Abstract
The RNA exosome is responsible for a wide variety of RNA processing and degradation reactions. The activity and specificity of the RNA exosome is thought to be controlled by a number of cofactors. Mtr4 is an essential RNA-dependent adenosine triphosphatase that is required for all of the nuclear functions of the RNA exosome. The crystal structure of Mtr4 uncovered a domain that is conserved in the RNA exosome cofactors Mtr4 and Ski2 but not in other helicases, suggesting it has an important role related to exosome activation. Rrp6 provides the nuclear exosome with one of its three nuclease activities, and previous findings suggested that the arch domain is specifically required for Rrp6 functions. Here, we report that the genetic interactions between the arch domain of Mtr4 and Rrp6 cannot be explained by the arch domain solely acting in Rrp6-dependent processing reactions. Specifically, we show that the arch domain is not required for all Rrp6 functions, and that the arch domain also functions independently of Rrp6. Finally, we show that the arch domain of Ski2, the cytoplasmic counterpart of Mtr4, is required for Ski2’s function, thereby confirming that the arch domains of these cofactors function independently of Rrp6.
INTRODUCTION
RNA maturation from the primary transcript to the mature active molecule includes a variety of modifications and cleavages. Mistakes at any step in the maturation process result in aberrant RNAs that are generally targeted for rapid degradation. The eukaryotic RNA exosome is one of the ribonucleases required for maturation of a variety of RNAs and for the degradation of aberrant RNAs. Specifically, the exosome promotes the processing of the 5.8S ribosomal RNA (rRNA), as well as small nucleolar RNA (snoRNA) and snRNA precursors. Moreover, the exosome degrades byproducts of gene expression, including the 5′ external transcribed spacer (5′ ETS) of pre-rRNA, introns and aberrant RNAs such as hypomodified transfer RNAs and messenger RNAs (mRNAs) that lack a stop codon (1–6).
The core of the eukaryotic RNA exosome is composed of nine subunits (7–11). Each of the nine subunits is essential, but catalytically inactive. Rather than being directly responsible for catalysis, they are critical for the exosome structure and are predicted to interact with substrate RNAs. Six of these subunits form a trimer of heterodimers that produce a channel whereby RNA can enter (PH ring) (8,10,12). Three ‘cap proteins’ have conserved putative RNA-binding domains that cluster around the entry site into the PH ring (7,13,14). The exosome core associates with two ribonuclease subunits, Rrrp44 and Rrp6. Rrp44 is on the opposite side of the channel at the putative RNA exit side of the PH ring (15,16). The Rrp44 subunit has both endonuclease and exonuclease domains, and provides the exosome with its two catalytic functions. Rrp6 is present only in a subset of the exosome complexes and is restricted to the nucleus. It thus provides the nuclear form of the exosome with a third catalytic site (7). The available exosome X-ray structures do not include Rrp6, and how it is oriented relative to the PH ring and the Rrp44 active sites is currently unknown.
The core subunits of the exosome are required for all exosome activities, but Rrp6 and a number of exosome cofactors are required for only a subset of its activities. Therefore, these cofactors appear to provide substrate specificity to the RNA exosome. Among the cofactors, Mtr4 and Ski2, collectively, are required for all exosome activities; Mtr4 is required for all of the nuclear functions, whereas Ski2 is required for all of the cytoplasmic functions. Mtr4 and Ski2 are related RNA-dependent adenosine triphosphatases (ATPases) (3,17,18). Mtr4 is also one of the subunits of the TRAMP complex, and as part of TRAMP, Mtr4 is thought to be involved in the degradation of aberrant transcripts (19,20). TRAMP is composed of Mtr4, a non-canonical poly(A) polymerase (either Trf4 or Trf5) and a putative RNA-binding protein (either Air1 or Air2) (19,20). Other exosome functions such as 5.8S rRNA maturation require Mtr4, but not the other TRAMP subunits (19).
The crystal structures of Mtr4 and Ski2 have been solved (17,21,22). This revealed that Mtr4 shares two RecA domains, a winged helix domain and a C-terminal domain with the extended family of Ski2-like RNA and DNA helicases that are involved in a variety of cellular processes (21,22). We will refer to these four domains as the core helicase domains. In addition to these conserved helicase features, the Mtr4 and Ski2 structures revealed two features that appear to be restricted to these two exosome-associated RNA helicases (17,21,22). First, analysis of surface-exposed residues revealed a large conserved area that surrounds the site where the 3′ end of the RNA emerges from the helicase core. The cap proteins of the exosome have a conserved surface similar in diameter, and we have speculated that the bottom of Mtr4 and Ski2 may form the binding site for the cap proteins of the exosome (21). The second Mtr4 feature that was resolved by the structure is a novel domain that is inserted into the winged helix domain and rises above the core helicase domains of Mtr4 on the RNA entry side (i.e. opposite to the conserved bottom surface). This ‘arch’ domain is conserved in all Mtr4 and Ski2 orthologs, but does adopt different conformations in the two published Mtr4 structures and in the Ski2 structure (17,21,22). The arch domain can be deleted from Mtr4 without any effect on its in vitro RNA-dependent ATPase or RNA-unwinding activity (21,22). In vivo, an mtr4-archless mutant complements an mtr4Δ but with a slow growth phenotype. The degradation of the 5′ ETS and the processing of the 5.8S rRNA are both defective in the mtr4-archless mutant (21). Comparison of the mtr4-archless defects with the phenotypes of strains lacking either Rrp44 endonuclease, Rrp44 exonuclease or Rrp6 exonuclease activities revealed a striking similarity between mtr4-archless and the rrp6Δ strains (21). We therefore previously suggested that the arch domain was required for Rrp6 function.
Several questions remain regarding exosome and Mtr4 function. What is the specific role of the arch domain of Mtr4 in terms of exosome-mediated processing and degradation? Is the arch restricted to recruiting Rrp6? To answer these questions we have analyzed the in vivo function of the Mtr4 arch domain in more detail. Here, we show that although the arch is necessary for specific Rrp6 functions, the arch has some functions independent of, and maybe redundant with, Rrp6. Additionally, although the arch domain is not required for the protein–protein interactions with the TRAMP complex subunit Trf5, the absence of the arch can affect Trf5 interaction. Finally, we further confirm that the arch domain functions beyond Rrp6, by performing the first in vivo analysis of the ski2-archless mutant.
MATERIALS AND METHODS
Strains, oligonucleotides and plasmids used are described in Supplementary Tables S1–S3. Most of the experiments were performed in the BY4721 strain background. For the experiments where MTR4 was over-expressed in the rrp6Δ, we used a W303 strain background because the growth defect of rrp6Δ is more pronounced in W303.
Yeast complementation assay
MTR4 and mtr4-archless plasmids were introduced into the mtr4Δ strain and the rrp6Δ mtr4Δ strain through the plasmid shuffle as described in Jackson et al. 2010 (21).
Growth suppression assay
W303 and the isogenic rrp6Δ strain Y765 were obtained from Michael Rosbash and transformed with 2µ plasmids encoding Mtr4 and archless Mtr4 (pAV716 and pAV717, respectively) with the URA3 selectable marker. Transformants were selected on plates lacking uracil (SC-URA). Single colonies were picked and grown overnight at 30°C. The liquid cultures were then serially diluted 1:5 and spotted onto SC-URA plates. Plates were then placed at 22°C, 30°C or 37°C.
Northern blot analysis
Single clones of each of the strains transformed with the MTR4 and mtr4-archless plasmid were streaked onto 5-Fluoroorotic Acid (5-FOA) plates. A patch of cells was grown in liquid overnight at 30°C in yeast peptone and dextrose liquid media (YPD). For the over-expression experiment, cells were streaked and grown in SC-URA plates and liquid media. Cultures were diluted to an OD600 of 0.2 in 40 ml of media and allowed to double twice. Cells were pelleted and frozen when they reached and OD600 of 0.8. Total RNA was isolated, loaded onto urea polyacrylamide gels and transferred to a nylon membrane. The membrane was probed with 5′- 32P-labeled nucleotides for the RNA-specific defects. As a loading control, the RNA subunit of the signal recognition particle (oAV224) was used.
Western blot analysis
Cells were obtained as explained in northern blot analysis, except only 20 ml of liquid media was used to grow the cells. Strains cured with 5-FOA were grown in YPD, and strains over-expressing Mtr4/Mtr4-archless were grown in SC-URA liquid media. Protein was obtained using the glass bead method (23), resolved in a 12% sodium dodecyl sulphate-polyacrylamide gel and transferred to a nitrocellulose membrane. The blot was analyzed using antibodies against Mtr4 (3) at a 1:5000 dilution or against the myc epitope (gifted by Eric J. Wagner) at a 1:1000 dilution. Antibodies against Pgk1 at a 1:10 000 dilution were used as a protein loading control (Molecular Probes).
Yeast two-hybrid (Y2H) assay
Yeast strain PJ69-4a was transformed with either pAV744 (Trf5) or pAV705 (empty vector). Transformants were selected on plates lacking leucine (SC-LEU). Single clones were streaked onto fresh SC-LEU plates. Likewise, PJ69-4α was transformed with pAV745 (Mtr4), pAV746 (Mtr4-archless) and pAV704 (empty vector) and streaked onto plates lacking tryptophan (SC-TRP). Single clones of transformants were mated with each respective tester strain. Briefly, PJ69-4a[pAV744] was crossed to PJ69-4α[pAV746] to determine whether Trf5 interacts with archless Mtr4. After mating, diploids were selected for in plates lacking both leucine and tryptophan (SC-LEU-TRP). Diploids were grown overnight at 30°C in liquid SC-LEU-TRP media. Liquid cultures were then serially diluted and spotted onto media lacking adenine and media lacking histidine (SC-ADE and SC-HIS, respectively) to screen for interaction as described previously in (24), and control media (SC-LEU-TRP). Growth was assessed at 4 and 8 days. SC-HIS plates containing 10 mM 3-aminotriazole (3-AT) were used. A positive control for the yeast two-hybrid (Y2H) method was assessed from the interaction between Mec3 and Rad17 DNA damage checkpoint proteins that form a complex. Negative controls involved Y2H analysis with tester proteins against an empty vector.
For β-gal readings, the diploid Y2H strains were grown overnight at 30°C, diluted to 0.15 OD600 the next day and allowed to double twice. β-Gal activity was quantitated using the Beta-Glo reagent (Promega) as per manufacturers instructions and a Synergy MX automated microplate reader.
His3-nonstop (His3-ns) growth assay
The His3-nonstop (His3-ns) growth assay was performed as previously described (25). Briefly, a ski2Δ (yAV517) strain was transformed with the His3-ns reporter (pAV188). His3-ns reporter lacks in-frame stop codons and encodes for the His3 protein (25). The ski2Δ [pAV188] cells were then transformed with pAV878 (SKI2), pAV879 (ski2-archless) or pRS415 (empty vector). The ski2Δ [pAV188] [pAV878/pAV879/pRS415] cells were serially diluted and replica plated onto plates with media lacking histidine and control plates to select for double transformants (i.e. SC-LEU-URA).
Synthetic lethality growth assay
yAV225 (dcp1-2/ski2Δ) was transformed with pAV876 (SKI2), pAV877 (ski2-archless) and pRS414 (empty vector with TRP maker). Transformations were plated at room temperature (20°C). Single colonies of the transformants were selected, serially diluted and pronged onto SC-TRP plates. Replica plates were placed at 20°C, 30°C and 37°C.
RESULTS
The arch domain of Mtr4 is not required for all Rrp6 functions
We have previously shown that mtr4-archless has RNA processing and degradation defects that resemble those in an rrp6Δ strain. An interesting hypothesis was that the arch domain of Mtr4 was required for all Rrp6 functions. To test whether all of the Rrp6 functions require the arch domain of Mtr4, we analyzed the snoRNA snR33. snoRNA processing requires the core exosome and Rrp6 in separate steps. In rrp6Δ strains, many snoRNAs are extended by three or four nucleotides. Some of these extra nucleotides are encoded in the snoRNA gene, whereas others are added by the TRAMP complex (2,5,26). In addition to these rrp6-specific short 3′ extensions, several different mutations in the core exosome, as well as rrp6Δ, lead to snoRNA species that carry longer encoded 3′ extensions, and some of these 3′ extended species are polyadenylated in a process that requires both TRAMP and Pap1 (2,5,26,27). To test whether either catalytic activity of Rrp44 was involved in snoRNA processing, we analyzed mutants with a point mutation in the Rrp44 exonuclease site (the rrp44-exo− mutant) or with a point mutation in the Rrp44 endonuclease site (the rrp44-endo− mutant). Figure 1 shows that rrp44-exo− accumulates long 3′ extended snR33 species, whereas the rrp44-endo− mutant has no defect in the maturation of snR33. Unlike what was seen for 5.8S rRNA processing and 5′ ETS degradation, the effect of mtr4-archless on processing of snR33 does not resemble the effect of rrp6Δ. The mtr4-archless strain does not accumulate snoRNAs with 2–3 nt extensions or long polyadenylated snoRNAs. We conclude that the arch domain of Mtr4 is only required for specific Rrp6 functions.
Figure 1.
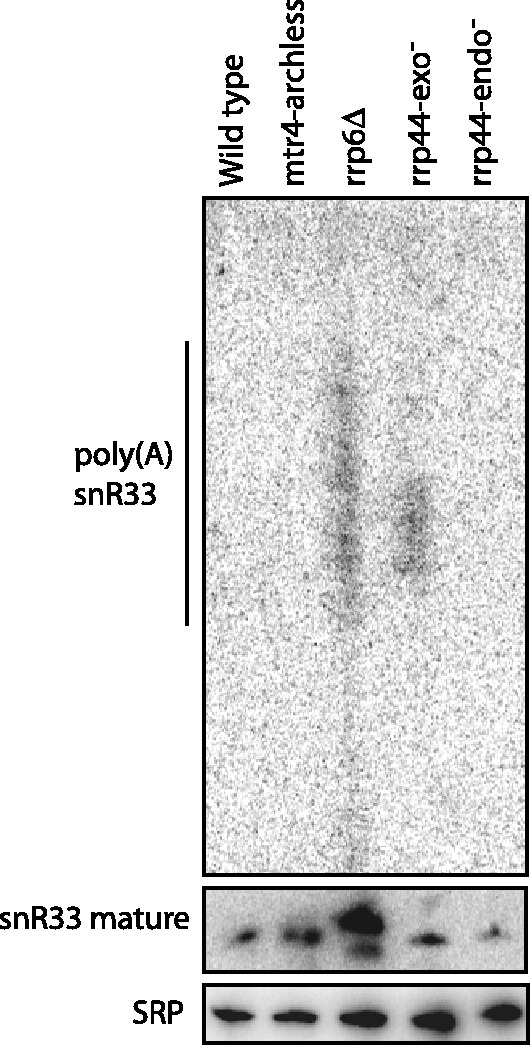
mtr4-archless and rrp6Δ have distinct effects on snoRNA processing. RNA was isolated from each of the strains indicated and analyzed by northern blotting of a urea polyacrylamide gel. The blot was perform using the probes complementary to the mature version of snR33 (middle panel), sequences downstream of the mature snR33 (top panel) and SRP for loading control (bottom panel), described in the oligonucleotide table.
Synthetic defects indicate that the arch domain of Mtr4 can function independently of Rrp6
To further resolve the relation between the functions of the arch domain and Rrp6, we used a genetic analysis and compared growth of the rrp6Δ and mtr4-archless mutants with the growth of an rrp6Δ mtr4-archless double mutant. To do this, we transformed isogenic mtr4Δ and mtr4Δ rrp6Δ strains that contained an MTR4 URA3 plasmid with LEU2 plasmids that contained either MTR4, mtr4-archless or an empty vector. These double transformants were then serially diluted and spotted onto 5-FOA-containing media to select for cells that had lost the MTR4 URA3 plasmid. Two observations suggest that the slow growth of mtr4-archless is not caused by a defect in Rrp6 activation. First, if the slow growth of mtr4-archless was caused by reduced activity of Rrp6, then rrp6Δ should lead to a growth defect at least as severe as mtr4-archless. We did not observe this, and instead, the mtr4-archless growth defect is much more severe than that of rrp6Δ (compare the 2nd and 4th rows of Figure 2). Second, the hypothesis that the arch is needed to assist Rrp6 predicts that deleting the arch in a strain already lacking Rrp6 would have no additional phenotypic effect. Instead, the rrp6Δ mtr4-archless double mutant grows much slower than either of the single mutants (compare row 5 of Figure 2 to rows 2 and 4). This synthetic growth defect indicates that the arch domain of Mtr4 and Rrp6 has overlapping functions, but that each can function independently of the other.
Figure 2.
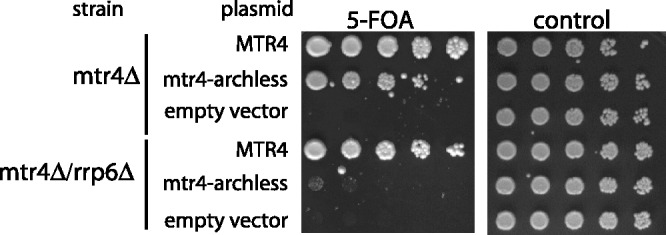
mtr4-archless and rrp6Δ have a synthetic growth defect. The mtr4Δ [MTR4, URA3] and mtr4Δ rrp6Δ [MTR4, URA3] strains were each transformed with plasmids encoding either Mtr4, Mtr4-archless or empty vector with the LEU2 selectable marker. Transformants were serially diluted and spotted onto 5-FOA media and control plate (SC-LEU-URA), and growth was assessed. Medium with 5-FOA selects for cells that have lost the URA3 plasmid (namely the WT copy of the MTR4). Therefore, only the MTR4 alleles that support viability will grow on 5-FOA.
We have previously shown that mtr4-archless and rrp6Δ result in similar 5.8S rRNA processing defects. Specifically, both these mutants accumulate a 5.8S rRNA processing intermediate that retains ∼30 nt of its 3′ extension. To test whether the synthetic growth defect of an rrp6Δ mtr4-archless mutant was related to this shared rRNA processing defect, we compared 5.8S rRNA processing in the double mutant with that in both single mutants by northern blot analysis. Figure 3 shows that the 5.8S rRNA defect in the double mutant is no more severe than in the single mutants. This is consistent with the suggestion that the arch domain and Rrp6 act in the same pathway, and indicates that the slower growth of the double mutant is not correlated with worsening of this processing defect. Overall, the results of the double mutant analysis suggest that the arch domain of Mtr4 and Rrp6 function in similar pathways, but there is not a complete overlap of these functions.
Figure 3.
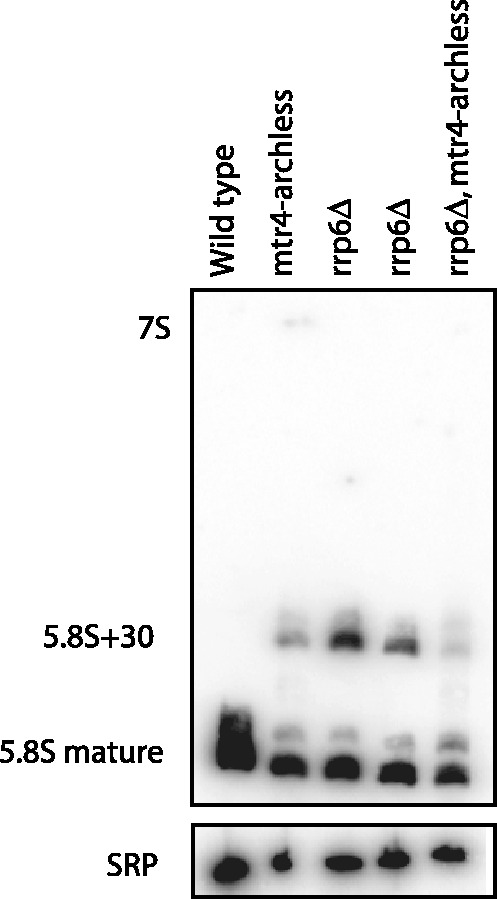
The 5.8S rRNA processing defects of mtr4-archless and rrp6Δ are not additive. RNA was isolated from each of the strains indicated and analyzed by northern blotting of a urea polyacrylamide gel. Two rrp6Δ strains are included; lane 3 is an rrp6Δ MTR4 strain, whereas lane 4 is an rrp6Δ mtr4Δ strain complemented with an MTR4 plasmid for direct comparison with lane 5. Northern blot was performed using the probes complementary to 5.8S rRNA and SRP for loading control.
Over-expression of the core domains of Mtr4 is sufficient to suppress the growth defect of rrp6Δ
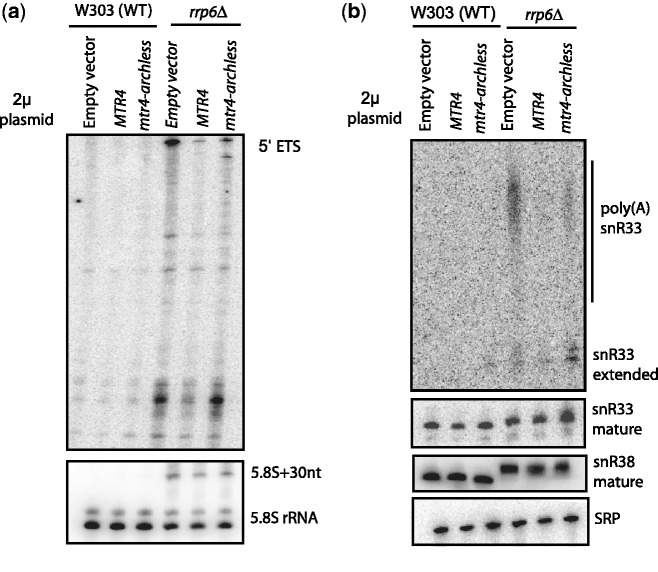
Previously, a high-copy suppressor screen showed that expressing Mtr4 from a high-copy plasmid reduced the growth defect of the rrp6Δ strain (28). To further understand the genetic interactions between RRP6 and MTR4, we analyzed whether this high-copy suppression required the arch domain of Mtr4. The high-copy suppressor screen was done in a different yeast background strain (W303) from the one used in our previous experiments (BY4742) because rrp6Δ in W303 results in a much more severe growth defect. Particularly, the rrp6Δ strain in the W303 background grows slowly at both 37°C and at room temperature. Thus, to fully understand the genetic interactions between rrp6Δ and mtr4-archless, we expressed either wild-type MTR4 or mtr4-archless from a high-copy plasmid in the wild-type and rrp6Δ W303 strain. Transformants were serially diluted, spotted onto plates and incubated at room temperature (20°C), 30°C and 37°C on media lacking uracil to select for transformants with the high-copy plasmid. Figure 4 shows that over-expression of wild-type and archless Mtr4 fully suppresses the growth defect of rrp6Δ at the non-permissive temperature, indicating that the arch domain is not necessary to restore the specific Rrp6-dependent functions that promote growth at the non-permissive temperature. The western blot in Figure 4b confirms that both Mtr4 and Mtr4-archless are over-expressed at similar levels in this yeast strain.
Figure 4.
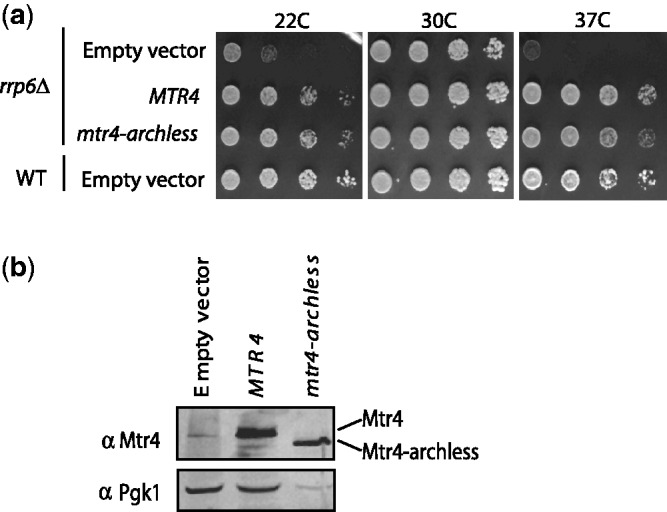
Over-expression of the core helicase domains of Mtr4 is sufficient to suppress the growth defect of an rrp6Δ mutant. (a) Wild-type and rrp6Δ strains in the W303 background strain were each transformed with 2µ (high-copy) plasmids encoding Mtr4, Mtr4-archless and empty vector. Transformants were serially diluted and spotted onto media selecting for the 2µ plasmid at the non-permissive temperatures, 20°C and 37°C, or at the permissive temperature 30°C. (b) Western blot analysis confirms that Mtr4 and Mtr4-archless are over-expressed at similar levels.
Over-expression of Mtr4 restores specific rrp6Δ RNA defects
It was previously reported that although over-expressing Mtr4 suppresses the growth defect in an rrp6Δ, it does not restore the defects in processing 5.8S rRNA and the snoRNA snR38. To determine whether other functions were restored in the rrp6Δ strain with each of the over-expressed Mtr4 versions, we performed northern blot analysis. Total RNA was isolated from transformants for each of these strains: rrp6Δ over-expressing Mtr4, rrp6Δ over-expressing Mtr4-archless and rrp6Δ with a high-copy empty vector. Additionally, control strains that had the wild-type RRP6 gene transformed with each of the high-copy plasmids (WT MTR4, mtr4-archless and empty vector) were analyzed. Over-expression of Mtr4 in the rrp6Δ strain did not affect the accumulation of 5.8S + 30 rRNA or snR38 with short 3′ extensions, as previously reported (29). However, in these same strains, the accumulation of longer polyadenylated forms of snR33 was almost completely suppressed (Figure 5b, compare fourth and fifth lanes), and accumulation of 5′ ETS degradation intermediates was partially suppressed (Figure 5a, compare fourth and fifth lanes). Over-expressing mtr4-archless reduced the rrp6Δ defects to a lesser extent than over-expressing wild-type Mtr4 (compare fifth and sixth lanes in Figure 5a and b). Overall, our results suggest that over-expressing Mtr4 in rrp6Δ does not affect the accumulation of snoRNA species with short 3′ extensions but does suppress the accumulation of longer 3′ extended species. Over-expressing mtr4- archless does so to a smaller extent but must suppress some other defects sufficiently to fully restore growth.
Figure 5.
Over-expression of Mtr4 restores specific rrp6Δ defects. RNA was isolated from the WT and the rrp6Δ strains transformed with high-copy plasmids (described in Figure 4). Northern blot was performed using the oligonucleotide probes against (a) 5′ ETS, 5.8S rRNA, (b) pre-snR33, mature snR33, snR38 and SRP described in the oligonucleotide table.
The arch domain of Ski2 is required to promote the cytoplasmic functions of the exosome
Although Ski2 and Mtr4 homology includes both the core helicase domains and the arch domain, the sequence similarity between the two arch domains is low. Moreover, it is not known whether the arch domain of Ski2 is functional. Like in Mtr4, in vitro activities of Ski2-archless are similar to the wild-type enzyme, including the capacity to form a Ski complex with Ski3 and Ski8 (17). Importantly, Ski2 function is independent of Rrp6, and therefore, if the arch domain of Ski2 is functional, it must function independently of Rrp6. Ski2 localizes to the cytoplasm, therefore only promoting the cytoplasmic functions of the exosome, which in turn are independent of the nuclear Rrp6. To investigate the role of the arch domain in one of the cytoplasmic functions of the exosome, we performed two different assays. First, Ski2 is required for one of the two redundant pathways for mRNA decay. Mutations that disrupt SKI2 function are synthetically lethal with mutations that inactivate the decapping enzyme. Therefore, we analyzed whether ski2-archless could rescue the synthetic lethality of the dcp1-2 ski2Δ strain, which contains a temperature-sensitive mutation that inactivates the decapping enzyme (30). As shown in Figure 6, the dcp1-2 ski2Δ synthetic lethality can not be rescued by the ski2-archless plasmid. Importantly, western blotting indicated that Ski2-archless is expressed at levels similar to wild-type Ski2.
Figure 6.
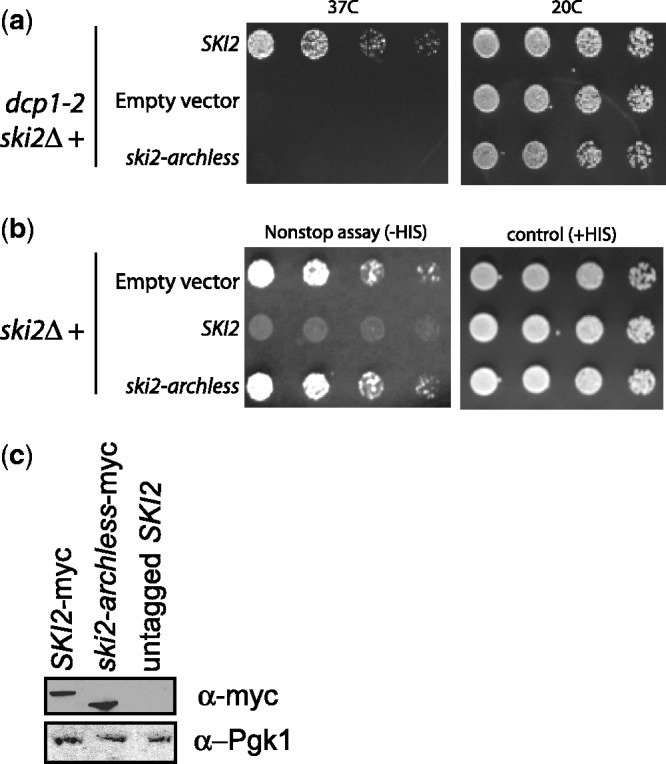
The arch domain of Ski2 is required for the degradation of normal and nonstop mRNAs by the exosome. (a) The dcp1-2 ski2Δ strain was transformed with SKI2, ski2-archless and empty plasmids and grown at either permissive or restrictive temperature (20°C and 37°C, respectively) to assay for defects in general mRNA decay. (b) The ski2Δ strain was transformed with his3-ns reporter and SKI2, ski2-archless and empty vector constructs. The nonstop assay plate lacks histidine, and growth indicates a defective nonstop decay pathway. In both assays, the growth from Ski2-archless resembles the empty vector control, indicating a defect in nonstop mRNA degradation by the exosome. (c) Western blotting with a myc antibody was used to show that Ski2-archless is expressed at similar levels to wild-type Ski2.
The second assay for Ski2 function is based on the preferential degradation of mRNAs that lack a stop codon (nonstop mRNAs) by the cytoplasmic exosome. A strain with SKI2 deleted in the chromosome was first transformed with the His3-ns reporter plasmid and then with either the SKI2, ski2-archless or empty vector plasmids. In cells with a defect in nonstop decay such as in a ski2Δ, the His3-ns reporter is stable, and such cells are able grow in media lacking histidine. Conversely, cells with a functional Ski2 quickly degrade the aberrant His3-ns mRNA, thereby unable to grow in media lacking the amino acid. The His3-ns growth assay in Figure 6 shows growth in cells expressing the Ski2-archless mutant in plates lacking histidine (non-stop assay, left panel). These two assays indicate that the arch domain of Ski2 is essential for both exosome-mediated functions, nonstop degradation and normal mRNA decay, consistent with the idea that arch domains can act independent of Rrp6 (31).
The arch domain of Mtr4 is not required for Trf5 interaction in vivo, but may modulate Trf5–Mtr4 interaction
Mtr4 is a subunit of the TRAMP complex but also has TRAMP-independent functions. Previous in vitro reconstitution and co-immunoprecipitation assays did not reveal a role for the arch domain in TRAMP complex formation (22,28). The TRAMP complex was initially identified because the Trf4 and Trf5 subunits were isolated in a Y2H screen using Mtr4 as bait (19). Therefore, we used this assay to test whether the arch domain of Mtr4 was required for Trf5 interaction in vivo. Trf5’s Mtr4-binding region was fused to the activation domain of the Gal4 transcription factor, whereas full-length Mtr4 and Mtr4-archless were independently fused to the Gal4 DNA-binding domain. Growth on plates lacking adenine or histidine indicated that there is positive interaction between Trf5 and Mtr4 even when the arch is removed (Figure 7). Thus, as seen in vitro, the arch domain is not required for in vivo interaction with Trf5. Surprisingly, the Mtr4-archless construct provided a more robust Y2H interaction than wild-type Mtr4. The activation of the HIS3 reporter gene in Y2H can be assayed semi-quantitatively on plates containing different concentrations of 3-AT, which is a competitive inhibitor of the His3 enzyme. Using this approach, the Y2H interaction between Trf5 and Mtr4 resulted in growth on plates containing 25 mM, but not 50 mM, 3-AT. Under identical conditions, the interaction between Trf5 and Mtr4-archless was sufficient to provide growth at up to 100 mM 3-AT (data not shown). Additionally, the LacZ reporter gene of the Y2H strains was activated 3-fold more by the Mtr4-archless–Trf5 interaction than the full-length Mtr4–Trf5 interaction (Figure 7). A stronger Y2H interaction can be caused by differences in expression level of the hybrid proteins or by a difference in the strength of the interaction. Using western blot analysis with anti-Mtr4 antibodies, we confirmed that the Gal4-AD-Mtr4 and Gal4-AD-Mtr4-archless proteins are expressed at similar levels, suggesting that the Mtr4-archless protein interacts better with Trf5 in the Y2H assay. As no change in TRAMP complex formation was observed in in vitro reconstitution or co-immunoprecipitation assays (22,28), the enhanced Y2H interaction is unlikely due to a direct change in binding affinity and may reflect involvement of additional proteins not stably associated with TRAMP.
Figure 7.
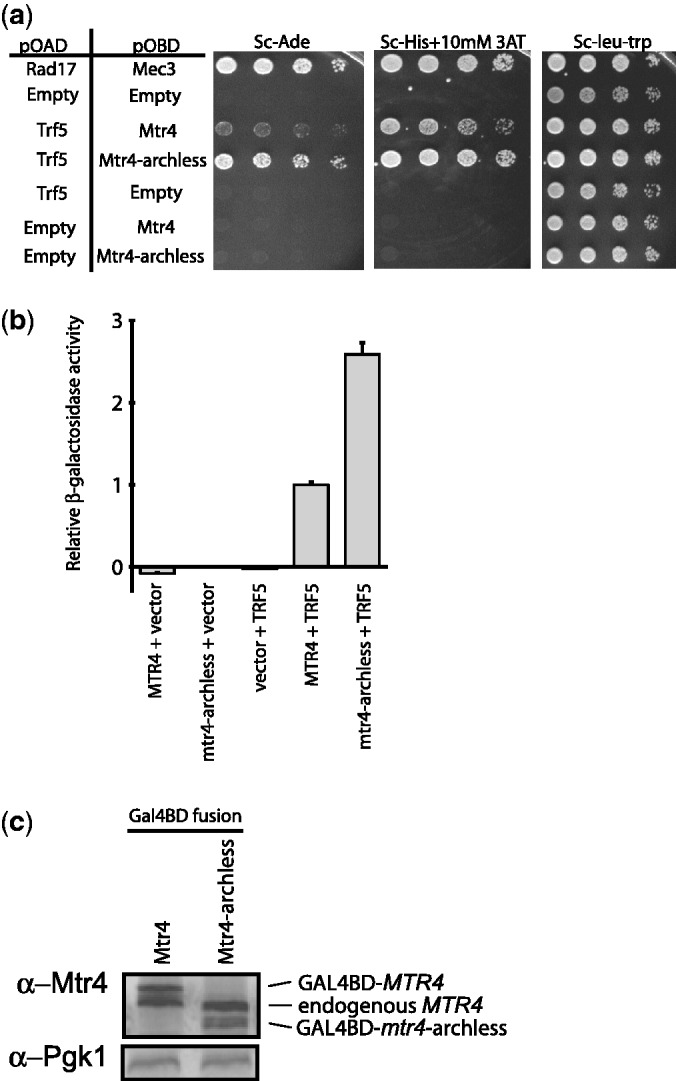
The arch domain of Mtr4 is not required for the Trf5–Mtr4 binding in vivo. (a) Y2H analysis was performed as explained in the ‘Materials and Methods’ section. Growth on SC-ADE and SC-HIS + 10 mM 3-AT indicates the physical interaction between the proteins tested. Empty vector were used against each of the proteins tested to control for autoactivation. (b) Quantitation of β-galactosidase expression. Plotted is the mean and standard deviation of triplicate cultures. The background reading obtained with empty vectors of pOAD and pOBD was subtracted. (c) Western blot analysis confirms the similar expression levels between Mtr4 and Mtr4-archless in the Y2H strains.
DISCUSSION
The crystal structure of Mtr4 revealed a novel arch domain (21,22). In vitro analysis of Mtr4 lacking this domain showed that the arch domain was not required for RNA-dependent ATPase activity, RNA helicase activity or TRAMP complex formation (21,22). However, initial in vivo experiments indicated that mtr4-archless had similar effects to rrp6Δ (21). Based on that observation, the arch domain was proposed to help make RNA substrate accessible to Rrp6, possibly by removal of the RNA from the core exosome (21). Here, we present a more complete description of the genetic interactions between mtr4-archless and rrp6Δ. Several in vivo effects can not be fully explained by the previous hypothesis, suggesting that the arch domain has additional functions independent of Rrp6.
First, we show that mtr4-archless and rrp6Δ have different effects on snoRNA maturation. The rrp6Δ mutant has previously been shown to accumulate distinct species of snoRNAs with 3′ extensions. rrp6Δ mutants accumulate snoRNAs with short extensions that can include a few A residues added by the TRAMP complex. Additionally, the rrp6Δ strain and other exosome mutants also accumulate some longer 3′ extended snoRNA species that can be polyadenylated (2,5). In contrast to rrp6Δ, the mtr4-archless mutation does not cause the accumulation of either species. Therefore, Rrp6 performs these functions without requiring the arch domain of Mtr4.
Second, we describe a synthetic growth defect between mtr4-archless and rrp6Δ, indicating that the arch domain of Mtr4 and Rrp6 can both function in the absence of each other. Specifically, if the sole function of the arch domain was to mediate Rrp6 function, deletion of the arch domain should have no further effect in rrp6Δ. The previously described 5.8S rRNA processing defect in rrp6Δ and mtr4-archless was no worse in the double mutant, consistent with the idea that the arch domain and Rrp6 act together in one pathway, but the more severe growth defect of the double mutant suggests that the arch domain has some other Rrp6-independent function.
Third, we show that over-expressing either full-length Mtr4 or Mtr4-archless can suppress the growth defect of rrp6Δ, suggesting that the function of the core helicase domains of Mtr4 also functionally overlaps with the function of Rrp6. Mtr4 was previously isolated as a high-copy suppressor of rrp6Δ, indicating that Mtr4 has some function that overlaps with that of Rrp6, and that Mtr4 can perform that function in the absence of Rrp6. We show that over-expression of Mtr4-archless also fully suppresses the growth defect of rrp6Δ. It was previously reported that Mtr4 over-expression did not suppress the accumulation of 5.8S + 30 rRNA or the snoRNAs with short TRAMP-generated 3′ extensions. We confirm this, but also show that Mtr4 does suppress some other processing defect of rrp6Δ, including the accumulation of snoRNAs with long poly(A) tails and 5′ ETS degradation intermediates. Mtr4-archless partially suppresses these same defects. This implies that not only do the functions of the arch domain and Rrp6 overlap but also the functions of the core helicase domains on Mtr4 and Rrp6 overlap.
Finally, we identified that, like Mtr4, the arch domain of Ski2 is important to promote exosome functions. Ski2-archless mutant is not able to stimulate normal and nonstop mRNA degradation by the exosome. This suggests that the arch domains of Mtr4 and Ski2 may have similar roles in assisting some function of the core exosome.
Overall, our genetic and molecular experiments indicate that Rrp6, the arch domain of Mtr4, and the core helicase domains of Mtr4 interact in different and possibly overlapping pathways that can not currently be explained by one simple model. Instead, we suggest that Mtr4 uses its arch domain and RNA-dependent ATPase activity promiscuously to modulate multiple RNA and protein interactions during RNA exosome processing and degradation rather than for one specific rearrangement.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3.
FUNDING
NIH [GM069900 and GM099790 to A.v.H.]; Welch foundation [AU1773 to A.v.H.]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Michael Rosbash for providing the W303 strain and the rrp6Δ strain derived from it, Patrick Linder for providing the anti-Mtr4 antibodies, Stan Fields for providing yeast two hybrid strains and plasmids, Eric J. Wagner’s laboratory for anti-myc antibodies, Sean Johnson’s laboratory for providing the SKI2 and ski2-archless plasmid in the Escherichia coli expression vector from which the yeast SKI2 constructs were derived and the van Hoof laboratory for comments on the work.
REFERENCES
- 1.Allmang C, Mitchell P, Petfalski E, Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000;28:1684–1691. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 5.van Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 2000;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Jia H, Jankowsky E, Anderson JT. Degradation of hypomodified tRNA(iMet) in vivo involves RNA-dependent ATPase activity of the DExH helicase Mtr4p. RNA. 2008;14:107–116. doi: 10.1261/rna.808608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′ –> 5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buttner K, Wenig K, Hopfner KP. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol. Cell. 2005;20:461–471. doi: 10.1016/j.molcel.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 9.Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 10.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 11.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′–>5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 12.Lorentzen E, Dziembowski A, Lindner D, Seraphin B, Conti E. RNA channelling by the archaeal exosome. EMBO Rep. 2007;8:470–476. doi: 10.1038/sj.embor.7400945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malet H, Topf M, Clare DK, Ebert J, Bonneau F, Basquin J, Drazkowska K, Tomecki R, Dziembowski A, Conti E, et al. RNA channelling by the eukaryotic exosome. EMBO Rep. 2010;11:936–942. doi: 10.1038/embor.2010.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mian IS. Comparative sequence analysis of ribonucleases HII, III, II PH and D. Nucleic Acids Res. 1997;25:3187–3195. doi: 10.1093/nar/25.16.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139:547–559. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- 16.Wang HW, Wang J, Ding F, Callahan K, Bratkowski MA, Butler JS, Nogales E, Ke A. Architecture of the yeast Rrp44 exosome complex suggests routes of RNA recruitment for 3′ end processing. Proc. Natl Acad. Sci. USA. 2007;104:16844–16849. doi: 10.1073/pnas.0705526104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halbach F, Rode M, Conti E. The crystal structure of S. cerevisiae Ski2, a DExH helicase associated with the cytoplasmic functions of the exosome. RNA. 2012;18:124–134. doi: 10.1261/rna.029553.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang S, Hitomi M, Hu YH, Liu Y, Tartakoff AM. A DEAD-box-family protein is required for nucleocytoplasmic transport of yeast mRNA. Mol. Cell. Biol. 1996;16:5139–5146. doi: 10.1128/mcb.16.9.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 20.Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson RN, Klauer AA, Hintze BJ, Robinson H, van Hoof A, Johnson SJ. The crystal structure of Mtr4 reveals a novel arch domain required for rRNA processing. EMBO J. 2010;29:2205–2216. doi: 10.1038/emboj.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weir JR, Bonneau F, Hentschel J, Conti E. Structural analysis reveals the characteristic features of Mtr4, a DExH helicase involved in nuclear RNA processing and surveillance. Proc. Natl Acad. Sci. USA. 2010;107:12139–12144. doi: 10.1073/pnas.1004953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jazwinski SM. Preparation of extracts from yeast. Methods Enzymol. 1990;182:154–174. doi: 10.1016/0076-6879(90)82015-t. [DOI] [PubMed] [Google Scholar]
- 24.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 25.Schaeffer D, Meaux S, Clark A, van Hoof A. Determining in vivo activity of the yeast cytoplasmic exosome. Methods Enzymol. 2008;448:227–239. doi: 10.1016/S0076-6879(08)02612-8. [DOI] [PubMed] [Google Scholar]
- 26.Grzechnik P, Kufel J. Polyadenylation linked to transcription termination directs the processing of snoRNA precursors in yeast. Mol. Cell. 2008;32:247–258. doi: 10.1016/j.molcel.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider C, Leung E, Brown J, Tollervey D. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res. 2009;37:1127–1140. doi: 10.1093/nar/gkn1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holub P, Lalakova J, Cerna H, Pasulka J, Sarazova M, Hrazdilova K, Arce MS, Hobor F, Stefl R, Vanacova S. Air2p is critical for the assembly and RNA-binding of the TRAMP complex and the KOW domain of Mtr4p is crucial for exosome activation. Nucleic Acids Res. 2012;40:5679–5693. doi: 10.1093/nar/gks223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abruzzi K, Denome S, Olsen JR, Assenholt J, Haaning LL, Jensen TH, Rosbash M. A novel plasmid-based microarray screen identifies suppressors of rrp6Delta in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:1044–1055. doi: 10.1128/MCB.01299-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobs Anderson JS, Parker RP. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown JT, Bai X, Johnson AW. The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA. 2000;6:449–457. doi: 10.1017/s1355838200991787. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.