Abstract
The nuclear hormone 1α,25-dihydroxyvitamin D3 (1α,25(OH)2D3 or 1,25D) regulates its target genes via activation of the transcription factor vitamin D receptor (VDR) far more specifically than the chromatin modifier trichostatin A (TsA) via its inhibitory action on histone deacetylases. We selected the thrombomodulin gene locus with its complex pattern of five VDR binding sites and multiple histone acetylation and open chromatin regions as an example to investigate together with a number of reference genes, the primary transcriptional responses to 1α,25(OH)2D3 and TsA. Transcriptome-wide, 18.4% of all expressed genes are either up-or down-regulated already after a 90 min TsA treatment; their response pattern to 1α,25(OH)2D3 and TsA sorts them into at least six classes. TsA stimulates a far higher number of genes than 1α,25(OH)2D3 and dominates the outcome of combined treatments. However, 200 TsA target genes can be modulated by 1α,25(OH)2D3 and more than 1000 genes respond only when treated with both compounds. The genomic view on the genes suggests that the degree of acetylation at transcription start sites and VDR binding regions may determine the effect of TsA on mRNA expression and its interference with 1α,25(OH)2D3. Our findings hold true also for other HDAC inhibitors and may have implications on dual therapies using chromatin modifiers and nuclear receptor ligands.
INTRODUCTION
Transcriptional regulation of genes is in most cases the balanced result of the activating function of transcription factors and the intrinsic repressive nature of chromatin (1). Nuclear receptors form a distinct family of transcription factors, many of which are activated by small lipophilic ligands (2). Vitamin D receptor (VDR) is a member of the nuclear receptor superfamily as it is activated by sub-nanomolar concentrations of its natural ligand 1α,25-dihydroxyvitamin D3 (1α,25(OH)2D3 or 1,25D) (3). VDR is able to associate with its genomic binding sites already in the absence of ligand and can form complexes with co-repressor proteins and chromatin modifying enzymes, such as histone deacetylases (HDACs), histone acetyltransferases (HATs), histone methyltransferases (KMTs) and histone lysine demethylases (KDMs) (4–6). The removal of acetyl groups from histones stabilizes their attraction to genomic DNA and keeps the respective genomic region repressed. In contrast to acetylation, there is a clear functional distinction between histone methylation marks, both concerning the exact histone residues as well as their degree of modification, such as mono-, di- or tri-methylation (7). For example, H3K4me3 and H3K27me3 are correlated with gene repression, whereas H3K9me1 and H4K20me1 are found primarily in active genes. When VDR is activated by binding of 1α,25(OH)2D3, the receptor’s ligand-binding domain changes its conformation, so that it is more favourable for the binding of co-activator proteins (8). Different classes of co-activators link the VDR either to HATs, which open chromatin locally, or to the basal transcriptional machinery containing RNA polymerase II (9). In this way, genes that are located in relative vicinity to genomic VDR binding sites can be specifically activated. A far more global, but less specific activation of genes can be achieved by inhibiting the enzymatic function of HDACs by small synthetic compounds, such as trichostatin A (TsA) (10,11). Interestingly, some of the genes coding for chromatin modifying enzymes, such as HDAC4, HDAC6 (12) and KDM6B (13), are VDR targets.
The classical, physiological role of 1α,25(OH)2D3 is the regulation of calcium and phosphate homeostasis and bone mineralization (14), but there is both epidemiological and pre-clinical evidence that VDR ligands also have anti-proliferative and immuno-modulatory actions (15,16). Therefore, 1α,25(OH)2D3 and its synthetic analogues are considered as therapeutic agents for the treatment and prevention of both hyper-proliferative and immunological diseases (17). HDAC inhibitors were used first as mood stabilizers and anti-epileptics (18), but within the last decade they were developed intensively towards the therapy of cancer and immune diseases (19–21). The idea to improve the specificity of gene regulatory and physiological action of HDAC inhibitors by a co-treatment with more specific drugs, such as nuclear receptor ligands, has already been followed since 15 years (22,23) and applied several times for VDR agonists (24–28). For example, successful clinical treatment of breast and prostate cancer had been reported for the combination of HDAC inhibitors with the anti-estrogen tamoxifen and the anti-androgen bicalutamide, respectively (29).
In a previous study, we related the anti-proliferative effect of 1α,25(OH)2D3 and TsA (30,31) on malignant and non-malignant human breast cell lines with the primary gene regulatory effect of the compounds on the gene family of cyclin-dependent kinase inhibitors (28). However, at that time the genome-wide view on VDR binding sites was missing, which was now made available by Ramagopalan et al. in human lymphoblastoids (32), by Meyer et al. in human colorectal cancer cells (33) and by us in human monocytic cells (34). The three studies reported between 1600 and 2800 genomic VDR locations that locate both up- and downstream of the transcription start sites (TSSs) of primary 1α,25(OH)2D3 target genes. For a few genes, such as cathelicidin anti-microbial peptide (CAMP), a single dominant VDR peak was found very close to the gene’s TSS, but the majority of 1α,25(OH)2D3 target genes showed a more complex arrangement of VDR binding sites: (i) a few VDR peaks in relative vicinity of a given TSS; (ii) one distal peak up to several 100 kb of the TSS; or (iii) a combination of both. The genomic locus of the thrombomodulin (THBD) gene displayed one of the most complex arrangements containing five VDR peak locations (34).
Mechanistic insight into the dynamics of transcriptional regulation that has been obtained during the last years for the VDR (12,35) and other nuclear receptors (36) indicates for a number of their target genes dynamic changes of mRNA expression levels in the order of 20–60 min. This dynamics relates to the cycling of nuclear proteins involved in transcriptional regulation through a deactivation, activation and initiation phase (37). These phases are characterized by the presence of different nuclear proteins that link the genomic regions of transcription factor binding sites with the respective TSS regions (38). For example, in the deactivation phase HDACs and co-repressor proteins play a central role (12). In this way HDAC inhibitors thus have an impact on the dynamics and timing of VDR target genes. Moreover, these studies emphasize that early time points in gene activation should be studied in more detail.
Therefore, we focused in this study our investigations on the individual and combined responses of genes to 1α,25(OH)2D3 and TsA at early time points. We found that the majority of the tested genes are either up- or down-regulated by a 90 min TsA treatment and confirmed this to be the case for 18.4% of all genes by microarray analysis. Based on their transcriptional response to 1α,25(OH)2D3 and TsA, genes can be sorted into multiple classes. The genome-wide view on these genes suggests that the degree of acetylation at TSSs and VDR binding regions may determine the effect of TsA on the mRNA expression and the possible interference with gene regulation by 1α,25(OH)2D3. Our findings hold true also for the HDAC inhibitor suberoylanilide hydroxamic acid (SAHA) and may have implications on dual therapies using chromatin modifiers and nuclear receptor ligands.
MATERIALS AND METHODS
Cell culture
THP-1 human monocytic leukemia cells (39) were grown in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS), 2 mM L-glutamine, 0.1 mg/ml streptomycin and 100 U/ml penicillin and the cells were kept at 37°C in a humidified 95% air/5% CO2 incubator. Prior to mRNA extraction, cells were grown overnight in phenol red-free medium supplemented with charcoal-stripped FCS. Then, cells were treated with solvent (0.2% ethanol or 0.02% DMSO), 100 nM 1α,25(OH)2D3, indicated concentrations of TsA (in most cases 300 nM), 3 µM SAHA or 1 mM valproic acid (VPA, all compounds from Sigma-Aldrich) for RNA extractions.
RNA extraction and cDNA synthesis
Total RNA was extracted using the Quick RNA Miniprep Kit (Zymo Research). cDNA synthesis was performed for 2 h at 37°C using 1 µg of total RNA as a template, 100 pmol oligo(dT)18 primers, 500 µM dNTPs, 40 U Ribolock Ribonuclease Inhibitor and 40 U MMuLV reverse transcriptase (Fermentas). Prior to real-time quantitative polymerase chain reaction (qPCR), the cDNA was diluted 10-fold.
Real-time quantitative polymerase chain reaction
qPCR reactions were performed using 250 nM of reverse and forward primers, 2 µl 1/10 diluted cDNA template and the ABsolute Blue Q-PCR SYBR Green Low ROX Mix (ABgene) in a total volume of 10 µl. In the PCR reaction the hotstart Taq polymerase was activated for 15 min at 95°C, followed by 40 amplification cycles of 15 s denaturation at 95°C, 15 s annealing at primer-specific temperatures (Supplementary Table S1) and 30 s elongation at 72°C and a final elongation for 5 min at 72°C. PCR product specificity was monitored using post-PCR melt curve analysis. Relative expression levels were determined with the comparative delta threshold cycle (delta-Ct) method. Amplification efficiencies of the primer pairs were taken into account and have been determined on the basis of a standard curve of a cDNA dilution series. Relative expression levels of the target genes were normalized to the three most stable of 10 tested internal reference genes (b2M, GAPDH, HPRT1). The expression stability of the reference genes was determined by the geNorm algorithm (40). Briefly, the arithmetic mean of replicated Ct values for each gene is transformed to a relative quantity (setting the sample with the highest expression as calibrator to 1), using the delta-Ct formula Q = EdeltaCt = E(calibratorCt – sampleCt) (Q = quantity sample relative to calibrator sample; E = amplification efficiency). For normalization, the relative quantities were divided by the normalization factor being the geometric mean of the three reference genes.
siRNA silencing
THP-1 cells were transfected with either non-specific control siRNA oligomers or specific siRNAs targeting VDR mRNA (41). The cells were grown 2–3 days in phenol red-free RPMI 1640 medium supplemented with 5% charcoal-stripped FCS, 2 mM L-glutamine, 0.1 mg/ml streptomycin and 100 U/ml penicillin. The transfection was performed in an Amaxa nucleofector (Lonza) using the Cell Line Nucleofector Kit V according to the manufacturer’s instructions with the following modifications. Per sample, 2 × 106 cells were harvested by centrifugation and re-suspended in 100 µl solution V. A total of 200 pmol of a mixture of three different siRNA oligonucleotides was added and the cell suspension was transferred to an electroporation cuvette (2 µM siRNA). The Nucleofector T-12 program was used for the nucleofection. Immediately after the pulse, 500 µl pre-warmed growth medium was added and the cell suspension was transferred to a microreaction tube. After an incubation at 37°C for 10 min the cells were transferred to pre-warmed 6-well plates containing 1 ml of medium per well (125 nM siRNA). The extent of the knock-down was determined on mRNA level after an incubation for 48 h including ligand or solvent treatment for 5 h.
Microarray analysis
Total RNA was checked for RNA integrity using an Experion electrophoresis station (Biorad) and analysed on Sentrix Human-12 v2 Expression BeadChips from Illumina at the Finnish Microarray Centre (Turku, Finland) using protocols recommended by the manufacturer. Raw and normalized microarray data are available at the Gene Expression Omnibus (GEO) under accession number GSE36323. Microarray data analysis was performed using R statistical software version 2.13 (R Dev Core) with associated libraries from Bioconductor project version 2.8. Data were normalized using VST transformation and RSN normalization used as the standard approach for Illumina arrays with the lumi package. Probe sets that were not linked to any known or predicted human gene were filtered out. Linear Models for Microarray Data (limma) package using linear model fitting for statistical testing with empirical Bayes variance smoothing procedure was applied to detection of differentially expressed genes. Obtained P-values were corrected for multiple testing using Benjamini–Hochberg false discovery rate procedure.
Chromatin immunoprecipitation
After treatment of THP-1 cells, nuclear proteins were cross-linked to DNA by adding formaldehyde directly to the medium to a final concentration of 1% and incubating at room temperature for 5 min on a rocking platform. Cross-linking was stopped by adding glycine to a final concentration of 0.125 M and incubating at room temperature for 5 min on a rocking platform. The cells were collected by centrifugation and washed twice with ice cold PBS. The cell pellets were re-suspended in 900 µl of lysis buffer (1% SDS, 10 mM EDTA, protease inhibitors, 50 mM Tris-HCl, pH 8.1) and the lysates were sonicated in a Bioruptor (Diagenode) to result in DNA fragments of 200–400 bp. Cellular debris was removed by centrifugation. Aliquots of 100 µl of the lysate were diluted 1:10 in chromatin immunoprecipitation (ChIP) dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 167 mM NaCl, protease inhibitors, 250 µg/ml BSA, 16.7 mM Tris-HCl, pH 8.1). 1.5 µg of anti-VDR antibody (sc-1008X, Santa Cruz), anti-H3K27ac (ab4729, Abcam) or non-specific IgG (12-370, Millipore) were bound to 60 µl protein A agarose beads (Millipore) in an overnight incubation at 4°C. The pre-formed bead–antibody complexes were then washed three times with ChIP dilution buffer and added to the chromatin aliquots. The samples were incubated for overnight at 4°C on a rotating platform to form and collect the immuno-complexes. The beads were washed sequentially for 4 min on a rotating platform with 1 ml of the following buffers: low salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl, pH 8.1), high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 500 mM NaCl, 20 mM Tris-HCl, pH 8.1) and LiCl wash buffer (0.25 M LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8.1). Finally, the beads were washed twice with 1 ml TE buffer (1 mM EDTA, 10 mM Tris-HCl, pH 8.0) and the immune complexes were eluted twice using 250 µl elution buffer (1% SDS, 100 mM NaHCO3) at room temperature for 15 min with rotation. The supernatants were combined and the immune complexes were reverse cross-linked at 65°C overnight in the presence of proteinase K (Fermentas) in a final concentration of 40 µg/ml. The DNA was isolated with the ChIP DNA Clean&Concentrator Kit (Zymo Research). Selected genomic regions containing VDR peaks were analysed by qPCR using equal DNA amounts of chromatin fragments, a SYBRGreen I master mix (Roche) and the genomic primers listed in Supplementary Table S2. The qPCR reactions were performed using the following profile: 10 min at 95°C, followed by 45 cycles of 20 s at 95°C, 15 s at 65°C and 15 s at 72°C and a final amplification step of 10 min at 72°C. The results were normalized with respect to input by using the formula 2−(ΔCp)*100, where ΔCp is Cp(input) – Cp(immunoprecipitated DNA) and Cp is the fractional cycle number.
Formaldehyde-assisted isolation of regulatory elements sequencing
FAIRE (formaldehyde-assisted isolation of regulatory elements) analysis was performed according to the protocol published by Giresi et al. (42). In short, THP-1 cells were cross-linked identically as for ChIP. After 5 min cross-linking and stopping with glycine the washed cell pellets were re-suspended and incubated sequentially in 2 ml of buffer L1, 2 ml of buffer L2 and 700 µl of buffer L3. The lysates were sonicated in a Bioruptor (Diagenode) to result in DNA fragments of 300–500 bp, and cellular debris was removed by centrifugation. Input samples were reverse cross-linked overnight at 65°C. The FAIRE samples and reverse cross-linked input samples were subjected to two sequential phenol/chloroform/isoamyl alcohol (25/24/1) extractions, re-suspended in 10 mM Tris-HCl (pH 7.4) and treated with 1 µl of RNase A (10 mg/ml) for 1 h at 37°C. The DNA was purified with the ChIP DNA Clean&Concentrator Kit (Zymo Research) and sequenced using a Solexa Gene Analyzer II platform at the Genomics Core Facility at the EMBL. For data analysis statistically significant peaks were identified using the Zinba program package version 1.06 by setting the mean fragment length at 200 bp and using other settings as recommended for FAIRE-seq in the Zinba web site (http://code.google.com/p/zinba/wiki/UsingZINBA) including peak refinement (43). Raw and normalized FAIRE-seq data are available at GEO under accession number GSE40075.
RESULTS
VDR binding and histone acetylation at the THBD locus
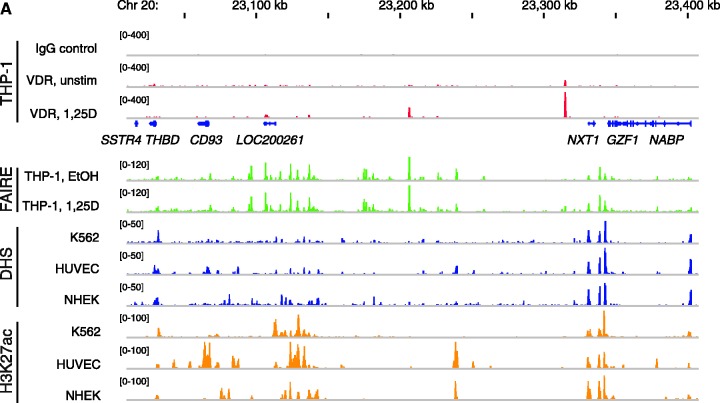
Throughout this whole study we used the human acute monocytic leukemia cell line THP-1 (39) as a model system. Being a cancer cell of the immune system, THP-1 is best suited for studies of genes with impact in cancer and the immune system. Using ChIP sequencing (ChIP-seq) analysis we have recently obtained a genome-wide view on VDR locations in these cells (34). One of the most complex loci identified in that study spreads over 400 kb and contains the seven genes somatostatin receptor 4 (SSTR4), THBD, CD93 molecule (CD93), hypothetical protein LOC200261 (LOC200261), nuclear transport factor 2-like export factor 1 (NXT1), glial cell line-derived neurotrophic factor-inducible zinc finger protein 1 (GZF1) and N-ethylmaleimide-sensitive factor attachment protein, beta (NAPB). Based on the microarray results from THP-1 cells that were treated for 4 h with 1α,25(OH)2D3 (34), THBD and CD93 are statistically significantly up-regulated VDR target genes within this locus. From the five VDR binding locations of the genomic locus, one shows up only in the absence of ligand, three are observed only in its presence and the most dominant peak is already present in unstimulated cells but strongly increases after 40 min treatment with 1α,25(OH)2D3 (Figure 1A). We performed FAIRE-seq analysis in THP-1 cells, which were treated for 100 min with EtOH or 1α,25(OH)2D3. This allows the identification of chromatin sites devoid of nucleosomes, roughly translating to the genome-wide localization of chromatin regions that are accessible to transcription factors at a given time and condition (44). All five VDR peaks are associated with a FAIRE-seq peak, although in some cases a significant amount of open chromatin was only observed after treatment with 1α,25(OH)2D3. For comparison, DNase hypersensitive sites (DHS) in K562, HUVEC and NHEK cells and histone 3 lysine 27 acetylation (H3K27ac) data from the same three cell lines obtained from the ENCODE project (45) are displayed. From all histone marks we selected H3K27ac, because this mark is most specific to separate active from poised enhancers (46). From all cellular models used for the ENCODE project the human myelogenous leukemia cell line K562 (47,48) resembles closest the THP-1 cells, but it is obvious that also the three other cellular models show for quite a number of sites a conserved signal. Interestingly, very most of the FAIRE-seq peaks from THP-1 cells were confirmed by both DHS and H3K27ac signals in at least one of the three ENCODE cell lines. With the exception of the SSTR4 gene, the TSSs of all seven genes overlap with FAIRE-seq, H3K27ac and DHS regions (Figure 1A). The very low expression of the SSTR4 gene was confirmed by qPCR in THP-1 cells (Supplementary Figure S1). However, in these cells also the LOC200261 gene is very low expressed, whereas the five other genes show 3- to 100-fold higher expression.
Figure 1.
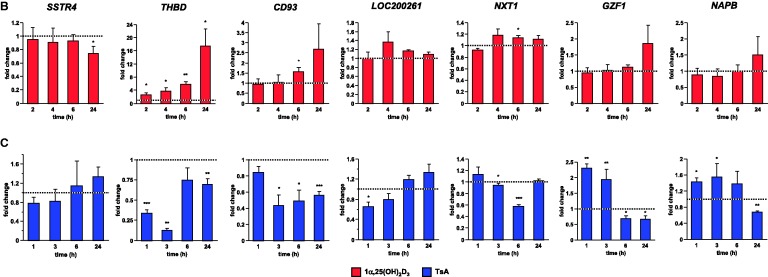
Genome view of the THBD locus and effects of 1α,25(OH)2D3 and TsA. (A) The peak tracks show data from a VDR ChIP-seq experiment in THP-1 cells (34) (IgG control in grey, VDR binding in unstimulated cells and VDR binding after 40 min 1α,25(OH)2D3 (1,25D) treatment in red), FAIRE-seq data from non-stimulated THP-1 cells (EtOH- and 1α,25(OH)2D3-treated for 100 min, green), and DHS-seq (blue) and H3K27ac ChIP-seq data from the ENCODE cell lines K562, HUVEC and NHEK (45) as a sign of open chromatin and histone acetylation. The gene structures are shown in blue. qPCR was performed to determine the relative changes of mRNA expression of all seven genes of the THBD locus in relation to the reference genes B2M, GAPDH and HPRT1 in THP-1 cells in response to incubation with 100 nM 1α,25(OH)2D3 (B) or 300 nM TsA (C) for the indicated times. The columns represent the means of three cell treatments and the bars indicate standard deviations. Two-tailed Student’s t-tests were performed to determine the significance of the mRNA induction by 1α,25(OH)2D3 or TsA in reference to solvent-treated cells (*P < 0.05; **P < 0.01; ***P < 0.001).
In summary, the genomic locus of the THBD gene displays a complex pattern of three VDR target genes, five VDR locations and multiple regions of histone acetylation and open chromatin. Due to the rather high level of conservation between the genome-wide chromatin marks, for additional genomic views (Figure 3A) the display of FAIRE-seq data from THP-1 cells and H3K27ac data from K562 cells was considered to be sufficient.
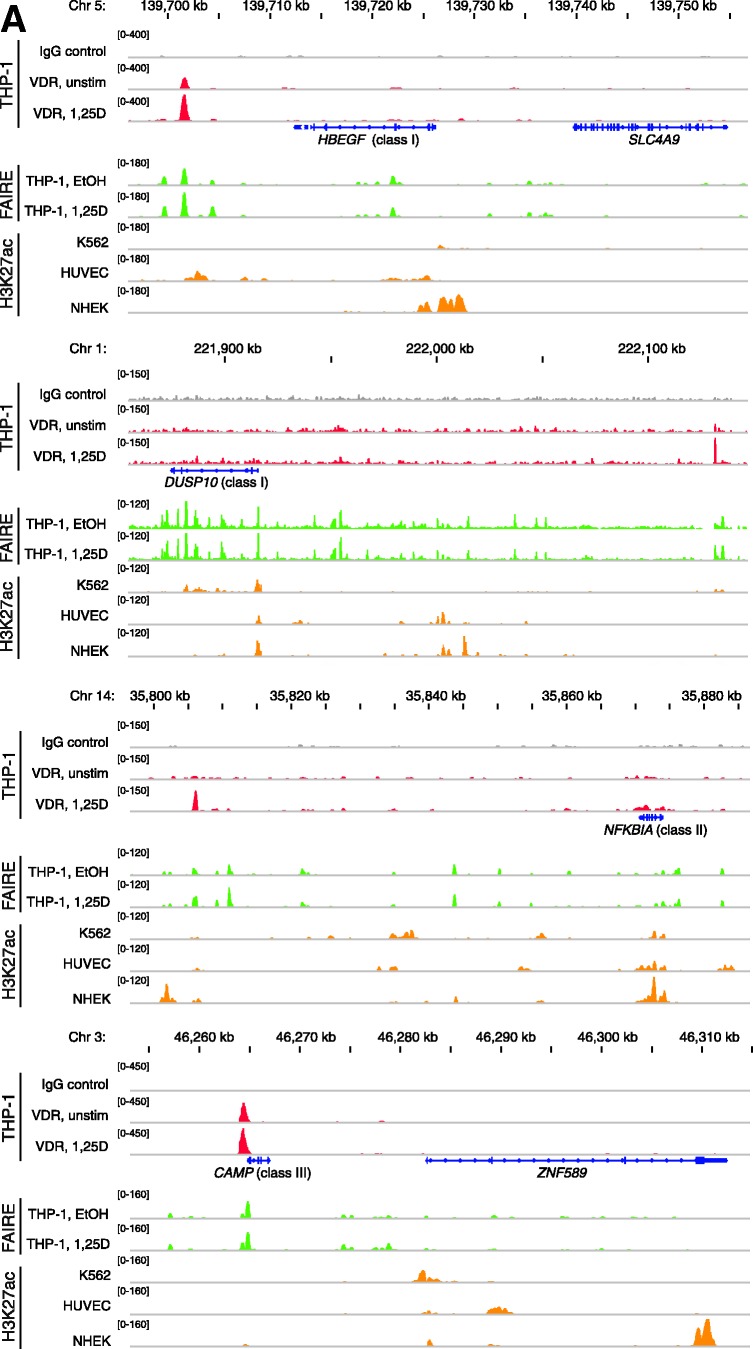
Figure 3.
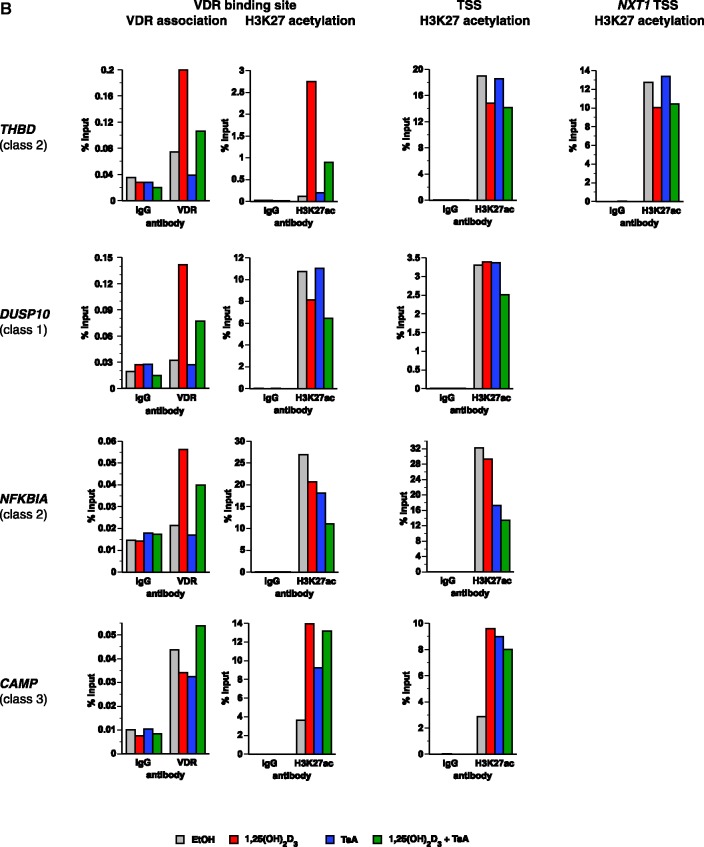
VDR association, histone acetylation and sites of open chromatin for selected loci. (A) Loci of the genes HBEGF, DUSP10, NFKBIA and CAMP. The peak tracks for the four different gene loci show data from a VDR ChIP-seq experiment in THP-1 cells (34) (IgG control in grey, VDR binding in unstimulated cells and VDR binding after 40 min 1α,25(OH)2D3 (1,25D) treatment in red), FAIRE-seq data from THP-1 cells (EtOH- and 1α,25(OH)2D3-treated for 100 min, green), and H3K27ac ChIP-seq data from the ENCODE (45) cell lines K562, HUVEC and NHEK (orange) as a sign of histone acetylation. The gene structures are shown in blue. (B) VDR association, H3K27 acetylation and unspecific IgG binding at VDR binding sites and TSS regions of four VDR target genes, determined by ChIP-qPCR after 40 min treatments of THP-1 cells with solvent, 1α,25(OH)2D3, TsA or the combination of both. Each graph is a representative of four independent experiments. Please note that the best VDR binding sites of the THBD and NFKBIA loci chosen for this assay are those close to the NXT1 and PSMA6 genes, respectively (Figures 1A and 3A).
Primary VDR target genes of the THBD locus
We tested by qPCR in THP-1 cells the response to 1α,25(OH)2D3 of all seven genes of the THBD locus 2–24 h after onset of treatment in reference to vehicle-treated samples (Figure 1B). The THBD gene was confirmed to be a prominent primary 1α,25(OH)2D3 target gene (2.7-, 3.7-, 5.8- and 17.4-fold induction after 2, 4, 6 and 24 h, respectively). In contrast, the six other genes of the investigated genomic locus are far less responsive: SSTR4 was 1.4-fold down-regulated after 24 h, whereas CD93 and NXT1 were 1.6- and 1.1-fold up-regulated after 6 h, respectively. The other genes showed no significant change in mRNA levels. For comparison, the known 1α,25(OH)2D3 target genes (34) dual specificity protein phosphatase 10 (DUSP10), nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (NFKBIA), CAMP and heparin-binding EGF-like growth factor (HBEGF) show a similar type of continuous up-regulation as observed for the THBD gene, whereas the genes cyclin-dependent kinase inhibitor 2D (CDKN2D), VDR and EIA binding protein p300 (EP300) served as negative control genes as in THP-1 cells they are not responsive to 1α,25(OH)2D3 (Supplementary Figure S2).
When the VDR gene was knocked down by the transfection of a mixture of three VDR-specific siRNAs, a subsequent significant decrease of their ligand response indicated the THBD gene as primary VDR target (Supplementary Figure S3). The genes DUSP10, NFKBIA, CAMP and HBEGF served again as positive references and CDKN2D and EP300 as negative controls. The results concerning the genes NXT1 and CD93 gene are not easy to interpret, since the effects of the VDR knock-down are at the borderline of statistical significance. Although the CD93 gene belongs to the same gene family as the THBD gene and may have resulted from a gene duplication (49), it is surprising that the THBD gene is far more responsive to 1α,25(OH)2D3. The genes GZF1 and NAPB still did not shown any response to 1α,25(OH)2D3 stimulation and VDR knock-down did not alter their expression levels, while due to their very low expression the genes SSTR4 and LOC200261 were not investigated with this assay.
Taken together, the THBD gene was confirmed as VDR target, for the genes NXT1 and CD93 only very low inductions by 1α,25(OH)2D3 were found, while the four other genes of the THBD locus are clearly not 1α,25(OH)2D3 targets.
Effect of TsA on genes of the THBD locus and other VDR targets
In order to study the impact of histone acetylation on the expression of VDR target genes, THP-1 cells were treated for 1–24 h with 300 nM TsA (Figure 1C). All seven genes of the THBD locus showed a rather individual profile in their response to the compound as measured by qPCR. The SSTR4 gene did not show any significant response to TsA treatment and the LOC200261 gene was only slightly down-regulated (1.5-fold) 1 h after onset of treatment, but one has to note that both genes are very low expressed in THP-1 cells (Supplementary Figure S1). In contrast, the THBD gene was down-regulated (2.9- and 7.7-fold) after 1 h and 3 h and stayed even after 24 h slightly repressed (1.4-fold). The CD93 gene was down-regulated (2.3-, 2.0- and 1.8-fold) after 3, 6 and 24 h of TsA treatment, whereas the NXT1 gene was only at time point 6 h prominently down-regulated (1.7-fold). The GZF1 gene was up-regulated (2.3- and 2.0-fold) at the early time points, but later down-regulated (1.4- and 1.5-fold). Similarly, the NAPB gene was up-regulated (1.4- and 1.6-fold) early, but after 24 h slightly down-regulated (1.5-fold). For comparison, the genes DUSP10, CDKN2D and HBEGF were up-regulated (1.7- to 2.5-fold) at early time points, but not later (Supplementary Figure S4). The NFKBIA gene was down-regulated (2.1- to 2.9-fold) within the first 3 h but nearly recovered to baseline after 6 h, whereas the EP300 gene was early down-regulated (1.3- and 2.5-fold after 1 and 3 h). The CAMP gene was not significantly affected by TsA treatment, whereas the VDR gene was slightly down-regulated (1.4-fold) after 24 h treatment.
Due to the rather high TsA concentration of 300 nM in the above experiment, we also tested TsA concentrations ranging from 15 to 500 nM at the early time point of 90 min (Supplementary Figure S5). It should be noted that even at the highest TsA concentrations no visible cytotoxic effects were observed within the time frame of the experiment (assessed by microscopy, data not shown). The resulting dose–response curves were in line with the results obtained with 300 nM TsA (Figure 1C and Supplementary Figure S4). The NXT1 gene is not responsive to TsA, whereas the genes CAMP, VDR and CD93 show a minor down-regulation (maximally 1.2-fold) at higher TsA concentrations. On the other hand, already low TsA concentrations suffice for the maximal down-regulation of EP300 (up to 2.0-fold) and for the up-regulation of the genes DUSP10 and NABP (up to 1.6-fold). Finally, a more direct dose-dependency across a wide range of TsA concentrations is seen for the up-regulation of the genes CDKN2D, HBEGF and GZF1 (up to 2.1- to 3.3-fold) and for the down-regulation of THBD and NFKBIA (up to 2.4-fold).
In order to obtain a transcriptome-wide view on the effects of TsA, we performed microarray experiments, where THP-1 cells were treated for 90 and 150 min with 300 nM TsA (Supplementary Figure S6). After 90 min treatment already 2161 genes were significantly regulated by TsA, representing 18.4% of all 11 775 expressed genes (Supplementary Table S3). After 150 min treatment nearly twice as many genes (4264, i.e. 36.2% of all) were found to be significantly regulated by TsA (Supplementary Table S3). Interestingly, the microarray confirms the TsA regulation measured by qPCR only of those genes that show significant effects already at lower TsA concentrations (Supplementary Figure S5), i.e. DUSP10, NFKBIA, CDKN2D, HBEGF, THBD, GZF1, NAPB and EP300.
In summary, the majority of the tested genes were either up- or down-regulated by a short-term TsA treatment. On a transcriptome-wide level this holds true for 18.4% of all genes. Regardless whether the tested genes responded already to lower TsA concentrations or only to higher amounts, all responses occurred at early time points.
Time-dependent transcriptome changes in response to 1α,25(OH)2D3 and TsA
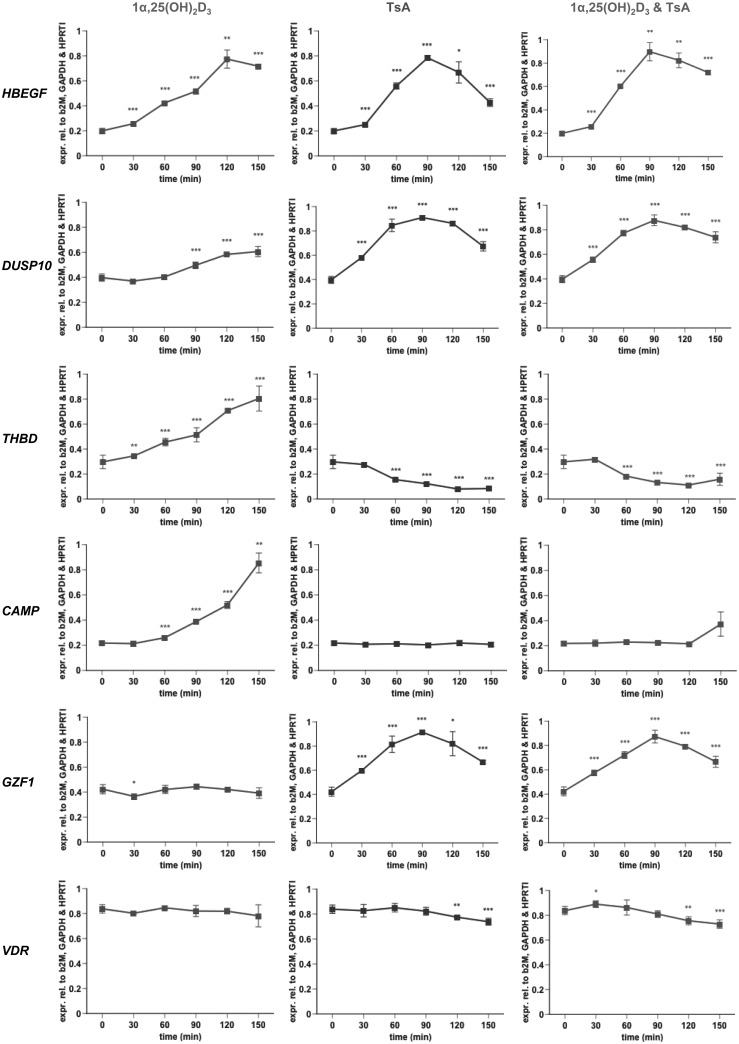
The experiments above indicated that time is an important parameter for the transcriptional response of genes to both 1α,25(OH)2D3 and TsA. Since our focus was on the primary response to both compounds, we performed time course experiments, in which THP-1 cells were treated with 1α,25(OH)2D3 and TsA, alone and in combination, every 30 min in a time frame of 0–150 min (Figure 2 and Supplementary Figure S7). By qPCR we tested the expression of the same 12 genes used in the previous experiments. Judging by their individual response to 1α,25(OH)2D3 and TsA, the 12 genes can be grouped into 5 classes. Class I contains the genes HBEGF and DUSP10, which are up-regulated by both 1α,25(OH)2D3 and TsA, class II is represented by the genes THBD and NFKBIA, which are up-regulated by 1α,25(OH)2D3 and down-regulated by TsA, and class III contains the CAMP gene, which is strongly up-regulated by 1α,25(OH)2D3 but does not respond to TsA treatment. The genes GZF1, CDKN2D and NAPB are representatives of class IV, which is characterized by no major response to 1α,25(OH)2D3 but an up-regulation by TsA. Finally, class V contains the genes VDR, CD93 and EP300, which show no major response to 1α,25(OH)2D3 within the investigated time frame but are down-regulated by TsA. The NXT1 gene cannot be classified non-ambiguously to any of the classes, since its response to both 1α,25(OH)2D3 and TsA is rather weak and transient. Interestingly, with the exception of HBEGF, no major difference could be detected for the response of the remaining 11 genes to the combined application of 1α,25(OH)2D3 and TsA in comparison to their response to TsA alone. This seems to suggest that at the early time points the effect of the treatment with TsA is far more dominant than that with 1α,25(OH)2D3.
Figure 2.
Early time course expression profiling of six representative genes. qPCR was performed to determine the relative changes of mRNA expression of selected genes normalized by the three reference genes B2M, GAPDH and HPRT1 in THP-1 cells in response to incubation with 100 nM 1α,25(OH)2D3 and 300 nM TsA, alone or in combination, over a time period of 150 min. A representative of at least three independent experiments, each performed in triplicate individual cell treatments, is shown. The data points represent the means of three cell treatments and the bars indicate standard deviations. Two-tailed Student’s t-tests were performed to determine the significance of the mRNA induction by the stimuli in reference to solvent-treated cells (*P < 0.05; **P < 0.01; ***P < 0.001).
Genes that respond neither to 1α,25(OH)2D3 nor TsA can be sorted into class VI. In principle there could be three additional classes of genes based on their response to 1α,25(OH)2D3 and TsA: genes that are down-regulated by 1α,25(OH)2D3 and either up-, down- or not regulated by TsA. These three classes are not represented by the 12 genes selected for qPCR analysis (Figure 2 and Supplementary Figure S7). Therefore, we performed at the end point of the time course series, 150 min, a microarray from THP-1 cells that were treated with 1α,25(OH)2D3 and TsA, alone and in combination, and compared them to the common solvent-treated control (Supplementary Figure S8 and Supplementary Table S3). The 4264 genes that were significantly regulated by TsA were already referred to above (Supplementary Figure S6). These TsA target genes show a large overlap of 3236 candidates with the 4253 genes that are regulated by the combination of 1α,25(OH)2D3 and TsA (Supplementary Figure S8). Notably, 1017 genes show a transcriptional response uniquely when cells are treated with both TsA and 1α,25(OH)2D3 but not when treated with TsA alone. Interestingly, only one of these 1017 genes, the gene leucine rich repeat containing 8 family, member A (LRRC8A), is a target of 1α,25(OH)2D3 alone (Supplementary Figure S8). All these numbers are high compared to the 27 genes that are regulated by 1α,25(OH)2D3 alone. Of the 25 up-regulated genes, 6 can be sorted into class I (up-regulation by TsA), 11 into class II (down-regulation by TsA) and 8 into class III (no response to TsA) (Supplementary Table S3). In contrast, class IV (up-regulation only by TsA alone) is represented by 2157 genes and class V (down-regulation only by TsA alone) by 2107 genes, while 7486 genes (63.6% of all expressed genes) belong to class VI (no response to neither of the 2 compounds). It should be noted that the genes DUSP10 and NFKBIA were not detected as 1α,25(OH)2D3 target genes in this microarray analysis, although we found them in THP-1 cells significantly responsive to 1α,25(OH)2D3 as measured by qPCR in this study and by microarray after 240 min treatment in a previous study (34). This may be in part due to the different time points in the two microarray experiments or the different locations of microarray probes compared to qPCR primers, which could lead to the detection of alternative transcript variants.
To get an alternative view on the results, we determined the subset of all expressed genes that show a significant difference between the treatment with TsA alone and the combined stimulation with 1α,25(OH)2D3. We identified 392 genes of which 225 were higher expressed in the presence of both compounds compared to a treatment with TsA alone (the HBEGF gene is one of them) and 167 were lower expressed (Supplementary Table S3). Interestingly, 200 of these 392 genes are responsive to TsA alone (94 up- and 106 down-regulated), two (HBEGF and zyxin (ZYX)) are 1α,25(OH)2D3 target genes (one up- and one down-regulated), while nearly half (190) are not significantly regulated by either single treatment. Interestingly, ZYX is already known to be a 1α,25(OH)2D3 and TSA target gene in prostate cancer cells (50).
Taken together, based on their transcriptional response to 1α,25(OH)2D3 and/or TsA, genes can be sorted into multiple classes. In short-term treatments, seeking for primary responses, TsA stimulates a far higher number of genes than 1α,25(OH)2D3 and dominates the outcome of the combined treatments. However, there is a subset of TsA target genes that can be modulated by 1α,25(OH)2D3 and even a larger number of additional genes become transcriptionally responsive only when treated with both compounds.
Linking the histone acetylation of TSS and VDR locations of a gene with its transcriptional response to 1α,25(OH)2D3 and TsA
The genomic loci of the VDR target genes HBEGF, DUSP10, NFKBIA and CAMP are displayed, like for the THBD locus (Figure 1A), with VDR ChIP-seq and FAIRE-seq data from THP-1 cells and H3K27ac data from K562 cells (Figure 3A). The regulation of these four genes by 1α,25(OH)2D3 seems to be far more straightforward than that of the THBD gene, since for each of the genes there is only one dominant VDR location in relative vicinity. Interestingly, the genes show different constellations of histone acetylation and open chromatin sites at their TSS and VDR locations. The HBEGF gene shows acetylation only at its TSS region but open chromatin at the VDR peak 10 kb downstream of the gene. The genes DUSP10 and NFKBIA display histone acetylation and open chromatin both at their TSS and the VDR binding regions 220 kb upstream and 60 kb downstream, respectively. In contrast, the CAMP gene shows open chromatin but no histone acetylation at its TSS, which is adjacent to a very strong VDR peak.
We conclude from these genomic views that in the cases of the genes HBEGF, DUSP10 and NFKBIA, where the TSS is acetylated, a treatment with TsA can influence the expression of the respective genes, but not in the case of the CAMP gene, where no TSS acetylation is observed. When in addition also the VDR binding sites are in regions of histone acetylation, such as for the genes DUSP10 and NFKBIA, TsA has the most dominant effect on mRNA transcription. In contrast, in cases of dominant VDR peaks without histone acetylation, a stimulation with 1α,25(OH)2D3 may interfere with the effects of TsA, as observed with the HBEGF gene, or even dominantly control the gene, as in the case of the CAMP gene.
Validation of VDR binding and H3K27 acetylation was performed by regular ChIP in THP-1 cells (Figure 3B). The TSS regions of the genes THBD, NXT1 and NFKBIA and the VDR binding site of the genes DUSP10 and NFKBIA showed far higher basal H3K27 acetylation than the TSS regions of the genes DUSP10 and CAMP and the VDR binding sites of the genes THBD (we selected here the major VDR peak, which is close to the NXT1 gene, see Figure 1A) and CAMP. With the exception of the TSS and the VDR binding site of the DUSP10 gene this confirms the indications obtained from the ENCODE cell lines (Figures 1A and 3A). Interestingly, 1α,25(OH)2D3 increased the acetylation at the TSS region of CAMP and at the VDR binding sites of THBD and CAMP, but decreased it at the TSS regions of THBD and NXT1 and at the VDR binding sites of DUSP10 and NFKBIA (which is close to the PSMA6 gene, see Figure 3A). Treatment with TsA enhanced histone H3K27 acetylation at the TSS region and VDR binding site of CAMP, but decreased it at the TSS region of NFKBIA and at the VDR binding site of NFKBIA. At the TSS regions of NFKBIA and CAMP the combined stimulation with both agents showed effects similar to that of TsA alone. However, at the TSS regions of THBD and NXT1 and the VDR binding sites of DUSP10, THBD and CAMP the combined stimulation rather resembled the effects observed with 1α,25(OH)2D3 alone. The VDR association suggested by our ChIP-seq data was confirmed at all four selected regions. 1α,25(OH)2D3 increased VDR binding at the sites of THBD, DUSP10 and NFKBIA and slightly decreased it at the site of CAMP, where high levels of VDR bind already in the absence of ligand. TsA treatment decreased VDR binding to the sites of THBD and CAMP and had no effects on those of DUSP10 and NFKBIA, while a combined stimulation with 1α,25(OH)2D3 and TsA reduced the increase of VDR association observed with 1α,25(OH)2D3 alone at the VDR binding sites of THBD, DUSP10 and NFKBIA. In line with the results obtained by ChIP-seq, no major recruitment of VDR to the five tested TSS regions could be observed (data not shown).
In summary, the degree of histone acetylation of TSS and VDR binding regions may determine the effect of TsA on mRNA expression and its influence on the regulation of the respective gene by 1α,25(OH)2D3. Regular ChIP essentially confirmed the VDR binding and acetylation at the selected sites and demonstrated that 1α,25(OH)2D3 and TsA have effects on VDR binding to its specific sites as well as on H3K27 acetylation to TSS regions and VDR binding sites.
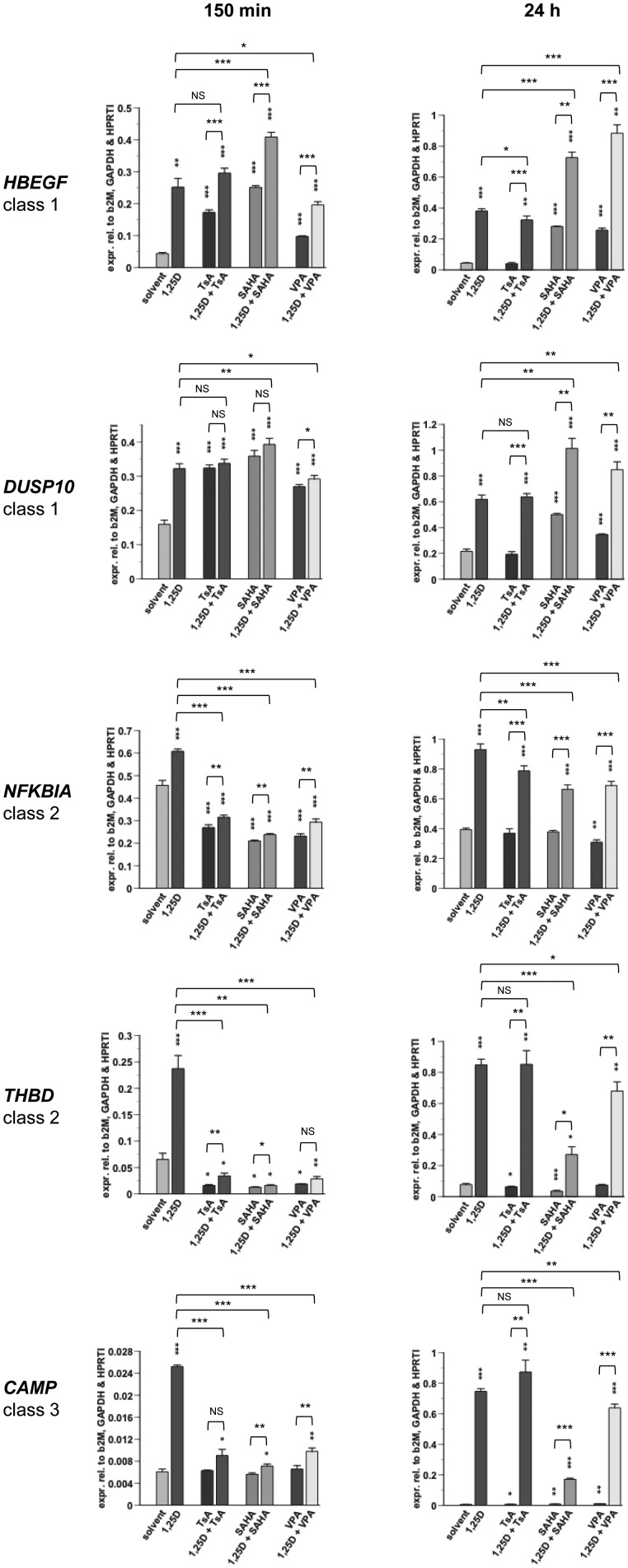
Physiological relevance of a dual treatment with 1α,25(OH)2D3 and HDAC inhibitors
In order to address the physiological relevance of a dual treatment with 1α,25(OH)2D3 and HDAC inhibitors, we compared the effects of TsA with those of SAHA and VPA that are in clinical trials for cancer treatment (51,52), and VPA is also on the market as an anti-convulsant in the therapy of epilepsy (53). In addition to an early time point (150 min), we treated the cells for a physiologically more relevant period of 24 h and determined by qPCR the expression of the genes HBEGF, DUSP10, NFKBIA, THBD and CAMP (Figure 4). We observed at 150 min only minor differences between the three HDAC inhibitors both when they were applied alone or in combination with 1α,25(OH)2D3. However, at the long-term treatment (24 h) gene-specific differences between the three HDAC inhibitors became obvious. The genes HBEGF and DUSP10 were up-regulated by TsA after 150 min (4.0- and 2.0-fold, respectively) but after 24 h their mRNA levels were already back to base-line levels. In contrast, the 150 min response to SAHA and VPA of the genes HBEGF (5.8- and 2.2-fold) and DUSP10 (2.3- and 1.7-fold) was maintained at 24 h (6.4- and 5.9-fold for HBEGF, and 2.3- and 1.6-fold for DUSP10). In contrast, the down-regulation of the NFKBIA gene by a short-term treatment with TsA, SAHA and VPA (1.7-, 2.0- and 2.0-fold, respectively) was lost or nearly lost after 24 h (1.1-, 1.0- and 1.3-fold, respectively), while for the THBD gene the down-regulation observed at 150 min (5.0-, 5.0- and 3.3-fold for TsA, SAHA and VPA, respectively) was reduced by a variable degree at 24 h to 1.3-, 2.0- and 1.0-fold inductions by TsA, SAHA and VPA, respectively. The CAMP gene did not respond to HDAC inhibition at 150 min but was slightly induced in long-term treatments (1.2-, 1.4- and 1.7-fold for TsA, SAHA and VPA, respectively). Interestingly, at the 24 h time point the 1α,25(OH)2D3 response was not any more dominated by HDAC inhibition. For the genes DUSP10, THBD and CAMP TsA did not even modulate the 1α,25(OH)2D3-mediated effects. However, SAHA and VPA significantly modulated the response of all five genes to 1α,25(OH)2D3. SAHA seems to be the most efficient in this respect: the 11.0-fold induction of THBD by 1α,25(OH)2D3 was reduced to 3.5-fold by the combination with SAHA, for the CAMP gene the increase of mRNA expression was reduced from 121.5-fold to 27.8-fold induction and for the NFKBIA gene from 2.4-fold to 1.7-fold, whereas for the HBEGF gene it increased from 8.7-fold induction to 16.6-fold and for the DUSP10 gene from 2.9-fold to 4.7-fold. These modulations were in the same direction as the effects of the short-term single treatments with the HDAC inhibitors (e.g. the THBD gene was down-regulated by HDAC inhibition for 150 min, while at 24 h the HDAC inhibitors repressed the response of the gene to 1α,25(OH)2D3).
Figure 4.
Short- and long-term effects of HDAC inhibition on VDR target genes. qPCR was performed to determine the relative changes of mRNA expression of selected genes normalized by the three reference genes B2M, GAPDH and HPRT1 in THP-1 cells in response to incubation with 100 nM 1α,25(OH)2D3 (1,25D), 300 nM TsA, 3 µM SAHA and 1 mM VPA, alone or in combination, for 150 min and 24 h. A representative experiment of two independent experiments, both performed in triplicate individual cell treatments, is shown. The data points represent the means of three cell treatments and the bars indicate standard deviations. Two-tailed Student’s t-tests were performed to determine the significance of the mRNA induction by the stimuli in reference to solvent-treated cells (*P < 0.05; **P < 0.01; ***P < 0.001).
Taken together, the effects of the three HDAC inhibitors are quite comparable, i.e. TsA can be replaced by the clinically more relevant compounds SAHA and VPA. For a long-term treatment SAHA seems to be the most effective compound and may be most suited for a clinical application.
DISCUSSION
In this study, we investigated the effects of the highly specific VDR agonist 1α,25(OH)2D3 and the globally acting chromatin modifier TsA on the regulation of their primary target genes. Being an inhibitor of enzyme activity, TsA can act within minutes. This is supported by the observation that chromatin modifications, such as the acetylation and deacetylation of histones, are dynamic and have been shown to change in the order of 20–60 min (35,36,54). Therefore, we focused in this study on early time points, such as 90–150 min, which represent primary effects of HDAC inhibition. In contrast, previous microarray analyses of TsA target genes in different human cancer cell lines measured 24 h after inhibitor treatment, i.e. at a far later time point (55–59). They found between 500 and 7000 target genes, which most likely represent to a large extent secondary and tertiary effects of the TsA treatment. However, these gene numbers are in the same order of magnitude of what we found in this study for genes that significantly change their expression already at an early time point. Even when we use a threshold of 1.4-fold for up- or down-regulation, our list reduces only to 177 genes after 90 min TsA treatment and to 679 genes at time point 150 min. This is far more than the number of genes responding within the same time frame to a stimulation with 1α,25(OH)2D3.
Previous TsA microarray studies (55–59) used concentrations of 300 nM to 5 µM, whereas in our TsA dose–response experiments as many as about half of all tested genes responded to an inhibitor concentration as low as 15 nM. This fits with previous studies in human breast cells, where also some genes responded to TsA concentration as low as 15 nM (28,60). Although this concentration is still above the Kd value of HDACs for the compound (61), we also found genes that responded only at concentrations of 100 nM TsA or higher. In our previous study (28) we had indications that at lower concentrations TsA seems to be less dominant over the effects of other compounds, such as 1α,25(OH)2D3, than at higher amounts, but we could not confirm this in THP-1 cells (data not shown). Interestingly, in human keratinocytes it was reported that a 24 h incubation with 661 nM TsA leads to synergistic effects of HDAC inhibition on the 1α,25(OH)2D3-mediated induction of the genes CAMP, CD14 and 24-hydroxylase (CYP24A1) (62), showing that the response to histone acetylation is cell type- and gene-specific.
The main mechanistic claim of this study is that genes, whose TSS region is not dominantly associated with H3K27 acetylation, are not likely to respond to TsA treatment. This is in line with a study, in which selected HATs and HDACs and the histone modifications H3K9Ac, H4K16Ac and H3K4me were profiled by ChIP-seq (63). Only active genes are shown to be associated with high levels of HATs, HDACs and histone acetylation and to respond to HDAC inhibitor treatment, whereas “poised” genes recruit far less HATs and HDACs and repressed genes no chromatin modifying enzymes at all. In THP-1 cells, the SSTR4 gene is an example for a silenced gene, which does not respond to TsA treatment. The other genes that were investigated in this study are already actively transcribed in non-treated THP-1 cells (Supplementary Figure S1 and data not shown). Furthermore we conclude that, if H3K27 acetylation is found on a VDR target gene’s TSS but not on the region of its VDR binding site, 1α,25(OH)2D3 has a chance to modulate the TsA response of the respective gene. In contrast, when both the TSS and the VDR binding site are controlled by H3K27 acetylation, the effects of 1α,25(OH)2D3 are most likely dominated by those of TsA. This also suggests that, at least in short-term treatments, a chromatin modifying compound, such as TsA, is more effective than a transcription factor modulating compound, such as 1α,25(OH)2D3, when both are addressing the same chromatin region. This observation is in agreement with comparable cases, where a direct effect on chromatin is in most cases more dominant than that of a transcription factor (64,65). From a mechanistic point of view this difference is obvious, since a 1α,25(OH)2D3 modulates the activity of only one transcription factor with a limited number of genomic binding sites, while TsA inhibits the activity of HDACs, which can affect every nucleosome in the whole genome. Moreover, since HDACs work in complex with co-repressor proteins (4,24,50), the modulation of HDAC activity by TsA affects not only a larger portion of the genome but also more nuclear proteins than 1α,25(OH)2D3.
The combined treatment with 1α,25(OH)2D3 and TsA provided a number of interesting results. From the perspective of the stimulation with TsA some 1000 additional genes responded to the treatment when 1α,25(OH)2D3 was used as a co-stimulant, while in parallel about the same number of genes ceased to respond, i.e. 1α,25(OH)2D3 changes the transcriptome profile of TsA by some 25%. We hypothesize that 1α,25(OH)2D3 can shift genes from the active state to the poised state rendering them un-responsive to TsA treatment and vice versa. In some cases at this early time point these shifts may be too small to detect on mRNA level activation or repression effects by the single treatments. We assume that the global effects of TsA allow several hundred VDR binding sites to contribute to the control of new target genes, i.e. the inhibition of HDACs by TsA has a similar effect than a pioneer factor. These binding sites can be either already known VDR peaks or new VDR locations that become accessible by the TsA-induced chromatin opening. In addition, from the 3236 TsA target genes that still respond to a co-treatment with both compounds, 200 significantly change their expression in the combined treatment. This means that although at a high TsA concentration of 300 nM the response of the transcriptome of a cell is dominated by the HDAC inhibitor, 1α,25(OH)2D3 has a modulatory effect on some TsA target genes. The VDR binding sites that control the 1α,25(OH)2D3 response of these genes should then not be under the control of H3K27 acetylation. This concept may be of general impact for dual therapies of cancer or immune diseases: the binding site of the transcription factor mediating the effects of the second compound, such as 1α,25(OH)2D3, should not be acetylated, in order to allow for a beneficial effect of the combinatorial therapeutic regimen. This also suggests that it may be advisable to use lower doses of HDAC inhibitors in dual treatments that aim for more specific responses. Since TsA will not be applied clinically, it is important that the same effects in dual treatments with 1α,25(OH)2D3 can also be obtained by more safe compounds, such as SAHA or VPA. For longer-term treatment, SAHA seems to be the most effective out of the three compounds. Therefore, SAHA may be even more promising than another dual therapy partner of 1α,25(OH)2D3, 9-cis retinoic acid, which is the ligand of the heterodimeric VDR partner protein retinoid X receptor (66).
We sorted VDR target genes into three classes based on whether the effects of TsA pointed into the same direction as 1α,25(OH)2D3, into opposite directions or whether TsA had no effect (Supplementary Table S3). The CAMP gene shows low basal expression compared to the other studied genes, which is in line with the missing H3K27 acetylation at its TSS and its non-responsiveness to TsA treatment. Thus, in the absence of the VDR ligand this gene seems to be in the poised state. The fact that VDR shows prominent binding to the site in the CAMP gene already in the absence of ligand suggests that the unliganded receptor may actively repress the gene possibly via the recruitment of HDACs. In contrast, the higher basal expression levels and responsiveness to HDAC inhibition, which we observed for other genes, indicate that they are actively transcribed already in non-treated THP-1 cells. In the case of the DUSP10 gene a stimulation with both 1α,25(OH)2D3 and TsA results in an up-regulation of mRNA levels. However, we could not observe any additive or synergistic effect of the two compounds, because the DUSP10 gene belongs to the majority of those on which TsA has a dominant effect at short time points. In case of the genes THBD and NFKBIA, a co-treatment with 1α,25(OH)2D3 and TsA even turned an up-regulated VDR target gene into a down-regulated gene. Interestingly, even the complex pattern of the THBD locus resulted in a rather simple response, where the H3K27 acetylation of the THBD TSS and of some VDR binding sites in the locus seems to dictate the outcome of the combined treatment. In contrast, HBEGF represents one of those genes where the response to 1α,25(OH)2D3 is further enhanced by a co-treatment with TsA, which is, to our interpretation, due to the dominant VDR peak without overlaying H3K27 acetylation. As discussed above for the CAMP gene, VDR is also recruited to this HBEGF site in the absence of 1α,25(OH)2D3 and actively represses the gene. This explains the rather strong ligand responsiveness of the HBEGF gene. Another important example of such a type of VDR target genes is CYP24A1, whose response to 1α,25(OH)2D3 was reported to be enhanced by TsA via the increase of histone acetylation and the decrease of histone methylation (67). However, in THP-1 cells the CYP24A1 gene is not very responsive to 1α,25(OH)2D3 (34). Similarly, we have shown previously that mRNA and promoter activity of the primary VDR target gene arachidonate 5-lipoxygenase (ALOX5) were strongly induced by HDAC inhibition and that TsA led to an increased binding of the transcription factors Sp1 and Sp3 to the ALOX5 promoter in Mono Mac 6 cells. However, the combined treatment of TsA with 1α,25(OH)2D3 and TGFβ reduced the strong mRNA induction observed with 1α,25(OH)2D3 and TGFβ alone (27,68).
In conclusion, the effective combination of genome-wide data, such as ChIP-seq for transcription factors and histone acetylation, with microarray analyses provides mechanistic insight into gene regulation. For most genes a combined treatment with 1α,25(OH)2D3 and TsA results in dominance of the chromatin modifier. Moreover, the number of responding genes rather increases than decreases, which not necessarily leads to a more focused response. This may have implications on possible therapeutic applications, such as the combination of a HDAC inhibitor with a nuclear receptor ligand.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Figures 1–8.
FUNDING
Academy of Finland, the Juselius Foundation (Finland) and Télévie (Belgium). Funding for open access charge: Juselius Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jussi Ryynänen for critical reading of the manuscript. Kind thanks to the Finnish Microarray and Sequencing Centre in Turku, Finland, and Genomics Core Facility at the EMBL in Heidelberg, Germany, for microarray and massive parallel sequencing services, respectively. S.S. and C.C. designed the project; S.S. performed the experiments; S.S., S.H. and C.C. analysed the data; and S.S. and C.C. wrote the article.
REFERENCES
- 1.Eberharter A, Becker PB. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 2002;3:224–229. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perissi V, Rosenfeld MG. Controlling nuclear receptors: the circular logic of cofactor cycles. Nat. Rev. Mol. Cell Biol. 2005;6:542–554. doi: 10.1038/nrm1680. [DOI] [PubMed] [Google Scholar]
- 3.Carlberg C, Molnár F. Current status of vitamin D signaling and its therapeutic applications. Curr. Top Med. Chem. 2012;12:528–547. doi: 10.2174/156802612799436623. [DOI] [PubMed] [Google Scholar]
- 4.Polly P, Herdick M, Moehren U, Baniahmad A, Heinzel T, Carlberg C. VDR-Alien: a novel, DNA-selective vitamin D3 receptor-corepressor partnership. FASEB J. 2000;14:1455–1463. doi: 10.1096/fj.14.10.1455. [DOI] [PubMed] [Google Scholar]
- 5.Abedin SA, Banwell CM, Colston KW, Carlberg C, Campbell MJ. Epigenetic corruption of VDR signalling in malignancy. Anticancer Res. 2006;26:2557–2566. [PubMed] [Google Scholar]
- 6.Nottke A, Colaiácovo MP, Shi Y. Developmental roles of the histone lysine demethylases. Development. 2009;136:879–889. doi: 10.1242/dev.020966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlberg C, Dunlop TW, Saramäki A, Sinkkonen L, Matilainen M, Väisänen S. Controlling the chromatin organization of vitamin D target genes by multiple vitamin D receptor binding sites. J. Steroid Biochem. Mol. Biol. 2007;103:338–343. doi: 10.1016/j.jsbmb.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 9.Lewis BA, Reinberg D. The mediator coactivator complex: functional and physical roles in transcriptional regulation. J. Cell Sci. 2003;116:3667–3675. doi: 10.1242/jcs.00734. [DOI] [PubMed] [Google Scholar]
- 10.Marks PA, Richon VM, Breslow R, Rifkind RA. Histone deacetylase inhibitors as new cancer drugs. Curr. Opin. Oncol. 2001;13:477–483. doi: 10.1097/00001622-200111000-00010. [DOI] [PubMed] [Google Scholar]
- 11.Monneret C. Histone deacetylase inhibitors. Eur. J. Med. Chem. 2005;40:1–13. doi: 10.1016/j.ejmech.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Malinen M, Ryynänen J, Heinäniemi M, Väisänen S, Carlberg C. Cyclical regulation of the insulin-like growth factor binding protein 3 gene in response to 1α,25-dihydroxyvitamin D3. Nucleic Acids Res. 2011;39:502–512. doi: 10.1093/nar/gkq820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pereira F, Barbáchano A, Silva J, Bonilla F, Campbell MJ, Muñoz A, Larriba MJ. KDM6B/JMJD3 histone demethylase is induced by vitamin D and modulates its effects in colon cancer cells. Hum. Mol. Genet. 2011;20:4655–4665. doi: 10.1093/hmg/ddr399. [DOI] [PubMed] [Google Scholar]
- 14.DeLuca HF. Overview of general physiologic features and functions of vitamin D. Am. J. Clin. Nutr. 2004;80:1689S–1696S. doi: 10.1093/ajcn/80.6.1689S. [DOI] [PubMed] [Google Scholar]
- 15.Ingraham BA, Bragdon B, Nohe A. Molecular basis of the potential of vitamin D to prevent cancer. Curr. Med. Res. Opin. 2008;24:139–149. doi: 10.1185/030079908x253519. [DOI] [PubMed] [Google Scholar]
- 16.Verstuyf A, Carmeliet G, Bouillon R, Mathieu C. Vitamin D: a pleiotropic hormone. Kidney Int. 2010;78:140–145. doi: 10.1038/ki.2010.17. [DOI] [PubMed] [Google Scholar]
- 17.Holick MF. Vitamin D: importance in the prevention of cancers, type 1 diabetes, heart disease, and osteoporosis. Am. J. Clin. Nutr. 2004;79:362–371. doi: 10.1093/ajcn/79.3.362. [DOI] [PubMed] [Google Scholar]
- 18.Lampen A, Carlberg C, Nau H. Peroxisome proliferator-activated receptor delta is a specific sensor for teratogenic valproic acid derivatives. Eur. J. Pharmacol. 2001;431:25–33. doi: 10.1016/s0014-2999(01)01423-6. [DOI] [PubMed] [Google Scholar]
- 19.Khan AN, Tomasi TB. Histone deacetylase regulation of immune gene expression in tumor cells. Immunol. Res. 2008;40:164–178. doi: 10.1007/s12026-007-0085-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee MJ, Kim YS, Kummar S, Giaccone G, Trepel JB. Histone deacetylase inhibitors in cancer therapy. Curr. Opin. Oncol. 2008;20:639–649. doi: 10.1097/CCO.0b013e3283127095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shuttleworth SJ, Bailey SG, Townsend PA. Histone deacetylase inhibitors: new promise in the treatment of immune and inflammatory diseases. Curr. Drug Targets. 2010;11:1430–1438. doi: 10.2174/1389450111009011430. [DOI] [PubMed] [Google Scholar]
- 22.Lin RJ, Nagy L, Inoue S, Shao W, Miller WH, Evans RM. Role of the histone deacetylase complex in acute promyelocytic leukemia. Nature. 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 23.Chang TH, Szabo E. Enhanced growth inhibition by combination differentiation therapy with ligands of peroxisome proliferator-activated receptor-gamma and inhibitors of histone deacetylase in adenocarcinoma of the lung. Clin. Cancer Res. 2002;8:1206–1212. [PubMed] [Google Scholar]
- 24.Rashid SF, Moore JS, Walker E, Driver PM, Engel J, Edwards CE, Brown G, Uskokovic MR, Campbell MJ. Synergistic growth inhibition of prostate cancer cells by 1α,25 Dihydroxyvitamin D3 and its 19-nor-hexafluoride analogs in combination with either sodium butyrate or trichostatin A. Oncogene. 2001;20:1860–1872. doi: 10.1038/sj.onc.1204269. [DOI] [PubMed] [Google Scholar]
- 25.Tavera-Mendoza LE, Quach TD, Dabbas B, Hudon J, Liao X, Palijan A, Gleason JL, White JH. Incorporation of histone deacetylase inhibition into the structure of a nuclear receptor agonist. Proc. Natl Acad. Sci. USA. 2008;105:8250–8255. doi: 10.1073/pnas.0709279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banwell CM, O'Neill LP, Uskokovic MR, Campbell MJ. Targeting 1α,25-dihydroxyvitamin D3 antiproliferative insensitivity in breast cancer cells by co-treatment with histone deacetylation inhibitors. J. Steroid Biochem. Mol. Biol. 2004;89–90:245–249. doi: 10.1016/j.jsbmb.2004.03.081. [DOI] [PubMed] [Google Scholar]
- 27.Klan N, Seuter S, Schnur N, Jung M, Steinhilber D. Trichostatin A and structurally related histone deacetylase inhibitors induce 5-lipoxygenase promoter activity. Biol. Chem. 2003;384:777–785. doi: 10.1515/BC.2003.086. [DOI] [PubMed] [Google Scholar]
- 28.Malinen M, Saramäki A, Ropponen A, Degenhardt T, Väisänen S, Carlberg C. Distinct HDACs regulate the transcriptional response of human cyclin-dependent kinase inhibitor genes to Trichostatin A and 1α,25-dihydroxyvitamin D3. Nucleic Acids Res. 2008;36:121–132. doi: 10.1093/nar/gkm913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thurn KT, Thomas S, Moore A, Munster PN. Rational therapeutic combinations with histone deacetylase inhibitors for the treatment of cancer. Future Oncol. 2011;7:263–283. doi: 10.2217/fon.11.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu M, Lee M-H, Cohen M, Bommakanti M, Freedman LP. Transcriptional activation of the Cdk inhibitor p21 by vitamin D3 leads to the induced differentiation of the myelomonocytic cell line U937. Genes Dev. 1996;10:142–153. doi: 10.1101/gad.10.2.142. [DOI] [PubMed] [Google Scholar]
- 31.Warrener R, Beamish H, Burgess A, Waterhouse NJ, Giles N, Fairlie D, Gabrielli B. Tumor cell-selective cytotoxicity by targeting cell cycle checkpoints. FASEB J. 2003;17:1550–1552. doi: 10.1096/fj.02-1003fje. [DOI] [PubMed] [Google Scholar]
- 32.Ramagopalan SV, Heger A, Berlanga AJ, Maugeri NJ, Lincoln MR, Burrell A, Handunnetthi L, Handel AE, Disanto G, Orton SM, et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome Res. 2010;20:1352–1360. doi: 10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer MB, Goetsch PD, Pike JW. VDR/RXR and TCF4/β-catenin cistromes in colonic cells of colorectal tumor origin: iImpact on c-FOS and c-MYC gene expression. Mol. Endocrinol. 2012;26:37–51. doi: 10.1210/me.2011-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heikkinen S, Väisänen S, Pehkonen P, Seuter S, Benes V, Carlberg C. Nuclear hormone 1α,25-dihydroxyvitamin D3 elicts a genome-wide shift in the locations of VDR chromatin accupancy. Nucleic Acids Res. 2011;39:9181–9193. doi: 10.1093/nar/gkr654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saramäki A, Diermeier S, Kellner R, Laitinen H, Väisänen S, Carlberg C. Cyclical chromatin looping and transcription factor association on the regulatory regions of the p21 (CDKN1A) gene in response to 1α,25-dihydroxyvitamin D3. J. Biol. Chem. 2009;284:8073–8082. doi: 10.1074/jbc.M808090200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 37.Carlberg C, Seuter S. Dynamics of nuclear receptor target gene regulation. Chromosoma. 2010;119:479–484. doi: 10.1007/s00412-010-0283-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carlberg C. The impact of transcriptional cycling on gene regulation. Transcription. 2010;1:13–16. doi: 10.4161/trns.1.1.11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsuchiya S, Yamabe M, Yamaguchi Y, Kobayashi Y, Konno T, Tada K. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1) Int. J. Cancer. 1980;26:171–176. doi: 10.1002/ijc.2910260208. [DOI] [PubMed] [Google Scholar]
- 40.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:R34. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toropainen S, Väisänen S, Heikkinen S, Carlberg C. The down-regulation of the human MYC gene by the nuclear hormone 1α,25-dihydroxyvitamin D3 is associated with cycling of corepressors and histone deacetylases. J. Mol. Biol. 2010;400:284–294. doi: 10.1016/j.jmb.2010.05.031. [DOI] [PubMed] [Google Scholar]
- 42.Giresi PG, Lieb JD. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements) Methods. 2009;48:233–239. doi: 10.1016/j.ymeth.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rashid NU, Giresi P, Ibrahim JG, Sun W, Lieb JD. ZINBA integrates local covariates with DNA-seq data to identify broad and narrow regions of enrichment, even within amplified genomic regions. Genome Biol. 2011;12:R67. doi: 10.1186/gb-2011-12-7-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.ENCODE-Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 46.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl Acad. Sci. USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lozzio CB, Lozzio BB. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood. 1975;45:321–334. [PubMed] [Google Scholar]
- 48.Klein E, Ben-Bassat H, Neumann H, Ralph P, Zeuthen J, Polliack A, Vánky F. Properties of the K562 cell line, derived from a patient with chronic myeloid leukemia. Int. J. Cancer. 1976;18:421–431. doi: 10.1002/ijc.2910180405. [DOI] [PubMed] [Google Scholar]
- 49.Greenlee MC, Sullivan SA, Bohlson SS. CD93 and related family members: their role in innate immunity. Curr. Drug Targets. 2012;9:130–138. doi: 10.2174/138945008783502421. [DOI] [PubMed] [Google Scholar]
- 50.Khanim FL, Gommersall LM, Wood VH, Smith KL, Montalvo L, O'Neill LP, Xu Y, Peehl DM, Stewart PM, Turner BM, et al. Altered SMRT levels disrupt vitamin D3 receptor signalling in prostate cancer cells. Oncogene. 2004;23:6712–6725. doi: 10.1038/sj.onc.1207772. [DOI] [PubMed] [Google Scholar]
- 51.Marks PA. Discovery and development of SAHA as an anticancer agent. Oncogene. 2007;26:1351–1356. doi: 10.1038/sj.onc.1210204. [DOI] [PubMed] [Google Scholar]
- 52.Michaelis M, Doerr HW, Cinatl J. Valproic acid as anti-cancer drug. Curr. Pharm. Des. 2007;13:3378–3393. [PubMed] [Google Scholar]
- 53.Bialer M, Yagen B. Valproic Acid: second generation. Neurotherapeutics. 2007;4:130–137. doi: 10.1016/j.nurt.2006.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Downs JA, Nussenzweig MC, Nussenzweig A. Chromatin dynamics and the preservation of genetic information. Nature. 2007;447:951–958. doi: 10.1038/nature05980. [DOI] [PubMed] [Google Scholar]
- 55.Suzuki H, Gabrielson E, Chen W, Anbazhagan R, van Engeland M, Weijenberg MP, Herman JG, Baylin SB. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat. Genet. 2002;31:141–149. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- 56.Lee HS, Park MH, Yang SJ, Jung HY, Byun SS, Lee DS, Yoo HS, Yeom YI, Seo SB. Gene expression analysis in human gastric cancer cell line treated with trichostatin A and S-adenosyl-L-homocysteine using cDNA microarray. Biol. Pharm. Bull. 2004;27:1497–1503. doi: 10.1248/bpb.27.1497. [DOI] [PubMed] [Google Scholar]
- 57.Chittur SV, Sangster-Guity N, McCormick PJ. Histone deacetylase inhibitors: a new mode for inhibition of cholesterol metabolism. BMC Genom. 2008;9:507. doi: 10.1186/1471-2164-9-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dannenberg LO, Edenberg HJ. Epigenetics of gene expression in human hepatoma cells: expression profiling the response to inhibition of DNA methylation and histone deacetylation. BMC Genom. 2006;7:181. doi: 10.1186/1471-2164-7-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roy S, Jeffrey R, Tenniswood M. Array-based analysis of the effects of trichostatin A and CG-1521 on cell cycle and cell death in LNCaP prostate cancer cells. Mol. Cancer Ther. 2008;7:1931–1939. doi: 10.1158/1535-7163.MCT-07-2353. [DOI] [PubMed] [Google Scholar]
- 60.Banwell CM, MacCartney DP, Guy M, Miles AE, Uskokovic MR, Mansi J, Stewart PM, O'Neill LP, Turner BM, Colston KW, et al. Altered nuclear receptor corepressor expression attenuates vitamin D receptor signaling in breast cancer cells. Clin. Cancer Res. 2006;12:2004–2013. doi: 10.1158/1078-0432.CCR-05-1218. [DOI] [PubMed] [Google Scholar]
- 61.Bantscheff M, Hopf C, Savitski MM, Dittmann A, Grandi P, Michon AM, Schlegl J, Abraham Y, Becher I, Bergamini G, et al. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat. Biotechnol. 2011;29:255–265. doi: 10.1038/nbt.1759. [DOI] [PubMed] [Google Scholar]
- 62.Schauber J, Oda Y, Büchau AS, Yun QC, Steinmeyer A, Zügel U, Bikle DD, Gallo RL. Histone acetylation in keratinocytes enables control of the expression of cathelicidin and CD14 by 1,25-dihydroxyvitamin D3. J. Invest. Dermatol. 2008;128:816–824. doi: 10.1038/sj.jid.5701102. [DOI] [PubMed] [Google Scholar]
- 63.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cheng C, Shou C, Yip KY, Gerstein MB. Genome-wide analysis of chromatin features identifies histone modification sensitive and insensitive yeast transcription factors. Genome Biol. 2011;12:R111. doi: 10.1186/gb-2011-12-11-r111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dawson MI, Xia Z. The retinoid X receptors and their ligands. Biochim. Biophys. Acta. 2012;1821:21–56. doi: 10.1016/j.bbalip.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luo W, Karpf AR, Deeb KK, Muindi JR, Morrison CD, Johnson CS, Trump DL. Epigenetic regulation of vitamin D 24-hydroxylase/CYP24A1 in human prostate cancer. Cancer Res. 2010;70:5953–5962. doi: 10.1158/0008-5472.CAN-10-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schnur N, Seuter S, Katryniok C, Rådmark O, Steinhilber D. The histone deacetylase inhibitor trichostatin A mediates upregulation of 5-lipoxygenase promoter activity by recruitment of Sp1 to distinct GC-boxes. Biochim. Biophys. Acta. 2007;1771:1271–1282. doi: 10.1016/j.bbalip.2007.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.