Abstract
Topoisomerase V (Topo-V) is the only member of a novel topoisomerase subtype. Topo-V is unique because it is a bifunctional enzyme carrying both topoisomerase and DNA repair lyase activities within the same protein. Previous studies had shown that the topoisomerase domain spans the N-terminus of the protein and is followed by 12 tandem helix–hairpin–helix [(HhH)2] domains. There are at least two DNA repair lyase active sites for apurinic/apyrimidinic (AP) site processing, one within the N-terminal region and the second within the C-terminal domain of Topo-V, but their exact locations and characteristics are unknown. In the present study, the N-terminal 78-kDa fragment of Topo-V (Topo-78), containing the topoisomerase domain and one of the lyase DNA repair domains, was characterized by structural and biochemical studies. The results show that an N-terminal 69-kDa fragment is the minimal fragment with both topoisomerase and AP lyase activities. The lyase active site of Topo-78 is at the junction of the fifth and sixth (HhH)2 domains. From the biochemical and structural data, it appears that Lys571 is the most probable nucleophile responsible for the lyase activity. Our experiments also suggest that Topo-V most likely acts as a Class I AP endonuclease in vivo.
INTRODUCTION
DNA topoisomerases are enzymes responsible for maintaining the topological state of DNA in the cell and are also involved in many other important cellular processes (1). They achieve this by relaxing/supercoiling, catenating/decatenating and knotting/unknotting DNA molecules (2). The importance of topoisomerases is emphasized by the fact that they are present in all domains of life (bacteria, archaea and eukarya) and help modulate several fundamental cellular processes, such as DNA replication, recombination and transcription (2,3). Topoisomerases create a transient break in either one or two strands of DNA, which is followed by either enzyme-bridged strand passage or controlled rotation of the DNA, thus modifying the DNA topology (3,4). At the end of the reaction, the DNA break is resealed and DNA is released from the protein. DNA topoisomerases are classified into two types based on whether they operate through a single- (Type I) or double-stranded (Type II) DNA break. Type I enzymes use the torsional energy stored in the supercoiled DNA to drive DNA relaxation, whereas Type II enzymes, in addition, require adenosine triphosphate for activity (4).
Type I topoisomerases are further subdivided into three subtypes, IA, IB and IC, based on sequence and structural similarities (5). Type IA and IB enzymes are well characterized with regard to their structure, DNA-binding mode, cleavage/religation mechanism and overall DNA relaxation mode (4). Methanopyrus kandleri topoisomerase V (Topo-V) is the most recently identified distinct topoisomerase and defines the Type IC subtype (6–8). Topo-V is found only in the archaeal Methanopyrus genus, which lives in deep hot vents in oceans (9). The enzyme is active at very high temperatures (122°C) and salt concentrations. Topo-V has a molecular mass of 112 kDa (984 amino acids) and is unique amongst topoisomerases owing to the presence of DNA relaxation and DNA repair lyase activities in the same polypeptide (10). DNA relaxation is carried out by the N-terminal domain (residues 1–268) (11), whereas the rest of the molecule is organized into 24 tandem helix–hairpin–helix (HhH) DNA-binding motifs arranged as 12 tandem (HhH)2 domains, some of which are involved in DNA repair lyase activity (8,12).
The topoisomerase domain of Topo-V has been well characterized by structural, biochemical and single-molecule studies. Structural studies of N-terminal fragments of Topo-V revealed a completely new fold with a different active site when compared with other topoisomerases, and recognized the need for conformational changes in the (HhH)2 domains to permit DNA binding (8,11). Furthermore, single-molecule studies confirmed that Topo-V relaxes DNA by a swiveling mechanism constrained by friction (13), similar to that used by Type IB topoisomerases (14). In contrast, the DNA repair lyase activity is not well understood or characterized. Previous biochemical studies had shown that Topo-V possesses apurinic/apyrimidinic (AP) lyase and deoxyribose-5-phosphate (dRP) lyase activities (10,12), which are important steps in the DNA base excision repair (BER) pathway. In BER, DNA lesions are processed by removing a damaged base, creating an abasic site, followed by cleavage of the DNA backbone, resulting in a gap that is later filled by a DNA polymerase (15). The enzymes that cleave an abasic site are classified into two groups based on their mechanism: Class I AP endonucleases (AP lyases) cleave 3′ to the AP site sugar via a β-elimination reaction, whereas Class II endonucleases cleave 5′ to the AP site sugar through a hydrolysis reaction (16,17). In Topo-V, there are at least two different DNA repair active sites, one located in the N-terminal 78-kDa fragment and the other in the 34-kDa C-terminal part of the protein (10,12), but the exact locations and mechanisms of action of these repair activities are unknown.
In the present study, we identified and characterized one of the DNA repair active sites of Topo-V. The N-terminal 78-kDa fragment of Topo-V (Topo-78), containing both DNA repair and relaxation activities, was studied by a combination of structural and biochemical methods. The minimal fragment containing AP lyase and dRP lyase activities was identified by deletion mutagenesis and corresponds to the N-terminal 69-kDa fragment spanning residues 1–599. Finer mapping showed that K566, K570 and K571 are needed for full AP lyase and dRP lyase activities. In addition, the 2.9-Å-resolution crystal structure of Topo-78 revealed the architecture of the lyase active site of Topo-78, which is located at the junction of the fifth and sixth (HhH)2 domains. The structure of this active site resembles the structure of other lyase-containing repair enzymes, such as Endonuclease III (EndoIII). Taken together, the studies indicate that the lyase activity occurs by a β-elimination reaction involving a catalytic Lys, and that Topo-V is a Class I AP endonuclease as well as a topoisomerase.
MATERIALS AND METHODS
Protein purification
DNA fragments coding for the Topo-V regions corresponding to residues 1–519 (Topo-61), residues 1–556 (Topo-65), residues 1–599 (Topo-69) and residues 1–640 (Topo-74) were polymerase chain reaction amplified from a plasmid containing the entire Topo-V sequence (12), adding 5′ NcoI and 3′ NdeI restriction sites, and cloned into the pET15b vector. Topo-78 (residues 1–685) and full-length Topo-V were purified as previously described (10,12). Lysine mutants were made by site-directed mutagenesis (QuikChange, Stratagene). For protein production, Topo-V was transformed into Escherichia coli BL21(DE3) pLysS cells, Topo-78 was transformed into E. coli BL21(DE3) cells and all other fragments were transformed into E. coli BL21 Rosetta (DE3) cells. Protein induction and purification were done according to previously reported protocols (11). Pure protein was concentrated and stored in 50 mM Tris (pH 8), 250 mM NaCl and 1 mM dithiothreitol (DTT), except for Topo-V, which required 1 M NaCl for solubility.
AP lyase and dRP lyase assays
AP lyase assays were done using the same DNA and a similar procedure as described before (16). The DNA strand containing uracil (U) was 3′-end labeled using [α-32P] dideoxyAdenosine Triphosphate and terminal deoxynucleotidyl transferase enzyme (USB). After heat-inactivating the labeling mix, it was annealed to a complementary strand and purified using a Bio-Rad Micro Bio-Spin 6 column. To produce the abasic site, the oligonucleotide was treated with uracil-DNA glycosylase (UDG) as previously described (16). The standard AP lyase assay contained 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (pH 7.5), 20 mM KCl, 0.5 mM ethylenediaminetetraacetic acid, 2 mM DTT, 500 nM abasic DNA (UDG treated) and 2.5 µM protein. Incubation was done at room temperature overnight to reduce degradation of substrate DNA. The gel was scanned using a STORM scanner (Molecular Dynamics 860), and bands were quantified using ImageQuantTL version 7.0 (GE Healthcare). Percentage of cleavage was calculated as follows: [intensity of product band/(intensity of substrate band + intensity of product band)] × 100. An overnight incubation was used because the reaction was carried out at suboptimal temperatures for Topo-V. dRP lyase assay was performed using a similar procedure as described before (10,12) In this case, a 34-bp DNA with U at the 16th position was used and pre-treated with UDG to prepare abasic DNA. The DNA was further treated with human AP endonuclease to cleave the sugar phosphate backbone at the abasic site, leaving a 5′-dRP moiety (17). Hundred nanomolar DNA substrate was then incubated with 100 nM of different Topo-V fragments at 60°C for 5 min. After this, the reaction products were stabilized by the addition of freshly prepared 20 mM NaBH4, followed by incubation on ice for 30 min. The samples were then heated at 75°C for 2 min, and the reaction products were separated by denaturing polyacrylamide gel and processed as mentioned earlier in the text. The intensity of product band in the DNA control lane was subtracted from all other lanes to account for the DNA degradation, and the percentage of DNA cleavage was calculated in the same manner as for AP lyase activity.
NaBH4 cross-linking experiments
The cross-linking experiments were performed using a similar procedure as mentioned before (10,12) using a 49-bp DNA with U at the 21st position. UDG-treated DNA (200 nM) was incubated with 200 nM protein and 1 mM NaBH4 on ice for 60 min, and the products were separated on a 10% sodium dodecyl sulphate polyacrylamide gel.
Crystallization, data collection and structure determination
The Topo-78 fragment was crystallized by vapor diffusion against 50 mM HEPES (pH 7), 0.1 M KCl, 0.01 M MgCl2 and 15% polyethylene glycol 400. Crystals appeared as tiny long clustered needles, with occasional appearance of single thicker (20–50 µm thick) crystals. The crystals were frozen using 20% glycerol as cryoprotectant. The thicker crystals diffracted to 2.9- to 3.1-Å resolution. Diffraction data were collected at the Life Science Collaborative Access Team station at the Advanced Photon Source in Argonne National Laboratory. Data collection and refinement statistics are shown in Supplemental Table S1. All data were processed and integrated using the program XDS (18) and scaled using the program SCALA (19). The 2.9-Å-resolution structure of Topo-78 was solved by molecular replacement (Phaser) (20) using Topo-61 [PDB ID: 2CSB (8)] as the search model. The missing part of the model was built by fitting the main-chain atoms of an (HhH)2 domain from Topo-61 on the electron density, one domain at a time, followed by model building in Coot (21) and refinement using Refmac5 (22). The appearance of side-chain density helped in the correct placement of each (HhH)2 domain. The electron density became weaker toward the C-terminal end of the protein, and despite several attempts, it was only possible to trace reliably until residue 612, although the construct had 685 residues and there was discontinuous density in the maps. The higher flexibility of the C-terminal end of the protein is also evident by the progressive increase in B-factors of the atoms as the chain length increases. Translation, libration and screw-rotation (TLS) refinement (23) was done during the later stages of refinement using six TLS groups, as suggested by the TLS server (24). The final Rfactor and Rfree for the model is 20.1% and 26.7%, respectively. The final model contains residues 3–612, four water molecules, three glycerol molecules and one Zn2+ ion. There was no Zn2+ ion present in the crystallization condition, but the coordinating residues (E161, H246 and E194) suggest Zn2+ as a plausible ligand. All figures were made using Pymol (25), and the electrostatic surfaces were calculated using the program APBS (26).
RESULTS AND DISCUSSION
Identification of the AP lyase active site of Topo-78
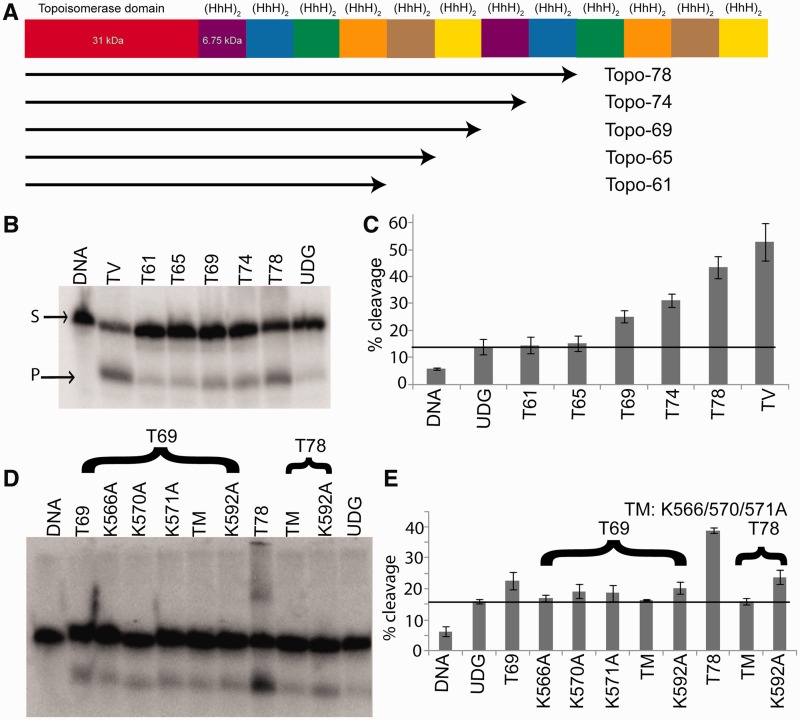
Previous studies had shown that Topo-78 has at least one DNA repair lyase active site located between residues 532 and 685 (10,12). To determine the exact location of this DNA repair active site, we created several deletion mutants, each with fewer number of (HhH)2 domains. The different fragments used for the repair assays included Topo-61 [residues 1–519, four (HhH)2 domains], Topo-65 [residues 1–556, five (HhH)2 domains], Topo-69 [residues 1–599, six (HhH)2 domains], Topo-74 [residues 1–640, seven (HhH)2 domains] and Topo-78 [residues 1–685, eight (HhH)2 domains] (Figure 1A). In addition, the full-length Topo-V (residues 1–982), with 12 (HhH)2 domains, was also examined. The fragments were designed based on an initial medium-resolution crystal structure of Topo-78 (see later in the text). All constructs were expressed in E. coli and purified to near homogeneity as described previously (11). While all the Topo-V fragments were soluble and active in 250 mM NaCl, full-length Topo-V required 1 M NaCl for solubility.
Figure 1.
AP lyase activity of the different Topo-V fragments. (A) Schematic representation of the Topo-V protein and the various fragments used in the present study. (B) AP lyase assay with different Topo-V fragments. The upper band corresponds to uncleaved DNA (S), whereas the lower band on the gel shows the product after cleavage of the DNA at the abasic site (P). (C) Graph showing quantification of the AP lyase activity for different Topo-V fragments (three replications). The partial cleavage of the DNA and UDG controls is due to the inherent instability of abasic DNA. Topo-69 is the shortest Topo-V fragment with AP lyase activity and indicates that the active site lies between residues 556 and 599. (D) Identification of the nucleophilic Lys involved in AP lyase activity of Topo-78. All the Lys residues in the region spanning 556–599 were mutated to Ala. (E) Graph showing AP lyase activity of different Lys mutants (three replications). The results from Topo-78 fragments show that mutating K566, K570 and K571 have an effect on AP lyase activity, whereas the K592 mutant does not show an effect. Error bars represent the standard deviation for three replications. The black line in (C) and (E) represents the basal activity due to the instability of DNA substrate. TM refers to the K566A, K570A, K571A triple mutant.
AP lyase assays using DNA with a single tetrahydrofuran (THF) abasic site, instead of a hemiacetal-containing abasic site, showed no activity, which is expected if Topo-V has a similar mechanism to the Class I AP endonucleases (17). To create the hemiacetal-containing abasic site, we used DNA with U at the targeted position and treated the DNA with UDG. The resulting DNA with a natural abasic site was used as the substrate for AP lyase assays. This abasic site–containing DNA was somewhat labile, especially when the reactions were carried out at a high temperature. To mitigate spontaneous DNA degradation, the AP lyase assays with Topo-V fragments were conducted at room temperature. The reaction products were resolved on a denaturing polyacrylamide gel, and typical results are shown in Figures 1B and C. Topo-69 is the shortest fragment with detectable AP lyase activity, whereas Topo-61 and Topo-65 showed no or little activity compared with the UDG-treated DNA control (Figure 1C). Interestingly, the AP lyase activity increases with an increase in the number of (HhH)2 domains (Figure 1C), a result reminiscent of the topoisomerase activity of Topo-V, where Topo-V fragments with more (HhH)2 domains showed higher DNA relaxation activities (11). It had been observed that the 34-kDa C-terminal fragment of Topo-V compacts DNA in electrophoretic mobility shift assays (12), suggesting that a role of the (HhH)2 domains is to bind DNA non-specifically. Thus, the increasing levels of AP lyase activity of Topo-V fragments with increasing number of (HhH)2 domains could be due to increased binding affinity of protein and DNA.
The AP lyase experiments with different Topo-V fragments suggested that the lyase active site is located between residues 556 and 599. The fact that there is no AP lyase activity using a THF-containing substrate suggested that Topo-V is similar to other DNA repair proteins that use a Lys residue for nucleophilic attack on the DNA backbone (17,27). To identify the catalytic residues involved in the AP lyase activity, all four Lys residues (K566, K570, K571 and K592) found in the region spanning residues 556–599 were mutated individually to Ala. These single mutants, as well a triple mutant (K566A/K570A/K571A), were made in the Topo-69 fragment. In addition, a triple mutant (K566A/K570A/K571A) and a single mutant (K592A) were made in the Topo-78 fragment. The results of AP lyase assays with these Lys mutants are summarized in Figure 1D and E. The triple mutant is devoid of AP lyase activity both in the Topo-69 and Topo-78 fragments. The K592A mutant is the most active among the single Lys mutants, whereas K566A, K570A and K571A have weaker activity compared with wild type. The observation that the triple mutants are devoid of activity suggested that one of the three Lys residues is the primary nucleophile in the β-elimination reaction. The low level of AP lyase activity observed with the single Lys mutants at 566, 570 and 571 suggests that these neighboring lysines can participate in or substitute partially for each other in the AP lyase reaction.
To confirm the presence of a second AP lyase active site, a K566A, K570A and K571A triple mutant in the full-length molecule was made. AP lyase activity assays show that this mutant retains robust AP lyase activity, although the first AP lyase active site was ablated (Supplemental Figure S1). This result confirms that Topo-V has at least two AP lyase active sites. Sequence analysis of the full-length protein did not reveal either a region with three lysines positioned as in the first active site or a single lysine in between two predicted (HhH)2 domains. Thus, the exact location of the second AP lyase active site remains unknown.
Identification of the dRP lyase active site of Topo-78
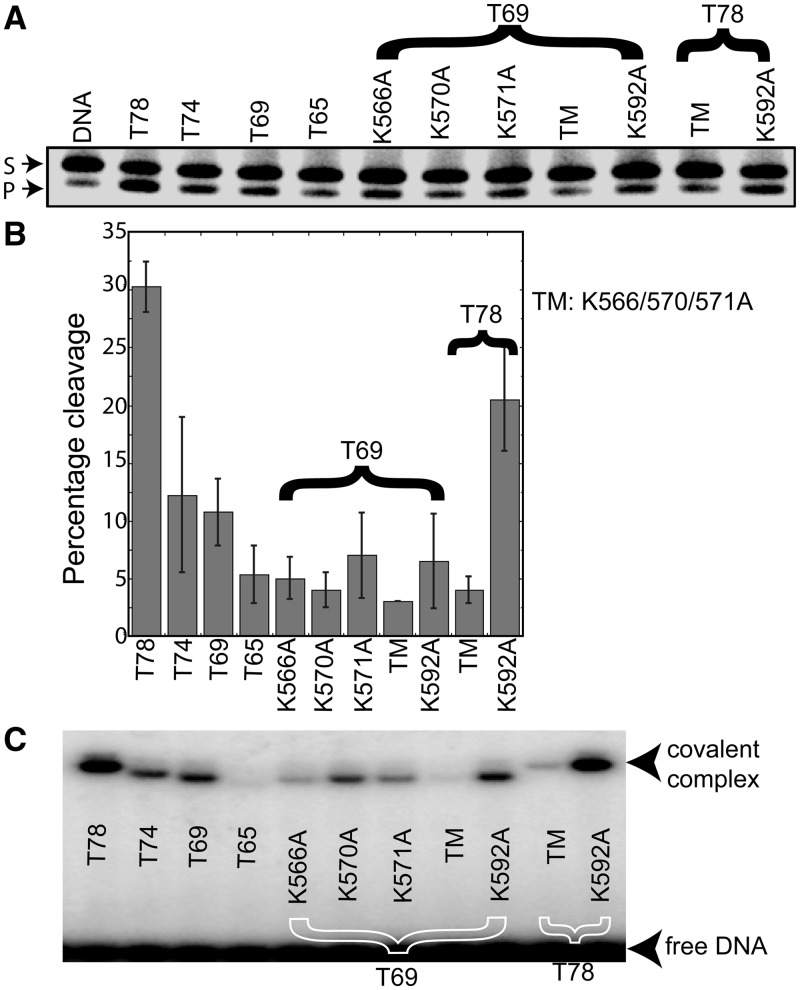
A dRP lyase assay was conducted with the Topo-V fragments to examine whether Topo-V uses the same active site for the AP lyase and dRP lyase activities (Figures 2A and B). The results show that Topo-65 has minimal dRP lyase activity, but the larger fragments, including Topo-78, Topo-74 and Topo-69, are active (Figure 2A and B). dRP lyase experiments were carried out with the lysine mutants to identify the nucleophile for the reaction. There is a substantial reduction in dRP lyase activity for the K566A/K570A/K571A triple mutant in both the Topo-69 and Topo-78 backbones. Individual alterations of Lys residues in the Topo-69 backbone shows that a lysine to alanine mutation at positions 566, 570, 571 and 592 has weak dRP lyase activity (Figure 2B). dRP lyase assay carried out in the Topo-78 backbone with a triple mutant and K592A single mutant clearly shows that the triple mutant is completely inactive and K592A retains significant activity compared with the wild-type Topo-78. Thus, these results prove that Topo-V uses the same active site for AP and dRP lyase activities, and that K566, K570 and K571 can probably partially substitute for each other owing to their proximity.
Figure 2.
dRP lyase assay with the different Topo-V fragments. (A) A 34-bp DNA containing a single U was pre-treated with UDG to generate an abasic site. The AP site-containing DNA was further treated with AP endonuclease to incise the DNA at the abasic site and used as the substrate for dRP lyase activity with different Topo-V fragments, as indicated. The lower band on the gel shows the product (P) after removal of the sugar phosphate at the abasic site, whereas the upper band illustrates the substrate (S). (B) Graph illustrating the quantification of the dRP lyase activity (%) of different Topo-V fragments. The results represent averages from four independent experiments. Comparison of lysine mutants on the Topo-69 and Topo-78 backbones shows that K566, K570 and K571 are required for dRP lyase activity, whereas K592 is not. Error bars represent the standard deviation for four replications. (C) NaBH4 cross-linking of the different Topo-V fragments with DNA. Topo-V fragments were incubated with abasic DNA and 1 mM NaBH4 to trap the Schiff base protein–DNA intermediate, as described under ‘Materials and Methods’ section. The results show that there is no DNA–protein complex formation for either the Topo-65 or the Lys triple mutant (K566A/570A/571A), although the single Lys mutants do form a DNA–protein complex. TM refers to the K566A, K570A, K571A triple mutant.
Analysis of a trapped protein–DNA covalent complex
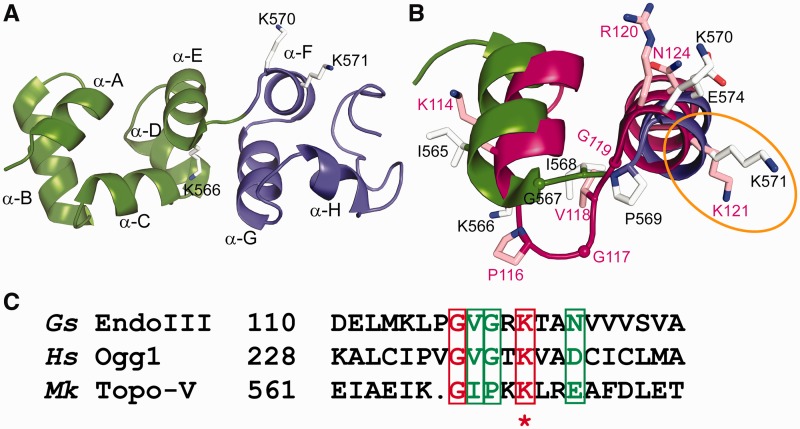
To further examine the active site and the nucleophile responsible for the AP lyase β-elimination reaction, cross-linking experiments by NaBH4 reduction were carried out using various deletion and lysine mutants (Figure 2C). This highly sensitive cross-linking technique relies on the ability of the C1′-aldehyde group of a base-less deoxyribose to react with a lysine γ-amine group of a protein to form a Schiff base protein–DNA complex, followed by reduction of the protein–DNA complex with NaBH4 (28). The results (Figure 2C) show that there is little or no covalent complex formation with Topo-65, whereas cross-linking is obvious with the Topo-78, Topo-74 and Topo-69 fragments. Experiments with single lysine mutants showed that K566A, K570A and K571A mutants form lesser amount of protein–DNA covalent complexes, whereas K592A retains a similar capacity as that of wild type in complex formation. The triple mutant failed to show covalent complex formation in both the Topo-69 and Topo-78 backbones, consistent with the idea that one of these three lysines can serve as a nucleophile. These results are consistent with that of the AP and dRP lyase assays described earlier in the text. Gel shift assays with DNA containing a THF abasic site showed that Topo-V fragments with a functional AP lyase active site bind to the DNA, showing that the abasic DNA binds specifically to the repair site and not to any of the other (HhH)2 domains in Topo-V (Supplemental Figure S2A). Covalent complex formation studies of Topo-V fragments containing a hemiacetal abasic DNA site clearly showed that the three lysines (K566, K570 and K571) are essential, as a triple mutant does not form a covalent complex with DNA (Supplemental Figure S2B). Only the triple mutant loses the activity to bind DNA and to form a covalent complex. Because all single mutants exhibit partial activity, it is very likely that all of them still retain the ability to bind DNA, although this was not tested directly. The experiments point to the fact that the three lysines are not only important for DNA binding but also for AP lyase activity. It is worth noting that K571 corresponds to K121 in Geobacillus stearothermophilus EndoIII, and that K121 was shown to be the Schiff base nucleophile responsible for the lyase activity of EndoIII (Figure 4C) (29).
Figure 4.
Superposition of the Topo-78 structure onto EndoIII [PDB ID: 1ORN (29)]. (A) Close-up view of (HhH)2 domains 5 (green) and 6 (blue), forming the AP lyase active site. The successive helices are named from A to J. K566 is the last residue of helix α-E, and K570 and K571 are the first two residues of helix α-F. (B) Comparison of the DNA repair active sites of Topo-78 (green and blue) and EndoIII (magenta). Residues K566, K570 and K571 of Topo-78 match L115, R120 and K121 of EndoIII respectively, with K114 immediately adjacent to K566. The location of these residues in the helices is very similar in both enzymes despite the longer loop connecting the two helices in EndoIII. (C) Sequence comparison of the HhH motifs from various proteins involved in DNA repair. The region for comparison was taken from Doherty et al. (30). Gs EndoIII, G. stearothermophilus EndoIII; Hs Ogg1, human 8-oxoguanine glycosylase; Mk Topo-V, M. kandleri Topo-V. Sequence analysis shows that the equivalent lysine of Topo-78, K571 (marked with an asterisk), is the nucleophile in all the proteins compared.
Overall structure of the Topo-78 fragment
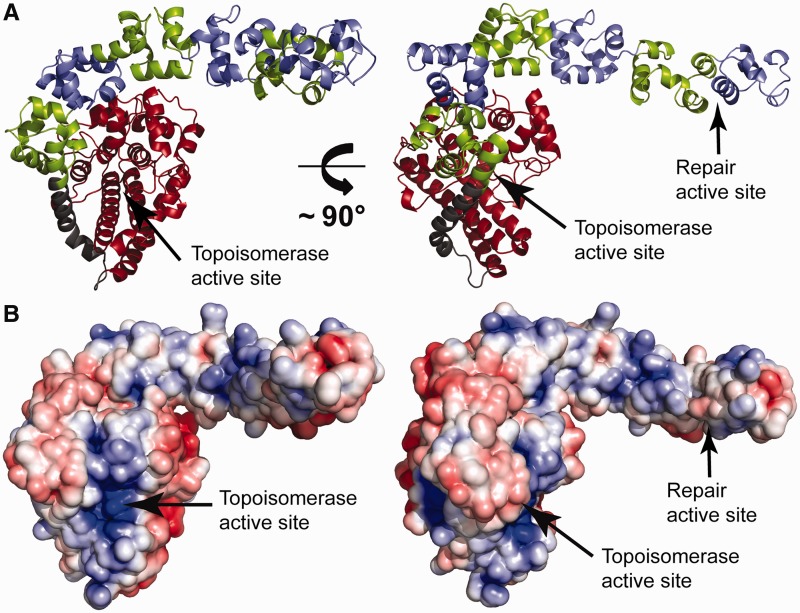
To gain further insight into the lyase reaction catalyzed by Topo-V, we determined the crystal structure of Topo-78. The structure of Topo-78 was solved to 2.9-Å resolution and shows that overall it is very similar to Topo-61, with a compact α-helix–rich N-terminal topoisomerase domain followed by six ordered (HhH)2 domains or 12 HhH motifs (Figure 3A). The seventh and eighth (HhH)2 domains are not visible in the structure, although it appears that the last two (HhH)2 domains can fold toward the topoisomerase domain. As in all the previous structures of Topo-V fragments, the first and second (HhH)2 domains wrap around the topoisomerase domain, whereas the remaining (HhH)2 domains extend away from the main body of the topoisomerase domain. In Topo-78, the sixth (HhH)2 domain folds slightly back over the fifth (HhH)2 domain (Figure 3A). Comparison of Topo-78 with other fragment structures shows that the topoisomerase domain superposes well (root-mean-square deviation (rmsd) of 0.6 Å and 0.5 Å for the main-chain atoms of the topoisomerase domain of Topo-61 (PDB ID: 2CSB) (8) and Topo-44 (PDB ID: 3M6Z) (11) respectively). The rmsd for the superposition of all common Topo-78 main-chain atoms on Topo-61 (PDB ID: 2CSB) is 1.1 Å and on Topo-44 (PDB ID: 3M6Z) is 1.4 Å. Superposition of only the (HhH)2 domains of the Topo-78 structure onto the common (HhH)2 domains of Topo-61 and Topo-44 yields an rmsd of 1.4 Å and 0.9 Å, respectively. The superpositions show that although the topoisomerase and (HhH)2 domains remain largely unchanged, their relative orientation is different, an observation that was made before (11). A surface charge representation of the Topo-78 fragment shows exposed positively charged patches on all the (HhH)2 domains (Figure 3B), as expected for this type of DNA-binding motif. Nevertheless, these positively charged patches do not form an obvious path for the DNA to bind. Instead, it indicates that either the (HhH)2 domains have to change relative orientation, or only selected (HhH)2 domains can bind DNA simultaneously, or the DNA has to follow a tortuous path around the (HhH)2 domains.
Figure 3.
Crystal structure of the Topo-78 fragment. (A) The diagram shows a model of the Topo-78 fragment, the first structure of a Topo-V fragment with both topoisomerase and DNA repair domains. Topo-78 is organized as a compact α-helix–rich topoisomerase domain (red), a long bent linker helix (grey) and six (HhH)2 domains (alternating green and blue). The (HhH)2 domains extend away from the topoisomerase domain; the last two (HhH)2 domains in the fragment are disordered and not visible in the structure. (B) Electrostatic surface representation of Topo-78. The orientation is the same as in (A). There is an accumulation of positive charge on the (HhH)2 domains, which could represent binding sites for DNA. The electrostatic potential was calculated with a dielectric constant of 78 for solvent and 2 for protein. The surface is colored with a blue to red gradient from +5 to −5 KbT/ec.
AP/dRP lyase active site
The AP/dRP lyase experiments showed that absence of the three lysines (K566, K570 and K571) has a deleterious effect on activity. These three lysines are in physical proximity (Figure 4B) and are located in the fifth and sixth (HhH)2 domains of Topo-78 (Figure 4A). K566 is the last residue of the second helix (α-E helix) of the fifth (HhH)2 domain, whereas K570 and K571 are the first two residues of the first helix (α-F helix) of the sixth (HhH)2 domain. A structure-based comparison with the crystal structure of EndoIII bound to DNA (PDB ID: 1ORN) (29) shows that residues K566, K570 and K571 correspond to residues L115, R120 and K121 of EndoIII, respectively (Figure 4B). K114 of EndoIII occupies a very similar position in the first helix of the HhH motif as K566 of Topo-V. Furthermore, comparisons with other DNA repair proteins shows that the only conserved amino acid in this region is the equivalent of K571, while K566 and K570 are not highly conserved (Figure 4C). Previous studies had shown that the dRP lyase activity of Topo-V proceeds through a β-elimination reaction, as a covalent protein–DNA complex could be trapped by NABH4 reduction (10). In the present study, we show that the AP lyase activity also proceeds through a β-elimination reaction, as the absence of the active site lysines (especially the triple mutant) leads to a strong reduction in AP lyase activity. This places Topo-V in the Class I AP lyase family, which cleaves 3′ to the DNA abasic site, leaving a 3′-α,β-unsaturated aldehyde and a deoxyribonucleoside 5′-phosphate group (17). Class I enzymes do not require divalent metal ions for activity, as is the case of Topo-V, as opposed to Class II AP endonucleases (eg. human AP endonuclease 1) that require metal ions and where the reaction proceeds by hydrolysis and not through a β-elimination reaction. This classification of Topo-V as a Class I AP lyase is also consistent with the observation that Topo-V cannot cleave THF-containing abasic DNA, whereas Class II enzymes can process this type of substrate. As both the AP lyase and dRP lyase reactions proceed through the same β-elimination mechanism, and AP lyase activity by Class I AP endonucleases does not produce a dRP substrate, it is most likely that in vivo Topo-V acts as an AP lyase. If that is the case, the α,β-unsaturated aldehyde left after AP lyase activity must be removed by a Class II endonuclease to produce a clean gap to be filled in by a DNA polymerase (17). The dRP lyase activity noted for Topo-V in vitro is the result of the common chemistry of AP and dRP lyase activities; when provided with a 5′-dRP–containing DNA substrate, Topo-V can remove the dRP moiety from DNA, as both AP and dRP lyase activities follow the β-elimination reaction (Figure 2).
Comparison of the Topo-78 structure with EndoIII bound to DNA
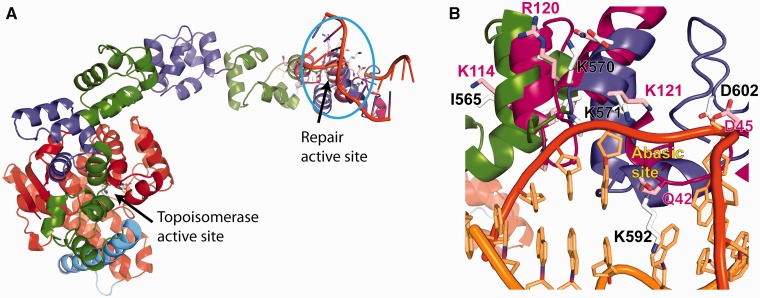
The HhH motif is a distinct class of DNA-binding motifs found in some DNA repair enzymes that has been implicated in non-sequence–specific DNA binding (30). In HhH motifs, protein–DNA interactions are mediated by hydrogen bonds between the DNA phosphate oxygens and the protein backbone nitrogens (30). The number of HhH motifs varies on different proteins. Most proteins contain a single HhH motif [e.g. EndoIII, Taq Pol I, AlkA (31–33)], whereas others such as Pol β have two spatially distant HhH motifs with different DNA-binding characteristics (30,34). In the case of Xeroderma Pigmentosa Complementation group F endonuclease, two tandem HhH motifs form an (HhH)2 domain, which in turn dimerizes to form an (HhH)2 dimer, but only one of the (HhH)2 domains in the dimer interacts with DNA (35). Topo-V is the only known protein where there is more than one (HhH)2 domain present in the same polypeptide, and the presence of 12 tandem (HhH)2 domains makes it a very unique DNA-binding protein. The (HhH)2 domains probably help enhance DNA binding in high-salt and high-temperature conditions (10). As HhH motifs are generally associated with proteins involved in BER of DNA (30), the repair active site region of Topo-78 crystal structure was compared with that of EndoIII bound to DNA (PDB ID: 1ORN) (29). The superposition (Figure 5A) shows that the cationic end of helix α-F and preceding loop of Topo-78 could make contacts with the phosphate backbone of DNA. In EndoIII, the protein contacts both the lesion- and non–lesion-containing DNA strands through separate α-helices (29). This is different from what is observed in other repair enzymes, such as hOgg1 (36) and AlkA (33), where the protein contacts only the backbone of the lesion-containing strand (29). A Topo-78/DNA model based on the EndoIII structure shows a short helix (α-H) inserted in between the two DNA strands. In the model, helix α-H and the preceding helix (α-G) in Topo-78 contact the lesion-containing DNA strand. It is not clear from the Topo-78 structure whether the protein contacts the non-lesion–containing DNA strand because the last two (HhH)2 domains are not visible in the present structure.
Figure 5.
A model of Topo-78 bound to DNA. (A) Superposition of Topo-78 structure onto EndoIII (magenta) bound to DNA (orange) (PDB ID: 1ORN (29)). The superposition was made based on the location of the helices with repair activity. Topo-78 shows helices contacting lesion-containing DNA. The invisible (HhH)2 domains of Topo-78 might also be involved in binding DNA. (B) Close-up view of Topo-78 (green and blue)–DNA (orange) interaction. In Topo-78, helix α-H inserts between both the DNA strands. Helices F, G, H, I and J are in positions to interact with the lesion-containing DNA strand, placing the catalytically important residues near the lesion. The catalytically important residues for both EndoIII (pink) and Topo-78 (grey) are shown as sticks. The position of the abasic site in the EndoIII structure is marked.
Repair enzymes that use the HhH motif can be either mono- or bifunctional glycosylases. The former ones only have glycosylase activity (e.g. AlkA, UDG), whereas the latter ones have glycosylase and AP lyase activities (e.g. hOgg1, EndoIII). In both cases, the HhH motif acts as a structural motif, which supports the highly bent DNA. The catalytically important residues of EndoIII were compared with the corresponding residues of Topo-78 based on the superposition (Figure 5B). These residues include K121, D45 and Q42 of EndoIII, which superimpose with K571, D602 and K592 of Topo-78. Q42 of EndoIII is responsible for flipping out the damaged base (29), and the same function is done by L125 in AlkA (33) and N149 in hOgg1 (37). The superposition of Topo-78 onto EndoIII shows that K592 of Topo-V occupies a similar position as that of Q42 of EndoIII. In EndoIII, D45 along with a water molecule activates the catalytic lysine for the nucleophilic attack on the DNA (29). The aspartate, along with the water molecule, deprotonates the catalytic lysine, forming an amine nucleophile that can then perform glycosidic bond cleavage and AP lyase activities (27,38). Analysis of the current model of Topo-78 reveals that D602 occupies a similar spatial position as D45 of EndoIII, although approaching from a different angle. Finally, based on sequence and structural comparisons, K121 of EndoIII matches K571 of Topo-V, and hence K571 is likely to be the catalytic nucleophile of Topo-V. Thus, the structural comparison shows that although Topo-V differs from EndoIII in terms of its overall structure and biological activities, the AP lyase active sites of both these proteins resemble each other.
Synergy between the topoisomerase and DNA repair activities of Topo-V
M. kandleri lives in hot vents deep in the ocean, and the elevated temperatures under these living conditions lead to an increased rate of formation of DNA lesions (10). Access of repair enzymes to the lesion sites may require melting of the duplex DNA by the action of a topoisomerase (1). In this context, the fusion of the topoisomerase and DNA lyase repair sites in Topo-V represents a good solution to the problem of needing the synergistic action of the two activities. The topoisomerase activity may help Topo-V to relax the supercoiled DNA and access the repair sites, leading to more efficient repair activity. The exact way that these two active sites coordinate their activities is unclear from the structure. In the conformation observed in the crystal structure, the two active sites are ∼70 Å apart. DNA binding will clearly lead to conformational changes, as the topoisomerase active site is buried and some movements are needed to expose the active site and assist DNA binding (8,11). This suggests that on DNA binding, the topoisomerase and DNA repair active sites could come closer together to interact, but this may not be needed because the local DNA changes required for efficient DNA repair activity may be the consequence of the overall alteration of the topological state of DNA. In other words, if the DNA is relaxed, this may provide better access to the lesion site without any direct interaction between the topoisomerase and repair active sites. As AP lyase activity creates a nick in the DNA that would lead to DNA relaxation, it is most likely that DNA relaxation activity precedes DNA repair. Overall, these observations and the fact that there are two DNA repair sites in Topo-V suggest that Topo-V may be primarily a DNA repair enzyme.
CONCLUSIONS
The present study identified that residues 1–599 constitute the shortest fragment of Topo-V with both topoisomerase and DNA repair (lyase) activities. Biochemical experiments show that K566, K570 and K571 can substitute for each other as the nucleophile for the AP and dRP lyase activities. Sequence and structural comparison of Topo-V with other DNA repair enzymes shows that K571 is highly conserved in all AP lyases with the HhH motif, and hence K571 is the most probable nucleophile of Topo-V, although the nearby lysines can partially substitute for K571 owing to their proximity. Superposition of Topo-78 onto EndoIII bound to DNA suggested a general mode of DNA binding, where the protein binds and bends the DNA drastically. Structural comparison with EndoIII also showed that two of its catalytically important residues (K121 and D45) have equivalent residues (K571 and D602, respectively) in Topo-V. The biochemical experiments classify Topo-V as a Class I AP endonuclease with a β-elimination reaction mechanism. The observations from the present study along with previous information on Topo-V suggest that Topo-V is most probably a DNA repair enzyme, which is fine-tuned by the topoisomerase domain for optimal activity in the extreme living habitat of M. kandleri.
ACCESSION NUMBERS
Coordinates for Topo-78 have been deposited in the PDB with accession number 4GFJ.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplemental Figures 1 and 2 and Supplementary References [39,40].
FUNDING
American Heart Association postdoctoral fellow [10POST2600325 to R.R.]. National Institutes of Health [R01GM51350 to A.M.]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge staff and instrumentation support from the Keck Biophysics Facility and the Center for Structural Biology at Northwestern University, and Life Science Collaborative Access Team station (LS-CAT) at the Advanced Photon Source (APS) at Argonne National Laboratory. LS-CAT was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor. Use of the APS is supported by the Department of Energy (DOE). Support from the R.H. Lurie Comprehensive Cancer Center of Northwestern University to the Structural Biology Facility is also acknowledged. We thank Alexei Slesarev for providing some of the Topoisomerase V plasmids.
REFERENCES
- 1.Vos SM, Tretter EM, Schmidt BH, Berger JM. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 2011;12:827–841. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 3.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 4.Baker NM, Rajan R, Mondragon A. Structural studies of type I topoisomerases. Nucleic Acids Res. 2009;37:693–701. doi: 10.1093/nar/gkn1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forterre P, Gribaldo S, Gadelle D, Serre MC. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89:427–446. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Slesarev AI, Stetter KO, Lake JA, Gellert M, Krah R, Kozyavkin SA. DNA topoisomerase V is a relative of eukaryotic topoisomerase I from a hyperthermophilic prokaryote. Nature. 1993;364:735–737. doi: 10.1038/364735a0. [DOI] [PubMed] [Google Scholar]
- 7.Forterre P. DNA topoisomerase V: a new fold of mysterious origin. Trends Biotechnol. 2006;24:245–247. doi: 10.1016/j.tibtech.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 8.Taneja B, Patel A, Slesarev A, Mondragon A. Structure of the N-terminal fragment of topoisomerase V reveals a new family of topoisomerases. EMBO J. 2006;25:398–408. doi: 10.1038/sj.emboj.7600922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huber R, Kurr M, Jannasch HW, Stetter KO. A novel group of abyssal methanogenic archaebacteria (Methanopyrus) growing at 110°C. Nature. 1989;342:833–834. [Google Scholar]
- 10.Belova GI, Prasad R, Kozyavkin SA, Lake JA, Wilson SH, Slesarev AI. A type IB topoisomerase with DNA repair activities. Proc. Natl Acad. Sci. USA. 2001;98:6015–6020. doi: 10.1073/pnas.111040498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajan R, Taneja B, Mondragon A. Structures of minimal catalytic fragments of topoisomerase V reveals conformational changes relevant for DNA binding. Structure. 2010;18:829–838. doi: 10.1016/j.str.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belova GI, Prasad R, Nazimov IV, Wilson SH, Slesarev AI. The domain organization and properties of individual domains of DNA topoisomerase V, a type 1B topoisomerase with DNA repair activities. J. Biol. Chem. 2002;277:4959–4965. doi: 10.1074/jbc.M110131200. [DOI] [PubMed] [Google Scholar]
- 13.Taneja B, Schnurr B, Slesarev A, Marko JF, Mondragon A. Topoisomerase V relaxes supercoiled DNA by a constrained swiveling mechanism. Proc. Natl Acad. Sci. USA. 2007;104:14670–14675. doi: 10.1073/pnas.0701989104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434:671–674. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 15.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 16.Prasad R, Beard WA, Strauss PR, Wilson SH. Human DNA polymerase beta deoxyribose phosphate lyase. Substrate specificity and catalytic mechanism. J. Biol. Chem. 1998;273:15263–15270. doi: 10.1074/jbc.273.24.15263. [DOI] [PubMed] [Google Scholar]
- 17.Piersen CE, McCullough AK, Lloyd RS. AP lyases and dRPases: commonality of mechanism. Mutat. Res. 2000;459:43–53. doi: 10.1016/s0921-8777(99)00054-3. [DOI] [PubMed] [Google Scholar]
- 18.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- 19.Collaborative Computational Project 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 20.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 22.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 23.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 24.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 25. The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC. [Google Scholar]
- 26.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCullough AK, Sanchez A, Dodson ML, Marapaka P, Taylor JS, Lloyd RS. The reaction mechanism of DNA glycosylase/AP lyases at abasic sites. Biochemistry. 2001;40:561–568. doi: 10.1021/bi002404+. [DOI] [PubMed] [Google Scholar]
- 28.Prasad R, Longley MJ, Sharief FS, Hou EW, Copeland WC, Wilson SH. Human DNA polymerase theta possesses 5'-dRP lyase activity and functions in single-nucleotide base excision repair in vitro. Nucleic Acids Res. 2009;37:1868–1877. doi: 10.1093/nar/gkp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fromme JC, Verdine GL. Structure of a trapped endonuclease III-DNA covalent intermediate. EMBO J. 2003;22:3461–3471. doi: 10.1093/emboj/cdg311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doherty AJ, Serpell LC, Ponting CP. The helix-hairpin-helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 1996;24:2488–2497. doi: 10.1093/nar/24.13.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thayer MM, Ahern H, Xing D, Cunningham RP, Tainer JA. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim Y, Eom SH, Wang J, Lee DS, Suh SW, Steitz TA. Crystal structure of Thermus aquaticus DNA polymerase. Nature. 1995;376:612–616. doi: 10.1038/376612a0. [DOI] [PubMed] [Google Scholar]
- 33.Hollis T, Ichikawa Y, Ellenberger T. DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. EMBO J. 2000;19:758–766. doi: 10.1093/emboj/19.4.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar A, Abbotts J, Karawya EM, Wilson SH. Identification and properties of the catalytic domain of mammalian DNA polymerase beta. Biochemistry. 1990;29:7156–7159. doi: 10.1021/bi00483a002. [DOI] [PubMed] [Google Scholar]
- 35.Newman M, Murray-Rust J, Lally J, Rudolf J, Fadden A, Knowles PP, White MF, McDonald NQ. Structure of an XPF endonuclease with and without DNA suggests a model for substrate recognition. EMBO J. 2005;24:895–905. doi: 10.1038/sj.emboj.7600581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 37.Banerjee A, Santos WL, Verdine GL. Structure of a DNA glycosylase searching for lesions. Science. 2006;311:1153–1157. doi: 10.1126/science.1120288. [DOI] [PubMed] [Google Scholar]
- 38.Krokan HE, Standal R, Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochem. J. 1997;(325 Pt 1):1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diederichs K, Karplus PA. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat. Struct. Biol. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
- 40.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.