Abstract
Progression to malignancy requires that cells overcome senescence and switch to an immortal phenotype. Thus, exploring the genetic and epigenetic changes that occur during senescence/immortalization may help elucidate crucial events that lead to cell transformation. In the present study, we have globally profiled DNA methylation in relation to gene expression in primary, senescent and immortalized mouse embryonic fibroblasts. Using a high-resolution genome-wide mapping technique, followed by extensive locus-specific validation assays, we have identified 24 CpG islands that display significantly higher levels of CpG methylation in immortalized cell lines as compared to primary murine fibroblasts. Several of these hypermethylated CpG islands are associated with genes involved in the MEK–ERK pathway, one of the most frequently disrupted pathways in cancer. Approximately half of the hypermethylated targets are developmental regulators, and bind to the repressive Polycomb group (PcG) proteins, often in the context of bivalent chromatin in mouse embryonic stem cells. Because PcG-associated aberrant DNA methylation is a hallmark of several human malignancies, our methylation data suggest that epigenetic reprogramming of pluripotency genes may initiate cell immortalization. Consistent with methylome alterations, global gene expression analysis reveals that the vast majority of genes dysregulated during cell immortalization belongs to gene families that converge into the MEK–ERK pathway. Additionally, several dysregulated members of the MAP kinase network show concomitant hypermethylation of CpG islands. Unlocking alternative epigenetic routes for cell immortalization will be paramount for understanding crucial events leading to cell transformation. Unlike genetic alterations, epigenetic changes are reversible events, and as such, can be amenable to pharmacological interventions, which makes them appealing targets for cancer therapy when genetic approaches prove inadequate.
INTRODUCTION
Cellular senescence is characterized by irreversible loss of cell proliferation, and is considered a powerful tumor suppressor mechanism to eliminate damaged cells prone to transformation (1,2). Cellular senescence has been shown to function as a barrier to somatic cell reprogramming (3), and to play a role in organismal aging (4). Features of senescence are often detected in pre-malignant lesions of mice and humans, but not in their respective invasive/malignant tumors. This suggests a role for senescent cells in the initiation and progression of tumorigenesis (2,5,6). For example, senescent cells are known to give rise to neighboring cancer cells through paracrine signaling with their microenvironment (7–9). Alternatively, cells may bypass or escape senescence and re-enter the cell cycle to become transformed following the inactivation of key components of the senescence-promoting pathways (2,4,10). Thus, investigating the underlying mechanisms of senescence bypass and immortalization may help elucidate tumor initiation and progression.
Despite obvious differences between human and mouse, key aspects of cellular senescence in the two species are well-conserved during evolution (11,12). Mouse models have been shown to provide mechanistic insights into the chain of events that leads to senescence bypass and immortalization of relevance to human carcinogenesis (2,13–15). Mouse embryonic fibroblasts (MEFs) are a validated model system to study senescence bypass and spontaneous immortalization (16,17). Primary MEFs have a limited growth capacity and undergo stress-induced senescence, presumably due to oxidative damage caused by standard cell culture conditions (10,18,19). Upon prolonged culturing and passaging, however, MEFs can override the senescence block and become spontaneously immortalized, partly due to mutation/deletion of the p53/p19Arf pathway (16,18,20).
The phenotypically normal Hupki (human p53 knock-in) mouse strain, harboring the humanized p53 gene (15), has extensively been used as a model system to study the underlying mechanisms of senescence bypass and immortalization (13,15,21,22). Using a library of several hundred immortalized cell lines, Whibley et al. (17) have demonstrated that MEFs derived from Hupki mice undergo senescence and become immortalized in the same way as their wild-type counterparts, i.e., embryonic fibroblasts from standard laboratory mice with a murine Trp53 gene. In both cases, immortalized cells typically harbor either a missense point mutation in the p53 gene leading to loss of p53 transactivation, or a biallelic deletion at the p19Arf locus. Surprisingly, however, more than half of the immortalized cell lines examined had neither a p53 mutation nor loss of the p19Arf activity (17). In some of these cell lines, genetic defects in tumor suppressor pathways, other than those affecting the p53/p19Arf axis, may contribute to escape or release from the senescence block into an immortal phenotype (17,23). Furthermore, active acquisition of epigenetic modifications, such as DNA methylation at regulatory CpG sites may interfere with the expression of key senescence genes, and pave the way to immortalization and cell transformation (23,24). To explore novel epigenetic/genetic routes that may modulate senescence bypass and immortalization, we have globally profiled DNA methylation in relation to gene expression in primary, senescent and immortalized fibroblasts derived from both the Hupki and wild-type mice. Using microarray-based approaches together with locus/gene-specific validation assays, we have constructed the whole methylome and transcriptome of MEFs at three distinct proliferation stages, including primary, senescent and immortalized cell lines.
MATERIALS AND METHODS
Isolation and immortalization of primary MEFs
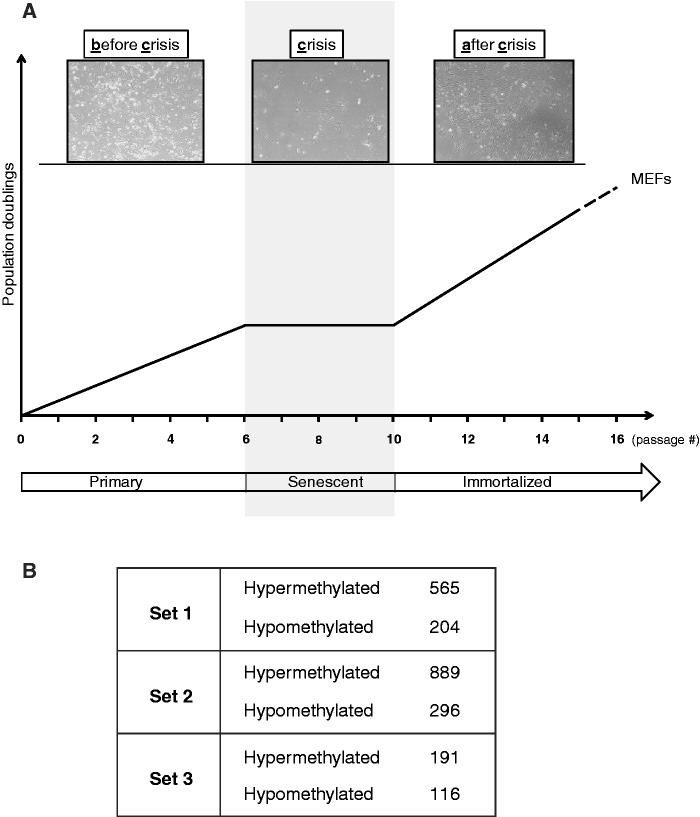
Primary MEFs were obtained from embryos derived from either Hupki (Sets 1, 2 and 3) or wild-type (Sets 5 and 7) mice according to standard procedures (15). Briefly, mouse embryos were harvested in utero at 13.5 days of gestation. Following the removal of the head and internal organs, embryo carcasses were roughly minced and digested for five times, 5 min each, with 0.25% trypsin at 37°C. Remaining tissue clumps were disaggregated by passage through a syringe equipped with an 18-gauge needle, and vigorous pipetting. Homogenous cell suspensions were then transferred to tissue culture flasks, and cultured in standard Dulbecco Modified Eagle’s Medium (DMEM, Irvine Scientific, Santa Ana, CA), supplemented with 10% fetal calf serum (FCS), non-essential amino acids and 0.1 mM β-mercaptoethanol. Following 1–3 days of incubation at 37°C, confluent cultures were trypsinized and frozen, at 2-3 × 106 cells per vial. To establish cell lines, cryopreserved embryonic fibroblasts (passage 0) were thawed and seeded onto T75 flasks containing DMEM supplemented with 10% FCS. At 80% confluence, the cells were trypsinized and re-seeded onto 6-well culture dishes or small flasks (12.5 cm2) at a density of ∼1 × 105 cells per well/flask in DMEM plus 10% FCS. Cultures were maintained under standard conditions (37°C, 5% CO2 in a humidified atmosphere), and passaged serially when reaching confluence [pass to: 1:3 or 1:4, as described previously (17)]. During crisis, which typically occurs by passage 5-6 and lasts up to 3 weeks, culture medium was changed twice a week. Following the crisis, over 95% of all cultures became spontaneously immortalized. For all experiments, cells were harvested at different passages before-, during- and after crisis, as outlined in Figure 1A, and genomic DNA/RNA was isolated according to standard procedures (17).
Figure 1.
Outline of the passaging protocol and identification of differentially methylated targets in immortalized MEFs. (A) MEFs were cultured in vitro as described in the text and harvested at low-passage (before crisis/primary), during crisis (crisis/senescent), and following immortalization (after crisis/immortalized). DNA/RNA was isolated according to standard procedures (17). (B) Aberrantly mCG islands were identified by MIRA-microarray analysis. Three independently established immortalized murine cell lines are shown (Set 1, Set 2 and Set 3). CpG islands were considered as hypermethylated when the immortalized cell line showed methylation peaks that were absent in all controls (primary MEFs). The minimum difference between immortalized and primary MEFs was ≥2-fold. For hypomethylated targets, reverse criteria were used. The numbers of methylation peaks, per each dataset, are shown.
Genome-wide DNA methylation profiling by MIRA-microarray analysis
We used the methylated CpG (mCG) island recovery assay (MIRA) in combination with microarray analysis (25,26) to globally profile DNA methylation in primary, senescent and immortalized MEFs. As a pull-down assay for enrichment of mCG islands, the MIRA is based on the ability of the methyl-CpG binding 2b (MBD2b) protein to bind methylated-CpG dinucleotides, while this reaction is enhanced in the presence of MBD3L1 protein (27). The MIRA-enriched- and input (non-enriched) DNA fractions were amplified by polymerase chain reaction (PCR), and subsequently labeled and hybridized to the Roche NimbleGen mouse methylation microarrays (Roche NimbleGen Inc., Indianapolis, IN) to interrogate the entire CpG-island battery of the mouse genome (see Supplementary Data) (25,26). The accession number for DNA methylation microarray data is GSE39034.
Single gene/locus DNA methylation analysis by COBRA and bisulfite sequencing
We verified the methylation status of individual target loci/genes identified by MIRA-microarray analysis using the conventional combined bisulfite restriction analysis (COBRA) (28), and sodium bisulfite sequencing (29). The primer sets used for PCR amplification of all the analysed targets are listed in Supplementary Table S1 (see Supplementary Data).
DNA methylation analysis of repetitive DNA elements
We have specifically determined the methylation status of major repetitive DNA elements, including long interspersed nuclear elements (LINE L1), intracisternal A particle long terminal repeat retrotransposons (IAP-LTR), and short interspersed nuclear elements (SINE B1) (30–32), in the genome of primary, senescent or immortalized MEFs using a bisulfite-based sequencing analysis (see Supplementary Data) (33).
Genome-wide gene expression profiling by Affymetrix-microarray analysis
We used the GeneChip® Mouse Genome 430 2.0 Array (Affymetrix Inc., Santa Clara, CA) to establish the global gene expression profile of MEFs at three different stages (before crisis, crisis, after crisis). Currently, this microarray platform is the most comprehensive mouse genome expression array, which enables interrogation of over 39 000 transcripts and variants from over 34 000 well-characterized mouse genes (see Supplementary Data). The accession number for gene expression microarray data is GSE39034.
Quantitative real-time PCR
Standard quantitative real-time-PCR (qRT-PCR) was used to confirm the transcriptional status of individual target genes identified by microarray analysis (34). The primer sets used for qRT-PCR are listed in Supplementary Table S1 (see Supplementary Data).
RESULTS
Genome-wide detection of aberrant DNA methylation following senescence bypass and immortalization
We have used the MIRA in combination with microarray analysis (25,26) to detect changes in the global methylation pattern of murine cells undergoing immortalization. MEFs were cultured in vitro as described in ‘Materials and Methods’ section (outlined in Figure 1A), and harvested at different passages (i.e., before crisis (bc): primary, during crisis (c): senescent and after crisis (ac): immortalized). Following pull-down of mCG islands by MIRA, the enriched- and input (non-enriched) DNA fractions were labeled, mixed and hybridized to the mouse CpG island plus promoter tiling arrays (Roche NimbleGen Inc.). Using standard algorithms for peak calling (see Supplementary Data), we have identified 24 CpG islands that were aberrantly methylated in three independent sets of immortalized MEF cell lines, but were methylation-free in the correspondent low-passage primary cells (Table 1). The identified CpG islands mapped to intergenic regions (21%), within gene bodies (38%), at promoter regions (29%) and near the 3′-end of genes (12%). Using a combination of the Ingenuity Pathway Analysis® (IPA®: v 9.0) and the Database for Annotation, Visualization and Integrated Discovery (DAVID: v 6.7) (35), we obtained gene ontology (GO) information for the annotated genes identified as hypermethylated targets in immortalized MEFs. Functional annotation analysis revealed that gene targets involved in intracellular signaling cascade (Rem 2, Rasd1, Wnk4, Adra1d, Mapk11, Plcd3; P < 0.007), small GTPase-mediated signal transduction (Rem2, Rasd1, Mapk11; P < 0.04), as well as neuropeptide signaling pathway (Eltd1, Lphn3; P < 0.09) were particularly enriched. Of significance, several genes involved in the MEK–ERK pathway, one of the most frequently disrupted pathways in cancer (36–39), showed progressive DNA hypermethylation as the cells bypassed senescence and became immortalized. Using reverse criteria for data analysis, we detected no significant loss of CpG island methylation across the three independently established immortalized cell lines relative to counterpart primary cells.
Table 1.
Hypermethylated targets detected in immortalized versus primary fibroblasts
| Target no. | Gene symbol | Description | CGI location | Position relative to gene | Strand | Enrichment ratio |
|---|---|---|---|---|---|---|
| 1 | chr19:59543322-59543961 | 6.40 | ||||
| 2 | Plcd3 | phospholipase C, delta 3 | chr11:102941383-102942047 | Intragenic | − | 5.99 |
| 3 | Wnk4 | WNK lysine deficient protein kinase 4 | chr11:101126459-101126913 | Intragenic | + | 5.09 |
| 4 | Adra1d | adrenergic receptor, alpha 1d | chr2:131371751-131372295 | Downstream | − | 5.01 |
| 5 | chr13:56854334-56854768 | 4.85 | ||||
| 6 | Phox2a | paired-like homeobox 2a | chr7:108969059-108969729 | Intragenic | + | 4.80 |
| 7 | Rem2 | Rad- and gem-related GTP-binding protein 2 | chr14:55098125-55098574 | Downstream | + | 4.78 |
| 8 | chr7:77514032-77515167 | 4.71 | ||||
| 9 | Mapk11 | mitogen-activated protein kinase 11 | chr15:88976796-88977465 | Intragenic | − | 4.67 |
| 10 | Sh2d5 | SH2 domain containing 5 | chr4:137806496-137806960 | Upstream | + | 4.25 |
| 11 | Cldn5 | claudin 5 | chr16:18777229-18778053 | Intragenic | + | 4.20 |
| 12 | Hoxc9 | homeobox C9 | chr15:102807421-102808173 | Upstream | + | 4.20 |
| 13 | Ntn3 | netrin 3 | chr17:24345612-24346041 | Upstream | − | 4.19 |
| 14 | Fbrsl1 | chr5:110815430-110815859 | Intragenic | − | 4.16 | |
| 15 | Ddn | dendrin | chr15:98637075-98637514 | Intragenic | − | 3.85 |
| 16 | Amh | anti-Mullerian hormone | chr10:80269858-80270292 | Downstream | + | 3.67 |
| 17 | Eltd1 | EGF, latrophilin seven transmembrane domain containing 1 | chr3:151100756-151101200 | Upstream | + | 3.57 |
| 18 | Rasd1 | RAS, dexamethasone-induced 1 | chr11:59777219-59777763 | Intragenica | − | 3.48 |
| 19 | Adra1d | adrenergic receptor, alpha 1d | chr2:131386864-131387318 | Upstream | − | 3.46 |
| 20 | Lphn3 | latrophilin 3 | chr5:81758777-81759233 | Intragenic | + | 3.38 |
| 21 | Akap5 | A kinase (PRKA) anchor protein 5 | chr12:77425710-77426441 | Upstream | + | 3.19 |
| 22 | Ccdc114 | coiled-coil domain containing 114 | chr7:53179579-53180028 | Upstream | + | 3.06 |
| 23 | chr11:21903730-21904098 | 2.96 | ||||
| 24 | chr15:102795737-102796201 | 2.86 |
aDownstream of Med9 gene.
Verification of aberrantly methylated targets by COBRA and bisulfite sequencing
To validate the DNA methylation microarray results, we have randomly selected and analysed several of the targets identified by MIRA-microarray analysis using both the COBRA and bisulfite DNA sequencing analysis (28,29). Representative results of the COBRA assay are shown in Figures 2C and 3C and Supplementary Figures S1–S4. Here, we have inspected the methylation status of CpG islands associated with the Plcd3 gene (target #2), Phox2a (target #6), Mapk11 (target #9), Cldn5 (target #11) and Rasd1 (target #18) genes. As shown in Table 1, these CpG-rich loci displayed enrichment ratios, by MIRA-microarray analysis, ranging from 5.99 to 3.48, respectively (on a linear scale, immortalized versus low-passage fibroblasts). COBRA analysis of the above-specified targets confirms the microarray results, indicating that novel epigenetic marks, i.e., CpG methylation, are established during cell immortalization (Figures 2A and 3A and Supplementary Figures S1A–S3A). Of note, these methylation changes were consistent in both the Hupki and wild-type immortalized MEFs (Supplementary Figures S3 and S4).
Figure 2.
Hypermethylated CpG island associated with the Phox2a locus. (A) Visualization of the NimbleGen array data for immortalized (in red) and primary MEFs (in green). Three independently established immortalized murine cell lines are shown (Set 1, Set 2 and Set 3). At this level of resolution, each peak corresponds to a CpG island. The methylation signal, obtained by the NimbleScan software, is plotted along the chromosome as a P-value on the y-axis (starting from 0 when P-value is 1). The P-value is derived from the Kolmogorov–Smirnov test comparing the log2 enrichment ratios between MIRA and input within a 750 bp window. (B) Snapshot of the UCSC Genome Browser indicating the location of the Phox2a gene. CpG islands are shown as green boxes. The locations of the H3K4me3, CTCF and Pol2-binding sites, are shown for low-passage MEFs, based on ENCODE/LICR published databases (http://genome.ucsc.edu/ENCODE/downloadsMouse.html). H3K4me3 and Pol2 signals are strong indicators of active promoters, whereas CTCF-binding sites are considered as a mark for potential insulator elements. (C) Genomic DNA from primary (1bc, 2bc, 3bc), senescent (2c, 3c) and immortalized (1ac, 2ac, 3ac) MEFs was treated with sodium bisulfite, and the CpG island within the Phox2a gene was amplified with gene-specific primers, and subjected to COBRA. Digested fragments on the gel are indicative of methylated BstUI restriction sites (5′-CGCG) within the CpG island. Mouse genomic DNA was methylated in vitro with the SssI methyltransferase, and served as positive control (Pos). (+) and (−) represent the presence and absence, respectively, of the BstUI restriction enzyme in the reaction mix. M = 100 bp ladder DNA marker. Given the limited amount of DNA available from sample 1c, bisulfite sequencing was performed only. (D) The extent of CpG methylation within the Phox2a CpG island was determined by sodium bisulfite sequencing in primary (1bc, 2bc, 3bc; left panel), senescent (1c, 2c, 3c; middle panel) and immortalized (1ac, 2ac, 3ac; right panel) MEFs. The sequencing results of up to 10 independent clones, and the respective percentage of methylation per sample are shown. Data were analysed using the web-based software, QUantification tool for Microarray Analysis (QUMA) (http://quma.cdb.riken.jp/). Open and closed circles represent unmethylated and mCG dinucleotides, respectively.
Figure 3.
Hypermethylated CpG island associated with the Mapk11 locus. (A) Visualization of the NimbleGen array data for immortalized (in red) and primary (in green) MEFs. Three independently established immortalized murine cell lines are shown (Set 1, Set 2 and Set 3). The methylation signal is plotted as a P-value on the y-axis, and was calculated as described in legend for Figure 2. (B) Snapshot of the UCSC Genome Browser indicating the location of the Mapk11 gene. The corresponding CpG island is shown as a green box together with the overlapping binding site for CTCF, and potential transcriptional initiation sites. (C) COBRA analysis of the mCG island associated with the Mapk11 gene in primary (1bc, 2bc, 3bc), senescent (2c, 3c) and immortalized (1ac, 2ac, 3ac) MEFs. Digested fragments on the gel are indicative of methylated BstUI restriction sites (5′-CGCG) within the CpG island (see, legend for Figure 2). (D) The extent of CpG methylation within the Mapk11 CpG island was determined by sodium bisulfite sequencing in primary (1bc, 2bc, 3bc; left), senescent (1c, 2c, 3c; middle) and immortalized (1ac, 2ac, 3ac; right) MEFs. The sequencing results of several independent clones, and the respective percentage of methylation per sample are shown.
To obtain a detailed methylation map for each CpG within the CpG islands, we also performed bisulfite sequencing on the targets analysed by COBRA. The sequencing results of the Phox2a-, Mapk11-, Rasd1- and Cldn5-associated CpG islands are shown in Figures 2D and 3D, and Supplementary Figures S1D and S2D. In all cases, we confirmed DNA methylation differences (up to 6-fold) between immortalized and primary MEFs, mirroring the differences observed in the microarray and COBRA analyses (Table 1). As shown in Figures 2D and 3D and Supplementary Figures S1D and S2D, aberrant methylation at the CpG islands is established mostly during the crisis stage, suggesting that DNA methylation of crucial genes involved in the maintenance of senescence may drive cells to the immortal phenotype. We did not detect any gene-specific CpG island that was hypomethylated across all the three immortalized cell lines. However, when less stringent criteria were used in the analysis (a methylation peak present in all primary cells but absent in two out of the three immortalized cell lines), we found several hypomethylated CpG islands common to two immortalized cell lines. Representative examples of hypomethylated targets are shown in Figure 4, which combines bisulfite sequencing results for the Neat1 gene (Figure 4A) and COBRA data for an unknown RIKEN cDNA 4833420G17 gene (Figure 4B) and the Hoxa3 gene (Figure 4C, bottom). As illustrated in Figure 4C, loss of methylation may occur at entire gene clusters, e.g., the Hoxa homeobox gene cluster that is known to be aberrantly methylated in a variety of human cancers (25,40,41).
Figure 4.
Hypomethylated CpG islands associated with the Neat1, RIKEN cDNA 4833420G17 and Hoxa3 locus. (A) Loss of CpG methylation within the Neat1 CpG island was determined by sodium bisulfite sequencing in primary (1bc, 2bc, 3bc; left panel), senescent (1c, 2c, 3c; middle panel) and immortalized (1ac, 2ac, 3ac; right panel) MEFs. The sequencing results of several independent clones and the respective percentage of methylation per sample are shown. (B) COBRA analysis of the hypomethylated CpG island associated with an unknown RIKEN cDNA 4833420G17 gene in primary (1bc, 2bc, 3bc), senescent (2c, 3c) and immortalized (1ac, 2ac, 3ac) MEFs. Digested fragments on the gel are indicative of methylated TaqI restriction sites (5'-TCGA) within the CpG island (see, legend for Figure 2). (C) Upper panel: hypomethylated CpG islands associated with the Hoxa cluster. Visualization of the NimbleGen array data for immortalized (ac, in red) and primary (bc, in green) MEFs. The methylation signal is plotted as a P-value on the y-axis. The location of the hypomethylated CpG island analysed by COBRA is shown as a snapshot of the UCSC Genome Browser. Lower panel: COBRA analysis of the hypomethylated CpG island associated with the Hoxa3 locus in primary (1bc, 2bc, 3bc), senescent (2c, 3c) and immortalized (1ac, 2ac, 3ac) MEFs. Digested fragments on the gel are indicative of methylated BstUI restriction sites (5'-CGCG) within the CpG island (see, legend for Figure 2).
Using a sodium bisulfite-based sequencing analysis (33), we have also investigated the methylation status of major repetitive DNA elements, including LINE L1, IAP-LTR and SINE B1, which constitute 18.78, 3.13 and 2.66%, respectively, of the mouse genome (30–32). As shown in Figure 5, DNA methylation at these repeat elements, in particular IAP-LTR and SINE B1 elements, was reduced in immortalized cell lines relative to primary MEFs, which indicates that genome-wide DNA methylation decreases with in vitro passaging.
Figure 5.
Methylation profiling in repetitive DNA elements in the mouse genome during immortalization. (A) Bisulfite sequencing of LINE L1, IAP-LTR and SINE B1 elements. Two independently established immortalized murine cell lines are shown (Set 5 and Set 7). bc = before crisis; ac = after crisis; 5 and 7 designate Set 5 and Set 7, respectively. Values in parentheses indicate percentages of mCG. (B) Quantification of mCG in major repetitive DNA elements in immortalized versus primary MEFs. Average results from two independently established immortalized murine cell lines are shown (Set 5 and Set 7). Fisher’s exact test was used to calculate the statistical significance of difference in mCpGs between immortalized and primary MEFs.
Gene expression profiling of primary, senescent and immortalized MEFs
To identify novel genes that are differentially expressed during immortalization, we have profiled the whole transcriptome of primary, senescent and immortalized MEFs using an Affymetrix-microarray platform (Affymetrix Inc.). Following standard bioinformatics analysis, we identified a large number of dysregulated genes across three independently established immortalized cell lines (Figure 6A). When we compared the expression profile of senescent versus primary fibroblasts (based on a log2 ratio of one = 2-fold difference), we detected 95 differentially expressed targets/probes, of which 34 were overexpressed (up to a log2 ratio of +5.54), and 61 were down-regulated (with a log2 ratio as low as −3.51) in all the three datasets. A much higher number of differentially expressed targets/probes (n = 313) was detected when immortalized cells were compared to low-passage primary fibroblasts (Figure 6B and Supplementary Table S2). Of the 313 identified targets, 80 were up-regulated (up to a log2 ratio of +6.07) and 233 were down-regulated (with a log2 ratio as low as −3.66). Interestingly, 50 of these probes/targets were common in the senescent and immortalized phenotype, suggesting a crucial role for these genes in circumventing senescence and initiating uncontrolled cell proliferation (Figure 6B).
Figure 6.
Genome-wide expression profiling of senescent and immortalized mouse fibroblasts versus counterpart primary cells and analysis of gene-set enrichment. (A) Global gene expression profile of immortalized MEFs was established using the Affymetrix-microarray analysis. Three independently established immortalized murine cell lines are shown (Set 1, Set 2 and Set 3). Differentially expressed genes were identified as described in Materials and Methods section. Genes with significantly different expression levels were selected by using a cutoff P-value of <0.05, and 2-fold change in the level of expression. (B) Venn diagram showing the number of differentially regulated transcripts in senescent (green) and immortalized (orange) mouse fibroblasts as compared to counterpart primary cells, based on gene expression microarray analysis. (C) GO analysis of the 95 differentially expressed transcripts (72 DAVID IDs) in senescent versus primary mouse fibroblasts, using DAVID Functional Annotation Clustering. The top GO categories with an enrichment score of ≥2 (x-axis) are listed in the y-axis. (D) GO analysis of the 313 differentially expressed transcripts (244 DAVID IDs) in immortalized versus primary mouse fibroblasts, using DAVID Functional Annotation Clustering. The top GO categories with an enrichment score of ≥2 (x-axis) are listed in the y-axis.
We have also investigated the functional relationships between genes that were differentially regulated in immortalized versus primary fibroblasts. Based on the IPA®, we found that these genes were mostly involved in cell signaling networks, cell cycle and cell morphology, embryonic and tissue development (Figure 6D) and were overrepresented compared to the senescent phenotype (Figure 6C). In agreement with the methylation data, we found that several pathways implicated in human cancer, such as the MEK–ERK, PI3K/Akt, Nfkb and Vegf pathway (36–39) were mostly affected upon immortalization (Figures 7A and 8). These findings are in agreement with those of a recent report in which major pathways involved in controlling senescence/immortalization were identified (42).
Figure 7.
Gene network analysis and relative quantification of gene expression by standard qRT-PCR. (A) Upon immortalization, genetic and epigenetic events may contribute to dysregulation of MAP kinase pathways. Upstream and downstream components of the p38-MAPK and MEK–ERK cascade are shown in pink. Network interactions were deduced from IPA® analysis and are illustrated in detail in Figure 8. Continuous line, direct interaction; broken line, indirect interaction. Red and green molecules represent up-regulated and down-regulated targets, respectively. Yellow node color is indicative of hypermethylation. Up-white-arrows represent up-regulation and down-white-arrows represent down-regulation. (B) Transcription levels of the Cldn5, Hoxc9, Wnk4, Mapk11, Rasd1, Adra1d and Akap5 genes following immortalization. Bars represent the mean normalized expression (±SD) of three independent immortalized cells lines (ac: 1ac & 2ac & 3ac; each in triplicate) relative to primary cells (bc: 1bc & 2bc & 3bc; each in triplicate). Data analysis was performed using the 2−ΔΔCt method (34) with Gapdh as reference gene. (C) The expression status of individual target genes identified by Affymetrix-microarray analysis was confirmed by standard qRT-PCR. Two up-regulated (Plf1 and Foxg1) and three down-regulated genes (Igf2, Hoxd10 and Hoxd13) were analysed in senescent (1c, 2c, 3c) and immortalized (1ac, 2ac, 3ac) samples according to the 2−ΔΔCt method (34). Expression data were normalized using an endogenous housekeeping gene as reference (Gapdh), and samples 1bc, 2bc, 3bc (RNA of primary fibroblasts) as calibrators within the respective group (fold ratio = 1). Data are expressed as mean ± SD of three replicates.
Correlation between DNA methylation and gene expression during senescence bypass and immortalization
To investigate the impact of DNA methylation on gene expression during senescence bypass and cell immortalization, we have individually quantified the transcription level of several of the hypermethylated genes identified by our DNA methylation analysis using standard qRT-PCR. The analysed targets are known to interact with key components of the MAP kinase pathways (Figure 7A), and are potentially relevant for the establishment of a malignant phenotype. Figure 7B shows the mean normalized expression levels of the Cldn5, Hoxc9, Wnk4, Mapk11, Rasd1, Adra1d and Akap5 genes in three independently immortalized cell lines (1ac, 2ac, 3ac) relative to primary cells (1bc, 2bc, 3bc). With the exception of the Akap5 gene, which is down-regulated, all the other targets show steady increase in transcription levels upon immortalization.
We have also selected several of the up-regulated and down-regulated genes detected by Affymetrix-chip analysis, and validated their relative expression by standard qRT-PCR. Representative qRT-PCR results are shown in Figure 7C. In line with microarray data (Supplementary Figure S5 and Supplementary Table S2), we confirmed that the Plf1 and Foxg1 genes were up-regulated, whereas the Hoxd10, Hoxd13 and Igf2 genes were down-regulated in immortalized MEFs (Figure 7C). Of significance, some of these genes encode proteins, which are relevant for MAPK cascades (Figures 7A and 8).
DISCUSSION
Bypass from senescence is widely recognized as the first step toward immortalization, which, in turn, may drive cells to malignancy (43). To gain insights into the epigenetic and genetic mechanisms involved in the senescence/immortalization switch, we have analysed the whole methylome and transcriptome of primary, senescent and immortalized MEFs, a well-established in vitro model of cell immortalization (16,17). Using a high-resolution genome-wide mapping technique (MIRA-chip) (25,26), followed by extensive single gene/locus-specific validation assays (COBRA and bisulfite sequencing) (28,29), we have identified 24 CpG islands that display aberrant DNA methylation in three independently established immortalized MEF cell lines as compared to counterpart low-passage primary cells (Table 1). Furthermore, we observed significant loss of CpG methylation at major repetitive DNA elements (Figure 5), a genome-wide epigenetic event often associated with genetic instability and cancer (44–46).
In immortalized murine fibroblasts, de novo DNA methylation marks preferentially genes that are involved in the development and function of the nervous system (Phox2a, target #6; Rem2, target #7; Cldn5, target #11; Ntn3, target #13; Amh, target #16; Adra1d, target #19; Akap5, target #21; IPA®, P = 9.75E-04 – 4.30E-02; Table 1). These and other developmental regulators (47% of the identified genes) are targets of the Polycomb repressive complexes, and often associate with bivalent modifications (H3K4me3 active marks and H3K27me3 inactive marks) in murine embryonic stem (ES) cells, as deduced by comparison of our data to published databases (47–49). Polycomb group (PcG) proteins are critical for the maintenance of stem cell identity and cell differentiation (47,50,51), and PcG-associated genes in ES cells are often the target of aberrant CpG methylation in cancer, thus, supporting the ‘stem cell origin’ model of transformation (52–54). The DNA methylation/PcG connection has been consistently observed in the epigenome of several human malignancies (55,56), and our findings suggest that epigenetic reprogramming of pluripotency genes may take place at early pre-neoplastic stages, once the cells acquire an infinite life span. Interestingly, GO analysis of the annotated hypermethylated targets (n = 19), identified in immortalized MEFs, confirmed a trend of enrichment for genes involved in intracellular signaling cascades, such as the MEK–ERK pathway, one of the most frequently altered genetic pathways in human cancer (Figures 7A and 8) (36–39). This finding reinforces the notion that alternative epigenetic mechanisms may contribute to the disruption of genetic pathways that are crucial for cell immortalization and growth, and tumor development (2,3,17,18,42).
To examine the potential impact of DNA methylation on gene expression during senescence bypass and immortalization, we have also constructed the whole transcriptome of primary, senescent and immortalized murine fibroblasts. A large number of differentially expressed targets/probes (n = 313) was identified in independently established immortalized MEF cell lines when compared to low-passage primary fibroblasts (Figure 6B and Supplementary Table S2). Apart from few known markers of senescence [i.e., Igfbp6, Ahr, Col1a1, Aldh1a2; reviewed in (42)], several of our identified transcripts are novel, and represent starting points for future research on transcriptional pathways that may regulate the acquisition of an infinite life span in somatic cells. We have further classified the function of the differentially expressed genes (crisis and after crisis versus before crisis phenotypes) by GO analysis. According to DAVID, a larger number of dysfunctional gene clusters was identified in the immortal phenotype, including genes involved in signaling, development, morphogenesis and neurogenesis (Figure 6C and D). The vast majority of these dysregulated genes converge into specific pathways, such as MEK/ERK, PI3K/Akt, NFkB and Vegf that are often impaired during tumorigenesis (Figure 8) (36–39). In addition, out of the 244 annotated DAVID IDs (from the 313 transcripts differentially expressed in immortalized MEFs), 41% (n = 101) are normally expressed in the brain, and play a role in neural development.
Figure 8.
Functionally related pathways of aberrantly methylated and differentially expressed genes in immortalized MEFs. Merging Networks were generated by Ingenuity Pathway Analysis® (IPA®: v 9.0), using the gene lists in Table 1 and Supplementary Table S2 (ac versus bc). The convergence of genes involved in the MEK–ERK pathway, one of the most frequently disrupted pathways in human cancer (36–39), is highly notable. Red and green nodes represent up-regulated and down-regulated genes, respectively. Yellow nodes are indicative of hypermethylated targets and white nodes show molecules that are not included in the datasets but interact with other components of the networks. Canonical pathways (CP: Molecular Mechanisms of Cancer) are outlined in blue.
We have also confirmed gene expression changes in several of the hypermethylated targets identified by our DNA methylation analysis using standard qRT-PCR. Apart from Akap5 and Hoxc9, which are aberrantly methylated at their promoter regions, the majority of the analysed targets contain intragenic CpG islands that become hypermethylated upon immortalization. These latter genes are all up-regulated in immortalized versus low-passage primary cells (Figure 7B). Our findings are in agreement with several reports indicating that intragenic methylation often correlates with gene up-regulation [reviewed in (57–61)]. Along with Cldn5, Wnk4, Mapk11 and Rasd1, several other targets are methylated within gene bodies (38%) rather than at canonical 5′-end promoters (29%) (Table 1), a trend consistently reported for different human epigenomes (62,63). Although the exact functional role of these intragenic CpG-rich elements remains to be determined, the general consent is that these elements constitute short independent transcriptional units and/or regulate alternative isoforms (62,63). A recent study has demonstrated a crucial role for the DNA-binding protein, CCCTC-binding factor (CTCF), in regulating alternative exon splicing by inhibiting Pol2 elongation dynamics (64). CTCF binding to the DNA occurs mostly intragenically (40–45%), and is inhibited by CpG methylation (64). In the present study, we have observed that about 70% of the methylated intragenic CpG islands are located at or adjacent to (within 1 kb) potential consensus elements for CTCF, when compared to published databases (Figures 2B and 3B, and Supplementary Figures S1 and S3) (http://genome.ucsc.edu/ENCODE/downloadsMouse.html). It is plausible that de novo methylation established during cell immortalization may impede the binding of the CTCF protein at these intragenic sites, thereby, resuming RNA polymerase extension and/or splicing. Alternatively, methylation-dependent CTCF release within gene bodies can predispose the DNA to an open chromatin configuration, which may lead to unusual gene activation, uncontrolled cell proliferation (i.e., immortalization), and cancer development (65,66).
A large body of evidence exists indicating that the Ras/Raf/ERK and the p38 MAP kinase pathways are two of the most relevant pathways involved in the activation of senescence as a fail-safe mechanism against oncogenic transformation and other damage in primary cells (42,67–69). By marked contrast, deregulated activation of the MEK/MAPK cascade can result in forced mitogenesis and transformation in immortal cells (67). These two totally opposite outcomes, cell cycle arrest and induction of proliferation, imposed by identical signal-transduction pathways are mostly cell context-dependent and dictated by the strength of the stimuli and the integrity of the senescence program (67). Interestingly, high-passage MEFs (PD 13), cultured according to the 3T3 protocol (as in this study), displayed elevated levels of activated p38MAPK (p38-P), compared to early passage MEF cells (PD 3), although apparently immortalized MEFs (PD 35) proliferate independently of the status of p38MAPK (70). In our study, although we did not directly analyse the activity (i.e. phosphorylation) of the kinases involved in the MAPK signaling cascades, all the indications are that these cascades are indeed activated during immortalization. Of note, we have shown that proliferin 1 (Plf1) a well-known component of the MAPK pathway is greatly up-regulated in senescent and immortal fibroblasts (up to 67-fold on linear scale; Supplementary Table S2). Plf1 modulates the activation of the MEK/ERK pathway and is transcriptionally regulated by β-catenin, which accumulates following activation of p38MAPK, via repression of GSK3β (Figure 7A) (71,72).
In conclusion, our global profiling of the methylome and transcriptome of primary, senescent and immortalized murine fibroblast cells shows abrogations of epigenetic and genetic pathways during cell immortalization, which closely resemble those found in human cancer (36,37,39,57–59). Despite species differences between human and mouse, identification of novel biomarkers of senescence bypass and immortalization in mouse models may help elucidate the underlying mechanisms of human carcinogenesis. If validated in vivo in humans, these biological markers can be used to predict cancer risk in early stage lesions, and help devise future strategies for cancer therapies, e.g., re-activation of key signaling effectors of senescence in tumor cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–5 and Supplementary Materials and Methods.
FUNDING
Funding for open access charge: American Cancer Society [RSG-11-083-01-CNE]; University of California Tobacco Related Disease Research Program [18KT-0040 and 20XT-0116 to A.B.].
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Reinhardt HC, Schumacher B. The p53 network: cellular and systemic DNA damage responses in aging and cancer. Trends Genet. 2012;28:128–136. doi: 10.1016/j.tig.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collado M, Serrano M. Senescence in tumours: evidence from mice and humans. Nat. Rev. Cancer. 2010;10:51–57. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banito A, Gil J. Induced pluripotent stem cells and senescence: learning the biology to improve the technology. EMBO Rep. 2010;11:353–359. doi: 10.1038/embor.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muller M. Cellular senescence: molecular mechanisms, in vivo significance, and redox considerations. Antioxid Redox Sign. 2009;11:59–98. doi: 10.1089/ars.2008.2104. [DOI] [PubMed] [Google Scholar]
- 5.Saab R. Senescence and pre-malignancy: how do tumors progress? Semin. Cancer Biol. 2011;21:385–391. doi: 10.1016/j.semcancer.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 6.Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguria A, Zaballos A, Flores JM, Barbacid M, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 7.Parrinello S, Coppe JP, Krtolica A, Campisi J. Stromal-epithelial interactions in aging and cancer: senescent fibroblasts alter epithelial cell differentiation. J. Cell Sci. 2005;118:485–496. doi: 10.1242/jcs.01635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coppe JP, Patil CK, Rodier F, Krtolica A, Beausejour CM, Parrinello S, Hodgson JG, Chin K, Desprez PY, Campisi J. A human-like senescence-associated secretory phenotype is conserved in mouse cells dependent on physiological oxygen. PLoS One. 2010;5:e9188. doi: 10.1371/journal.pone.0009188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coppe JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu. Rev. Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.d'Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat. Rev. Cancer. 2008;8:512–522. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- 11.Hahn WC, Weinberg RA. Modelling the molecular circuitry of cancer. Nat. Rev. Cancer. 2002;2:331–341. doi: 10.1038/nrc795. [DOI] [PubMed] [Google Scholar]
- 12.Itahana K, Campisi J, Dimri GP. Mechanisms of cellular senescence in human and mouse cells. Biogerontology. 2004;5:1–10. doi: 10.1023/b:bgen.0000017682.96395.10. [DOI] [PubMed] [Google Scholar]
- 13.Clarke AR, Hollstein M. Mouse models with modified p53 sequences to study cancer and ageing. Cell Death Differ. 2003;10:443–450. doi: 10.1038/sj.cdd.4401188. [DOI] [PubMed] [Google Scholar]
- 14.Anisimov VN, Ukraintseva SV, Yashin AI. Cancer in rodents: does it tell us about cancer in humans? Nat. Rev. Cancer. 2005;5:807–819. doi: 10.1038/nrc1715. [DOI] [PubMed] [Google Scholar]
- 15.Luo JL, Yang Q, Tong WM, Hergenhahn M, Wang ZQ, Hollstein M. Knock-in mice with a chimeric human/murine p53 gene develop normally and show wild-type p53 responses to DNA damaging agents: a new biomedical research tool. Oncogene. 2001;20:320–328. doi: 10.1038/sj.onc.1204080. [DOI] [PubMed] [Google Scholar]
- 16.Wadhwa R, Sugihara T, Taira K, Kaul SC. The ARF-p53 senescence pathway in mouse and human cells. Histol. Histopathol. 2004;19:311–316. doi: 10.14670/HH-19.311. [DOI] [PubMed] [Google Scholar]
- 17.Whibley C, Odell AF, Nedelko T, Balaburski G, Murphy M, Liu Z, Stevens L, Walker JH, Routledge M, Hollstein M. Wild-type and Hupki (human p53 knock-in) murine embryonic fibroblasts: p53/ARF pathway disruption in spontaneous escape from senescence. J. Biol. Chem. 2010;285:11326–11335. doi: 10.1074/jbc.M109.064444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Odell A, Askham J, Whibley C, Hollstein M. How to become immortal: let MEFs count the ways. Aging (Albany NY) 2010;2:160–165. doi: 10.18632/aging.100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat. Cell Biol. 2003;5:741–747. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lundberg AS, Hahn WC, Gupta P, Weinberg RA. Genes involved in senescence and immortalization. Curr. Opin. Cell Biol. 2000;12:705–709. doi: 10.1016/s0955-0674(00)00155-1. [DOI] [PubMed] [Google Scholar]
- 21.Besaratinia A, Pfeifer GP. Applications of the human p53 knock-in (Hupki) mouse model for human carcinogen testing. FASEB J. 2010;24:2612–2619. doi: 10.1096/fj.10-157263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng L, Hollstein M, Xu Y. Ser46 phosphorylation regulates p53-dependent apoptosis and replicative senescence. Cell Cycle. 2006;5:2812–2819. doi: 10.4161/cc.5.23.3526. [DOI] [PubMed] [Google Scholar]
- 23.Decottignies A, d'Adda di Fagagna F. Epigenetic alterations associated with cellular senescence: a barrier against tumorigenesis or a red carpet for cancer? Semin. Cancer Biol. 2011;21:360–366. doi: 10.1016/j.semcancer.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 24.Yasaei H, Gilham E, Pickles JC, Roberts TP, O'Donovan M, Newbold RF. Carcinogen-specific mutational and epigenetic alterations in INK4A, INK4B and p53 tumour-suppressor genes drive induced senescence bypass in normal diploid mammalian cells. Oncogene. 2012 doi: 10.1038/onc.2012.45. Mar 12 (doi:10.1038/onc.2012.45; epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 25.Tommasi S, Karm DL, Wu X, Yen Y, Pfeifer GP. Methylation of homeobox genes is a frequent and early epigenetic event in breast cancer. Breast Cancer Res. 2009;11:R14. doi: 10.1186/bcr2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tommasi S, Kim SI, Zhong X, Wu X, Pfeifer GP, Besaratinia A. Investigating the epigenetic effects of a prototype smoke-derived carcinogen in human cells. PLoS One. 2010;5:e10594. doi: 10.1371/journal.pone.0010594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang CL, Jin SG, Pfeifer GP. MBD3L1 is a transcriptional repressor that interacts with methyl-CpG-binding protein 2 (MBD2) and components of the NuRD complex. J. Biol. Chem. 2004;279:52456–52464. doi: 10.1074/jbc.M409149200. [DOI] [PubMed] [Google Scholar]
- 28.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodier JL, Ostertag EM, Du K, Kazazian HH., Jr A novel active L1 retrotransposon subfamily in the mouse. Genome Res. 2001;11:1677–1685. doi: 10.1101/gr.198301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martens JH, O'Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 33.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 35.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 36.Steelman LS, Franklin RA, Abrams SL, Chappell W, Kempf CR, Basecke J, Stivala F, Donia M, Fagone P, Nicoletti F, et al. Roles of the Ras/Raf/MEK/ERK pathway in leukemia therapy. Leukemia. 2011;25:1080–1094. doi: 10.1038/leu.2011.66. [DOI] [PubMed] [Google Scholar]
- 37.Whittaker S, Marais R, Zhu AX. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29:4989–5005. doi: 10.1038/onc.2010.236. [DOI] [PubMed] [Google Scholar]
- 38.Chin L, Garraway LA, Fisher DE. Malignant melanoma: genetics and therapeutics in the genomic era. Genes Dev. 2006;20:2149–2182. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- 39.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 40.Ordway JM, Bedell JA, Citek RW, Nunberg A, Garrido A, Kendall R, Stevens JR, Cao D, Doerge RW, Korshunova Y, et al. Comprehensive DNA methylation profiling in a human cancer genome identifies novel epigenetic targets. Carcinogenesis. 2006;27:2409–2423. doi: 10.1093/carcin/bgl161. [DOI] [PubMed] [Google Scholar]
- 41.Strathdee G, Holyoake TL, Sim A, Parker A, Oscier DG, Melo JV, Meyer S, Eden T, Dickinson AM, Mountford JC, et al. Inactivation of HOXA genes by hypermethylation in myeloid and lymphoid malignancy is frequent and associated with poor prognosis. Clin. Cancer Res. 2007;13:5048–5055. doi: 10.1158/1078-0432.CCR-07-0919. [DOI] [PubMed] [Google Scholar]
- 42.Fridman AL, Tainsky MA. Critical pathways in cellular senescence and immortalization revealed by gene expression profiling. Oncogene. 2008;27:5975–5987. doi: 10.1038/onc.2008.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boehm JS, Hahn WC. Understanding transformation: progress and gaps. Curr. Opin. Genet. Dev. 2005;15:13–17. doi: 10.1016/j.gde.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 44.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 45.Kulis M, Esteller M. DNA methylation and cancer. Adv. Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 46.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat. Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 47.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 48.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rugg-Gunn PJ, Cox BJ, Ralston A, Rossant J. Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proc. Natl Acad. Sci. USA. 2010;107:10783–10790. doi: 10.1073/pnas.0914507107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vermeulen L, de Sousa e Melo F, Richel DJ, Medema JP. The developing cancer stem-cell model: clinical challenges and opportunities. Lancet Oncol. 2012;13:e83–e89. doi: 10.1016/S1470-2045(11)70257-1. [DOI] [PubMed] [Google Scholar]
- 53.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat. Rev. Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 54.Glinsky GV. Genomic models of metastatic cancer: functional analysis of death-from-cancer signature genes reveals aneuploid, anoikis-resistant, metastasis-enabling phenotype with altered cell cycle control and activated Polycomb Group (PcG) protein chromatin silencing pathway. Cell Cycle. 2006;5:1208–1216. doi: 10.4161/cc.5.11.2796. [DOI] [PubMed] [Google Scholar]
- 55.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ballestar E, Esteller M. Epigenetic gene regulation in cancer. Adv. Genet. 2008;61:247–267. doi: 10.1016/S0065-2660(07)00009-0. [DOI] [PubMed] [Google Scholar]
- 57.Baylin SB, Jones PA. A decade of exploring the cancer epigenome – biological and translational implications. Nat. Rev. Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 59.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat. Rev. Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 61.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 62.Illingworth R, Kerr A, Desousa D, Jorgensen H, Ellis P, Stalker J, Jackson D, Clee C, Plumb R, Rogers J, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gomes NP, Espinosa JM. Gene-specific repression of the p53 target gene PUMA via intragenic CTCF-Cohesin binding. Genes Dev. 2010;24:1022–1034. doi: 10.1101/gad.1881010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Recillas-Targa F, de la Rosa-Velazquez IA, Soto-Reyes E. Insulation of tumor suppressor genes by the nuclear factor CTCF. Biochem. Cell Biol. 2011;89:479–488. doi: 10.1139/o11-031. [DOI] [PubMed] [Google Scholar]
- 67.Lin AW, Barradas M, Stone JC, van Aelst L, Serrano M, Lowe SW. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 1998;12:3008–3019. doi: 10.1101/gad.12.19.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu J, Woods D, McMahon M, Bishop JM. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 1998;12:2997–3007. doi: 10.1101/gad.12.19.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Debacq-Chainiaux F, Boilan E, Dedessus Le Moutier J, Weemaels G, Toussaint O. p38(MAPK) in the senescence of human and murine fibroblasts. Adv. Exp. Med. Biol. 2010;694:126–137. doi: 10.1007/978-1-4419-7002-2_10. [DOI] [PubMed] [Google Scholar]
- 70.Iwasa H, Han J, Ishikawa F. Mitogen-activated protein kinase p38 defines the common senescence-signalling pathway. Genes Cells. 2003;8:131–144. doi: 10.1046/j.1365-2443.2003.00620.x. [DOI] [PubMed] [Google Scholar]
- 71.Glazer RI, Wang XY, Yuan H, Yin Y. Musashi1: a stem cell marker no longer in search of a function. Cell Cycle. 2008;7:2635–2639. doi: 10.4161/cc.7.17.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cuadrado A, Nebreda AR. Mechanisms and functions of p38 MAPK signalling. Biochem. J. 2010;429:403–417. doi: 10.1042/BJ20100323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.