarized in Table 1 clearly indicate that the individual proteins are monomeric in solution, and their distance distribution functions p(r) display skewed profiles typical for elongated particles (Figure 5B). From the MM and volume (V) estimates (Table 1), both binary complexes MetRS:Arc1p and GluRS:Arc1p have a 1:1 stoichiometry, and the ternary complex MetRS:GluRS:Arc1p has a 1:1:1 stoichiometry. The radii of gyration (R g) and maximum diameters (D max) of the binary and ternary complexes are larger than those of the individual proteins, and the p(r) functions suggest highly anisometric structures of these complexes. For the pentameric complex, the increase of the apparent MM estimated from the forward scattering values is fully compatible with the addition of the two tRNAs to the 1:1:1 ternary protein complex. This indicates, in agreement with the above EMSA results, that the pentameric complex contains all the three proteins and the two tRNA molecules. However, very surprisingly, the geometrical parameters of the pentameric complex are significantly smaller than those of the ternary complex. The distance distribution of the pentameric complex (Figure 5) is a bell-shaped function typical for compact globular bodies. These data unambiguously point to a collapse of the quaternary structure of the MetRS:GluRS:Arc1p complex upon the binding of the tRNA mixture, preferentially of tRNAMet and tRNAGlu.
Figure 5.
Scattering curves and distance distribution functions. (A) Experimental scattering curves and the scattering from the structural models; the experimental data are shown in black (with error bars in grey), the ab initio fit for each data set (and the final rigid body refinement for the pentameric complex) in colours corresponding to the structures presented. (B) Distance distribution (p(r)) functions: 1, Pentameric complex MetRS:GluRS:Arc1p:tRNAs (grey-dashed line: ab intio fit, purple: SASREF refined rigid body fit); 2, Trimeric complex of MetRS:GluRS:Arc1p (yellow); 3, GluRS:Arc1p (lilac); 4, MetRS:Arc1p complex (yellow); 5, GluRS (red) the largest of the monomeric complexes; 6, MetRS (blue) slightly smaller than GluRS but both are noticeably bigger then Arc1p; 7, Arc1p (green) is the smallest of the proteins in the complex.
Table 1.
Overall structural parameters calculated from SAXS
| Parameter | Source | Arc1p | GluRS | MetRS | MetRS:Arc1p | GluRS:Arc1p | Ternary complex | Pentameric complex |
|---|---|---|---|---|---|---|---|---|
| R g (Å) | Guiner | 49 ± 2 | 50 ± 1 | 50 ± 2 | 62 ± 2 | 65 ± 2 | 97 ± 4 | 60 ± 6 |
| I(0) | Guiner | 146 ± 10 | 280 ± 20 | 270 ± 30 | 360 ± 30 | 490 ± 40 | 830 ± 80 | 880 ± 90 |
| MW | Literature (18) | 43 | 84.5* | 85.5 | 128.5 | 127.5* | 214.5* | 239.5* |
| Calculated from I(0) relative to BSA | 39 ± 4 | 75 ± 8 | 72 ± 8 | 97 ± 10 | 130 ± 15 | 217 ± 20 | 234 ± 25 | |
| Volume (Å3) | DAMMIN | 110 900 | 211 300 | 203 100 | 293 000 | 333 900 | 699 600 | 406 200 |
| No. of residues | Literature (15) | 376 | 724 | 751 | 1127 | 1100 | 1851 | 1999 |
Shown are R g and I(0) of the monomers as well as of the complexes, the resulting molecular weights as well as the volumes resulting from the ab initio reconstructions. Corresponding references are shown in brackets. A star (*) indicates that the protein contains a His tag.
In the next step, the low-resolution shapes of all the constructs were determined ab initio using the program DAMMIN, representing the particle as a collection of interconnected beads (29). The independently determined shapes of the individual proteins and of the ternary and pentameric complexes are presented in Supplementary Data (Supplementary Figure S3A–E). A visual comparison of these models strongly suggests that the elongated shapes of all the three proteins do not significantly change upon complex formation. This observation opens a possibility of the advanced multiphase ab initio modelling using the program MONSA (29,30). Here, the ternary complex is represented by three ‘phases’ (types of beads), belonging to Arc1p, MetRS and GluRS, and the model is constructed in such a way that the measured data from the individual proteins, binary and ternary complexes are simultaneously fitted by a single model. Several independent runs yielded similar models fitting the experimental data very well (Figure 5A); importantly, the multiphase fits were not worse than those from the individual shape reconstructions further confirming that no significant shape changes took place upon complex formation. A typical MONSA reconstruction in Figure 4A and B reveals a distinct star-like assembly of the elongated individual proteins (represented by beads of different colours).
The above multiphase modelling requires that the conformations of the free subunits are preserved in the complexes. This is the case for the trimeric but definitely not for the pentameric complex. Attempts to perform a four-phase MONSA modelling including the tRNA moiety to fit all the scattering data sets including that of the pentameric complex were unsuccessful. This indicates that the shape of the protein moiety changes significantly in the pentameric complex compared with all the other constructs. The shape of the pentameric complex reconstructed ab initio from its scattering pattern (Figure 4C and D, and Figure 5A) appears as a globular particle, significantly more compact than the star-shaped trimeric complex. This result fully agrees with the above analysis of the overall parameters and further corroborates the finding that the binding of the two tRNAs prompts a drastic compaction of the Arc1p-complex.
Molecular modelling
To further rationalize the obtained low-resolution shapes, molecular modelling of the Arc1p-complexes was performed utilizing the available high-resolution structures and homology models. For the trimeric complex, the structures of the N-termini and the homologues of the C-termini were manually fitted into the ab initio MONSA envelope of the trimeric complex using biochemical and structural data known for the complex as additional positioning constraints (see Materials and Methods section). The positioning of the fragments within the appropriate envelopes (Figure 4A and B) demonstrates that the reconstructed volume can accommodate all the individual components. Indeed, the high-resolution structures fit well within the shape whereby the individual domains interact with each other as previous studies have shown (6,13–15). The arms of the ab initio envelope appear to be slightly longer compared to the overlaid structures, which can be explained by the potential flexibility of the highly extended trimeric complex.
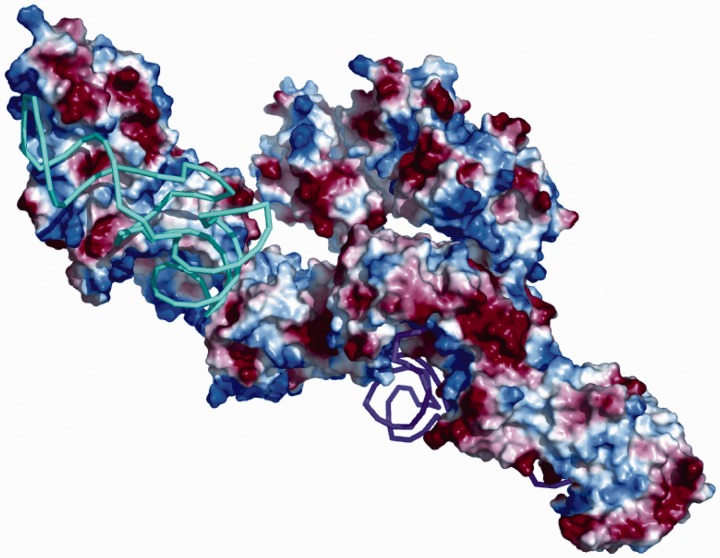
Also for the pentameric complex structural homologues of the C-termini and the known structures of the N-termini were used for the initial manual fitting in the same way as it was done for the trimeric complex. The surface charges of the individual proteins, in particular of the binding sites between the tRNAs, the synthetases and the C-terminal domain of Arc1p, were taken into account while fitting the domains into the ab initio model of the pentameric complex. A tentative model thus constructed was further refined by rigid body modelling of the positions of the components using the program SASREF (24) while keeping the major protein–RNA interfaces. The refined model is displayed together with the ab initio shape in Figure 4C and D. The surface charge of the pentameric complex analysed with PyMOL is presented in Figure 6 in the same orientation as in Figure 4C. Here, the synthetases show positive surface charge at the binding site of the negatively charged tRNAs, and, interestingly, the C-terminal domain of Arc1p also has positive charges where the tRNAs can bind.
Figure 6.
Pentameric complex with surface charges of the individual proteins (blue: positive, red: negative). The proteins are at the same positions as in Figure 4C.
DISCUSSION
The analysis of the SAXS data has provided us with the ab initio models of both the trimeric and the pentameric complexes, displayed in Figure 4. Without the tRNAs the protein components of the Arc1p-complex appear to point in different directions and the proteins themselves look stretched with none or only very few interactions between their individual domains. There is still some empty volume within the overall shapes left after fitting the structure homologues inside the surface of the trimeric complex. This can be explained by the fact that we did not find structural homologues with the same number of amino acids as our S. cerevisiae proteins. All the individual components have a number of residues missing Arc1p 82 (residues 122–204), GluRS 36 (residues 181–205 and 711–723) and MetRS 47 (residues 705–751). Potential flexibility of the extremely extended trimeric complex may also play a role, but, overall, the extended shapes of the proteins can be well matched by the available high-resolution portions. The fidelity of the obtained model of the ternary complex is especially high given that it fits simultaneously six independently measured SAXS patterns.
The binding of the two tRNAs causes a significant compaction, literally a collapse, of the pentameric complex compared to the trimeric. The complex with the tRNAs is very compact, and even its apparent (Porod) volume is smaller than the volume without the tRNAs. This is explained by two reasons. First, the pentameric complex is a heterogeneous molecule (consisting of two components with significantly different X-ray contrast, proteins and RNA).These density fluctuations lead to additional scattering at higher angles and thus a smaller effective volume. Second and perhaps most important, the volume as seen by SAXS corresponds to the volume of the unbound solvent excluded by the molecule. The very extended trimeric complex is able to exclude more solvent (because of potential flexibility, higher hydration and electrostatic interactions) than the rather compact and rigid pentameric complex. This considerable volume compaction indicates substantial conformational changes of all three proteins when the tRNAs are bound. The arrangement of the individual domains inside the pentameric shape is not as straightforward as it is for the trimeric complex. It is not possible to assign a particular part of the shape to a specific protein, because of the dramatic volume compaction of the pentameric complex. However, by using the results of previous studies (6,13–15) and the surface charges of the specific proteins as additional constraints, we can derive a sensible model for the structural assembly within the pentamer. The structures of the homologues fit nearly perfectly inside the pentameric envelope resulting from SAXS experiments. The fact, that there are some minor parts of the structures outside the ab initio shape (shown in grey), may be a consequence of the relatively low sequence identities between the S. cerevisiae proteins and the structural homologues, ranging from 23% to 45.3%.
The compaction observed upon addition of tRNA indicates the protein components of the ternary complex undergo substantial conformational changes upon the formation of the pentameric complex. The most plausible cause of these changes are electrostatic interactions between the proteins and the tRNA. Indeed, the analysis of the charge distribution of the synthetases and of Arc1p within their TRBDs show positive-charged surfaces, where the tRNAs can bind (Figure 6). These interactions, given the extended shapes of the free proteins, may lead to significant compaction upon the binding of the synthetases to their cognate tRNA.
In the present work we report a totally unexpected effect, namely a drastic compaction of a protein complex upon tRNA binding. A compaction effect of such magnitude, literally a collapse, has, to our knowledge, not been reported before for nucleoprotein complexes. These impressive rearrangements of the subunits are most probably due to the surface charges of the individual protein domains. Electrostatic repulsion of the positively charged tRNA-binding domains of the synthetases as well as Arc1p may cause the star-like arrangement of the proteins in the trimeric complex. In the pentamer, on the other hand, the negatively charged tRNA backbones will induce a mutual contraction of these domains. The flexible links connecting the domains facilitate these rearrangements.
In yeast as well as in eukaryotic cells, it is important to have an effective translation machinery connected to a high degree of complex formation, because of the size and complexity of the cell. The affinity of binding the tRNAs is increased in the pentameric complex compared to the synthetase–tRNA complex alone, which shows the importance of the pentameric complex for the yeast cell. The aminoacylation of the tRNA is faster and more effective and therewith the whole translation machinery can be faster. Also the specificity to the tRNA is higher, because of the additional binding sites to the cofactor Arc1p. The observed structural results have therefore clear functional implications in the context of the efficiency of the translational mechanism.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3, Supplementary Methods and Supplementary References [20,44].
FUNDING
EU grant 3D-repertoire [LSHG-CT-2005-512028]; BMBF grant SYNC-LIFE [05K10YEA]. Funding for open access charge: EMBL.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Sebastian Glatt (SCB unit, EMBL Heidelberg) for consulting and technical advice with the EMSA assay.
REFERENCES
- 1.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Giegé R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kisselev LL, Wolfson AD. Aminoacyl-tRNA synthetases from higher eukaryotes. Prog. Nucleic Acids Res. Mol. Biol. 1994;48:83–142. doi: 10.1016/s0079-6603(08)60854-5. [DOI] [PubMed] [Google Scholar]
- 4.Harris CL, Kolanko CJ. Aminoacyl-tRNA synthetase complex in Saccharomyces cerevisiae . Biochem. J. 1995;309:321–324. doi: 10.1042/bj3090321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karanasios E, Simos G. Building arks for tRNA: structure and function of the Arc1p family of non-catalytic tRNA-binding proteins. FEBS Lett. 2010;584:3842–3849. doi: 10.1016/j.febslet.2010.08.023. [DOI] [PubMed] [Google Scholar]
- 6.Simader H, Hothorn M, Köhler C, Basquin J, Simos G, Suck D. Structural basis of yeast aminoacyl-tRNA synthetase complex formation revealed by crystal structures of binary sub-complexes. Nucleic Acids Res. 2006;34:3968–3979. doi: 10.1093/nar/gkl560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cahuzac B, Berthonneau E, Birlirakis N, Guittet E, Mirande M. A recurrent RNA-binding domain is appended to eukaryotic aminoacyl-tRNA synthetases. EMBO J. 2000;19:445–452. doi: 10.1093/emboj/19.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klipcan L, Levin I, Kessler N, Moor N, Finarov I, Safro M. The tRNA- induced conformational activation of human mitochondrial phenylalanyl-tRNA synthetase. Structure. 2008;16:1095–1104. doi: 10.1016/j.str.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 9.Yadavalli SS, Klipcan L, Zozulya A, Banerjee R, Svergun D, Safro M, Ibba M. Large-scale movement of functional domains facilitates aminoacylation by human phenylalanyl-tRNA synthetase. FEBS Lett. 2009;583:3204–3208. doi: 10.1016/j.febslet.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Lee SW, Cho BH, Park SG, Kim S. Aminoacyl-tRNA synthetase complexes: beyond translation. J. Cell Sci. 2004;117:3725–3734. doi: 10.1242/jcs.01342. [DOI] [PubMed] [Google Scholar]
- 11.Robinson JC, Kerjan P, Mirande M. Macromolecular assemblage of aminoacyl-tRNA synthetases: quantitative analysis of protein-protein interactions and mechanism of complex assembly. J. Mol. Biol. 2000;304:983–994. doi: 10.1006/jmbi.2000.4242. [DOI] [PubMed] [Google Scholar]
- 12.Hausmann CD, Ibba M. Aminoacyl-tRNA synthetase complexes: molecular multitasking revealed. FEMS Microbiol. Rev. 2008;32:705–721. doi: 10.1111/j.1574-6976.2008.00119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simos G, Sauer A, Fasiolo F, Hurt EC. A conserved domain within Arc1p delivers tRNA to aminoacyl-tRNA synthetases. Mol. Cell. 1998;1:235–242. doi: 10.1016/s1097-2765(00)80024-6. [DOI] [PubMed] [Google Scholar]
- 14.Karanasios E, Simader H, Panayotou G, Suck D, Simos G. Molecular determinants of the yeast Arc1p-aminoacyl-tRNA synthetase complex assembly. J. Mol. Biol. 2007;374:1077–1090. doi: 10.1016/j.jmb.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Galani K, Grosshans H, Deinert K, Hurt EC, Simos G. The intracellular location of two aminoacyl-tRNA synthetases depends on complex formation with Arc1p. EMBO J. 2001;20:6889–6898. doi: 10.1093/emboj/20.23.6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frechin M, Kern D, Martin RP, Becker HD, Senger B. Arc1p: anchoring, routing, coordinating. FEBS Lett. 2010;584:427–433. doi: 10.1016/j.febslet.2009.11.037. [DOI] [PubMed] [Google Scholar]
- 17.Simos G, Segref A, Fasiolo F, Hellmuth K, Shevchenko A, Mann M, Hurt EC. The yeast protein Arc1p binds to tRNA and functions as a cofactor for the methionyl- and glutamyl-tRNA synthetases. EMBO J. 1996;15:5437–5448. [PMC free article] [PubMed] [Google Scholar]
- 18.Deinert K, Fasiolo F, Hurt EC, Simos G. Arc1p organizes the yeast aminoacyl-tRNA synthetase complex and stabilizes its interaction with the cognate tRNAs. J. Biol. Chem. 2001;276:6000–6008. doi: 10.1074/jbc.M008682200. [DOI] [PubMed] [Google Scholar]
- 19.Graindorge J-S, Senger B, Tritch D, Simos G, Fasiolo F. Role of Arc1p in the modulation of yeast glutamyl-tRNA synthetase activity. Biochemistry. 2005;44:1344–1352. doi: 10.1021/bi049024z. [DOI] [PubMed] [Google Scholar]
- 20.Simader H, Suck D. Expression, purification, crystallization and preliminary phasing of the heteromerization domain of the tRNA-export and aminoacylation cofactor Arc1p from yeast. Acta. Crystallograph. F Struct. Biol. Cryst. Commun. 2006;62:346–349. doi: 10.1107/S1744309106005823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meinnel T, Blanquet S. Maturation of Pre-tRNA by Escherichia coli RNase P specified by a Guanosine of the 5′-Flanking Sequence. J. Biol. Chem. 1995;270:15908–15914. doi: 10.1074/jbc.270.26.15908. [DOI] [PubMed] [Google Scholar]
- 22.Hellman LM, Fried MG. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2007;2:1849–1861. doi: 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roessle MW, Klaering R, Ristau U, Robrahn B, Jahn D, Gehrmann T, Konarev P, Round A, Fiedler S, Hermes C, et al. Upgrade of the small-angle X-ray scattering beamline X33 at the European Molecular Biology Laboratory, Hamburg. J. Appl. Crystallogr. 2007;40:190–194. [Google Scholar]
- 24.Round AR, Franke D, Moritz S, Huchler R, Fritsche M, Malthan D, Klaering R, Svergun DI, Roessle M. Automated sample-changing robot for solution scattering experiments at the EMBL Hamburg SAXS station X33. J. Appl. Crystallogr. 2008;41:913–917. doi: 10.1107/S0021889808021018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petoukhov MV, Konarev PV, Kikhney AG, Svergun DI. ATSAS 2.1 – towards automated and web-supported small-angle scattering data analysis. J. Appl. Crystallogr. 2007;40:223–228. [Google Scholar]
- 26.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS – a Windows-PC based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 27.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- 28.Porod G. General theory. In: Glatter O, Kratky O, editors. Small-Angle X-Ray Scattering. London: Academic Press; 1982. pp. 17–51. [Google Scholar]
- 29.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small angle scattering. J. Appl. Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Svergun DI, Nierhaus KH. A map of protein-rRNA distribution in the 70 S Escherichia coli ribosome. J. Biol. Chem. 2000;275:14432–14439. doi: 10.1074/jbc.275.19.14432. [DOI] [PubMed] [Google Scholar]
- 32.Wriggers W. Using Situs for the integration of multi-resolution structures. Biophys. Rev. 2010;2:21–27. doi: 10.1007/s12551-009-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svergun DI, Barberato C, Koch MHJ. CRYSOL – a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- 34.Petoukhov MV, Svergun DI. Global rigid body modelling of macromolecular complexes against small-angle scattering data. Biophys. J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simader H, Hothorn M, Suck D. Structures of the interacting domains from yeast glutamyl-tRNA synthetase and tRNA-aminoacylation and nuclear-export cofactor Arc1p reveal a novel function for an old fold. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1510–1519. doi: 10.1107/S0907444906039850. [DOI] [PubMed] [Google Scholar]
- 36.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL Workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 37.Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T. The SWISS-MODEL repository and associated resources. Nucleic Acid Res. 2009;37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acid Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modelling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 40.Peitsch MC. Protein modelling by e-mail. Bio/Technology. 1995;13:658–660. [Google Scholar]
- 41.Renault L, Kerjan P, Pasqualato S, Menetrey J, Robinson JC, Kawaguchi S, Vassylyev DG, Yokoyama S, Mirande M, Cherfils J. Structure of the EMAPII domain of human aminoacyl-tRNA synthetase complex reveals evolutionary dimer mimicry. EMBO J. 2001;20:570–578. doi: 10.1093/emboj/20.3.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sherlin LD, Bullock TL, Newberry KJ, Lipman RS, Hou YM, Beijer B, Sproat BS, Perona JJ. Influence of transfer RNA tertiary structure on aminoacylation efficiency by glutaminyl and cysteinyl-tRNA synthetases. J. Mol. Biol. 2000;299:431–446. doi: 10.1006/jmbi.2000.3749. [DOI] [PubMed] [Google Scholar]
- 43.Nakanishi K, Ogiso Y, Nakam T, Nureki O. Structural basis for anticodon recognition by methionyl-tRNA synthetase. Nat. Struct. Mol. Biol. 2005;12:931–932. doi: 10.1038/nsmb988. [DOI] [PubMed] [Google Scholar]
- 44.Combet C, Blanchet C, Geourjon C, Deléage G. NPS@: network protein sequence analysis. Trends Biochem. Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.