Abstract
Folding of RNA molecules into their functional three-dimensional structures is often supported by RNA chaperones, some of which can catalyse the two elementary reactions helix disruption and helix formation. Hfq is one such RNA chaperone, but its strand displacement activity is controversial. Whereas some groups found Hfq to destabilize secondary structures, others did not observe such an activity with their RNA substrates. We studied Hfq’s activities using a set of short RNAs of different thermodynamic stabilities (GC-contents from 4.8% to 61.9%), but constant length. We show that Hfq’s strand displacement as well as its annealing activity are strongly dependent on the substrate’s GC-content. However, this is due to Hfq’s preferred binding of AU-rich sequences and not to the substrate’s thermodynamic stability. Importantly, Hfq catalyses both annealing and strand displacement with comparable rates for different substrates, hinting at RNA strand diffusion and annealing nucleation being rate-limiting for both reactions. Hfq’s strand displacement activity is a result of the thermodynamic destabilization of the RNA through preferred single-strand binding whereas annealing acceleration is independent from Hfq’s thermodynamic influence. Therefore, the two apparently disparate activities annealing acceleration and duplex destabilization are not in energetic conflict with each other.
INTRODUCTION
Due to rugged folding landscapes, especially large RNA molecules tend to get trapped in non-native conformations that need to be resolved to enable refolding of the molecules into their native and functional structures (1–3). Besides the rescue of misfolded RNAs, cells also need to ensure refolding between functional conformations, e.g. in the context of riboswitches (4). RNA refolding can be broken down into two elementary reactions—the disruption of an existing duplex and the annealing of one of these strands to a third, complementary strand. Duplex disruption involves the loss of enthalpy due to the breaking of hydrogen bonds and a partial de-stacking of bases resulting in an increase of Gibbs free energy of between 0.9 kcal/mol and 3.4 kcal/mol per base-pair (5). Depending on the length and GC-content of the helix, rate constants of duplex dissociation are often too small to ensure refolding at biological relevant time frames in the absence of auxiliary factors (6). Annealing on the other hand is a spontaneous reaction due to the significant gain of enthalpic energy during base-pair formation. This reaction is restricted by RNA strand diffusion, the activation energy necessary for the formation of the first few base-pairs in the encounter complex and the entropic penalty caused by conformational restriction and ion uptake from the bulk solution into the RNA’s ion atmosphere (7–10). Importantly, during refolding events helix destabilization and annealing occur in a concerted fashion, in the way that a maximal possible number of base-pairs is maintained during replacement of one of the original strands through a third, competing strand (11–14).
To support correct RNA folding, cells have developed several different strategies, among which is the utilization of specialized proteins (15,16). Among these are RNA chaperones, such as Escherichia coli StpA and HIV-1 NCp7, which destabilize duplexes independently of external energy and thus enable the RNA molecule to find its functional structure in a second folding attempt. Instead of containing a common consensus sequence or fold, these proteins facilitate chaperone activity via a variety of RNA binding motifs which only share the concentrated localization of positively charged amino acids (6,17). RNA chaperone–RNA interactions are often of electrostatic and/or hydrophopic nature and transient which seems to be an underlying feature of the activity (18). Another class of proteins, which only accelerate annealing, are called RNA annealer proteins. Also in this group electrostatic and transient interactions with the RNA are dominant and essential, as shown by the example of the HIV-1 Tat fragment Tat(44-61) (19). HIV-1 Tat is an early viral regulator whose best described function is the trans-activation of transcription through specific RNA binding, but which has been implied in many other viral processes some of which involve RNA molecules (20). Besides catalysing the strand displacement reaction, many RNA chaperones have been found to also accelerate the annealing reaction. The immense diversity and the ATP-independence of RNA chaperone and annealing activities pose the question of the energy source and the molecular mechanism of helix disruption and annealing.
A sequence-dependent RNA chaperone seems to be the hexameric E. coli Hfq. The Sm-like protein is an important post-transcriptional regulator that facilitates the base-pair formation of several mRNAs and their cognate non-coding RNAs and thus affects the transcript’s translation and stability (21). The restructuring of RNA secondary structure seems to be based on Hfq’s ability to accelerate annealing and catalyse strand displacement. However, only some groups were able to show an Hfq-induced structural alteration in mRNAs or non-coding RNAs (22–24) whereas other groups did not detect an influence of Hfq on RNA secondary structure (25–27).
Here, we show that Hfq’s annealing acceleration and helix disruption activities depend on Hfq’s binding preference toward AU-rich sequences as opposed to the substrate’s thermodynamic stability. Interestingly, Hfq can catalyse strand displacement of substrates with higher GC-contents when a hairpin is attached to the sequence that serves as an Hfq-binding platform. Hfq-catalysed strand displacement and thermodynamic destabilization of the substrate are interrelated. However, the observed rate constant of strand displacement does not correlate with the extent of the destabilization. Thus, the bottle-neck for strand displacement is not the strand opening, but the formation of the new duplex involving the competitor strand. Furthermore, using Hfq and the HIV-1 Tat-derived peptide Tat(44-61) we demonstrate that annealing acceleration is separate from the proteins’ thermodynamic influence on duplex stability and thus annealing acceleration is not in energetic conflict with helix destabilization or strand displacement.
MATERIALS AND METHODS
RNAs, peptides and proteins
RNAs were ordered at Eurogentec, Microsynth or Dharmacon, dissolved in H2O and stored at −20°C or −80°C (for long-term storage). Sequences are listed in Table 1. All Tat-derived peptides Tat(44-61) and scr1-3 were a generous gift from Peter Steinlein (Research Institute for Molecular Pathology, Vienna, Austria). They were lyophilized and stored at −20°C. Prior to use peptides were dissolved in Tat peptide buffer (30 mM Tris-HCl pH 7, 30 mM NaCl, 1 mM DTT). Purification of the protein Hfq was carried out as described elsewhere (28,29). Hfq concentrations in this article exclusively refer to the hexamer form of the protein (Hfq6).
Table 1.
Sequences and GC-contents of RNA substrates that were used in this study
| RNA | GC-content [%] | Sequences |
|---|---|---|
| 21R | 38.1 | 5’-AUGUGGAAAAUCUCUAGCAGU-3’ |
| 3’-UACACCUUUUAGAGAUCGUCA-5’ | ||
| JM1 | 4.8 | 5’-AAUUUAAUGUUUUAUUUAUUA-3’ |
| 3’-UUAAAUUACAAAAUAAAUAAU-5’ | ||
| J1h | 5’-UUAAUUUAAUGUUUUAUUUAUUAGGGUUUAUGGCUGUUCGCCAUUU–3’ | |
| JM2 | 14.3 | 5’-AAUUUAAUGUUUUAGUUACUA-3’ |
| 3’-UUAAAUUACAAAAUCAAUGAU-5’ | ||
| J2h | 5’-UUAAUUUAAUGUUUUAGUUACUAGGGUUUAUGGCUGUUCGCCAUUU–3’ | |
| JM3 | 23.8 | 5’-AAUUCAAUGUUUUAGUUACUG-3’ |
| 3’-UUAAGUUACAAAAUCAAUGAC-5’ | ||
| J3h | 5’-UUAAUUCAAUGUUUUAGUUACUGGGGUUUAUGGCUGUUCGCCAUUU–3’ | |
| JM4 | 33.3 | 5’-AACUCAAUGUCUUAGUUACUG-3’ |
| 3’-UUGAGUUACAGAAUCAAUGAC-5’ | ||
| J4h | 5’-UUAACUCAAUGUCUUAGUUACUGGGGUUUAUGGCUGUUCGCCAUUU–3’ | |
| JM6 | 52.4 | 5’-GACUCAGUGUCAGAGUCACUG-3’ |
| 3’-CUGAGUCACAGUCUCAGUGAC-5’ | ||
| J6h | 5’-UUGACUCAGUGUCAGAGUCACUGGGGUUUAUGGCUGUUCGCCAUUU–3’ |
The base-paired form of 21R is referred to as ds21R throughout the article while the single-strand (first line) is called 21R+. For the JM series, JM always refers to the double-strand and J (first line) and M (second line) are the single-strands. The RNAs termed J1h-J4h and J6h contain an additional 3′hairpin binding-platform (separated by GGGUUU from the base-pairing region) as well as a 5′-UU overhang. The nucleotides base-pairing with each other to form the stem of the hairpin are underlined.
FRET assays
Combined annealing and strand displacement assays were carried out and analysed as described by Rajkowitsch and Schroeder (2007) (26) using a GENios Pro™ microplate reader and applying the following changes. Concentrations of donor and acceptor dye-labeled RNAs were 10 nM, the unlabeled competitor strand was added at a final concentration of 100 nM. Annealing data were fitted to the following second-order reaction equation using Prism 5 (GraphPad Software, San Diego, CA):
| (1) |
(where Y, normalized FRET index; Aann, amplitude of annealing; kann, observed rate constant for annealing; t, time). FRET signals describing strand displacement were fit to a mono-exponential decay:
| (2) |
(where ASD, amplitude of strand displacement; kSD, observed rate constant for strand displacement).
UV melting
Complementary RNAs were annealed by heating them to 95°C for 2 min and subsequent slow cooling and the resulting double-strand was gel-purified. For each experiment 500 nM dsRNA was dissolved in a total volume of 500 µl containing 10 mM sodium cacodylate buffer pH 6.8 (HCl), 0.5 mM EDTA and 10 mM NaCl (Tat-derived peptides) or 50 mM sodium phosphate buffer pH 8, 0.5 mM EDTA and 50 mM NaCl (Hfq). Quartz cuvettes with 10 mm path length were used.
Temperature-dependent absorption experiments were conducted on a ‘Cary’Bio100 spectrophotometer equipped with a thermoelectrically controlled multi-cell holder.
Following a 5 min equilibration time, samples were heated from 15°C to 95°C with a rate of 0.3°C/min. Absorption values at 260 nm A260 nm were recorded every minute. From the obtained absorbance values for 260 nm versus temperature curves the folded fraction α(T) and the melting temperature Tm were calculated using a baseline approach (30). For the 21 base-pair long helix in JM4h the melting temperature was determined as the second peak of the derivative dA260 nm/dT. This method was sufficiently exact for this substrate as Tm, JM4h was identical with Tm,JM4 (Table 2).
Table 2.
Protein-dependent annealing and strand displacement rate constants and melting temperatures of RNAs in the absence and presence of protein Annealing rates kann of all RNA substrates in the absence of protein were between 0.005 and 0.008 s−1
| Protein | RNA | Annealing and strand displacement assay |
UV melting analysis |
||
|---|---|---|---|---|---|
| + Protein |
RNA only | + Protein | |||
| kann [s−1] | kSD [s−1] | Tm [°C] | Tm [°C] | ||
| Hfqa | JM1 | 0.028 ± 0.004 | 0.028 ± 0.004 | 44.7 ± 0.3 | 36.6 ± 3.6 |
| JM1h | 0.031 ± 0.004 | 0.027 ± 0.006 | n.d. | n.d. | |
| JM2 | 0.033 ± 0.008 | 0.024 ± 0.004 | 52.5 ± 0.4 | 50.9 ± 0.7 | |
| JM2h | 0.029 ± 0.007 | 0.027 ± 0.002 | n.d. | n.d. | |
| JM3 | 0.026 ± 0.006 | No SD | 56.2 ± 0.2 | 56.6 ± 0.2 | |
| JM3h | 0.030 ± 0.005 | 0.024 ± 0.003 | n.d. | n.d. | |
| JM4 | 0.032 ± 0.006 | No SD | 64.8 ± 0.2 | 65.3 ± 0.3 | |
| JM4h | 0.033 ± 0.007 | 0.031 ± 0.009 | 65.0 ± 0.3b | 64.4 ± 0.6b | |
| JM6 | No ann acc | No SD | 76.0 ± 0.1 | 76.4 ± 0.4 | |
| JM6h | No ann acc | No SD | n.d. | n.d. | |
| Tat(44-61)c | JM1 | 0.036 ± 0.096 | No SD | n.d. | |
| 21R | 0.032 ± 0.004 | No SD | 58.3 ± 0.3 | 71.7 ± 0.3 | |
| Scr1 | 21R | 0.022 ± 0.002 | n.d. | 68.4 ± 0.6 | |
| Scr2 | 21R | 0.014 ± 0.003 | n.d. | 73.0 ± 0.4 | |
| Scr3 | 21R | 0.016 ± 0.003 | n.d. | 69.6 ± 0.1 | |
Strand displacement was not measureable in the absence of protein. Values are means ± standard deviations of at least three measurements.
No ann acc, no acceleration of annealing; No SD, no strand displacement; n.d., not determined.
a100 nM Hfq (ann and SD assay), 750 nM Hfq (UV melting).
bMelting transition of the 21 base-pair long helix.
c1 µM Tat(44-61) (ann and SD assay with JM1), 300 nM Tat(44-61) and scr1-3 (ann and SD assay with 21R), 2 µM Tat(44-61) and scr1-3 (UV melting).
Microscale thermophoresis
1 µM of Cy5-labeled single-stranded or double-stranded RNA (except Cy5-J2: 200 nM) was incubated for 5 min at room temperature with different concentrations of protein or peptide in either Hfq-binding buffer (50 mM Na-phosphate buffer pH 8, 50 mM NaCl, 2 mM MgCl2, 0.5 mM EDTA, 0.05 mg/ml BSA) or Tat peptide-binding buffer (50 mM Tris-HCl pH 7, 30 mM NaCl, 3 mM MgCl2, 0.5 mg/ml BSA). The reactions were transferred to standard-treated capillaries and loaded onto the capillary holder of the NanoTemper microscale thermophoresis (MST) device. A primary capillary scan determined the exact position of the capillaries on the holder and gave information on fluorescence quenching as well as the solubility of the peptide or peptide–RNA complex. Thermophoresis experiments were carried out with an infrared laser intensity of 0.7 V (Hfq) or 0.8 V (Tat peptide). Obtained fluorescence signals were normalized to the fluorescence at time point 0. Normalized fluorescence versus time curves were analysed using the NanoTemper analysis software to obtain Fnorm,Tjump [User Manual Monolith NT.015, www.nanotemper-technologies.com (4 October 2012, date last accessed)]. Fnorm,Tjump versus Tat peptide concentration was fit to a Hill equation for multiple binding sites:
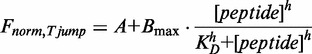 |
(3) |
(where A, offset; Bmax, scaling factor; KD, dissociation constant; h, Hill factor).
RESULTS
Design of RNA constructs of increasing thermal stability
To test the ability of our three model proteins to anneal and open up double-stranded RNAs with different thermal stabilities in a simplified system, we designed perfectly complementary 21mers with increasing GC-content (Table 1). Cy5 fluorescent dyes were attached to the 5′ ends of the + strands (J1-J4 and J6), whereas the – strands (M1–M4 and M6) were 5′-labeled with a Cy3 dye. To observe ‘pure annealing’ (without the necessity to open up internal base-pairs before duplex formation), all RNAs were designed devoid of internal secondary structures. However, this was only possible for RNAs with a GC-content of up to around 50%. The J6 and M6 RNAs each form five internal base-pairs which need to be unfolded to allow annealing of the complementary strands and thus reduce the calculated JM6 duplex stability (Supplementary Table S1).
The RNAs termed J1h–J4h and J6h are based on the respective J1–J4 and J6 RNAs and form 21 base-pairs with the respective M1–M4 and M6 strands. They contain an additional 3′hairpin binding-platform (separated by GGGUUU from the base-pairing region) as well as a 5′-UU overhang.
Assuming an axial rise of 2.79 Å per base-pair, an annealed and labeled 21 base-pair long JM duplex allows for an estimated FRET efficiency of EFRET =0.35 (Förster radius, R0 = 53 Å) (31). The actual determined EFRET was 0.22 which can be explained by a sub-optimal relative orientation of the fluorophors due to the twisted helix and stacking of the fluorophors on the terminal bases (32). Indeed, we observed a stabilization of the JM duplexes through the 5′-attached fluorophors by an average value of 3.6° which indicates stacking (Supplementary Figure S3).
In our coupled, FRET-based annealing and strand displacement assay, we measured the kinetics of duplex formation (phase I) and the strand displacement through a competitor RNA (phase II) for each JM pair by monitoring the FRET signal (Figure 1A). The annealing rates of all substrates were between 0.005 s−1 and 0.008 s−1 with no significant correlation between GC-content/internal structures and observed rate constants. These values are in good agreement with annealing rate constants reported by Pörschke and Eigen (1971) (10). Also, as expected for the bimolecular reaction, the observed annealing rate of JM1 was linearly dependent on the squared initial RNA concentration (Supplementary Figure S1). Strand displacement was not observed for any of the RNA duplexes in the absence of protein.
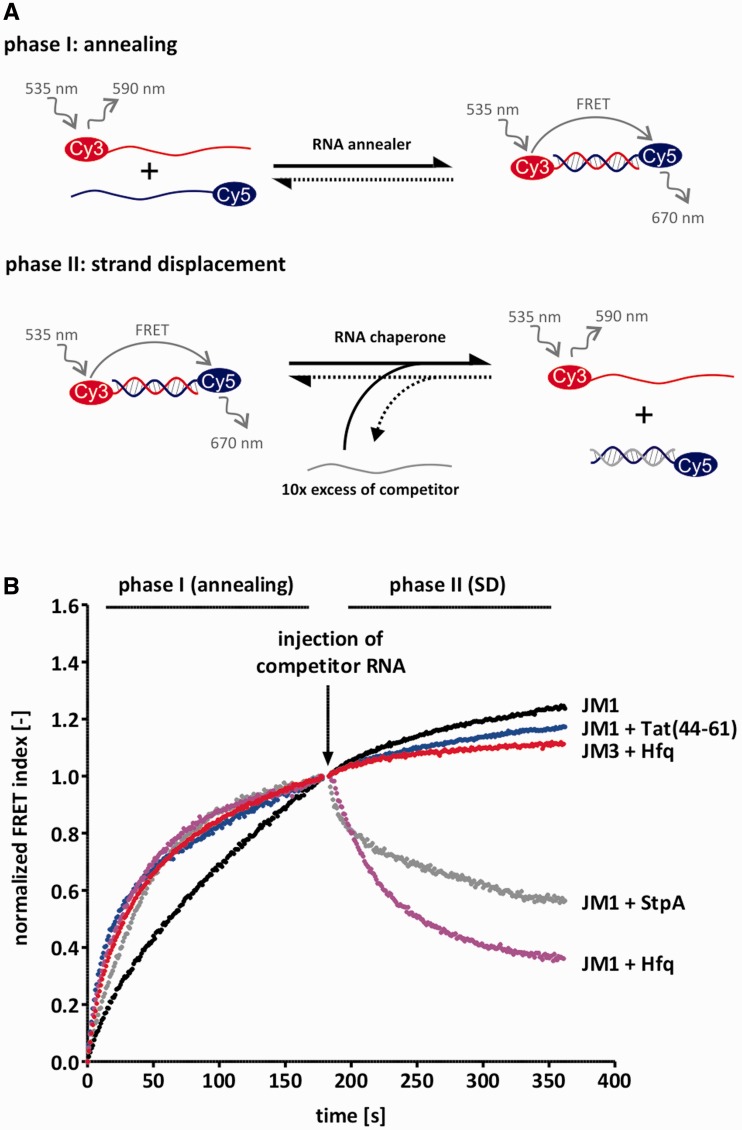
Figure 1.
Tat(44-61) and Hfq catalyse strand displacement and/or annealing. (A) Scheme of the FRET-based annealing and strand displacement assay. In phase I, the kinetics of annealing are tested by mixing 10 nM of two complementary RNA strands which are 5′-labeled with a Cy5 or a Cy3 dye, respectively, and monitoring their fluorescent signals. Duplex formation allows for FRET which is therefore a measure of the fraction of annealed double-strands. Phase II is started by the injection of a 10-fold excess of competitor RNA which resembles one of the strands from phase I, but is unlabeled. In the presence of a protein with helix destabilizing activity, the pre-formed duplex is opened up so that the competitor strand can invade and as a result, the FRET signal decreases. (B) 1 µM Tat(44-61), 100 nM Hfq and 1 µM StpA (serving as a positive control), were tested in this assay using the JM1 RNA substrate. For better visual comparability, the calculated FRET index was normalized between 0 and 1 (phase I) or to 1 only (phase II). While all three proteins accelerated annealing of JM1, only StpA and Hfq showed strand displacement activity with this substrate. Interestingly, Hfq did not catalyse strand displacement of the substrate JM3 which has a higher GC-content than JM1. The JM3 ‘RNA only’ curve is very similar to the JM1 curve and is thus not shown.
Hfq and Tat(44-61)—proteins with different RNA annealing and strand displacement activities
We measured Hfq-catalysed annealing and strand displacement rates using our FRET-based assay and 100 nM Hfq6 which corresponds approximately to the concentration at which the protein’s annealing activity reaches a plateau (Supplementary Figure S2). Consistent with the reports from other groups, Hfq’s annealing and strand displacement activities were strongly dependent on the RNA sequence used (Figure 1, Table 2). Hfq accelerated annealing of the RNAs JM1–JM4 by about 4- to 5-fold. Annealing of the GC-rich RNA pair JM6, however, was not accelerated by Hfq. Interestingly, the observed reaction rates of annealing in the presence of Hfq did not decrease gradually with increasing GC-content. Instead, the accelerated rate constants were comparable for substrates with a GC-content of 4.8–33.3%. At GC-contents higher than these values, annealing acceleration was not detectable. In agreement with previous studies, Hfq was active in our strand displacement assay dependent upon the substrate used. Hfq readily exchanged strands of the least stable and AU-rich duplexes, JM1 and JM2. Observed reaction rates were 0.028 s−1 and 0.024 s−1. With increasing GC-content, however, the strand displacement ability of Hfq was lost. In contrast to annealing acceleration, the cut-off for strand displacement was a GC-content of around 23.8% (the GC-content of JM3). Importantly, the attachment of a stem-loop structure to the 3′ end of the J3 and J4 strands (J3h and J4h, Table 1) allowed for strand displacement of the JM3h and JM4h substrates (Table 2).
The peptide Tat(44-61) efficiently accelerated annealing of our JM1 and 21R substrate, but did not have strand displacement activity (Figure 1B, Table 2). The annealing activity of the full-length protein and its fragment Tat(44-61) has been tested in different assays and with different substrates (19,33). Despite earlier reports (34,35), Doetsch et al. did not observe a duplex destabilization activity of the Tat protein and its fragment Tat(44-61) (19,33,36,37). We conclude that the Tat peptide accelerates annealing in a sequence-independent fashion and is thus a useful tool for the investigation of this activity.
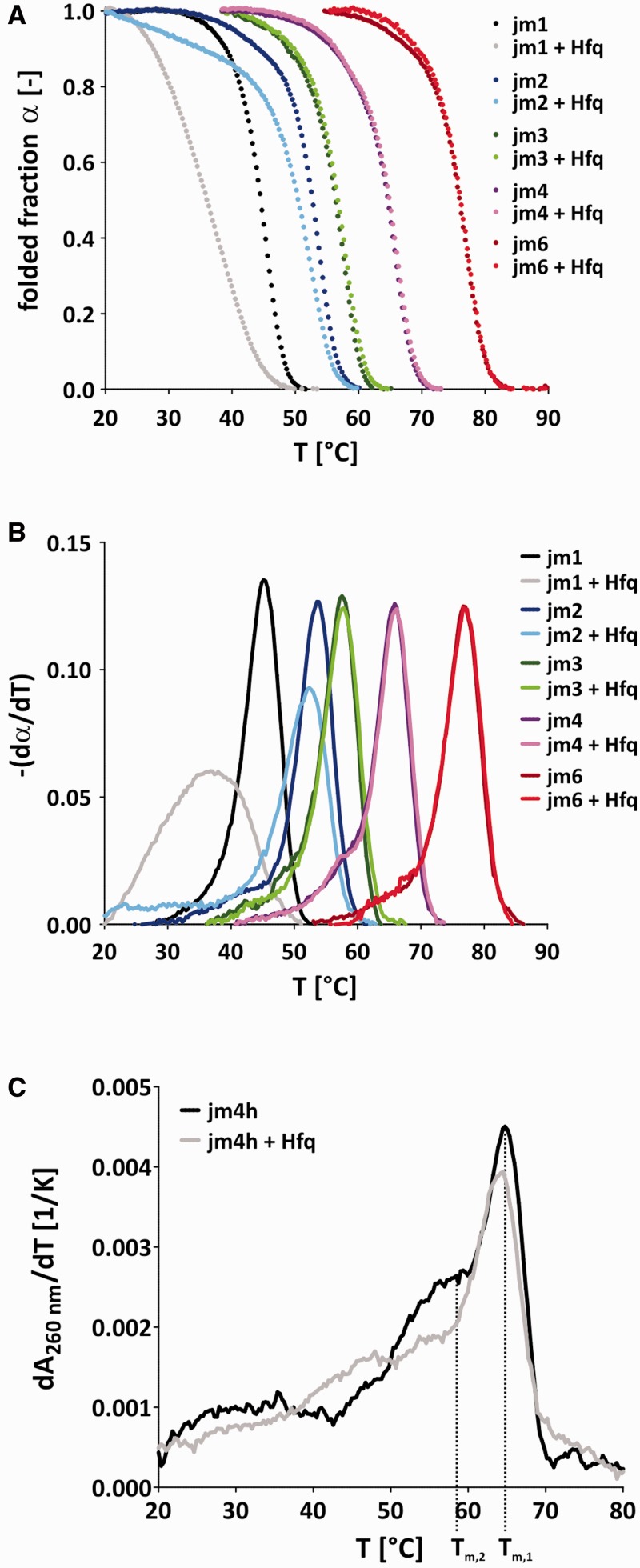
The RNA chaperone activity of Hfq is qualitatively correlated with the thermodynamic destabilization of the RNA substrates
It is believed that an RNA chaperone-catalysed strand displacement event is due to the thermodynamic destabilization of a pre-formed helix, allowing for invasion of a competitor strand (16). To further study this hypothesis, we tested Hfq together with different RNA substrates in UV melting experiments. Hfq lowered the melting temperature of the JM1 and JM2 substrates whereas the melting temperatures of JM3, JM4 and JM6 in the presence of Hfq were identical to the ones measured in the absence of the protein within the error range (Table 2). Although the melting temperature difference of JM2 in the presence and absence of Hfq ΔTm,JM2 was only marginal with about −1.6°, the influence of Hfq on the RNA’s melting behavior was very obvious from the α(T) versus T plot (double-stranded fraction versus temperature) and its derivative (Figure 2A and B). In contrast, Tm,JM1+Hfq was significantly lower than Tm,JM1 with a difference of about −8.1°. Consistent with the calculated melting temperatures, a significant influence of Hfq on the shape of the melting curves of the substrates JM3, JM4 and JM6 was not visible (Figure 2A and B). Because Hfq-catalysed annealing and strand displacement of the JM4h substrate, we also carried out UV melting experiments on this substrate (Figure 2C, Table 2). The resulting melting curve is a superimposition of the melting profiles of the 21 base-pair long helix and the attached hairpin. Interestingly, while Hfq destabilized the hairpin significantly, its influence on the melting temperature of the 21 base-pair duplex was much less pronounced. The melting temperatures of the duplex in the presence and absence of the protein are identical within the error range, but the derivative dA260nm/dT shows a slight destabilization of the 21-mer duplex by Hfq.
Figure 2.
Hfq destabilizes the JM1 and JM2 duplex while it does not affect substrates with a higher GC-content. (A) UV melting experiments were carried out with gel-purified JM1-4 and JM6 double-strands in the presence or absence of 750 nM Hfq. The folded fraction α(T) = folded(T) / (folded(T) + unfolded(T)) was calculated from the melting transitions using a baseline approach. (B) The negative first derivatives of α often make small changes in the melting curve (e.g. biphasic melting behavior) more obvious. Hfq clearly shifted the melting curves of JM1 and JM2 toward lower temperatures, while its influence on the other three substrates JM3, JM4 and JM6 was insignificant. (C) The first derivative dA260 nm/dT of UV melting profiles of the jm4h substrate in the presence and absence of Hfq. The JM4h substrate displays two main melting transitions- one for the 21 base-pair long duplex (Tm,1) and one for the attached hairpin (Tm,2). Hfq clearly shifts Tm,2 toward lower temperatures while the influence on Tm,1 is comparably small.
Notably, despite the fact that the thermodynamic destabilization through Hfq was larger for JM1 than for JM2 and JM4h, the Hfq-catalysed strand displacement rates as measured with the FRET-based assay were identical for both substrates (Table 2).
Annealing activities of Hfq and Tat(44-61) and thermodynamic stabilization of the RNA double-strand are distinct activities
We have often observed that proteins which accelerate annealing also stabilize RNA double-strands thermodynamically. Hfq accelerated annealing of JM3 and JM4 and we therefore wondered whether we would observe an Hfq-induced thermodynamic stabilization of RNA substrates JM3 and JM4 in our UV melting experiments. However, Hfq did not influence the melting profile or temperature of these two RNA substrates (Table 2 and Figure 2). These data indicate that the kinetic influence of Hfq on RNA base-pair formation does not depend on its thermodynamic influence.
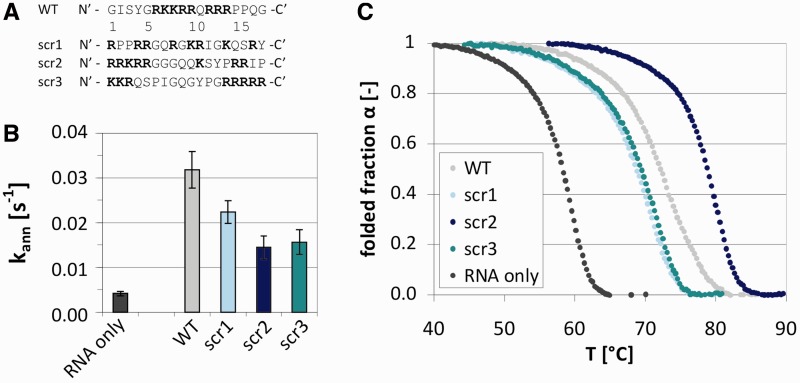
More evidence that an annealing acceleration is not directly correlated with thermodynamic stabilization comes from the Tat fragment Tat(44-61) and the derived scrambled peptides scr1-3 (Figure 3A). Tat(44-61) accelerated annealing of the RNA 21R by about 7-fold (Figure 3B, Table 2). Furthermore, it increased the melting temperature of the same substrate in a concentration-dependent manner (Figure 3C and Supplementary Figure S4). The three scrambled peptides also accelerated annealing of 21R, although not as effectively as the wildtype peptide (Figure 3B, Table 2). Scr1 was more active than the peptides scr2 and scr3. The activities of the two latter were identical within the error range. In the UV melting assays, however, scr2 stabilized the double-strand most efficiently whereas scr1 and scr3 showed the least thermodynamic stabilization of the RNA duplex. Therefore, the magnitude of annealing acceleration did not correlate with the extent of thermodynamic stabilization.
Figure 3.
Annealing acceleration activity and double-strand stabilization activity of Tat-derived peptides do not correlate. (A) Sequences of scrambled peptides scr1-3, based on Tat(44-61) wildtype sequence (WT). (B) Observed annealing constants kann of scr1-3 as measured using the FRET-based annealing assay. (C) The Tat peptide and the scrambled peptides increased the stability of double-stranded 21R RNA (and thus the melting temperature) as was measured with UV melting. α(T) = folded(T) / (unfolded(T) + folded(T)).
Differential annealing and strand displacement activities of the chaperone proteins can be partially explained by variations in RNA substrate binding
A theoretical model for the influence of a small ligand on a nucleic acid’s melting temperature was put forward by Crothers (1971) (38). According to this model a melting temperature increase can be roughly explained by a preferential binding of the ligand to double-stranded RNA (KD,ss > KD,ds) or the presence of more binding sites on the double-strand. Conversely, a Tm decrease is due to the preferential binding of the ligand to the single-stranded form (KD,ss < KD,ds) or the availability of more binding sites on the single-strand relative to the double-stranded form.
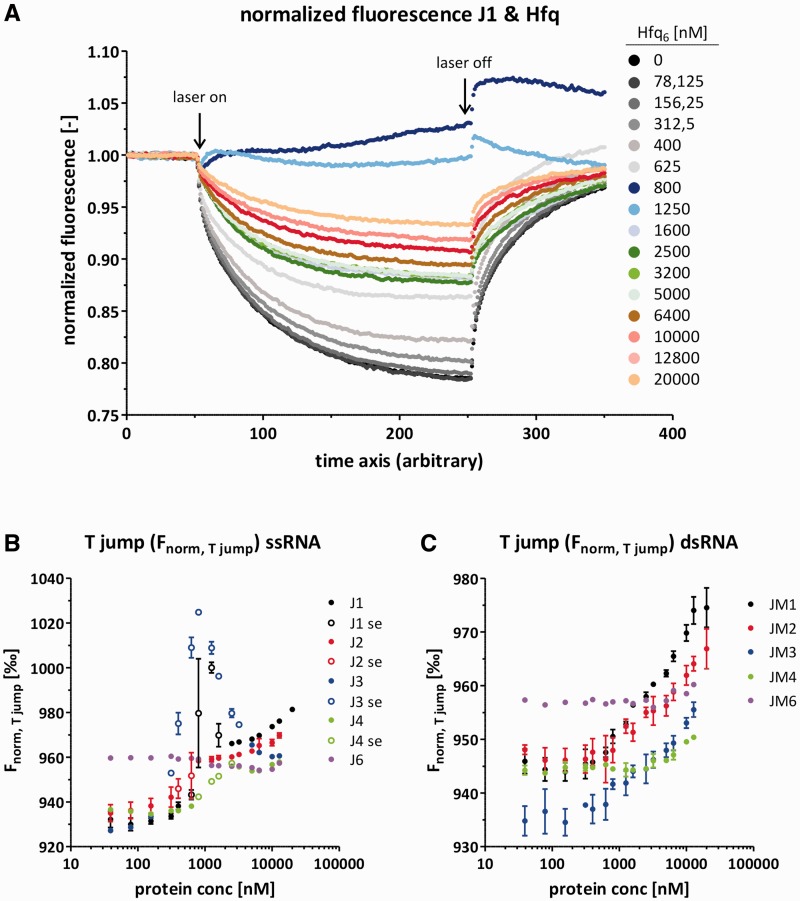
We tested Hfq binding to single-stranded and double-stranded RNAs of the JM series using MST as an alternative to gel shift assays (39). This technique monitors the average influence of the used protein on the RNA’s thermophoretic mobility, but is unable to discriminate Hfq monomers from multimers or protein binding from an RNA structural change. Therefore, we compared the obtained data qualitatively instead of quantitatively. Nevertheless, these data explain both the differential influence of Hfq on annealing and strand displacement as well as on the melting temperature of the JM RNAs. No significant Hfq-induced change in the T jump signal was found for J6 and JM6 and, thus, no binding to J6 or JM6 was detected under the applied conditions (Figure 4B and C, Supplementary Figure S6E). This correlates well with Hfq’s missing influence on JM6 annealing, strand displacement and the RNA’s UV melting profile. In contrast, the J1 and J2 single-stranded RNAs both interacted with Hfq. The corresponding double-stranded RNAs JM1 and JM2 also interacted with Hfq, although the Hfq-induced alteration of the T jump signal was less severe suggesting the affinity to dsRNA may be reduced compared with ssRNA (Figure 4B and C, Supplementary Figure S6A and B). This differential binding of double- and single-stranded RNA explains the decrease of the melting temperature of JM1 and JM2 according to Crothers (1971). Hfq binding to JM3 and JM4 was even weaker while Hfq interacted significantly with the single-strands J3 and J4 (Figure 4B and C, Supplementary Figure S6C and D).
Figure 4.
Hfq binds single- and double-stranded RNAs as well as RNAs of different GC-contents with different affinities as determined with MST. (A) Representative normalized fluorescence curves of an MST experiment using Cy5-J1 and a titration series of Hfq. Importantly, the RNA molecules move from warm to cold areas as indicated by the fluorescence decrease after the laser has been switched on. Notably, at Hfq6 concentrations of about 800–1600 nM the contrary behavior (molecules moving toward warm areas) or a superimposing effect was visible. (B) From the MST experiments the T jump signal Fnorm,T jump was derived for all single-stranded RNAs. Unfilled circles represent values that stem from curves showing the superimposing effect (se). (C) From the MST experiments the T jump signal Fnorm, T jump was derived for all double-stranded RNAs.
The thermophoresis curves for all single-stranded RNAs are slightly difficult to interpret, which is due to a superimposing effect that was visible at Hfq6 concentrations of around 800–1600 nM, depending on the single-stranded RNA used (Figure 4A, Supplementary Figure S6A–D). At these proteins concentrations, which happened to be approximately equimolar to the RNA concentration, we also observed a stronger quenching effect (Supplementary Figure S5B). Judging from the capillary scan which was carried out before each MST measurement, aggregation or strong protein binding to the capillary can probably be excluded as it would have yielded a different peak shape (Supplementary Figure S5A) [Nano Temper MST Starting Guide—Monolith NT.115, www.nanotemper-technologies.com (4 October 2012, date last accessed)]. Gel shift assays showed that at sub-equimolar Hfq–RNA concentrations only very few RNA molecules were bound while at concentrations above equimolar Hfq–RNA concentrations all RNA molecules were in a complex with Hfq. The RNA–Hfq interaction appears to be transient, although Hfq binding was highly cooperative (Supplementary Figure S7). The high cooperativity might be due to a switch in Hfq’s oligomerization state which would be in line with data recently reported by Panja and Woodson (2012) (40). Such a switch of the Hfq’s oligomerization state might also explain the change in the protein’s thermophoretical behavior.
Our Hfq-binding data are in good agreement with previous studies that describe a preference of Hfq for AY-rich sequences (41–43). The single-strands J1, M1, J2 and M2 are rich in adenine and pyrimidines (mostly uridine) while in the substrates with higher GC-contents the AU/C stretches are interspersed by a G more frequently (Table 1). Also the preference for single-stranded RNA has been suggested before (44,45).
Determined by MST measurements, the Tat peptide bound both single-stranded and double-stranded RNAs 21R+ and ds21R with dissociation constants and Hill factors that are identical to each other within the error range: KD,ssRNA = 6.3 ± 0.5 µM, KD,dsRNA = 7.4 ±0.9 µM (see Supplementary Data). The contradiction with Crother’s theoretical model might be explained with the technical difficulties one encounters when measuring binding constants for the Tat peptide and different RNAs. Due to the small size and high charge of the peptide as well as the low binding energy of the non-specific peptide–RNA interaction the most commonly used techniques to determine dissociation constants such as gel mobility shift assays, quenching methods, filter binding or isothermal calorimetry were not applicable to our system (see Supplementary Data). MST seemed to work best for this system. However, especially when measuring Tat peptide binding to single-stranded RNA we observed a superimposing effect of uncertain origin (Supplementary Figure S11). Therefore, the determined KDs may not be exact enough to illustrate the differential binding of the peptide to single- and double-stranded RNA. Also, the double-stranded RNA probably offers more peptide-binding sites than the single-stranded RNA which is due to an increased charge density. This might be sufficient to explain the peptide-conferred duplex stabilization despite similar KDs for single- and double-strand binding.
DISCUSSION
GC-content dependence of Hfq’s strand displacement and annealing activities
In the past, contradictions arose as to whether Hfq is able to open up secondary structures or not. Although several groups reported a structural alteration in mRNAs or non-coding RNAs (22–24) others were not able to detect any helix destabilization activity of Hfq with their substrates (25–27). Our results help to explain the discrepancies of these reports. Hfq only catalysed strand displacement of 21mers with overall GC-contents of up to about 14%. Notably, longer RNAs with a higher overall GC-content might still be destabilized locally if they harbor at least one Hfq-binding motif or longer AU-rich stretch which is sufficiently bound by Hfq. Such an effect was indeed visible in our artificial JM2 substrate. The UV melting profile of this RNA in the presence of Hfq showed a ‘shoulder’ which we interpret as the opening of the eight base-pair long AU-stretch on one end while the other end which is interspersed by Gs and Cs was melted less efficiently (Figure 2, Table 1). Also, we found that the addition of a hairpin structure flanked by several uridines to the 3′ end of the J3 and J4 strands (yielding J3h and J4h) allowed for strand displacement of JM3h and JM4h (Tables 1 and 2). Furthermore, Hfq destabilized the 21 base-pair region of JM4h thermodynamically while JM4’s melting temperature was not influenced (Figure 2). This artificial hairpin with three upstream uridines might mimic a natural one that has been shown to improve Hfq binding to its RNA substrate (46). We conclude that nucleic acid structures or sequences that are bound by Hfq can sequester Hfq to regions the protein would bind only weakly in the absence of the auxiliary sequence and thus aid destabilization of these duplexes. The limiting factor for helix destabilization is thus not so much the thermodynamic stability of the duplex as Hfq’s affinity for the substrate or parts of it. Indeed, it has been reported that certain single-stranded or hairpin Hfq-binding sites improve the base-pairing of mRNAs and their cognate non-coding RNA even when located further away from the region (42,46–48). The fact that Hfq did not catalyse strand displacement of the JM6h substrate despite possibly being able to bind it via the 3′ hairpin, probably owes to the high stability of JM6 and the resulting low frequency of temperature-induced duplex breathing.
Mechanism of Hfq-catalysed strand displacement
While RNA helicases rely on ATP hydrolysis to catalyse the disruption of stable RNA base-pairs, RNA chaperones function without the consumption of energy-rich substances. According to one model, the ends of a double-stranded RNA ‘breath’, that is they are in a dynamic equilibrium of opening and closing base-pairs. Due to its increased binding affinity toward single-stranded RNA relative to the double-strand, the chaperone sequesters the accessible strand and thus drives the equilibrium toward the unfolded RNA form (16). Prerequisites for the strand displacement reaction are therefore a sufficiently high temperature to ensure duplex breathing as well as a certain affinity of the RNA chaperone for the single-strands.
Our data recorded with Hfq fit well to this model. They suggest that Hfq catalyses strand displacement via the sequestration of a single-strand of a breathing duplex and thus allows for invasion of a competitor strand. There is a clear correlation between preferred single-strand binding (as measured with MST) and strand displacement activity (as measured in our FRET-based assay, Table 2). However, according to MST experiments, Hfq also interacted stronger with J3 and J4 than with JM3 and JM4, but did not catalyse strand displacement of these substrates in our FRET-based assay. Notably, Hfq influenced the behavior of JM3 and JM4 in MST to a smaller extent than the behavior of JM1 and JM2, indicating weaker binding of Hfq to JM3 and JM4. We therefore think that in the case of JM3 and JM4, Hfq is not associated to these double-strands long enough to catch the event of strand opening.
Competitor strand diffusion and annealing constitute the rate-limiting steps of the strand displacement reaction
A surprising and striking observation we report here is that the extent of duplex destabilization as measured with UV melting does not correlate with the observed rate constant of strand displacement. The measured rate constants kobs,SD were identical within the error range for all tested substrates. Furthermore, the rate constants for annealing acceleration kobs,ann equaled the strand displacement rate constants kobs,SD (Table 2). We recently reported the same phenomenon for E. coli StpA (49). This indicates that the opening of just a few base-pairs of the pre-formed duplex is sufficient for the strand displacement reaction. As soon as enough base-pairs are disrupted to allow for competitor strand invasion, the replacement of the respective strand is fast so that the overall thermodynamic stability of the strand is negligible. Therefore, duplex destabilization is not the bottle-neck of the strand displacement reaction. The rate-limiting factors for strand displacement are diffusion of the competitor RNA to the breathing duplex as well as the nucleation event of the competitor strand annealing to its complementary strand.
Coexistence of two apparently disparate activities, RNA annealing and strand displacement, in one protein
Hfq and other chaperones such as StpA and NCp7 catalyse both annealing acceleration and strand displacement, two activities that at first sight seem energetically and mechanistically disparate. For NCp7 a local separation of helix destabilization and annealing acceleration within the protein has been suggested (50,51). While the strand displacement activity seems to be conferred mainly by the protein’s zinc fingers, annealing acceleration as well as the duplex stabilizing activity have been located to the basic stretches.
Our Tat-derived peptides, which stabilized the ds21R double-strand more efficiently than other peptides, did not accelerate annealing to a stronger extent—the two activities did not correlate quantitatively (Figure 3). Therefore, in mechanistic terms Tat(44-61) influences the RNA substrate in two distinct ways of which one causes the stabilization of the double-strand (which is a thermodynamic effect) whereas the other accelerates the annealing reaction (which is a kinetic effect). According to Crothers (1971), the double-strand stabilization is a result of stronger peptide binding to the duplex when compared with the single-stranded form. The acceleration of annealing on the other hand is due to the selection of an annealing-competent conformation of the RNA single-strand through Tat(44-61) as we recently reported (19).
In the case of Hfq, the strand displacement reaction is catalysed by preferred binding of the single-stranded RNA and thermodynamic stabilization of the latter. The kinetic effect of annealing acceleration is believed to be due to the co-localization of complementary RNA strands. Indeed, Hfq harbors two RNA binding sites (a proximal and a distal face) which can bind RNAs simultaneously. Furthermore, each of the six monomers has the potential to interact with 1–3 nt (52–54). Therefore, annealing acceleration and strand displacement can be explained through different mechanisms that are energetically not in conflict with each other.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–11 and Supplementary References [40,55–59].
FUNDING
University of Vienna and the Austrian Science Fund FWF through the Special Research Program (SFB17) on ‘Modulators of RNA fate and function’ [F1703 to R.S.]; DK W1207 RNA Biology (to M.D.); Lise Meitner [M1157-B12 to B.F.]. Funding for open access charge: Austrian Science Fund FWF [F1703].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLDEGEMENTS
We are grateful to Dr Peter Steinlein (Research Institute for Molecular Pathology, Vienna) for supplying us with Tat peptides and to Dr Euripedes De Almeida Ribeiro (Max F. Perutz Laboratories, Vienna) for advice concerning the Hfq purification. We also want to thank Prof. Dr Harald Schwalbe (Johann Wolfgang Goethe University, Frankfurt am Main) for giving us the possibility to carry out the MST measurements in his laboratory.
REFERENCES
- 1.Thirumalai D, Woodson SA. Kinetics of folding of proteins and RNA. Acc. Chem. Res. 1996;29:433–439. [Google Scholar]
- 2.Russell R, Zhuang X, Babcock HP, Millett IS, Doniach S, Chu S, Herschlag D. Exploring the folding landscape of a structured RNA. Proc. Natl Acad. Sci. USA. 2002;99:155–160. doi: 10.1073/pnas.221593598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solomatin SV, Greenfeld M, Chu S, Herschlag D. Multiple native states reveal persistent ruggedness of an RNA folding landscape. Nature. 2010;463:681–684. doi: 10.1038/nature08717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM. Functional complexity and regulation through RNA dynamics. Nature. 2012;482:322–330. doi: 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freier SM, Kierzek R, Jaeger JA, Sugimoto N, Caruthers MH, Neilson T, Turner DH. Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl Acad. Sci. USA. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herschlag D. RNA chaperones and the RNA folding problem. J. Biol. Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 7.Craig ME, Crothers DM, Doty P. Relaxation kinetics of dimer formation by self complementary oligonucleotides. J. Mol. Biol. 1971;62:383–401. doi: 10.1016/0022-2836(71)90434-7. [DOI] [PubMed] [Google Scholar]
- 8.Draper DE, Grilley D, Soto AM. Ions and RNA folding. Annu. Rev. Biophys. Biomol. Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- 9.Proctor DJ, Ma H, Kierzek E, Kierzek R, Gruebele M, Bevilacqua PC. Folding thermodynamics and kinetics of YNMG RNA hairpins: specific incorporation of 8-bromoguanosine leads to stabilization by enhancement of the folding rate. Biochemistry. 2004;43:14004–14014. doi: 10.1021/bi048213e. [DOI] [PubMed] [Google Scholar]
- 10.Pörschke D, Eigen M. Co-operative non-enzymic base recognition. 3. Kinetics of the helix-coil transition of the oligoribouridylic–oligoriboadenylic acid system and of oligoriboadenylic acid alone at acidic pH. J. Mol. Biol. 1971;62:361–381. doi: 10.1016/0022-2836(71)90433-5. [DOI] [PubMed] [Google Scholar]
- 11.Coutts SM. Thermodynamics and kinetics of G-C base pairing in the isolated extra arm of serine-specific transfer RNA from yeast. Biochim. Biophys. Acta. 1971;232:94–106. doi: 10.1016/0005-2787(71)90494-1. [DOI] [PubMed] [Google Scholar]
- 12.Uhlenbeck OC, Borer PN, Dengler B, Tinoco I. Stability of RNA hairpin loops: A 6 -C m -U 6. J. Mol. Biol. 1973;73:483–496. doi: 10.1016/0022-2836(73)90095-8. [DOI] [PubMed] [Google Scholar]
- 13.Furtig B, Wenter P, Pitsch S, Schwalbe H. Probing mechanism and transition state of RNA refolding. ACS Chem. Biol. 2010;5:753–765. doi: 10.1021/cb100025a. [DOI] [PubMed] [Google Scholar]
- 14.LeCuyer KA, Crothers DM. Kinetics of an RNA conformational switch. Proc. Natl Acad. Sci. USA. 1994;91:3373–3377. doi: 10.1073/pnas.91.8.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Bläsi U, Schroeder R. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- 16.Woodson SA. Taming free energy landscapes with RNA chaperones. RNA Biol. 2010;7:677–686. doi: 10.4161/rna.7.6.13615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cristofari G, Darlix JL. The ubiquitous nature of RNA chaperone proteins. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:223–268. doi: 10.1016/s0079-6603(02)72071-0. [DOI] [PubMed] [Google Scholar]
- 18.Doetsch M, Schroeder R, Fürtig B. Transient RNA-protein interactions in RNA folding. FEBS J. 2011;278:1634–1642. doi: 10.1111/j.1742-4658.2011.08094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doetsch M, Furtig B, Gstrein T, Stampfl S, Schroeder R. The RNA annealing mechanism of the HIV-1 Tat peptide: conversion of the RNA into an annealing-competent conformation. Nucleic Acids Res. 2011;39:4405–4418. doi: 10.1093/nar/gkq1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brigati C, Giacca M, Noonan DM, Albini A. HIV Tat, its TARgets and the control of viral gene expression. FEMS Microbiol. Lett. 2003;220:57–65. doi: 10.1016/S0378-1097(03)00067-3. [DOI] [PubMed] [Google Scholar]
- 21.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geissmann TA, Touati D. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moll I, Leitsch D, Steinhauser T, Blasi U. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep. 2003;4:284–289. doi: 10.1038/sj.embor.embor772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arluison V, Hohng S, Roy R, Pellegrini O, Regnier P, Ha T. Spectroscopic observation of RNA chaperone activities of Hfq in post-transcriptional regulation by a small non-coding RNA. Nucleic Acids Res. 2007;35:999–1006. doi: 10.1093/nar/gkl1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brescia CC, Mikulecky PJ, Feig AL, Sledjeski DD. Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA. 2003;9:33–43. doi: 10.1261/rna.2570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajkowitsch L, Schroeder R. Coupling RNA annealing and strand displacement: a FRET-based microplate reader assay for RNA chaperone activity. Biotechniques. 2007;43:304–310. doi: 10.2144/000112530. [DOI] [PubMed] [Google Scholar]
- 27.Hopkins JF, Panja S, McNeil SA, Woodson SA. Effect of salt and RNA structure on annealing and strand displacement by Hfq. Nucleic Acids Res. 2009;37:6205–6213. doi: 10.1093/nar/gkp646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayer O, Rajkowitsch L, Lorenz C, Konrat R, Schroeder R. RNA chaperone activity and RNA-binding properties of the E. coli protein StpA. Nucleic Acids Res. 2007;35:1257–1269. doi: 10.1093/nar/gkl1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beich-Frandsen M, Vecerek B, Sjöblom B, Bläsi U, Djinovic-Carugo K. Structural analysis of full-length Hfq from Escherichia coli. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011;67:536–540. doi: 10.1107/S174430911100786X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mergny JL, Lacroix L. Analysis of thermal melting curves. Oligonucleotides. 2003;13:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- 31.Coban O, Lamb DC, Zaychikov E, Heumann H, Nienhaus GU. Conformational heterogeneity in RNA polymerase observed by single-pair FRET microscopy. Biophys. J. 2006;90:4605–4617. doi: 10.1529/biophysj.105.078840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iqbal A, Arslan S, Okumus B, Wilson TJ, Giraud G, Norman DG, Ha T, Lilley DM. Orientation dependence in fluorescent energy transfer between Cy3 and Cy5 terminally attached to double-stranded nucleic acids. Proc. Natl Acad. Sci. USA. 2008;105:11176–11181. doi: 10.1073/pnas.0801707105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herschlag D, Khosla M, Tsuchihashi Z, Karpel RL. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J. 1994;13:2913–2924. doi: 10.1002/j.1460-2075.1994.tb06586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuciak M, Gabus C, Ivanyi-Nagy R, Semrad K, Storchak R, Chaloin O, Muller S, Mely Y, Darlix JL. The HIV-1 transcriptional activator Tat has potent nucleic acid chaperoning activities in vitro. Nucleic Acids Res. 2008;36:3389–3400. doi: 10.1093/nar/gkn177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo X, Kameoka M, Wei X, Roques B, Gotte M, Liang C, Wainberg MA. Suppression of an intrinsic strand transfer activity of HIV-1 Tat protein by its second-exon sequences. Virology. 2003;307:154–163. doi: 10.1016/s0042-6822(02)00068-5. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, Ma X, Yeh YS, Zhu Y, Daugherty MD, Frankel AD, Musier-Forsyth K, Barbara PF. Comparative analysis of RNA/protein dynamics for the arginine-rich-binding motif and zinc-finger-binding motif proteins encoded by HIV-1. Biophys. J. 2010;99:3454–3462. doi: 10.1016/j.bpj.2010.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boudier C, Storchak R, Sharma KK, Didier P, Follenius-Wund A, Muller S, Darlix JL, Mely Y. The mechanism of HIV-1 Tat-directed nucleic acid annealing supports its role in reverse transcription. J. Mol. Biol. 2010;400:487–501. doi: 10.1016/j.jmb.2010.05.033. [DOI] [PubMed] [Google Scholar]
- 38.Crothers DM. Statistical thermodynamics of nucleic acid melting transitions with coupled binding equilibria. Biopolymers. 1971;10:2147–2160. doi: 10.1002/bip.360101110. [DOI] [PubMed] [Google Scholar]
- 39.Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 2010;1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- 40.Panja S, Woodson SA. Hexamer to monomer equilibrium of E. coli Hfq in solution and its impact on RNA annealing. J. Mol. Biol. 2012;417:406–412. doi: 10.1016/j.jmb.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lorenz C, Gesell T, Zimmermann B, Schoeberl U, Bilusic I, Rajkowitsch L, Waldsich C, von Haeseler A, Schroeder R. Genomic SELEX for Hfq-binding RNAs identifies genomic aptamers predominantly in antisense transcripts. Nucleic Acids Res. 2010;38:3794–3808. doi: 10.1093/nar/gkq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soper TJ, Woodson SA. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA. 2008;14:1907–1917. doi: 10.1261/rna.1110608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Folichon M, Arluison V, Pellegrini O, Huntzinger E, Regnier P, Hajnsdorf E. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res. 2003;31:7302–7310. doi: 10.1093/nar/gkg915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Franze de Fernandez MT, Hayward WS, August JT. Bacterial proteins required for replication of phage Q ribonucleic acid. Pruification and properties of host factor I, a ribonucleic acid-binding protein. J. Biol. Chem. 1972;247:824–831. [PubMed] [Google Scholar]
- 45.Moller T, Franch T, Hojrup P, Keene DR, Bachinger HP, Brennan RG, Valentin-Hansen P. Hfq. A bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 46.Ishikawa H, Otaka H, Maki K, Morita T, Aiba H. The functional Hfq-binding module of bacterial sRNAs consists of a double or single hairpin preceded by a U-rich sequence and followed by a 3' poly (U) tail. RNA. 2012;18:1062–1074. doi: 10.1261/rna.031575.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc. Natl Acad. Sci. USA. 2010;107:9602–9607. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Panja S, Woodson SA. Hfq proximity and orientation controls RNA annealing. Nucleic Acids Res. 2012;40:8690–8697. doi: 10.1093/nar/gks618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doetsch M, Gstrein T, Schroeder R, Fürtig B. Mechanisms of StpA-mediated RNA remodeling. RNA Biol. 2010;7:96–104. doi: 10.4161/rna.7.6.13882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams MC, Rouzina I, Wenner JR, Gorelick RJ, Musier-Forsyth K, Bloomfield VA. Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc. Natl Acad. Sci. USA. 2001;98:6121–6126. doi: 10.1073/pnas.101033198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hargittai MR, Gorelick RJ, Rouzina I, Musier-Forsyth K. Mechanistic insights into the kinetics of HIV-1 nucleocapsid protein-facilitated tRNA annealing to the primer binding site. J. Mol. Biol. 2004;337:951–968. doi: 10.1016/j.jmb.2004.01.054. [DOI] [PubMed] [Google Scholar]
- 52.Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl Acad. Sci. USA. 2009;106:19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schumacher MA, Pearson RF, Moller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fender A, Elf J, Hampel K, Zimmermann B, Wagner EG. RNAs actively cycle on the Sm-like protein Hfq. Genes Dev. 2010;24:2621–2626. doi: 10.1101/gad.591310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 56.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cann JR. Theory and practice of gel electrophoresis of interacting macromolecules. Anal. Biochem. 1996;237:1–16. doi: 10.1006/abio.1996.0193. [DOI] [PubMed] [Google Scholar]
- 58.Walter NG, Burke JM. Fluorescence assays to study structure, dynamics, and function of RNA and RNA-ligand complexes. Methods Enzymol. 2000;317:409–440. doi: 10.1016/s0076-6879(00)17027-2. [DOI] [PubMed] [Google Scholar]
- 59.Hammack A, Chen YL, Pearce JK. Role of dissolved salts in thermophoresis of DNA: lattice-Boltzmann-based simulations. Phys. Rev. E Stat. Nonlin. Soft. Matter. Phys. 2011;83:031915. doi: 10.1103/PhysRevE.83.031915. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.