Abstract
The first crystal structure of human telomeric DNA in complex with the natural alkaloid berberine, produced by different plant families and used in folk medicine for millennia, was solved by X-ray diffraction method. The G-quadruplex unit features all-parallel strands. The overall folding assumed by DNA is the same found in previously reported crystal structures. Similarly to previously reported structures the ligand molecules were found to be stacked onto the external 5′ and 3′-end G-tetrads. However, the present crystal structure highlighted for the first time, the presence of two berberine molecules in the two binding sites, directly interacting with each tetrad. As a consequence, our structural data point out a 2:1 ligand to G-tetrad molar ratio, which has never been reported before in a telomeric intramolecular quadruplex structure.
INTRODUCTION
The 3′-end single-stranded overhang of human telomeric DNA is currently considered an interesting target in new anticancer therapy strategies. This is due to its ability to form, in the presence of stabilizing agents, such as monovalent cations, non-canonical G-quadruplex folds (Scheme 1a), which in their turn, can inhibit the activity of telomerase. This enzyme is involved in the cell life cycle and is over-expressed in most of the cancer cells. This imbalance has an impact on the recruitment of telomere-associated proteins required for the capping and maintenance of telomeric ends (1–3).
Scheme 1.
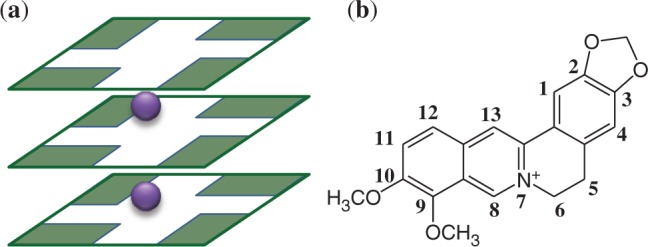
(a) Schematic representation of three guanine tetrads and monovalent cations as found in G-quadruplex telomeric DNA (b) Berberine (Ber) molecule drawing.
Many efforts have recently been devoted to the development of new G-quadruplex stabilizing molecules as potential anticancer drugs. Compounds found to be active in the binding to the telomeric G-quadruplex include anthraquinones, acridines, quinacridines, porphyrins, perylenes and the natural macrocyclic molecule telomestatin (3,4). Most of them are charged positively and contain a large aromatic surface.
The nuclear magnetic resonance and X-ray structures of a few telomeric quadruplex-small molecule complexes in the presence of K+ ions have already been reported and studied (5–11), and these all feature a quadruplex parallel stranded topology (12). In almost all of these cases, only one molecule of ligand was observed stacked onto an external G-tetrad (5,6,9,11). Isoquinoline alkaloids, such as berberine (Ber) (Scheme 1b), possess structural features similar to those mentioned earlier, and constitute an important class of natural products, produced by different plant families, and used in Ayurvedic, Chinese and Middle-Eastern folk medicine for millennia. Moreover, they possess striking biological and pharmacological activities, and have an emerging role in contemporary biomedical research. In the past few years, several papers have been published discussing the binding properties of the natural benzophenantridine alkaloids berberine and berberine-derivatives toward G-quadruplex structures, illustrating again that the properties and behavior of these compounds are becoming a key area of study and interest (13–18).
Biophysical and biochemical studies have shown that these substances bind to G-quadruplex structures formed by the human telomeric, the c-myc oncogene and other biologically relevant G-rich sequences (19–21). In the case of berberine, a stacking interaction with 1:1 binding stoichiometry has been observed and documented, with a high binding affinity for a 22-mer human telomeric sequence in K+ containing solution (22). Moreover, the induction of an antiparallel G-quadruplex in a 21-mer human telomeric sequence has been evidenced by circular dicroism measurements also without the addition of monovalent cations (23).
Isoquinoline alkaloids show a notable efficiency in telomerase inhibition and, most importantly, they are also selective toward G-quadruplex over duplex DNA (20). Furthermore, semi-synthetic derivatives of berberine, especially those 9-O substituted, which contain charged aza-aromatic terminal groups, show an even higher specificity and have an increased ability over berberine to inhibit telomerase, suggesting that they might become lead compounds for the development of new anticancer drugs (23,24).
The X-ray diffraction analysis of berberine complexed with double helix DNA d(CGTACG) was recently reported (25). However, to the best of our knowledge, no atomic detail structural data have been published up till now on the adduct formed by berberine or berberine-derivatives with the G-quadruplex. Thus, to shed more light on the binding features of this natural alkaloid, we solved the structure of the berberine/d[TAG3(T2AG3)3] complex (referred as Ber/h-Tel23 in the text later).
MATERIALS AND METHODS
The d[TAG3(T2AG3)3] sequence was purchased from Jena Bioscience (Jena, Germany), HPLC purified berberine chloride was purchased from Sigma-Aldrich Co. Ltd., USA. The purified oligonucleotide was dissolved to a final concentration of 4 mM, in a solution containing 20 mM potassium cacodylate buffer at pH 6.5 and 50 mM potassium chloride, and then heated to 368 K for 5 min. It was subsequently allowed to cool overnight to room temperature to induce the quadruplex formation.
A search for crystallization conditions was performed using a home-made crystallization screen based on the one described by Campbell and Parkinson (26). The DNA quadruplex–drug complex was crystallized at 296 K using the sitting drop vapour diffusion method. One microliter of the 4 mM solution of the human telomeric sequence d[TAG3(T2AG3)3] was mixed with 1 µL of a 2 mM solution of berberine in water, and with 2 µL of a solution containing the following: 1.8 M ammonium sulphate, 0.2 M KCl, 0.05 M Li2SO4 and 0.05 M potassium cacodylate with pH = 6.5. Drops were equilibrated against this latter solution. Yellow hexagonal crystals of the complex appeared after 2 weeks.
Data collection on crystals of the quadruplex–drug complex was performed at 100 K, using as cryoprotectant a solution analogous to the crystallization solution but with an increased concentration of ammonium sulphate (1.9 M) and 30% glycerol. The berberine–quadruplex complex crystallizes in the space group P6, with unit cell parameters a = 65.620 Å and c = 41.81 Å.
Data for the complex were collected at the X12 beamline (EMBL, DESY, Hamburg) to a maximum resolution of 2.4 Å, using a wavelength of 1.000 Å. Data were integrated using the program mosflm (27,28) (and scaled with SCALA from the CCP4 program package (29). Unit cell parameters and data collection statistics are given in Table 1.
Table 1.
Summary of data collection and atomic model refinement statistics
| Data collection | |
| Wavelength (Å) | 1.000 |
| Space group | P6 |
| Cell dimension (Å) | a = 65.620, c = 41.81 |
| Limiting resolution (Å) | 2.3 (2.42–2.3) |
| Unique reflections | 4674 |
| Rsym | 0.088 (0.68) |
| Multiplicity | 22.2 (22.3) |
| Completeness overall (%) | 100.0 (100.0) |
| <I/σ(I)> | 24.9 (1.1) |
| Refinement | |
| Resolution range (Å) | 30.0–2.4 |
| Unique reflections, working/free | 4128/188 |
| Rfactor (%) | 21.5 |
| Rfree (%) | 25.2 |
| RMSD bonds(Å) | 0.008 |
| RMSD angles (°) | 2.451 |
Values in parentheses are for the highest resolution shell.
The structure of the complex was solved by molecular replacement using the program Molrep (30). Coordinates of the complex of the same DNA sequence with a tetra-substituted naphthalene diimide (9) (PDB code: 3CDM) were used as a search model, after deleting atoms belonging to ligand and solvent molecules.
Fo-Fc electron density maps revealed a clear density for the drug molecules and they were introduced and refined at full occupancy. Two spherical densities were also present in the Fo-Fc electron density maps inside the G-quadruplex channel that could be interpreted as potassium ions. The model was refined using the program Refmac5 (31) from the CCP4 package (29). Manual rebuilding of the model was performed using the program Coot (32). The crystal packing analysis was performed with the program Mercury (33). Coordinates of the complex have been deposited with the Protein Data Bank (PDB access number 3R6R).
RESULTS
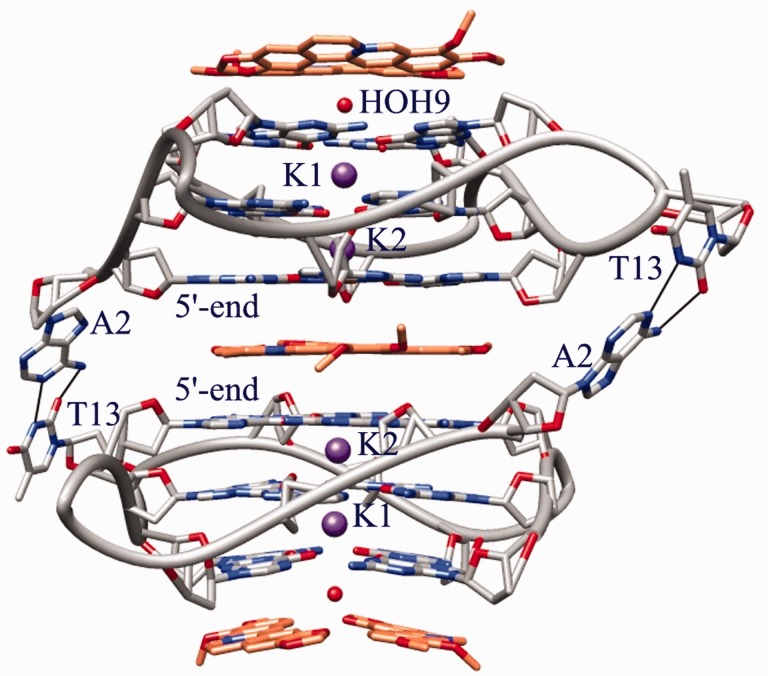
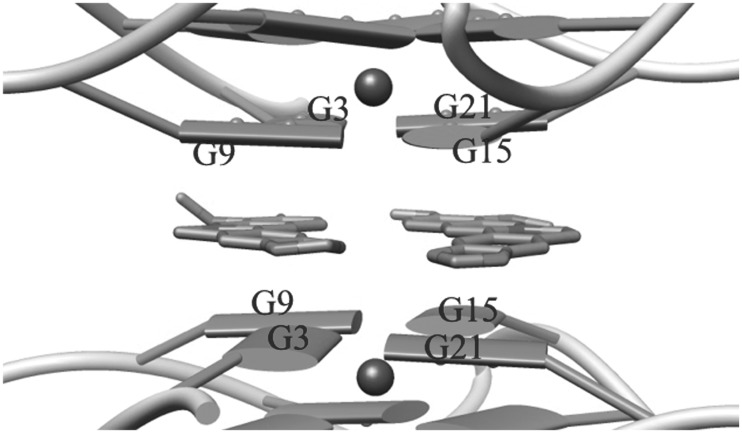
The complex crystallizes in the P6 space group, with each asymmetric unit containing an intramolecular G-quadruplex in complex with berberine in a DNA/ligand molar ratio of 1:3. The overall DNA topology is of parallel type and quite similar to those described for the native 22-mer intramolecular quadruplex (34), and also for the h-Tel23 in complex with a tetra-substituted naphthalene diimide (9,11). The analysis of the crystal structure (Figure 1) shows a parallel orientation of all strands and a square planar arrangement of the G-tetrads, which were observed 3.2 Å apart from each other and featuring an average 29° twist between each successive step. Monovalent K+ cations, featuring an antiprismatic coordination environment, are positioned between adjacent G-tetrad, with an average distance of 2.7 Å from the O6 carbonyl atoms of the guanine residues (with maximum and minimum bond lengths of 3.0 and 2.7 Å, respectively). All three loops protrude from the stacked tetrad platforms, with the adenine in each TTA link intercalated between the two thymines. In contrast to the structure of h-Tel23 in complex with the naphthalene diimide, no ligand molecules intercalate in the loops. These loops have the same distinctive conformation, as has been found in the native 22-mer structure (34). The crystal packing features dimers of symmetry related (2-fold axis) intramolecular G-quadruplexes connected by the A2 and T13 residues belonging to different DNA monomers (Figure 1). They are linked by Watson and Crick type H-bonds, as found for the structures of the native 22-mer and the diimide/h-Tel23 complex (9,11,34). As a consequence, the two 5′-ends define a binding site where two coplanar Ber molecules are stacked, and lying 3.4 Å apart from both the 5′-end tetrads (Figure 2). Each Ber molecule interacts with a pair of adjacent guanines per tetrad. As a result of this placement and of the 2-fold symmetry relationship featuring the binding site, each ligand molecule actually interacts with the same couple of guanine from both the tetrads: G3/G9 in one case and G15/G21 in the other (Figure 2).
Figure 1.
DNA topology and binding sites in the Ber/h-Tel23 adduct (Ber molecules related by C2 pseudosymmetry are omitted for clarity).
Figure 2.
5′-end binding site for the Ber molecules (the other couple of Ber molecules, related by C2 pseudosymmetry axis, is omitted for clarity).
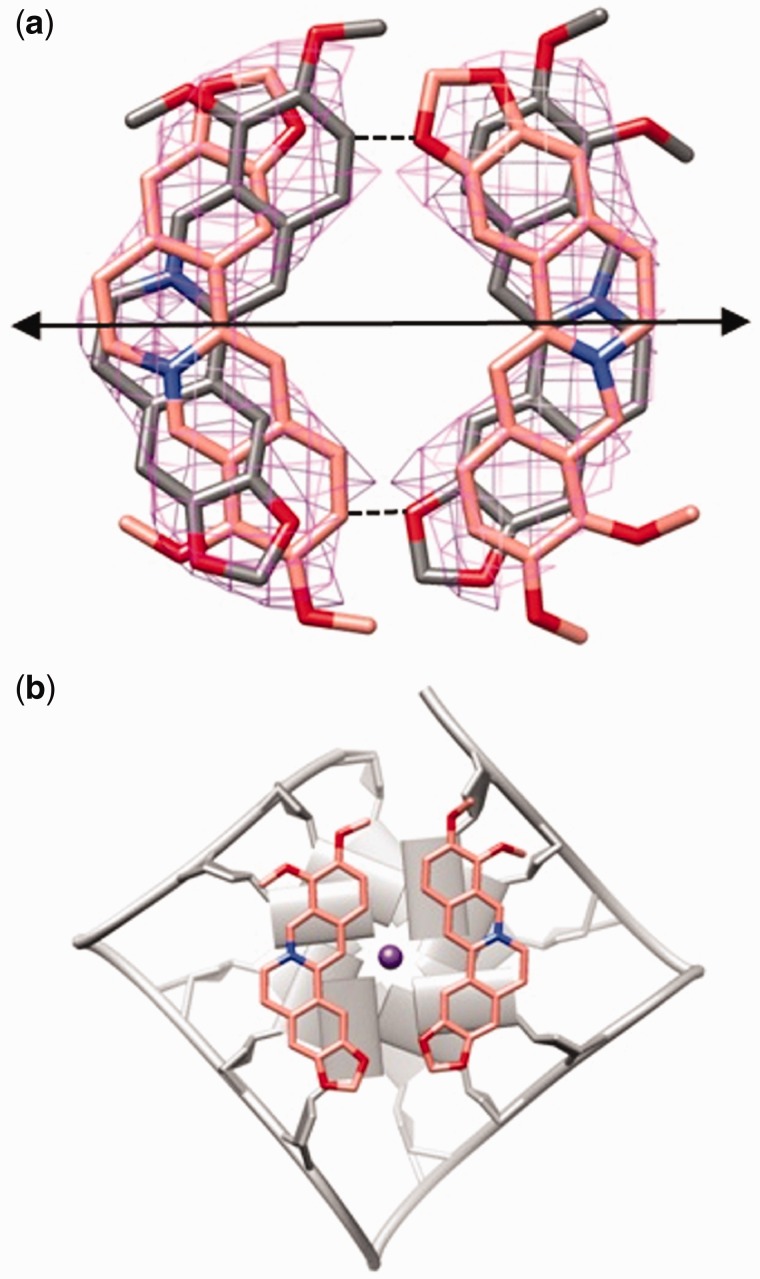
The 2-fold crystallographic axis, relating the two G-quadruplexes, in addition, bisects the Ber molecules, that are disordered and possessing the dyad axis only as a pseudo-symmetry (Figure 3a). Only the pair formed by the combination of Ber molecules with the metoxy groups pointing in the same direction is allowed in the case of the 5′-end binding site. With reference to Figure 3a, the pair can be formed either by the two pink or by the two gray Ber molecules. Certainly, the ligand pair in the binding site cannot be formed by Ber molecules with methoxy groups pointing in the opposite direction, due to the short, not allowed contacts that would otherwise be formed between the molecules belonging to the pair (Figure 3a, dashed lines). Interestingly, Ber molecules in each couple interact with each other through van der Waals interactions and define an approximate four-sided polygon, which leaves a free central channel, the one occupied by the monovalent ions (Figure 3b). It is worth noting that, as clearly shown in Figure 3b, the distance between the berberine carbon atoms C3 and C10 (9.3 Å) almost corresponds to the distance between the N9 nitrogen atoms of the guanine residues of the interacting pairs (9.6 Å), namely G3/G9 and G15/G21. Thus, the isoquinoline system of each Ber molecule is very well overlapped on the purinic moieties belonging to the G3/G9 and G15/G21 couples.
Figure 3.
(a) Disordered Ber molecules in the 5′-end binding sites. Only the pair of molecules featured by the same color is really present in the binding site. The position of the crystallographic 2-fold axis, relating the disordered Ber molecules, is also shown. Skeleton and Fo-Fc omit map for each Ber contoured at 1.5σ level. (b) Top view of a couple of Ber molecules stacked onto the 5′-end G-tetrads: (the other couple of Ber molecules, related by C2 pseudosymmetry, is omitted for clarity).
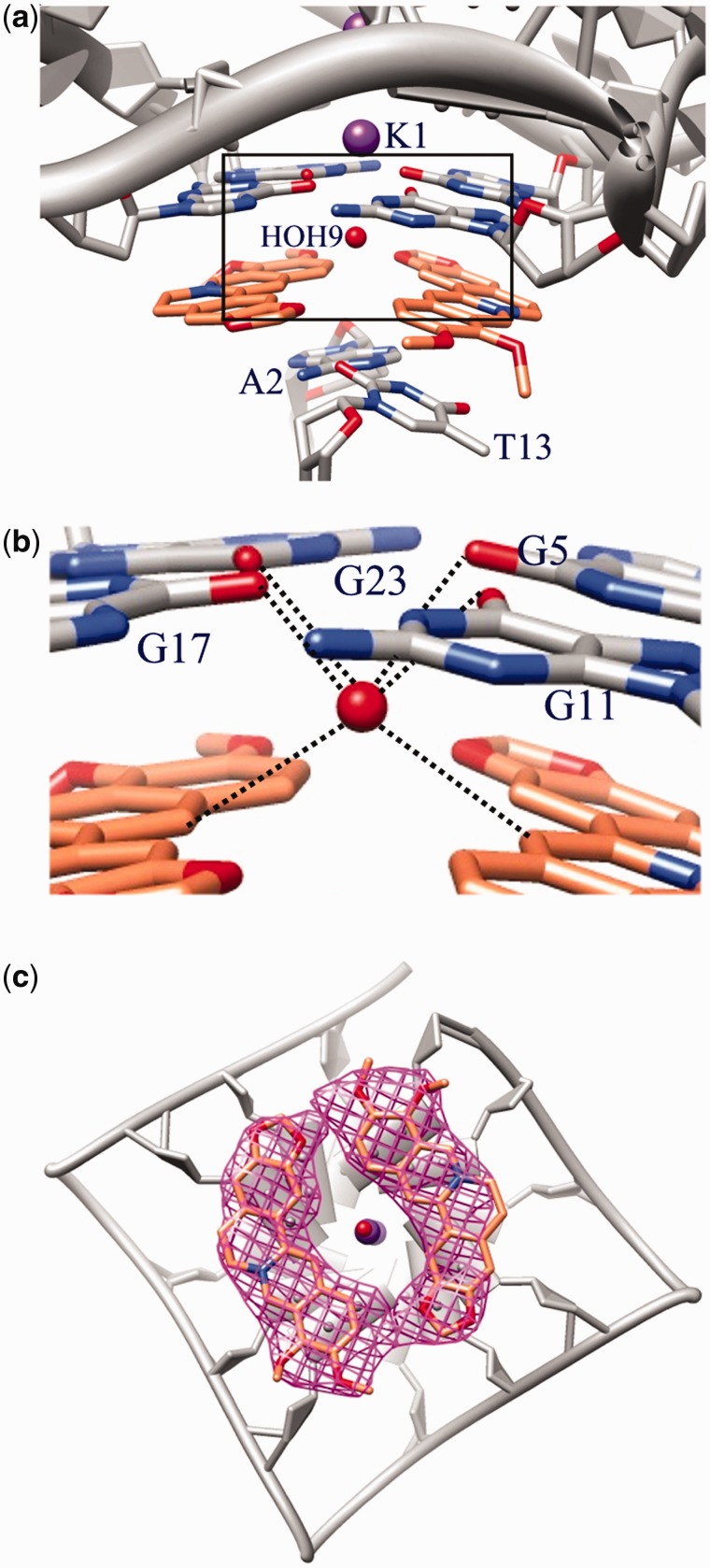
Two further Ber molecules are stacked on the 3′-end, about 3.4 Å apart from the tetrad. This binding site is completed by the A2–T13 base pair from a different G-quadruplex dimer, symmetry related by a 3-fold axis (Figure 4a). As well as for the 5′-end binding site, each Ber molecule is at stacking distance with only two adjacent guanines (G5/G11 and G17/G23, respectively; Figure 4b). Also, in this case, the isoquinoline moiety perfectly overlaps onto the base pair. However, unlike what occurs for the 5′-end binding site, the two stacked ligand molecules form a dihedral angle of 19.7°, so maximizing the stacking interaction with the 3′-end tetrad, which is bent as well, with a dihedral angle of 13.9° between the planes defined by the G5/G11 and G17/G23 pairs.
Figure 4.
(a) Binding site for the Ber molecules at the 3′-end G-tetrad; (b) H-bond contacts established by the HOH9 water molecule in the 3′-end binding site; (c) top view of the two Ber molecules stacked on to the 3′-end G-tetrad: skeleton and Fo-Fc omit map for each Ber contoured at 1.5σ level.
It is to be noted that the absence of a planarity of the 3′-end tetrad is a common feature for the intramolecular telomeric G-quadruplex and it is present in all the to date known crystal structure of both native and complexed DNA, where the tetrad is slightly funnel-shaped (dihedral angles between adjacent guanines ranging from 12° to 18°) (9,11,34). Instead, the strong interaction established in the here described structure between the ligand molecules and the 3′-end tetrad, makes the guanines in each pair (G5/G11 or G17/G23) indeed coplanar (dihedral angles falling in the range 5–6°).
In a similar way to the ligands positioned on the 5′-end, the two Ber molecules define a four-sided polygon, but here their methoxy groups point in the opposite direction (Figure 4c). The molecules are in contact through CH…O H-bonds (C…O distance 2.6 Å; Figure 4b). The open central channel is again visible, and occupied by a disordered water molecule, which lies 3.3 Å far from the nearest potassium ion. This water molecule forms H-bonds with the O6 carbonyl oxygen from the guanine residues (O…O mean distance 3.0 Å) and CH…O interactions (C…O mean distance 3.2 Å) with the Ber molecules contributing to fix their orientation. Instead the symmetry featuring the 5′-end binding site, as well as the absence of water molecules, could explain the disorder observed for the ligands in this binding site.
DISCUSSION
The structure of the complex here reported identifies two different binding sites, each containing two ligand molecules stacked side-by-side onto the tetrad. The overall 3:1 binding stoichiometry here found differs from the ligand to quadruplex molar ratio previously reported in literature for the adducts formed in diluted K+ solution between Ber derivatives and the 22-mer telomeric DNA. These complexes exhibit a 1:1 binding stoichiometry (19–24) with DNA, which is present in solution as a mixture of hybrid1 and hybrid2 topology.
The two Ber molecules stacked on the tetrads represent a significant novelty when compared with the structural data available to date regarding unimolecular G-quadruplex formed by telomeric DNA. Indeed, a ligand to tetrad ratio different from 1:1 has only been previously observed in two cases. The first refers to the X-ray structure of tetramolecular adduct daunomycin/d(TGGGGT), where three ligand molecules on a G-tetrad were found (35), and the second describes an acridine derivative adduct with the TERRA RNA (7). The latter complex is characterized by a 2:1 ligand to G-tetrad molar ratio stoichiometry, but the ligand gives rise to a 1:1 molar ratio when complexed with telomeric DNA in an all parallel arrangement (7). The stoichiometric ratio greater than 1:1 found for the adducts of (7) and (35) is, most likely, favored by the folding topology of the target and ultimately on how the loops connect the tetrads. In these complexes, as well as in our X-ray solved complex where G-DNA is in parallel conformation, loops connect opposite ends of the quadruplex leaving the tetrads completely available for binding. Among the foldings reported to date for telomeric intramolecular G-quadruplex, the parallel is the only one featuring both external tetrads which are not hindered by the adenine and thymine residues. Instead, the hybrid type, typical of diluted K+ containing solutions, presents partially not accessible tetrads. Because for some time the parallel G-quadruplex has been considered to be exclusively distinctive of the solid state and to be induced by the crystal packing forces, one could doubt its biological relevance. However, recent literature has reported that under crowding conditions telomeric sequences might co-exist as an equilibrium mixture of different kinds of arrangements, also including the parallel one (36–38). In addition, while the antiparallel G-quadruplex conformation is induced by a number of compounds (39), ligands are known which are able to induce, in solution, the parallel arrangement of telomeric DNA (40). Thus, the question about the most likely possible form of the telomeric DNA sequence in vivo or at least in experimental settings simulating the in vivo condition (41–44), is still strongly debated and researchers are far from reaching a convergence of opinions. Moreover, it has been demonstrated that when working with significantly longer telomeric sequences than those normally used in experimental studies (in the range 21–26 nt), the very same ligand molecule can give rise to complexes featuring a ligand to DNA molar ratio that is different from those found in monomer forming sequences (45). This behavior would suggest that the quadruplex units along a multimer structure are not completely independent.
The three-dimensional arrangement of the full length telomeric overhang can lead the G-tetrads of not consecutive G-quadruplex units into stacking distance. This allows the formation of additional putative binding sites besides the quadruplex–quadruplex interfaces of consecutive units. The driving forces of the interactions that are established to these interfaces are mainly of electrostatic and van der Waals types, not affected by the fact that the facing ends are of 3′–5′, 3′–3′ or 5′–5′ type (12). Therefore, given the nature of these interactions, the two molecules of Ber, that in our experiments interact with the 5′-ends of two short independent G-quadruplex units (23 nucleotide bases), may establish in vivo stacking interactions onto 3′ or 5′ G-tetrads of non-consecutive G-quadruplexes belonging to long telomeric sequences. Indeed, it has been proposed that telomeric DNA and RNA might afford different higher-order arrangements of parallel-stranded G-quadruplexes (12). These include the 5′–5′ stacking mode we have experimentally observed, and also the 3′–3′ and the 3′–5′ one.
The presence of two Ber molecules stacked side-by-side onto the tetrad is due, most likely, to how small the molecule is compared with other ligands whose telomere/ligand structure is reported in literature (5,6,9). As observed in the Results section, the Ber-molecule gives rise, both in 3′- and 5′-end binding site, to an optimal dimensional match with a pair of guanines, so maximizing the π-stacking interaction between the aromatic moieties and leaving room in the binding site to lodge a second molecule of ligand. The really strong interaction can also be inferred from the coplanarity of the G5/G11 and G17/G23 pairs and from the modified conformation of the 3′-end tetrad with respect to the till now reported intramolecular telomeric G-quadruplex structures, at least in the solid state.
The size of the Ber molecule may also explain its G-quadruplex over duplex selectivity (46). In fact, while the two Ber molecules can be easily located on the telomeric tetrad, the results obtained from the X-ray diffraction analysis of the d(CGTACG)-Ber complex point out that this planar ligand is too wide to give rise to a canonical intercalation in the ds-DNA, and is forced to intercalate at the junction of two consecutive distorted double helices (25).
Summarizing, this study presents the first X-ray structure of the adduct formed between Ber and the telomeric DNA, and provides a key to interpret the observed selectivity of the ligand toward the G-quadruplex compared with the DNA double helix. The solved structure highlights a stoichiometric ratio that is different from the one observed in diluted solution for the adducts with the hybrid type folding. In particular, it shows the presence of two Ber molecules stacked side-by-side onto the tetrad. These data, together with the recent literature that considers the parallel conformation as one of the biologically relevant folding topology for telomeric DNA, show new possibilities for ligand to DNA quadruplex binding, that have never been considered up to now. In our opinion, this interaction mode provides an important clue for interpreting binding data obtained with experimental settings which simulate the in vivo conditions.
ACCESSION NUMBERS
PDB accession number 3R6R.
FUNDING
Italian Ministero dell’Università e della ricerca (MIUR). Funding for open access charge: FUNZ. BILMAYA MED 1.12.05.01.
Conflict of interest statement. None declared.
REFERENCES
- 1.Balasubramanian S, Neidle S. G-quadruplex nucleic acids as therapeutic targets. Curr. Opin. Chem. Biol. 2009;13:345–353. doi: 10.1016/j.cbpa.2009.04.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neidle S. Human telomeric G-quadruplex: The current status of telomeric G-quadruplexes as therapeutic targets in human cancer. FEBS J. 2010;277:1118–1125. doi: 10.1111/j.1742-4658.2009.07463.x. [DOI] [PubMed] [Google Scholar]
- 3.Ou TM, Lu YJ, Tan JH, Huang ZS, Wong KY, Gu LQ. G-Quadruplexes: targets in anticancer drug design. ChemMedChem. 2008;3:690–713. doi: 10.1002/cmdc.200700300. [DOI] [PubMed] [Google Scholar]
- 4.Monchaud D, Teulade-Fichou MP. A Hitchhiker's guide to G-quadruplex ligands. Org. Biomol. Chem. 2008;6:627–636. doi: 10.1039/b714772b. [DOI] [PubMed] [Google Scholar]
- 5.Campbell NH, Karim NHA, Parkinson GN, Gunaratnam M, Petrucci V, Todd AK, Vilar R, Neidle S. Molecular basis of structure-activity relationships between salphen metal complexes and human telomeric DNA quadruplexes. J. Med. Chem. 2012;55:209–222. doi: 10.1021/jm201140v. [DOI] [PubMed] [Google Scholar]
- 6.Campbell NH, Parkinson GN, Reszka AP, Neidle S. Structural basis of DNA quadruplex recognition by an acridine drug. J. Am. Chem. Soc. 2008;130:6722–6724. doi: 10.1021/ja8016973. [DOI] [PubMed] [Google Scholar]
- 7.Collie GW, Sparapani S, Parkinson GN, Neidle S. Structural basis of telomeric RNA quadruplex-acridine ligand recognition. J. Am. Chem. Soc. 2011;133:2721–2728. doi: 10.1021/ja109767y. [DOI] [PubMed] [Google Scholar]
- 8.Gavathiotis E, Heald RA, Stevens MFG, Searle MS. Drug recognition and stabilisation of the parallel-stranded DNA quadruplex d(TTAGGGT)4 containing the human telomeric repeat. J. Mol. Biol. 2003;334:25–36. doi: 10.1016/j.jmb.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 9.Parkinson GN, Cuenca F, Neidle S. Topology conservation and loop flexibility in quadruplex-drug recognition: crystal structures of inter- and intramolecular telomeric DNA quadruplex-drug complexes. J. Mol. Biol. 2008;381:1145–1156. doi: 10.1016/j.jmb.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 10.Parkinson GN, Ghosh R, Neidle S. Structural basis for binding of porphyrin to human telomeres. Biochemistry. 2007;46:2390–2397. doi: 10.1021/bi062244n. [DOI] [PubMed] [Google Scholar]
- 11.Collie GW, Promontorio R, Hampel SM, Micco M, Neidle S, Parkinson GN. Structural basis for telomeric G-quadruplex targeting by naphthalene diimide ligands. J. Am. Chem. Soc. 2012;134:2723–2731. doi: 10.1021/ja2102423. [DOI] [PubMed] [Google Scholar]
- 12.Phan AT. Human telomeric G-quadruplex: structures of DNA and RNA sequences. FEBS J. 2010;277:1107–1117. doi: 10.1111/j.1742-4658.2009.07464.x. [DOI] [PubMed] [Google Scholar]
- 13.Bhadra K, Kumar GS. Isoquinoline alkaloids and their binding with DNA: calorimetry and thermal analysis applications. Mini Rev. Med. Chem. 2010;10:1235–1247. doi: 10.2174/13895575110091235. [DOI] [PubMed] [Google Scholar]
- 14.Kuo C-L, Chi C-W, Liu T-Y. The anti-inflammatory potential of berberine in vitro and in vivo. Cancer Lett. 2004;203:127–137. doi: 10.1016/j.canlet.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Maiti M, Kumar GS. Molecular aspects on the interaction of protoberberine, benzophenanthridine, and aristolochia group of alkaloids with nucleic acid structures and biological perspectives. Med. Res. Rev. 2007;27:649–695. doi: 10.1002/med.20087. [DOI] [PubMed] [Google Scholar]
- 16.Maiti M, Kumar GS. Polymorphic nucleic acid binding of bioactive isoquinoline alkaloids and their role in cancer. J. Nucleic Acids. 2009;2010:pii: 593408. doi: 10.4061/2010/593408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satou T, Akao N, Matsuhashi R, Koike K, Fujita K, Nikaido T. Inhibitory effect of isoquinoline alkaloids on movement of second-stage larvae of Toxocara canis. Biol. Pharm. Bull. 2002;25:1651–1654. doi: 10.1248/bpb.25.1651. [DOI] [PubMed] [Google Scholar]
- 18.Singhal KC. Anthelmintic activity of berberine hydrochloride against Syphacia obvelata in mice. Indian J. Exp. Biol. 1976;14:345–347. [PubMed] [Google Scholar]
- 19.Bhadra K, Suresh KG. Interaction of berberine, palmatine, coralyne, and sanguinarine to quadruplex DNA: A comparative spectroscopic and calorimetric study. Biochim. Biophys. Acta. 2011;1810:485–496. doi: 10.1016/j.bbagen.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 20.Franceschin M, Rossetti L, D'Ambrosio A, Schirripa S, Bianco A, Ortaggi G, Savino M, Schultes C, Neidle S. Natural and synthetic G-quadruplex interactive berberine derivatives. Bioorg. Med. Chem. Lett. 2006;16:1707–1711. doi: 10.1016/j.bmcl.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Ma Y, Ou TM, Hou JQ, Lu YJ, Tan JH, Gu LQ, Huang ZS. 9-N-Substituted berberine derivatives: Stabilization of G-quadruplex DNA and down-regulation of oncogene c-myc. Bioorg. Med. Chem. Lett. 2008;16:7582–7591. doi: 10.1016/j.bmc.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 22.Arora A, Balasubramanian C, Kumar N, Agrawal S, Ojha RP, Maiti S. Binding of berberine to human telomeric quadruplex—spectroscopic, calorimetric and molecular modeling studies. FEBS J. 2008;275:3971–3983. doi: 10.1111/j.1742-4658.2008.06541.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhang WJ, Ou TM, Lu YJ, Huang YY, Wu WB, Huang ZS, Zhou JL, Wong KY, Gu LQ. 9-Substituted berberine derivatives as G-quadruplex stabilizing ligands in telomeric DNA. Bioorg. Med. Chem. 2007;15:5493–5501. doi: 10.1016/j.bmc.2007.05.050. [DOI] [PubMed] [Google Scholar]
- 24.Ma Y, Ou TM, Tan JH, Hou JQ, Huang SL, Gu LQ, Huang ZS. Synthesis and evaluation of 9-O-substituted berberine derivatives containing aza-aromatic terminal group as highly selective telomeric G-quadruplex stabilizing ligands. Bioorg. Med. Chem. Lett. 2009;19:3414–3417. doi: 10.1016/j.bmcl.2009.05.030. [DOI] [PubMed] [Google Scholar]
- 25.Ferraroni M, Bazzicalupi C, Bilia AR, Gratteri P. X-Ray diffraction analyses of the natural isoquinoline alkaloids Berberine and Sanguinarine complexed with double helix DNA d(CGTACG) Chem. Commun. 2011;47:4917–4919. doi: 10.1039/c1cc10971e. [DOI] [PubMed] [Google Scholar]
- 26.Campbell NH, Parkinson GN. Crystallographic studies of quadruplex nucleic acids. Methods. 2007;43:252–263. doi: 10.1016/j.ymeth.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data, Joint CCP4 + ESF-EAMCB. Newsletter Protein Crystallogr. 1992;26 [Google Scholar]
- 28.Leslie AGW, Powell HR. Processing diffraction data with Mosflm. In: Read RJ, Sussman JL, editors. Evolving Methods for Macromolecular Crystallography. Vol. 245. Springer: Physics and Astronomy NATO Series; 2007. pp. 41–51. [Google Scholar]
- 29.Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 30.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 31.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 32.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macrae CF, Bruno IJ, Chisholm JA, Edgington PR, McCabe P, Pidcock E, Rodriguez-Monge L, Taylor R, Van De Streek J, Wood PA. Mercury CSD 2.0—new features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008;41:466–470. [Google Scholar]
- 34.Parkinson GN, Lee MPH, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 35.Clark GR, Pytel PD, Squire CJ, Neidle S. Structure of the first parallel DNA quadruplex-drug complex. J. Am. Chem. Soc. 2003;125:4066–4067. doi: 10.1021/ja0297988. [DOI] [PubMed] [Google Scholar]
- 36.Hänsel R, Löhr F, Foldynová-Trantírková S, Bamberg E, Trantírek L, Dötsch V. The parallel G-quadruplex structure of vertebrate telomeric repeat sequences is not the preferred folding topology under physiological conditions. Nucleic Acids Res. 2011;39:5768–5775. doi: 10.1093/nar/gkr174. and references cited therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heddi B, Phan AT. Structure of human telomeric DNA in crowded solution. J. Am. Chem. Soc. 2011;133:9824–9833. doi: 10.1021/ja200786q. [DOI] [PubMed] [Google Scholar]
- 38.Martino L, Pagano B, Fotticchia I, Neidle S, Giancola C. Shedding light on the interaction between TMPyP4 and human telomeric quadruplexes. J. Phys. Chem. B. 2009;113:14779–14786. doi: 10.1021/jp9066394. [DOI] [PubMed] [Google Scholar]
- 39.Georgiades SN, Abd Karim NH, Suntharalingam K, Vilar R. Interaction of metal complexes with G-quadruplex DNA. Angew. Chem.-Int. Edit. 2010;49:4020–4034. doi: 10.1002/anie.200906363. [DOI] [PubMed] [Google Scholar]
- 40.Nicoludis JM, Barrett SP, Mergny JL, Yatsunyk LA. Interaction of human telomeric DNA with N-methyl mesoporphyrin IX. Nucleic Acids Res. 2012;40:5432–5447. doi: 10.1093/nar/gks152. and references cited therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cang X, Sponer J, Cheatham TE., III Insight into G-DNA structural polymorphism and folding from sequence and loop connectivity through free energy analysis. J. Am. Chem. Soc. 2011;133:14270–14279. doi: 10.1021/ja107805r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dao NT, Haselsberger R, Michel-Beyerle ME, Phan AT. Following G-quadruplex formation by its intrinsic fluorescence. FEBS Lett. 2011;585:3969–3977. doi: 10.1016/j.febslet.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 43.Petraccone L, Malafronte A, Amato J, Giancola C. G-quadruplexes from human telomeric DNA: how many conformations in PEG containing solutions? J. Phys. Chem. B. 2012;116:2294–2305. doi: 10.1021/jp209170v. [DOI] [PubMed] [Google Scholar]
- 44.Xue Y, Liu JQ, Zheng KW, Kan ZY, Hao YH, Tan Z. Kinetic and thermodynamic control of G-quadruplex folding. Angew. Chem. Int. Edit. 2011;50:8046–8050. doi: 10.1002/anie.201101759. [DOI] [PubMed] [Google Scholar]
- 45.Cummaro A, Fotticchia I, Franceschin M, Giancola C, Petraccone L. Binding properties of human telomeric quadruplex multimers: a new route for drug design. Biochimie. 2011;93:1392–1400. doi: 10.1016/j.biochi.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 46.Bessi I, Bazzicalupi C, Richter C, Jonker HRA, Saxena K, Sissi C, Chioccioli M, Bianco S, Bilia AR, Schwalbe H, et al. Spectroscopic, molecular modeling, and NMR-spectroscopic investigation of the binding mode of the natural alkaloids Berberine and Sanguinarine to human telomeric G-quadruplex DNA. ACS Chem. Biol. 2012;7:1109–1119. doi: 10.1021/cb300096g. [DOI] [PubMed] [Google Scholar]