Abstract
16S ribosomal RNA gene (rDNA) amplicon analysis remains the standard approach for the cultivation-independent investigation of microbial diversity. The accuracy of these analyses depends strongly on the choice of primers. The overall coverage and phylum spectrum of 175 primers and 512 primer pairs were evaluated in silico with respect to the SILVA 16S/18S rDNA non-redundant reference dataset (SSURef 108 NR). Based on this evaluation a selection of ‘best available’ primer pairs for Bacteria and Archaea for three amplicon size classes (100–400, 400–1000, ≥1000 bp) is provided. The most promising bacterial primer pair (S-D-Bact-0341-b-S-17/S-D-Bact-0785-a-A-21), with an amplicon size of 464 bp, was experimentally evaluated by comparing the taxonomic distribution of the 16S rDNA amplicons with 16S rDNA fragments from directly sequenced metagenomes. The results of this study may be used as a guideline for selecting primer pairs with the best overall coverage and phylum spectrum for specific applications, therefore reducing the bias in PCR-based microbial diversity studies.
INTRODUCTION
Understanding microbial diversity has been the ambition of scientists for decades. Because diversity analysis by cultivation is problematic for a significant fraction of Bacteria and Archaea, culture-independent surveys have been developed. In the past, the most commonly used approach was cloning and sequencing of the 16S ribosomal RNA gene (rDNA) using conserved broad-range PCR primers (1). With the advent of massive parallel sequencing technologies, direct sequencing of PCR amplicons became feasible (2–4). In 2006, Roche’s 454 GS 20 pyrosequencing (5) became the first high-throughput sequencing technology to be successfully applied for large scale biodiversity analysis and was key to uncovering the ‘rare biosphere’ (6). The continuous development of the technology, offering read lengths of up to 1000 bp nowadays, further improved throughput and resolution of 16S rDNA sequencing (7). Since then, additional high-throughput sequencing technologies have become commercially available. The attractiveness of Illumina (8) lies in the reduced per base costs and comparatively high sequencing depth (9), despite having short read lengths. While the major advantage of Ion Torrent (10) are its relatively low cost and rapid sequencing speed. Furthermore, Pacific Bioscience (PacBio) now employs the ‘single-molecule real-time’ (SMRT) sequencing technology, designed to achieve average read lengths of more than 3000 bp (11). For a detailed review of sequencing technologies please refer to Loman et al. (12). There is no doubt that the rapid development of sequencing technologies has opened a new dimension in biodiversity analysis, but the diversity of technologies also adds complexity to the experimental design of a study.
The most critical step for accurate rDNA amplicon analysis is still the choice of primers (4,13). Using suboptimal primers, or more precisely, primer pairs, can lead to under-representation (14) or selection against single species or even whole groups (15–17). Using inappropriate primers consequently leads to questionable biological conclusions (17–19).
In this study, 175 broad range 16S rDNA primers and 512 primer pairs were investigated in silico with respect to overall coverage and phylum spectrum for Bacteria and Archaea as well as amplicon length. Primer sequences were compared with all 376 437 16S/18S rDNA sequences available in the SILVA non-redundant reference database (SSURef NR) release 108 (20). For consistency, all primers were renamed according to the primer nomenclature suggested by Alm et al. (21). Two pairs of bacterial PCR primers were selected for empirical evaluation at the field station Helgoland Roads (North Sea). Finally, the obtained results were compared with diversity estimates from previous metagenome studies (22).
MATERIALS AND METHODS
Primer nomenclature
Primers were renamed according to Alm et al. (21). Each name is composed of seven dash-separated parts, describing: the target gene, the rank of the target group, the target group, the target position within the gene, the primer version, the target strand and the length of the primer. For illustration, the seven parts comprising the primer name ‘S-D-Bact-0338-a-A-18’ are to be interpreted as follows:
An indication of the target gene. In this case, ‘S’ for small subunit rDNA (S);
An indication of the largest taxonomic group targeted by the PCR primer. For example, ‘D’ for domain level;
An abbreviated description, limited to three to five letters, of the specific taxonomic or phylogenetic group targeted by the primer. For example, ‘Bact’ for the domain Bacteria;
A four-digit number indicating the 5′ position of the sense strand. For example, ‘0338’ stands for start position 338 in the Escherichia coli system of nomenclature (23);
A single lowercase letter indicating the version of the probe. For example ‘a’ for a first version;
A single uppercase letter indicating whether the probe sequence is identical to the DNA sense strand (S) or to the antisense (A) strand; and
A number indicating the length of the PCR primer. 18 bases in the example.
Nomenclature for in silico evaluation
In this study, the term ‘coverage’ refers to the percentage of matches for a given taxonomic path. Taxonomic paths were considered ‘not covered’ if their coverage was below 50%. The term ‘phylum spectrum’ refers to the number of matched phyla. For example, if a primer or primer pair covers the majority of all phyla it is described as having a ‘large phylum spectrum’.
Selection of primers
A total of 175 forward and reverse 16S rDNA primers were chosen for the in silico evaluation. Primer sequences were either obtained from a literature survey or provided by the SILVA user community in response to a poll on the ARB/SILVA mailing list in January 2012 (Supplementary Table S1). Only primers with an overall coverage above 75% for either Bacteria or Archaea were considered for primer pair analysis. All primers are available in probeBase, a comprehensive online database for rRNA-targeted oligonucleotides, at www.microbial-ecology.net/probebase/ (24).
Selection criteria for primer pairs
Primer pairs were chosen according to annealing temperatures, overall coverage of variable regions and amplicon length. Annealing temperatures were calculated with OligoCalc (25). Primer pair combinations with annealing temperature differences of less than 5°C were accepted as pairs. Suitable primer pairs were organized into three different groups (Supplementary Table S8): Group Short (Group S) generates 100–400 bp fragments. Group Middle (Group M) generates 400–1000 bp fragments. Group Long (Group L) generates fragments ≥1000 bp. A total of 512 primer combinations were evaluated. The best 30 bacterial primer pairs in each group and all archaeal primer pairs with a combined overall coverage >70% were analyzed in detail.
In silico evaluation of primers and primer pairs
Primer evaluation was based on two datasets: Firstly, the non-redundant SILVA Reference database (release SSURef 108 NR) containing 376 437 sequences. The SILVA SSURef 108 NR was prepared from all SSU sequences longer than 1200 bp for Bacteria and Eukaryota and longer than 900 bp for Archaea. Sequences are required to have a SINA (26) alignment quality value better than 50 (20). Redundant sequences were removed by clustering with UCLUST (27) using a 99% identity criterion. A second SSU database was prepared from the Global Ocean Survey (GOS) (28,29) metagenomes using the SILVA pipeline. Alignment was attempted with SINA for all GOS reads and all sequences with an alignment quality of at least 30 and a minimum length of 300 were retained, yielding a dataset of 10 945 sequences. Taxonomic classifications for each read were applied as described below.
Primer matching was executed using the probe match function of the ARB PT server (30) at two levels of stringency, allowing zero or one mismatch, respectively. For each primer and stringency level the database entries were separated into three groups: (i) matches; (ii) mismatches; and (iii) unknown. The match status was considered to be unknown if no sequence data was available at the match position of the respective primer. Furthermore, only sequences corresponding to the primer at the intended position where considered to be matches. From these numbers, coverage was computed as the matched fraction of entries either matches or mismatches, excluding entries for which the match status was unknown. Individual coverages were computed for all taxa. When computing the combined coverage of forward and reverse primer pairs, an entry was considered to have unknown match status if the match status for either of the two primers was unknown. Likewise, the pair was only considered to be a match if both primers matched at the intended match position.
Detailed information for each analysed primer and primer pair are provided in the Supplementary Material Online (single primer: Supplementary Tables S2–S7; primer pairs: Supplementary Tables S9–S38). All scripts and SQL queries as well as database dumps are available online at www.arb-silva.de/download/archive/primer_evaluation.
Sampling site and collection of water samples
Sample collection was carried out as part of the ‘multi omic’ approach of the MIMAS (Microbial Interaction in MArine Systems) project (www.mimas-project.de). Surface water was collected on 11 February 2009 and weekly from 31 March 2009 until October 2009. Water samples (total volume 360 l) from the Kabeltonne site at Helgoland Roads in the North Sea (54°11.18′N, 7°54.00′E) were collected at a depth of 0.5 m and processed immediately at the Biological Station Helgoland. The water was pre-filtered through a 10 μm and a 3 µm pore-size filter. For harvesting a 0.2 -μm-pore-size filter was used. At each time point 10 l and 15 l of seawater were filtered onto 8 filters for genomic DNA extraction. All filters were stored at −80°C until future usage. Details can be found in Teeling et al. (22). In this study, 16S rDNA pyrotag analysis with Roche’s 454 FLX Titanium technology was performed using samples from: 11 February 2009, 7 April 2009 and 14 April 2009. Results from 16S rDNA diversity analysis gained from metagenome studies of the same sampling dates (22) were used for comparison.
DNA extraction
Genomic DNA was directly extracted from filters as described in Zhou et al. (31) with the following modifications: all extraction steps were performed with 50 µl proteinase K (10 mg/ml), and after isopropanol precipitation, pelleted nucleic acids were obtained by centrifugation at 50 000g for 30 min at room temperature. The genomic DNA was stored at −20°C until PCR amplification and metagenomic sequencing were carried out.
Amplification
Per sample, two separate PCR reactions were performed in order to test two bacterial primer pairs for 16S rDNA amplification. Primer pairs were: (i): S-D-Bact-0341-b-S-17, 5′-CCTACGGGNGGCWGCAG-3′ (32), and S-D-Bact-0785-a-A-21, 5′-GACTACHVGGGTATCTAATCC-3 (32); and (ii): S-D-Bact-0008-a-S-16, 5′-AGAGTTTGATCMTGGC-3′ (33), and S-D-Bact-0907-a-A-20, 5′-CCGTCAATTCMTTTGAGTTT-3′ (34). The reaction was carried out in 50 µl volumes containing 0.3 mg/ml BSA (Bovine Serum Albumin), 250 µM dTNPs, 0.5 µM of each primer, 0.02 U Phusion High-Fidelity DNA Polymerase (Finnzymes OY, Espoo, Finland) and 5x Phusion HF Buffer containing 1.5 mM MgCl2. The following PCR conditions were used: initial denaturation at 95°C for 5 min, followed by 25 cycles consisting of denaturation (95°C for 40 s), annealing (2 min) and extension (72°C for 1 min) and a final extension step at 72°C for 7 min. Annealing temperature for primer pair (i) was set at 55°C and for (ii) at 44°C. PCR products were purified with a QiaQuick PCR purification kit (QIAGEN, Hilden, Germany). The quantity and quality of the extracted DNA were analysed by spectrophotometry using an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and by agarose gel electrophoresis. The PCR products were stored at −20°C for sequencing.
Sequencing
The pyrosequencing reactions were performed at LGC Genomics GmbH, Berlin, Germany. All sequencing reactions were based upon FLX—Titanium chemistry (Roche/454 Life Sciences, Branford, CT 06405, USA; www.454.com) and all methods were performed using the manufacturers’ protocol. Briefly, genomic DNA from metagenome studies (22) as well as PCR-amplified DNA fragments were checked for quality on a 2% agarose gel. 500 ng of each sample was then used for the sequencing library. In a minor modification to the protocol, no size selection of the fragments was performed. The fragments were subjected to end repair and polishing. An extra A was added to the ends of the fragments and the Roche Rapid Library adaptors were ligated on to the fragments as described in the Roche Rapid Library Preparation Manual for GS FLX Titanium Series, October 2009, Rev. January 2010 (Roche/454 Life Sciences, Branford, CT 06405, USA; www.454.com). After subsequent emulsion PCR the fragment libraries were processed and sequenced according to the Roche protocols. The resulting sequences were processed using the standard Roche software for base calling, trimming of adaptors and quality trimming (Genome Sequencer FLX System Software Manual version 2.3, Roche/454 Life Sciences, Branford, CT 06405, USA; www.454.com). For PCR-amplified DNA fragments, per sample two distinct PCR reactions were sequenced on 1/8 of picotiter plate (PTP). Raw data were stored as FNA file. Sequences were submitted to INSDC (EMBL-EBI/ENA, Genbank, DDBJ) with accession number ERP001031. For metagenomics two full PTPs per sample were sequenced. Metagenome sequences were published by the MIMAS project (22) and can be obtained from INSDC with accession number ERP001227.
Identification and taxonomic classification of 16S rDNA fragments
Unassembled sequence reads from both SSU rRNA gene PCR amplicons (pyrotags) and metagenome sequencing were preprocessed (quality control and alignment) by the bioinformatics pipeline of the SILVA project (20). Briefly, reads shorter than 200 nt or with more than 2% of ambiguities or more than 2% of homopolymers were removed. Remaining reads from amplicons and metagenomes were aligned against the SSU rDNA seed of the SILVA database release 108 (www.arb-silva.de/documentation/background/release-108/) (20) using SINA (26). Unaligned reads were not considered in downstream analysis to eliminate non 16S rDNA sequences.
Remaining PCR amplicons were separated based on the presence of aligned nucleotides at E. coli positions of the respective primer binding sites instead of searching for the primer sequences itself. This strategy is robust against sequencing errors within the primer signatures or incomplete primer signatures. This separation strategy works because the amplicon size of one primer pair is significant longer, with overhangs on both 3′ and 5′ site, compared with the amplicon of the second primer pair. With this approach the need for barcoding during combined sequencing of 16S pyrotags derived from different PCR reactions on the same PTP lane was avoided. FASTA files for each primer pair of the separated samples are available online at www.arb-silva.de/download/archive/primer_evaluation.
Reads of the filtered and separated 16S pyrotag datasets as well as metagenomes were dereplicated, clustered and classified on a sample by sample basis. Dereplication (identification of identical reads ignoring overhangs) was done with cd-hit-est of the cd-hit package 3.1.2 (www.bioinformatics.org/cd-hit) using an identity criterion of 1.00 and a wordsize of 8. Remaining sequences were clustered again with cd-hit-est using an identity criterion of 0.98 (wordsize 8). The longest read of each cluster was used as a reference for taxonomic classification, which was done using a local BLAST search against the SILVA SSURef 108 NR dataset (www.arb-silva.de/projects/ssu-ref-nr/) using blast-2.2.22+ (http://blast.ncbi.nlm.nih.gov/Blast.cgi) with default settings. The full SILVA taxonomic path of the best BLAST hit was assigned to the reads if the value for (percentage of sequence identity + percentage of alignment coverage)/2 was at least 93. In the final step, the taxonomic path of each cluster reference read was mapped to the additional reads within the corresponding cluster plus the corresponding replicates (as identified in the previous analysis step) to finally obtain (semi-) quantitative information (number of individual reads representing a taxonomic path). Raw output data are available in the Supplementary Material in Supplementary Tables S48–S50.
Adjustment of the total number of sequence reads to smaller subsets by random re-sampling
Sequencing depth may infringe on the comparability of the resulting taxonomic resolution. To verify that the results derived from the 16S pyrotags were not an artefact of deep sequencing, the total number of 16S pyrotags was reduced until roughly equal amounts of classified pyrotags and classified metagenome reads remained for each sample. Three subsets of each 16S pyrotag sample were adjusted by withdrawing equal amounts of sequences randomly without replacement. Raw output data are available in the Supplementary Material Online (Supplementary Tables S51–S52). An analogue approach was described in Gilbert et al. (35).
RESULTS AND DISCUSSION
In silico evaluation of 16S rDNA primers
The overall coverage of 175 single primers was evaluated for all three domains of life (Supplementary Table S1). Additionally for Bacteria and Archaea the phylum spectrum was investigated with respect to zero and one mismatch (Supplementary Tables S2–S5). Eukaryota are only considered on domain level (Supplementary Tables S5–S6). A total of 122 single primers passed the 50% overall coverage threshold with 31, 51 and 1 primer(s) specific for the domain Archaea (A), Bacteria (B) and Eukaryota (E), respectively. At one-mismatch-stringency the total number increased to 150 eligible primers.
For Archaea, primers S-D-Arch-0519-a-A-19 (A: 91.3%, B: 0.1%, E: 1.0%) and S-D-Arch-0787-a-A-20 (A: 87.4%, B: 7.8%, E: 0.0%) stand out. This is in line with a recent study by Wang and Qian (15). The highest overall coverage and specificity for the domain Bacteria was detected for the primers S-D-Bact-1061-a-A-17 (A: 2.9%, B: 96.4%, E: 0.0%) and S-D-Bact-0564-a-S-15 (A: 16.3%, B: 96.0%, E: 0.0%). Furthermore, 39 primers show relatively high overall coverage for more than one domain. For instance, S-*-Univ-0515-a-S-19 (A: 54.5%, B: 95.4%, E: 92.2%) detects all three domains and S-D-Bact-0787-b-A-20 (A: 89.9%, B: 90.6%, E: 0.0%) targets Bacteria and Archaea as recently reported (36).
It has previously been asserted (15) that the primers S-*-Univ-0789-a-S-18 (A: 86.1%, B: 6.8%, E: 0.0%) and S-*-Univ-0906-a-S-17 (A: 83.7%, B: 0.3%, E: 76.8%) target Bacteria and Archaea. Contrary to this, with only 6.8% and 0.3% overall coverage of the domain Bacteria, but 86.1% and 83.7% overall coverage of the domain Archaea, respectively, our results confirm the original intention of both primers to be specific for the domain Archaea (37,38). However, if one mismatch is tolerated, S-*-Univ-0789-a-S-18 (A: 96.0%, B: 93.0%, E: 0.0%) targets Archaea and Bacteria. S-*-Univ-0906-a-S-17 (A: 93.2%, B: 49.8%, E: 0.0%) still fails to pass our 50% threshold.
The primer sequence of S-*-Univ-0779-a-S-20 (A: 0.0%, B: 0.0%, E: 0.0%) is misspelled in Wang and Qian (15). Allowing one mismatch increases the overall coverage to A: 64.8%, B: 6.8%, E: 77.6% and indicates that the correct primer sequence targets Archaea and Eukaryota.
A direct comparison of our results with the studies of Huws et al. (39) and Baker et al. (14) is not possible, as the overall coverage of the primers is not given. Nossa et al. (1) restricted their evaluation to a single habitat. Walter et al. (36) analysed a total of only four primers.
In respect to detailed phylum coverage (Supplementary Tables S2–S5) it should be noted that the numbers of sequences present in a phylum affects the values for phylum coverage. If the majority of a small phylum (e.g. Caldiserica with 61 sequences) is targeted, the coverage will be higher than for a member rich phylum (e.g. Firmicutes with 84 910 sequences). Similar effects occur for phyla in which only a small number of sequences contain sequence information at the primer position of interest.
In silico evaluation of primer pairs
When combining forward and reverse primers, the bias of single primers can accumulate. To minimize the overall bias, primers with similar overall coverage and phylum spectrum must be used. Using the 75% overall coverage criterion, 86 single primers qualify for primer pair analysis. In order to get suitable combinations for the different sequencing technologies, primer pairs were organized into three groups according to their amplicon length (Supplementary Table S8). Group S(mall) could be of particular interest for Illumina (8) and Ion Torrent (10) sequencing. Primer pairs of Group M(iddle) are suitable for Roche’s 454 (40) technology. Group L(arge) primer pairs are useful for sequencing methods such as PacBio (11) as well as for creating classical clone libraries. A total of 512 primer combinations were evaluated. Again, the focus of this evaluation was Archaea and Bacteria. Eukaryota are only considered on domain level.
Assuming that a standard PCR can tolerate up to two mismatches between the primer and its target (1), results with one mismatch are also taken into account. However, it should be noted that a primer mismatch can result in a biased picture of the bacterial diversity (41) and preferential amplification might lead to under-representation of important members of a community (14,41).
In silico evaluation of primer pairs suitable for Illumina and Ion Torrent sequencing (Group S)
Only 12 archaeal primer pairs have an overall coverage above 70%. The best results with an overall coverage of 76.8% are obtained with S-D-Arch-0349-a-S-17/S-D-Arch-0519-a-A-16 (A: 76.8%, B: 0.0%, E: 0.0%) (Supplementary Table S9). This pair generates an amplicon length of 185 bp which spans the hypervariable (HV) region three. The evaluation revealed that it misses five out of eight phyla: Ancient Archaeal Group (AAG), GoC-Arc-109-D0-C1-M0, Korarchaeota, Marine Hydrothermal Vent Group 2 (MHVG-2) and Nanoarchaeota. The remaining three archaeal phyla are detected (Crenarchaeota, Marine Hydrothermal Vent Group 1 (MHVG-1) and Euryarchaeota). With one mismatch allowed, overall coverage for Archaea increases to A: 91.0%, B: 0.0%, E: 0.1% now covering additionally Korarchaeota and MHVG-2 (Supplementary Table S10). However, in the case of Korarchaeota detailed analysis of the primer target position revealed a 3′ end mismatch of the forward primer, which is known to affect amplification. Nanoarchaeota and AAG show three mismatches. Moreover, PCR has to tolerate up to four mismatches of the forward primer to amplify GoC-Arc-109-D0-C1-M0. In summary, S-D-Arch-0349-a-S-17/S-D-Arch-0519-a-A-16 generates short amplicons, has a comparatively high overall coverage by detecting up to four out of eight archaeal phyla and excellent domain specificity. Hence, this primer pair shows the most promising results for Illumina and Ion Torrent sequencing.
For Bacteria, the primer pair S-D-Bact-0341-b-S-17/S-D-Bact-0515-a-A-19 (A: 0.0%, B: 91.2%, E: 0.0%) has the highest overall coverage (Supplementary Table S11). Detailed analysis reveals that 10 phyla are not detected (Armatimonadetes, Chlamydiae, Caldiserica, Hyd24-12, GOUTA4, Kazan-3B-28, SM2F11, as well as Candidate divisions WS6, OP11, TM7 and OD1). If one mismatch is tolerated some Archaea (A: 44.6%, B: 96.7%, E: 0.2%) as well as seven additional phyla are detected (Supplementary Table S12), but amplification of Candidate divisions OP11 and WS6 as well as Armatimonadetes remains unlikely. In all three cases, the mismatch position of the forward primer is located at the 3′ end. For Candidate divisions OP11 and WS6, the reverse primer would need to tolerate three mismatches. These findings are in line with the conclusions of Baker and Cowan (42), who claim that no domain-specific primer exists or can be designed that matches all bacterial 16S rDNA sequences.
The best candidate for the domain Bacteria is S-D-Bact-0564-a-S-15/S-D-Bact-0785-b-A-18. This primer pair has a slightly lower overall coverage for Bacteria (A: 14.6%, B: 89.0%, E: 0.0%) compared with the previous candidate but only fails to detect four bacterial phyla (Chloroflexi, Elusimicrobia, BHI80-139 and Candidate division OP11). With one allowed mismatch (A: 57.1%, B: 95.2%, E: 0.0%), only Candidate division OP11 sequences remain undetected due to a 3′ end mismatch of both primers. Please note that one mismatch may also lead to amplification of archaeal 16S rDNA sequences. Based on the promising phylum spectrum we are in favour of this primer pair in comparison to the previous described S-D-Bact-0341-b-S-17/S-D-Bact-0515-a-A-19. In summary, S-D-Bact-0564-a-S-15/S-D-Bact-0785-b-A-18 generates an amplicon of 253 bp covering the fourth HV region and satisfies with a high overall coverage and reasonably good domain specificity. Hence, it is recommended for Bacteria.
Two primer pairs target the domains Bacteria and Archaea: S-D-Arch-0519-a-S-15/S-D-Bact-0785-b-A-18 (A: 88%, B: 89.1%, E: 0.7%) and S-D-Arch-0519-a-S-15/S-D-Bact-0785-a-A-21 (A: 86.5%, B: 87.1%, E: 0.0%). Within the bacterial domain, those two primer pairs cover 49 out of 59 phyla. The coverage for Chlamydiae, Caldiserica, Chloroflexi, SM2F11, Kazan-3B-28, BHI80-139 and Candidate divisions WS6, OP11, TM7 and OD1 is below 50%. If one mismatch is tolerated, seven additional phyla are detected and overall coverage increases for S-D-Arch-0519-a-S-15/S-D-Bact-0785-b-A-18 (A: 94.9%, B: 95.1%, E: 1.6%) and S-D-Arch-0519-a-S-15/S-D-Bact-0785-a-A-21 (A: 94.6%, B: 94.8%, E: 0.7%). Amplification of Candidate divisions WS6, TM7 and OP11 remains unlikely. The mismatch position of S-D-Arch-0519-a-S-15 is located at the 3′ end in case of Candidate divisions WS6 and TM7. For Candidate division OP11, both reverse primers show a 3′ end mismatch. For Archaea, each primer pair fails to detect four out of eight phyla (AAA, MHVG-1 and MHVG-2 and Nanoarchaeota), which is reduced to one (Nanoarchaeota) if one mismatch is allowed. The continuous failure of primers to detect Nanoachaeota is not surprising, due to the majority of Archaea-specific primers being designed prior to the discovery of the Nanoarchaeota (14). Detailed analysis of the mismatch positions reveals one internal mismatch for AAA, MHVG-1 and MHVG-2 but three mismatches for Nanoarchaeota. Addition of Nanoarchaeota-specific primers (43) is recommended. Previous evaluation showed S-P-Nano-0008-a-S-16 and S-P-Nano-1390-a-A-17 to be highly specific for Nanoarchaeota (Supplementary Table S2). Note that these primers generate almost full-length sequences. In summary, both primer pairs can be recommended for amplification. They generate amplicons specific for Bacteria and Archaea with an average length of 278 bp that spans the HV region four.
In silico evaluation of primer pairs suitable for sequencing technologies like Roche’s 454 (Group M)
No archaeal-specific primer pair achieves a full phylum spectrum (Supplementary Table S15). S-D-Arch-0519-a-S-15/S-D-Arch-1041-a-A-18 (A: 76.6%, B: 0.0%, E: 0.0%) shows the best results with respect to a relatively high overall coverage coupled with a high domain specificity. This primer pair covers two out of eight phyla (Crenarchaeota and Euryarchaeota), but the phylum spectrum increases remarkably to six detected phyla if one mismatch is allowed (A: 92.8%, B: 0.0%, E: 0.0%). Detection of the four additional phyla (AAG, Korarchaeota, MHVG I and MHVG II) is likely due to a middle mismatch position in the reverse primer. Amplification of GoC-Arc-109-D0-C1-M0 and Nanoarchaeaota remains challenging due to more than one mismatch. In summary, S-D-Arch-0519-a-S-15/S-D-Arch-1041-a-A-18 is the most suitable primer pair with a 540 bp amplicon spanning HV regions 4-6 and excellent domain specificity. The frequent use of HV region six in diversity analysis makes this pair particularly interesting for comparative analysis (35,44,45).
For the domain Bacteria, several domain-specific primer pairs attain high overall coverage, but 27 out of 30 fail to detect more than 10 phyla (Supplementary Table S17). The three best pairs are S-D-Bact-0341-b-S-17/S-D-Bact-1061-a-A-17 (A: 0.0%, B: 91.9%, E: 0.0%), S-D-Bact-0564-a-S-15/S-*Univ-1100-a-A-15 (A: 8.0%, B: 92.7%, E: 0.0%) and S-D-Bact-0341-b-S-17/S-D-Bact-0785-a-A-21 (A: 0.5%, B: 86.2%, E: 0.0%). Although the first two show higher overall coverage, the latter exhibits a larger phylum spectrum. S-D-Bact-0341-b-S-17/S-D-Bact-0785-a-A-21 only fails to detect seven bacterial phyla (Hyd24-12, GOUTA4, Armatimonadetes, Chloroflexi, BHI80-139 and Candidate divisions OP11 and WS6). If one mismatch is tolerated (A: 64.6%, B: 94.5%, E: 0.1%), amplification of four additional phyla is likely (Chloroflexi, BHI80-139, Hyd24-12 and GOUTA4). However, some archaeal sequences are also detected. Detailed analysis reveals that only the coverage for Candidate division OP11 remains below the 50% threshold (Supplementary Table S18). Besides four mismatches for the reverse primer, the mismatch positions in both primers are located towards the 3′ end. Moreover, amplification of Armatimonadetes and Candidate division WS6 is unlikely due to the 3′ end mismatch position of the forward primer. Although not covering the complete phylum spectrum, the pair S-D-Bact-0341-b-S-17/S-D-Bact-0785-a-A-21 shows the best combination of domain and phylum coverage and can thus be recommended for 464 bp amplicons covering the HV regions 3–4.
S-D-Bact-0785-a-S-18/S-*-Univ-1392-a-A-15 (A: 72.3%, B: 74.1%, E: 0.0%) qualifies as a suitable primer pair for Bacteria and Archaea. With no mismatches it only fails to detect Nanoachaeota and expands to full archaeal phylum spectrum if one mismatch is tolerated. Detailed analysis revealed that none of the mismatch positions are located towards the 3′ end, which should allow amplification. For Bacteria, an overall coverage of 76.3% is achieved but this pair fails to detect nine phyla (Chloroflexi, SM2F11, HDB-SIOH1705, BD1-5, EM19, BHI80-139, Candidate divisions OP11, SR1, OD1 as well as Epsilonproteobacteria). Allowing one mismatch results in an increased overall coverage (A: 79.0%, B: 86.1%, E: 1.3%) and the additional detection of six phyla due to internal mismatches. Only the coverage of HDB-SIOH17005, SM2F11 and Candidate division OP11 remains below the 50% threshold. In summary, with an amplicon length of 608 bp and detection of HV regions 5–8 this primer pair qualifies to target Bacteria and Archaea.
This detailed evaluation also demonstrates that reverse and forward primers with individual high coverage do not automatically qualify as an optimal primer pair. For instance, S-D-Bact-0347-a-S-19 (A: 0.0%, B: 86.1%, E: 0.0%) and S-D-Bact-0785-a-A-19 (A: 8.5%, B: 86.4%, E: 0.0%) have been designed and approved by the Human Microbiome Project for analysing the foregut microbiome (1). Based on promising results within the human habitat, they suggested that this primer pair may be a good candidate to access the bacterial diversity in any habitat (1). However, our evaluation reveals a lower overall coverage of A: 0.0%, B: 76.5%, E: 0.0% and detection of only 25 out of 59 bacterial phyla if they act as a primer pair. Even if one mismatch is allowed (A: 0.0%, B: 90.6%, E: 0.0%) this primer pair still fails to detect 17 phyla (Armatimonadetes, Chlamydiae, Dictyoglomi, Planctomycetes, Verrucomicrobia, Spirochaetes, Lentisphaerae, HDB-SIOH1705, LD1-PA38, NPL-UPA2, Hyd24-12 and SM2F11, as well as Candidate divisions OP11, WS6, BRC1, OD1, WS3 and OP3).
In silico evaluation of primer pairs suitable for sequencing technologies such as PacBio SMRT or classical clone libraries (Group L)
For fragments >1000 bases we could not find an archaeal primer pair with both an overall coverage of over 70% and a satisfying phylum spectrum (Supplementary Table S21). The majority detects only the two sequence-rich phyla, Crenarchaeota and Euryarchaeota. S-D-Arch-0349-a-S-17/S-*-Univ-1392-a-A-15 (A: 65.8%, B: 0.0%, E: 0.0%) has the highest overall coverage. Detailed analysis revealed that this pair fails to detect six out of eight phyla (AAG, GoC-Arc-109-D0-C1-M0, Korarchaeota, MHVG-1, MHVG-2 and Nanoarchaeota) (Supplementary Table S21). Although performance increases slightly when one mismatch is allowed (A: 76.0%, B: 0.0%, E: 0.1%), the coverage for three phyla (AAG, GoC-Arc-109-D0-C1-M0 and Nanoarchaeota) remains below 50% (Supplementary Table S22). In addition, a 3′ mismatch of the forward primer hampers amplification of Korarchaeota. In summary, this primer pair cannot be recommended. Similar results are obtained for the other archaeal primer pairs of Group L.
The bacterial primer pairs show more satisfying results (Supplementary Table S23). S-D-Bact-0008-c-S-20/S-D-Bact-1391-a-A-17 (A: 0.1%, B: 78.0%, E: 0.0%) has a high overall coverage and detects 55 out of 59 phyla. The four phyla with below-threshold coverage are Chlamydiae, WCHB1-60, Candidate division SR1 and OP11. If one mismatch is allowed, overall coverage increases to A: 0.1%, B: 86.2%, E: 0.0% and Candidate division OP11 is now likely to be detected due to an internal mismatch. S-D-Bact-0008-c-S-20/S-D-Bact-1046-a-A-19 (A: 0.0%, B: 81.3%, E: 0.0%) achieves the highest overall coverage but fails to detect eight phyla (S2R-29, SM2F11, Chlamydiae, Thermotogae, WCHB1-60, Kazan-3B-28, EM19, Candidate division OP11 and Epsilonproteobacteria). Remarkably, this is mostly compensated if one mismatch is allowed. However, amplification of some sequences belonging to Candidate division OP11 and WHCBI-60 is unlikely due to 3′ end mismatches. Moreover, the reverse primer fails to detect SM2F11 due to two mismatches of which one is located towards the 3′ end. Chlamydiae remains undetected due to three internal mismatches of the forward primer. The promising results and excellent domain specificity of both primer pairs are depreciated by the fact that they only span HV regions 1–6 and 1–8, respectively. Nevertheless, if an amplicon length of <1400 bp is sufficient we are in favour of both primer pairs.
For nearly full-length sequences (>1400 bp) we recommend S-D-Bact-0008-a-S-16/S-D-Bact-1492-a-A-16 (A: 0.2%, B: 77.1%, E: 0.0%). This domain-specific primer pair spans HV regions 1–9 and covers 52 out of 59 bacteria phyla. The missing phyla are: GAL08, Kazan-3B-28, Chlamydiae, Dictyoglomi, WCHB1-60, MVP-21 and Caldiserica. One mismatch (A: 0.2%, B: 86.8%, E: 0.0%) allows additional detection of Caldiserica and Dictyoglomi due to an internal mismatch. The remaining five phyla have either more than two mismatches or, in case of Chlamydiae, the forward primer has a 3′ end mismatch. In the past, S-D-Bact-0008-a-S-16/S-D-Bact-1492-a-A-16, which is commonly known as GM3/GM4, has been intensively used for clone library-based studies from different habitats (46–48). Thus plenty of data for comparative analysis are available. However, the high number of sequences originally obtained with the GM3/GM4 pair is also likely to have artificially inflated the coverage values we obtained. Ideally, sequences obtained with a given primer should be excluded when evaluating that same primer.
In silico re-evaluation of primer pairs using a PCR free metagenome database
The majority of the sequences in specialized 16S/18S rDNA databases such as SILVA (20), greengenes (49) or RDP II (50) are a result of prior PCR amplification. In order to calibrate our previous analysis, re-evaluation of the results using the publicly available Global Ocean Sampling (GOS) database was performed. The initial GOS dataset consisted of 6.3 billion bp of Sanger sequence reads (28) and has recently been augmented by samples from the Atlantic and Indian Oceans (51). Although it is limited to the marine habitat, it is the most comprehensive dataset that provides a reasonable amount of relatively long fragments necessary for primer evaluation.
A total of 10 685 16S/18S rDNA sequences were extracted from the GOS dataset. 95% of the reads range between 900 bp and 1200 bp in length; the average length was 1053 bp. However, the bacterial fraction was dominant, consisting of 9965 sequences, compared with only 290 archaeal and 439 eukaryotic 16S and 18S sequences, respectively. Thus the results for Archaea and Eukaryota are uncertain and should only be seen as complementary information. In addition to the limited number of sequences, only a subset of phyla is present. For example, for Archaea 288 sequences belong to Crenarchaeota (63 sequences) and Euryarchaeota (225 sequences). The remaining two sequences could be assigned to AAG and MHVG-1, respectively. For Korarchaeota, GoC-Arc-109-D0-C1-M0, MHVG-2 and Nanoarchaeota, no sequences are present.
For the domain Bacteria, the 9956 reads span 28 out of 59 phyla. The majority belong to Actinobacteria (1006 sequences), Bacteroidetes (785 sequences), Cyanobacteria (805 sequences) and Proteobacteria (6655 sequences). Other member rich phyla such as Firmicutes (167 sequences) and Acidobacteria (29 sequences) are only present in low numbers. The lack of a full phylum spectrum clearly limits the re-evaluation and prevents direct comparisons with our previous results. The much lower and also varying number of sequences in the respective target regions affects the results as well. Furthermore primer pairs of Group L had to be excluded from the re-evaluation due to the lack of sufficient numbers of long sequences.
In the previous evaluation for Group S, the archaeal primer pair S-D-Arch-0349-a-S-17/S-D-Arch-0519-a-A-16 (A: 76.8%, B 0.0%, E: 0.0%) was proposed as a suitable pair for amplicon sequencing of <400 bases. Re-evaluation based on the GOS dataset again yielded the highest overall coverage (A: 74.5%, B: 0.0%, E: 1.2%) and excellent domain specificity. The recommended bacterial primer pair S-D-Bact-0564-a-S-15/S-D-Bact-0785-b-A-18 (A: 0.0%, B: 83.4%, E: 0.0%) also performs well. Tolerating one mismatch still confirms domain specificity (A: 10.6%, B: 86.2%, E: 0.0%). Unfortunately, detailed comparison on phylum level proved difficult. For example, within the SILVA database, 84 910 Firmicutes sequences of sufficient length are present and 91.8% of these are covered by S-D-Bact-0564-a-S-15/S-D-Bact-0785-b-A-18. Using the GOS dataset, only two sequences from Firmicutes are available.
Promising trends could also be observed for the two primer pairs targeting both, Archaea and Bacteria. In particular, S-D-Arch-0519-a-S-15/S-D-Bact-0785-b-A-18 stands out with high overall coverage (A: 76.5%, B: 83.4%, E: 1.9%), which increases slightly if one mismatch is allowed (A: 81.8%, B: 86.5%, E: 1.9%).
For Group M, only 32 sequences of sufficient length were available to re-evaluate the recommended archaeal primer pair S-D-Arch-0519-a-S-15/S-D-Arch-1041-a-A-18. Thus the Archaea primer pairs were excluded from further validation.
With on average 2600 available bacterial sequences for re-evaluating Group M, the conditions were slightly better. As in the previous evaluation, several primer pairs show high overall coverage: S-D-Bact-0564-a-S-15/S-*-Univ-1100-a-A-15 proves its suitability with a high domain-specific and overall coverage (A: 0.0%, B: 76.2%, E: 0.0%). Overall coverage for Bacteria increases up to 80.2%, if one mismatch is tolerated (A: 2.3%, B: 80.2%, E: 0.0%). In contrast, S-D-Bact-0341-b-S-17/S-D-Bact-1061-a-A-17 (A: 0.0%, B: 58.9%, E: 0.0%) fails to match the previous results, which could be a consequence of the specific dataset. Even allowing one mismatch does not achieve satisfying results (A: 0.0%, B: 64.8%, E: 0.0%). At first glance, similar results were obtained for S-D-Bact-0341-b-S-17/S-D-Bact-0785-a-A-21 (A: 0.0%, B: 43.1%, E: 0.0%). However, considering one mismatch the overall coverage significantly increased to A: 58.2%, B: 70.9%, E: 0.0%.
The re-evaluation of the primer pairs based on the GOS dataset (Supplementary Tables S27–S38) shows that, despite the relatively large dataset size, it still lacks resolution power, especially when considering a specific gene. Unfortunately, the data obtained by other large scale projects, such as the Earth Microbiome Project (52), is of little use for primer evaluation due to their cost effective, but length-limited sequencing strategy. Due to the inherent risk of creating chimeric sequences we would not consider assembly a solution to this limitation. Should the error rate of long read sequencing technologies such as PacBio be significantly reduced, data from metagenomic studies relying on these technologies would become a valuable resource for revisiting the primer sensitivity issue. In summary, if a sufficient amount for metagenomic 16S rDNA sequences were available, the previous primer pair recommendations could be confirmed.
Experimental evaluation of the primer pair S-D-Bact-0341-b-S-17/S-D-Bact-0785-a-A-21
The primer pair S-D-Bact-0341-b-S-17/S-D-Bact-0785-a-A-21 (Group M) was applied to DNA extracted from a time series of three marine environmental samples at Helgoland Roads. For simplification, we will refer to the obtained reads as ‘16S pyrotags’. In the course of the MIMAS project, metagenomic analysis was performed using marine samples from the same site and time points (22). The results from the metagenomic-based diversity studies are used to evaluate the accuracy of each primer pair by comparing the taxonomic classifications.
On average, 59 700 sequences were obtained per sampling occasion, of which 52 400 could be assigned as 16S pyrotags (88.4%) (Supplementary Table S39). The relatively high loss is due to the stringent quality checks used for the identification and taxonomic classification of 16S rDNA fragments. In contrast, metagenome analysis resulted on average in 2 109 000 sequences (22) per sampling occasion, but only 1600 sequences (0.1%) qualified as 16S rDNA gene fragments.
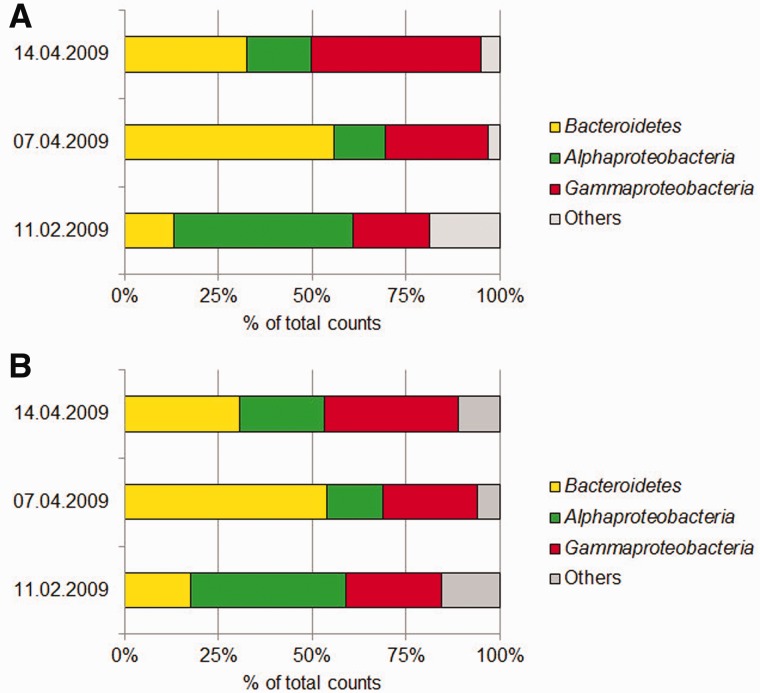
The results of the 16S pyrotag analysis show that the bacterial community is dominated by Alphaproteobacteria, Bacteroidetes and Gammaproteobacteria (Figure 1A and Supplementary Table S40). According to the in silico evaluation, for primer pair S-D-Bact-0341-b-S-17/S-D-Bact-0785-a-A-21 high coverage of these three groups are expected (Bacteroidetes: 89.2%, Alphaproteobacteria: 81.4%, Gammaproteobacteria: 90.6%). Allowing one mismatch the overall coverage increases to up to 95% for each group. The results from the 16S pyrotags also revealed a succession of the relative abundances. Bacteroidetes peaked on 7 April 2009, but were still abundant on 14 April 2009. For Alphaproteobacteria more sequences could be detected in winter on 11 February 2009. In contrast, the relative abundance of Gammaproteobacteria increased on 14 April 2009. The same trends were observed in the metagenomes (Figure 1B and Supplementary Table S41) (22). To verify that the results derived from the 16S pyrotags are not an artefact of deep sequencing, the total number of reads was adjusted to smaller subsets of around 2000 sequences by random re-sampling. Detailed analysis of these re-sampled subsets confirmed the results (Supplementary Table S42).
Figure 1.
Taxonomic distribution of 16S rRNA gene sequences gained from a time series of three different surface water samples at Helgoland Roads in the North Sea, (A) 16S pyrotags generated from PCR and sequenced with Roche’s 454 pyrosequencing (relative abundance, percentage of total counts) (B) 16S sequences gained from metagenome studies (relative abundance, percentage of total counts).
16S pyrotag analysis provides an enhanced resolution up to the group or genus level. Six relatively abundant taxonomic groups and genera (Formosa, Polaribacter, SAR11 clade surface 1, NAC11-7 lineage, Reinekea and SAR92 clade) have been examined in detail (Supplementary Figure S1A and Supplementary Table S43). Noteworthy is the Formosa peak on 7 April 2009 and the presence of Reinekea only on 14 April 2009. Both results were supported by diversity studies from the corresponding metagenomes (Supplementary Figure S1B and Supplementary Table S44). Again, the re-sampled 16S pyrotag subsets confirmed that the results are not an artefact of deep sequencing (Supplementary Table S45). In addition, it is interesting to note that corresponding metaproteome studies described in Teeling et al. (22) reflect the same succession of the bacterial community on the protein level.
Considering the in silico evaluation, S-D-Bact-0341-b-S-17/S-D-Bact-0785-a-A-21 should fail to detect SAR 11 clade surface 1 (0.7%). However, experimental evaluation clearly shows that the primer pair is able to amplify this taxonomic group. This can be explained by the increased coverage of up to 97% if one mismatch is allowed. A closer look at the primer target position of the reverse primer reveals an internal mismatch position towards the 5′ end. The results demonstrate that S-D-Bact-0341-b-S-17/S-D-Bact-0785-a-A-21 provides a good representation of the bacterial diversity down to genus and group level and illustrates that an internal mismatch towards the 5′ end can be tolerated by standard PCR.
To test the assumption that a suboptimal primer pair might result in a biased picture of the bacterial diversity, S-D-Bact-0008-a-S-16/S-D-Bact-0907-a-A-20 was applied to the same samples. This primer pair was chosen due to its relatively high overall coverage (A: 0.0%, B: 75.1%, E: 0.0%) but distinctly lower phylum spectrum. Based on the in silico evaluation it should fail to detect 18 bacterial phyla (Aquificae, BD1-5, BHI80-139, Chlamydiae, Dictyoglomi, EM19, Lentisphaerae, SM2F11, Thermotogae, Tenericutes, Verrucomicrobia, WCHB1-60 and Candidate divisions TM7, WS6, OD1, SR1 and OP11). With relatively high coverage of Bacteroidetes (77.6%), Alphaproteobacteria (71.3%) and Gammaproteobacteria (80.5%) in silico evaluation and experimental data confirm that this primer pair is able to detect the same dominant taxonomic groups (Supplementary Figure S2 and Supplementary Table S46). However, in comparison with the 16S pyrotags generated with S-D-Bact-0341-b-S-17/S-D-Bact-0785-a-A-21 and metagenome studies Alphaproteobacteria appear to be more abundant throughout all samples. Bacteroidetes, on the other hand, are under-represented. A similar bias can be found on the group level (Supplementary Figure S3 and Supplementary Table S47). Use of this primer pair indicates a higher relative abundance of Alphaproteobacteria SAR11 clade surface 1 as well as NAC11-7 lineage on 7 April 2009 and 14 April 2009. In turn, particularly the genus Formosa is less prominent. This is in line with the results from the in silico evaluation, which shows that S-D-Bact-0008-a-S-16/S-D-Bact-0907-a-A-20 only detects 52.9% of the Formosa sequences. Even one allowed mismatch results only in an increase of 9% up to 61.9%. A closer look reveals a mismatch of the reverse primer towards the 3′ end for several Formosa sequences.
Although S-D-Bact-0008-a-S-16/S-D-Bact-0907-a-A-20 is able to detect all major groups, a bias in the relative abundances as well as community structure is clearly confirmed by the experimental data (Figure 1 and Supplementary Figures S1–S3). This supports our assumption that the overall coverage need always to be considered in combination with the phylum spectrum. Detailed analysis of the mismatch position should also be taken into account. Nevertheless, the experimental results strongly indicate that in silico evaluation can serve as a guideline for choosing the most suitable primer pair.
CONCLUSIONS
The advent of new sequencing methods has been a paradigm shift for molecular ecology and especially microbial diversity analysis using marker genes. The rapid adoption of the new techniques caused a backlog in proper evaluation of the primers used for diversity surveys. Our study shows that even commonly used single primers exhibit significant differences in overall coverage and phylum spectrum. Consequently, primer pairs need to be carefully selected to avoid accumulative bias. Out of the 175 primers and 512 primer pairs checked, only 10 can be recommended as broad range primers. Although none of them are perfect, and especially for the Archaea we recommend the design of additional primers, the experimental validation shows that a good combination of primers approximate PCR-free metagenomic approaches with respect to community structure and relative abundances. The experimental results confirm that single internal mismatches, when located towards the 5′ end, are tolerated in the amplification process. Re-inspection of the primers using GOS metagenomes was found to be a reasonable approach for determining possible primer bias in the public rDNA repositories. However, the incomplete phylum spectrum as well as the comparatively small dataset size with respect to 16S rRNA genes in the GOS metagenomes did not allow for an in-depth re-evaluation. For example, Group M primer pair S-D-Bact-0341-b-S-17/S-D-Bact-0785-a-A-21, which we recommended based on the SSURef 108 NR results, fails to detect major groups in the GOS dataset, yet excels in the experimental evaluation. This demonstrates the validity of using comprehensive, non-redundant datasets like the SILVA SSURef 108 NR for detailed evaluation of probes and primers. We would like to note that the SILVA project has prepared an online service for this purpose at www.arb-silva.de/search/testprime, which is modelled after our evaluation method and allows inspection of per-taxon coverages for individual primer pairs. Furthermore, all primers, including bibliographic information and information on specificity and overall coverage, have been added to probeBase. The availability of the evaluated primers in a central and publically accessible repository plus the online primer evaluation tool should facilitate the search for, and the evaluation and selection of, suitable primers in future studies.
AVAILABILITY
Supplementary files are available at www.arb-silva.de/download/archive/primer_evaluation/.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–52 and Supplementary Figures 1–3.
FUNDING
Max Planck Society and the Federal Ministry of Education and Research Germany [03F0480D]. Research in the lab of M.H. is funded through grants from the Austrian Research Fund [Y277-B03]; the European Research Council [Starting Grant EVOCHLAMY 281633]. Funding for open access charge: Max Planck Society.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge Jack A. Gilbert (Argonne National Laboratory, Argonne, IL, USA), Bernhard M. Fuchs (Max Planck Institute, Bremen, Germany) and Christine Klockow for critical discussion of this manuscript. E. Karamehmedovic and M. Meiners for helping with the laboratory work. G. Gerds and A. Wichels from the Alfred Wegner Institute (Bremerhaven, Germany) for supporting and performing the water sampling. Hannah Marchant, Elizabeth Robertson, Mira Okshevsky and Mario Schimak for critical reading of the manuscript.
REFERENCES
- 1.Nossa C.W., Oberdorf W.E., Yang L., Aas J.A., Paster B.J., Desantis T.Z., Brodie E.L., Malamud D., Poles M.A., Pei Z. Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. WJG. 2010;16:4135–4144. doi: 10.3748/wjg.v16.i33.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medini D., Serruto D., Parkhill J., Relman D.A., Donati C., Moxon R., Falkow S., Rappuoli R. Microbiology in the post-genomic era. Nat. Rev. Microbiol. 2008;6:419–430. doi: 10.1038/nrmicro1901. [DOI] [PubMed] [Google Scholar]
- 3.Mardis E.R. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Armougom F., Raoult D. Exploring microbial diversity using 16S rRNA high-throughput methods. J. Comput. Sci. Syst. Biol. 2009;2:74–92. [Google Scholar]
- 5.Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sogin M.L., Morrison H.G., Huber J.A., Mark Welch D., Huse S.M., Neal P.R., Arrieta J.M., Herndl G.J. Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc. Natl Acad. Sci. USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Z., Lozupone C., Hamady M., Bushman F.D., Knight R. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 2007;35:e120. doi: 10.1093/nar/gkm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5:433–438. doi: 10.1517/14622416.5.4.433. [DOI] [PubMed] [Google Scholar]
- 9.Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Huntley J., Fierer N., Owens S.M., Betley J., Fraser L., Bauer M., et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rothberg J.M., Hinz W., Rearick T.M., Schultz J., Mileski W., Davey M., Leamon J.H., Johnson K., Milgrew M.J., Edwards M., et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 11.Eid J., Fehr A., Gray J., Luong K., Lyle J., Otto G., Peluso P., Rank D., Baybayan P., Bettman B., et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 12.Loman N.J., Misra R.V., Dallman T.J., Constantinidou C., Gharbia S.E., Wain J., Pallen M.J. Performance comparison of benchtop high-throughput sequencing platforms. Nat. Biotechnol. 2012;30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- 13.Schloss P.D., Gevers D., Westcott S.L. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker G.C., Smith J.J., Cowan D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods. 2003;55:541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y., Qian P.Y. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS ONE. 2009;4:e7401. doi: 10.1371/journal.pone.0007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tringe S.G., Hugenholtz P. A renaissance for the pioneering 16S rRNA gene. Curr. Opin. Microbiol. 2008;11:442–446. doi: 10.1016/j.mib.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 17.Hamady M., Knight R. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome. Res. 2009;19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersson A.F., Lindberg M., Jakobsson H., Bäckhed F., Nyrén P., ngstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS ONE. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z., DeSantis T.Z., Andersen G.L., Knight R. Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res. 2008;36:e120. doi: 10.1093/nar/gkn491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pruesse E., Quast C., Knittel K., Fuchs B.M., Ludwig W., Peplies J., Glöckner F.O. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alm E.W., Oerther D.B., Larsen N., Stahl D.A., Raskin L. The oligonucleotide probe database. Appl. Environ. Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teeling H., Fuchs B.M., Becher D., Klockow C., Gardebrecht A., Bennke C.M., Kassabgy M., Huang S., Mann A.J., Waldmann J., et al. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science. 2012;336:608–611. doi: 10.1126/science.1218344. [DOI] [PubMed] [Google Scholar]
- 23.Brosius J., Palmer M.L., Kennedy P.J., Noller H.F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl Acad. Sci. USA. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loy A., Maixner F., Wagner M., Horn M. probeBase – an online resource for rRNA-targeted oligonucleotide probes: new features 2007. Nucleic Acids Res. 2007;35:D800–D804. doi: 10.1093/nar/gkl856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kibbe W.A. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 2007;35:W43–W46. doi: 10.1093/nar/gkm234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pruesse E., Peplies J., Glöckner F.O. SINA: accurate high throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 28.Rusch D.B., Halpern A.L., Sutton G., Heidelberg K.B., Williamson S., Yooseph S., Wu D., Eisen J.A., Hoffman J.M., Remington K., et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS ONE. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yooseph S., Sutton G., Rusch D.B., Halpern A.L., Williamson S.J., Remington K., Eisen J.A., Heidelberg K.B., Manning G., Li W., et al. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ludwig W., Strunk O., Westram R., Richter L., Meier H., Kumar Y., Buchner A., Lai T., Steppi S., Jobb G., et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou J., Bruns M.A., Tiedje J.M. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herlemann D.P.R., Labrenz M., Juergens K., Bertilsson S., Waniek J.J., Anderrson A.F. Transition in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011;5:1571–1579. doi: 10.1038/ismej.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muyzer G., de Waal E.C., Uitterlinden A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muyzer G., Brinkhoff T., Nuebel U., Santegoeds C., Schaefer H., Waver C. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Akkermans A.D.L., van Elsas J.D., de Bruijn F.J., editors. Molecular Microbial Ecology Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 1–27. [Google Scholar]
- 35.Gilbert J.A., Field D., Swift P., Thomas S., Cummings D., Temperton B., Weynberg K., Huse S., Hughes M., Joint I., et al. The taxonomic and functional diversity of microbes at a temperate coastal site: a 'multi-omic' study of seasonal and diel temporal variation. PLoS ONE. 2010;5:e15545. doi: 10.1371/journal.pone.0015545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walters W.A., Caporaso J.G., Lauber C.L., Berg-Lyons D., Fierer N., Knight R. PrimerProspector: de novo design and taxonomic analysis of barcoded PCR primers. Bioinformatics. 2011;27:1159–1161. doi: 10.1093/bioinformatics/btr087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barns S.M., Fundyga R.E., Jeffries M.W., Pace N.R. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc. Natl Acad. Sci. USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reysenbach A.L., Pace N.R. Robb F.T., Place A.R., editors. Archaea: A Laboratory Manual—Thermophiles. CSHLP. Protocol 16. 1995:101–107. [Google Scholar]
- 39.Huws S.A., Edwards J.E., Kim E.J., Scollan N.D. Specificity and sensitivity of eubacterial primers utilized for molecular profiling of bacteria within complex microbial ecosystems. J. Microbiol. Methods. 2007;70:565–569. doi: 10.1016/j.mimet.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 40.Droege M., Hill B. The genome sequencer FLX system–longer reads, more applications, straight forward bioinformatics and more complete data sets. J. Biotechnol. 2008;136:3–10. doi: 10.1016/j.jbiotec.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 41.Sipos R., Székely A.J., Palatinszky M., Révész S., Márialigeti K., Nikolausz M. Effect of primer mismatch, annealing temperature and PCR cycle number on 16S rRNA gene-targetting bacterial community analysis. FEMS Microbiol. Ecol. 2007;60:341–350. doi: 10.1111/j.1574-6941.2007.00283.x. [DOI] [PubMed] [Google Scholar]
- 42.Baker G.C., Cowan D.A. 16 S rDNA primers and the unbiased assessment of thermophile diversity. Biochem. Soc. Trans. 2004;32:218–221. doi: 10.1042/bst0320218. [DOI] [PubMed] [Google Scholar]
- 43.Huber H., Hohn M.J., Rachel R., Fuchs T., Wimmer V.C., Stetter K.O. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature. 2002;417: 63–67. doi: 10.1038/417063a. [DOI] [PubMed] [Google Scholar]
- 44.Huse S.M., Dethlefsen L., Huber J.A., Mark Welch D., Relman D.A., Sogin M.L. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCliment E.A., Nelson C.E., Carlson C.A., Alldredge A.L., Witting J., Amaral-Zettler L.A. An all-taxon microbial inventory of the Moorea coral reef ecosystem. ISME J. 2012;6:309–319. doi: 10.1038/ismej.2011.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abed R.M., Al-Thukair A., de Beer D. Bacterial diversity of a cyanobacterial mat degrading petroleum compounds at elevated salinities and temperatures. FEMS Microbiol. Ecol. 2006;57:290–301. doi: 10.1111/j.1574-6941.2006.00113.x. [DOI] [PubMed] [Google Scholar]
- 47.Liebner S., Harder J., Wagner D. Bacterial diversity and community structure in polygonal tundra soils from Samoylov Island, Lena Delta, Siberia. Int. Microbiol. 2008;11:195–202. [PubMed] [Google Scholar]
- 48.Schauer R., Bienhold C., Ramette A., Harder J. Bacterial diversity and biogeography in deep-sea surface sediments of the South Atlantic Ocean. ISME J. 2010;4:159–170. doi: 10.1038/ismej.2009.106. [DOI] [PubMed] [Google Scholar]
- 49.DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cole J.R., Chai B., Farris R.J., Wang Q., Kulam-Syed-Mohideen A.S., McGarrell D.M., Bandela A.M., Cardenas E., Garrity G.M., Tiedje J.M. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007;35:D169–D172. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yooseph S., Nealson K.H., Rusch D.B., McCrow J.P., Dupont C.L., Kim M., Johnson J., Montgomery R., Ferriera S., Beeson K., et al. Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature. 2010;468:60–66. doi: 10.1038/nature09530. [DOI] [PubMed] [Google Scholar]
- 52.Gilbert J.A., Meyer F., Jansson J., Gordon J., Pace N., Tiedje J., Ley R., Fierer N., Field D., Kyrpides N., et al. The Earth Microbiome Project: meeting report of the “1st EMP meeting on sample selection and acquisition” at Argonne National Laboratory October 6th 2010. Stand. Genomic Sci. 2010;3: 249–253. doi: 10.4056/aigs.1443528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.