Abstract
Human cytomegalovirus (HCMV) miRNAs are important for regulation of viral infection and evasion of host immune responses. Unfortunately, the importance of HCMV miRNAs cannot be addressed in vivo due to the species specificity of CMVs. Rhesus CMV (RhCMV) infection of rhesus macaques provides an important model system for HCMV pathogenesis due to the genetic similarity between the viruses. In this report, seventeen RhCMV miRNAs were identified using Next Generation Sequencing. In fibroblasts, RhCMV miRNAs associate with Argonaute proteins and display several patterns of expression, including an early peak in expression followed by decline and accumulation throughout infection. Additionally, RhCMV encodes an HCMV miR-US5-2 homologue that targets the 3’ UTR of RhCMV US7. Finally, examination of salivary gland tissue from infected animals revealed the presence of a subset of viral miRNAs. This study highlights the importance of the RhCMV model system for evaluating the roles of CMV miRNAs during viral infection.
Keywords: RhCMV, HCMV, miRNA, endothelial cells, salivary gland, miR-US5-2
Introduction
Human cytomegalovirus (HCMV) is an ubiquitous pathogen that causes persistent, but clinically benign, infections in healthy individuals. Severe disease can occur in immunocompromised patients such as organ transplant recipients or individuals co-infected with HIV (Clarke et al., 1996; Rowshani et al., 2005). In addition, HCMV is the leading congenitally-transmitted viral infection (Pereira et al., 2005). Animal models play a crucial role in understanding host/pathogen interactions, but studies of HCMV pathogenicity and immune evasion have been hampered due to the strict species specificity of CMVs (McGeoch, Dolan, and Ralph, 2000). Although rodent CMVs are useful models for virus infection, these viruses share limited genetic homology with HCMV, especially in genes non-essential for growth in tissue culture (McGeoch et al., 1995; Rawlinson, Farrell, and Barrell, 1996; Vink, Beuken, and Bruggeman, 2000). In contrast, rhesus CMV (RhCMV), which is ubiquitous in rhesus macaque (Macaca mulatta) colonies, has extensive homology with HCMV at the genome level (Baskin, 1987; Hansen et al., 2003; Kuhn et al., 1999; Lockridge et al., 1999; Pitcher et al., 2002; Vogel et al., 1994). Most RhCMV infections are asymptomatic, but in immunocompromised animals the virus causes similar patterns of infection and pathology and exhibit humoral and cellular immune responses closely matching those of HCMV (Baskin, 1987; Kaur et al., 2003; Kuhn et al., 1999; Lockridge et al., 1999; Picker et al., 2004; Pitcher et al., 2002; Price et al., 2008). Thus, infection of rhesus macaques with RhCMV allows for the study of CMV pathogenicity, immune evasion and persistence in a relevant non-human primate system.
MicroRNAs (miRNAs) are a class of ~22 nucleotide (nt) non-coding RNAs involved in post-transcriptional regulation of target genes. pri-miRNAs are capped and polyadenylated transcripts which can contain one or several ~80nt hairpins that are further processed by the cell to form mature miRNAs (Cai, Hagedorn, and Cullen, 2004). These hairpins are cleaved first in the nucleus by the RNase III enzyme Drosha to produce a ~60nt pre-miRNA, and then transported to the cytoplasm by Exportin5 (Yi et al., 2003). The pre-miRNA hairpin is further processed by a second RNase III enzyme Dicer, which releases the double-stranded miRNA (Hutvagner et al., 2001; Lee et al., 2002). A single strand of the mature miRNA is incorporated into the multi-protein RNA-induced silencing complex (RISC) that associates with the target mRNAs, usually within the mRNA 3’ untranslated region (UTR) (Hammond et al., 2000). This interaction results in gene silencing through either mRNA degradation or translational repression (Bartel, 2009; Guo et al., 2010; Lim et al., 2005). miRNAs are found in most multicellular eukaryotes and play important regulatory roles in many cellular processes, including development, apoptosis and cancer (Farazi et al., 2010; Lima et al., 2011; Tiscornia and Izpisua Belmonte, 2011). DNA viruses, mostly of the Herpesvirus family, also encode miRNAs. To date, over 230 viral miRNAs have been identified (Grundhoff and Sullivan, 2011; Tuddenham and Pfeffer, 2011). HCMV encodes at least 14 miRNAs (Grey et al., 2005; Pfeffer et al., 2004) several of which target cellular genes involved in cell cycle regulation and the immune response to viral infection (Grey et al., 2010; Stern-Ginossar et al., 2007). In addition, some HCMV miRNAs also target viral genes (Grey et al., 2007; Murphy et al., 2008; Stern-Ginossar et al., 2009; Tirabassi et al., 2011), which could be important for limiting viral gene expression during persistence, latency and/or during reactivation. miRNAs are non-immunogenic, and as such may be an excellent mechanism to modulate host and viral gene expression in situations of viral latency. Unfortunately, these hypotheses are difficult to address for HCMV due to the lack of a relevant model system.
While the miRNAs encoded by HCMV, mouse CMV (MCMV) and rat CMV (RCMV) have been reported (Buck et al., 2007; Dolken et al., 2007; Grey et al., 2005; Meyer et al., 2011; Pfeffer et al., 2004), the identification and expression of RhCMV miRNAs is unknown. In this study we examined the expression of RhCMV miRNAs in rhesus fibroblasts, endothelial cells and tissues. We found that RhCMV encodes multiple miRNAs with different abundance and kinetics of expression as well as a positional and functional homologue of an HCMV miRNA. These studies provide the basis for examining the role of CMV miRNAs during pathogenesis and persistence in a relevant non-human primate model system.
Results
RhCMV encodes multiple miRNAs that are clustered in distinct regions of the viral genome
To identify RhCMV-encoded miRNAs, RNA isolated from virus-infected fibroblasts at three days post infection (dpi) was analyzed by Next Generation Sequencing (NGS). A total of 14,186,475 reads were made from the RhCMV-infected fibroblasts, with over 75 percent of the sequences mapping to miRBase (Table 2). 88.9 percent of these sequences were within 20 to 24 nucleotides in length. Of the 2,307,330 unmapped sequences, 44,949 (0.02%) mapped to hairpin structures within the RhCMV genome. The remainder of the unmapped sequences likely represent non-coding RNAs, unrecognized rhesus macaque miRNAs or RNA degradation products.
Table 2.
Distribution of small RNAs from RhCMV-infected rhesus fibroblasts
| # of sequences | % of sequences | # of unique sequences | % of unique sequences | |
|---|---|---|---|---|
| Raw | 14,186,475 | 100 | 790,102 | 100 |
| Mappable | 13,068,450 | 92.12 | 40,665 | 5.15 |
| Mapped (Total) | 10,773,233 | 75.94 | 16,439 | 2.08 |
| Unmapped (Total) | 2,307,330 | 16.26 | 25,063 | 3.17 |
| Mapped to RhCMV genome | 44,949 | 0.32 | 190 | 0.02 |
Seventeen potential RhCMV miRNAs were identified by NGS based on sequence length, copy number, mapping to the RhCMV genome and formation of stable hairpins. Consistent with standard nomenclature, the miRNAs are identified according to the closest open reading frame on the same strand. The viral miRNAs ranged in size from 18 to 23 nucleotides and were detected at copy numbers from 3 to 42705. Secondary structure predictions (LC Sciences, (Zuker, 2003)) demonstrate that the potential RhCMV miRNAs could fold into the hairpin structures typical of pre-miRNAs (Figure 1). Table 3 shows the predominant consensus sequence for each miRNA. Variability in the 3’ ends of the miRNAs was common, whereas any sequences detected with variability in the 5’ end for each miRNA was only a minor species (data not shown). Similar 3’ and 5’ variability has been noted with many other herpesviruses (Amen and Griffiths, 2011; Meyer et al., 2011; Riley, Rabinowitz, and Steitz, 2010; Zhu et al., 2010).
Figure 1. RhCMV pre-miRNA stem-loop structures.
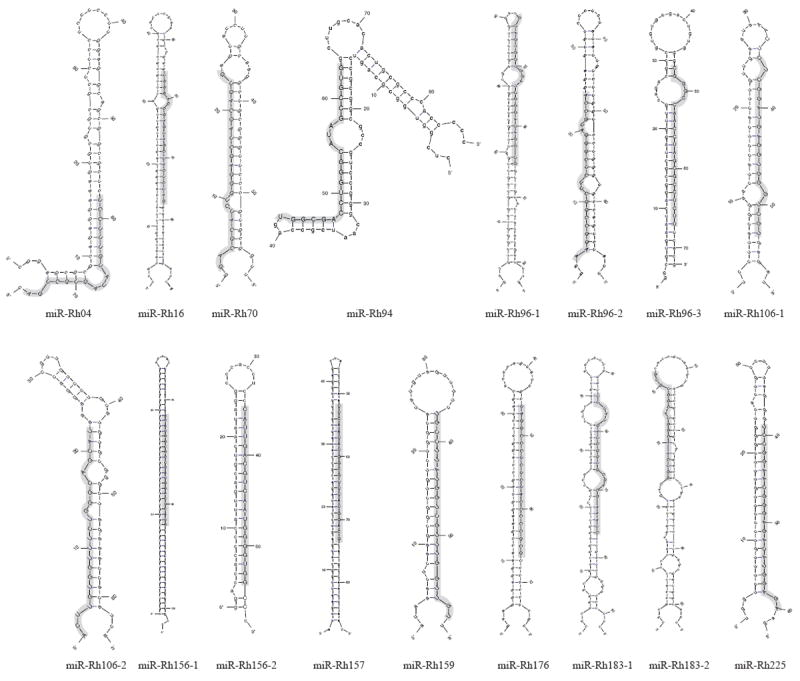
Stem-loop structures were predicted using the mfold program (Zuker, 2003). Mature miRNA sequences are highlighted in grey and depicted in capitalized letters.
Table 3.
RhCMV miRNAs identified from infected rhesus fibroblasts
| miRNA | Sequence | Length | Copy Number of isoform | Copy Number of all isoforms | Strand | Starta |
|---|---|---|---|---|---|---|
| miR-Rh04 | TCCTTTGTATAGTGGTGA | 18 | 3 | 3 | - | 3278 |
| miR-Rh16 | TGGGACCGAGTAGCTACTCTAG | 22 | 18253 | 42705 | - | 13078 |
| miR-Rh70 | GACGCATCTGGGAGCTAGCAGTG | 23 | 6 | 15 | - | 55945 |
| miR-Rh94 | TGGCGACCTGGGCATAGCCGTG | 22 | 3 | 3 | + | 87960 |
| miR-Rh96-1 | GCAGTGCTGGATCGGGACAGGCG | 23 | 3 | 3 | + | 88610 |
| miR-Rh96-2 | AAGGCCGGCTTCTCGGGATGCCA | 23 | 3 | 3 | + | 89430 |
| miR-Rh96-3 | CGGGCCAACCTAGCGCCTCTT | 21 | 17 | 49 | + | 89771 |
| miR-Rh106-1 | AACTCTCGGATGAGGTGGGTCGT | 23 | 3 | 3 | + | 102704 |
| miR-Rh106-2 | ACTTGTGGTTTCTGGGCAGCAT | 22 | 83 | 200 | + | 103662 |
| miR-Rh156-1 | ATATGGCGTCTATGGACTCATG | 22 | 8 | 16 | - | 162033 |
| miR-Rh156-2 | CATTGACGTCAATGGGGTGGCC | 22 | 9 | 29 | - | 161426 |
| miR-Rh157 | TTCAATATGGCGTTTATGGACT | 22 | 3 | 3 | + | 162076 |
| miR-Rh159 | TGCCTTATGGTCGTTTGGTTGT | 22 | 4 | 4 | + | 164314 |
| miR-Rh176 | TAGGCCCTCTCTACCCCCGCGT | 22 | 13 | 16 | - | 180692 |
| miR-Rh183-1 | TTTATGATCGGTGCGACGAAATC | 23 | 366 | 842 | + | 187856 |
| miR-Rh183-2 | GACGGTCAACGGTACGGTGTG | 21 | 3 | 3 | + | 191178 |
| miR-Rh225 | TCCGGAACGGGAGACATGGAGA | 22 | 58 | 264 | + | 216779 |
miRNA start sites refer to the 68-1 strain of RhCMV
The genomic locations of the 17 RhCMV miRNAs are depicted in Figure 2. Similar to HCMV, the RhCMV miRNAs are generally distributed across the viral genome with major clusters in two regions. One cluster of three miRNAs is located near the IE region of the genome (encoded by the Rh156 exons) and includes miR-Rh156-1, miR-Rh156-2 and miR-Rh157. Intriguingly, similar to the rodent CMVs, a second cluster exists either close to or within the viral origin of replication (nucleotides 88161 to 90525 (Hansen et al., 2003)) including miR-Rh94, miR-Rh96-1, miR-Rh96-2 and miR-Rh96-3. Although these miRNAs are clustered, each miRNA is encoded within their own hairpin structure (Figure 1). Comparison of the miRNA sequences of HCMV and RhCMV did not reveal significant sequence homology between the miRNAs of these viruses, with the exception of miR-Rh183-1 which is a positional homologue of HCMV miR-US5-2 and encodes a similar seed sequence (see below).
Figure 2. Genomic organization of RhCMV miRNAs.
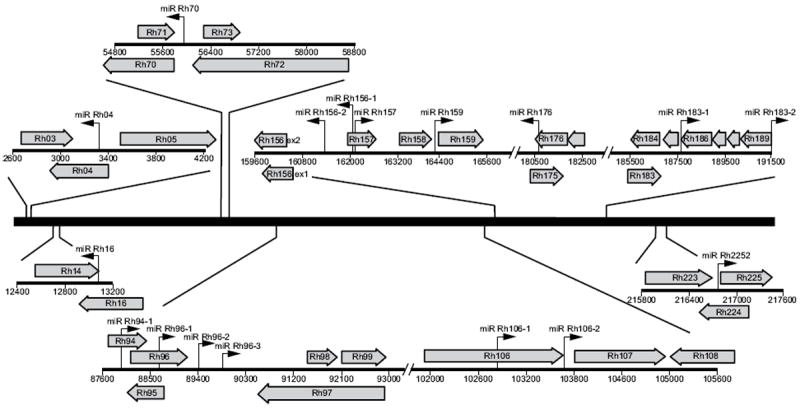
The RhCMV miRNAs are depicted as black arrows with the corresponding nomenclature above. Right-facing arrows indicate miRNAs encoded on the sense strand, while left-pointing arrows indicate miRNAs encoded on the antisense strand. Large grey arrows represent annotated open reading frames as per Hansen et al. (Hansen et al., 2003).
RhCMV miRNAs are differentially expressed in cells
To confirm the expression of RhCMV miRNAs during infection, we designed primer and probe sets for miRNA-specific, stem-loop real time PCR for 16 of the 17 predicted miRNAs. Using this technique, we were able to detect 13 rhesus-specific miRNAs. We were not able to reliably detect miR-Rh156-1, miR-Rh157 and miR-Rh-159. The miR-Rh156-1 assay detected product in the mock samples at levels comparable to infected cells, suggesting the presence of a cellular product with similar 3’ sequence (stem-loop primers are designed based on the 3’ sequence for each miRNA). Additionally, two of the viral miRNAs identified by deep sequencing, miR-Rh157 and miR-Rh159, were detected at copy numbers below the reliable limit of detection by the Real-time PCR assay. We were unable to successfully design an assay to detect miR-Rh04, which may be due to the shorter size of the miRNA.
Expression of each RhCMV miRNA was examined over a time-course of infection to determine their temporal expression patterns. In fibroblasts, HCMV miRNAs are undetectable at very early times in infection but increases in abundance throughout infection (Grey et al., 2005; Murphy et al., 2008). In contrast, RhCMV miRNAs display three distinct expression patterns (Figure 3). The first pattern of expression is similar to that observed for HCMV miRNAs, where miR-Rh16, miR-Rh176 and miR-Rh183-1 accumulated to high copy number over the 120 hour time-course. Similarly, miR-Rh96-1, miR-Rh156-2 and miR-Rh225 also accumulated, but were much less abundant. The second pattern of expression is exemplified by miR-Rh70, miR-Rh96-2, miR-Rh96-3, miR-Rh106-1and miR-Rh183-2 where copy number per cell peaked between 24-48 hours followed by a considerable decline in miRNA copy number. Finally, miR-Rh106-2 expression peaked at the earliest times tested followed by a decline over the remainder of infection
Figure 3. Kinetics of RhCMV miRNA expression in fibroblasts.
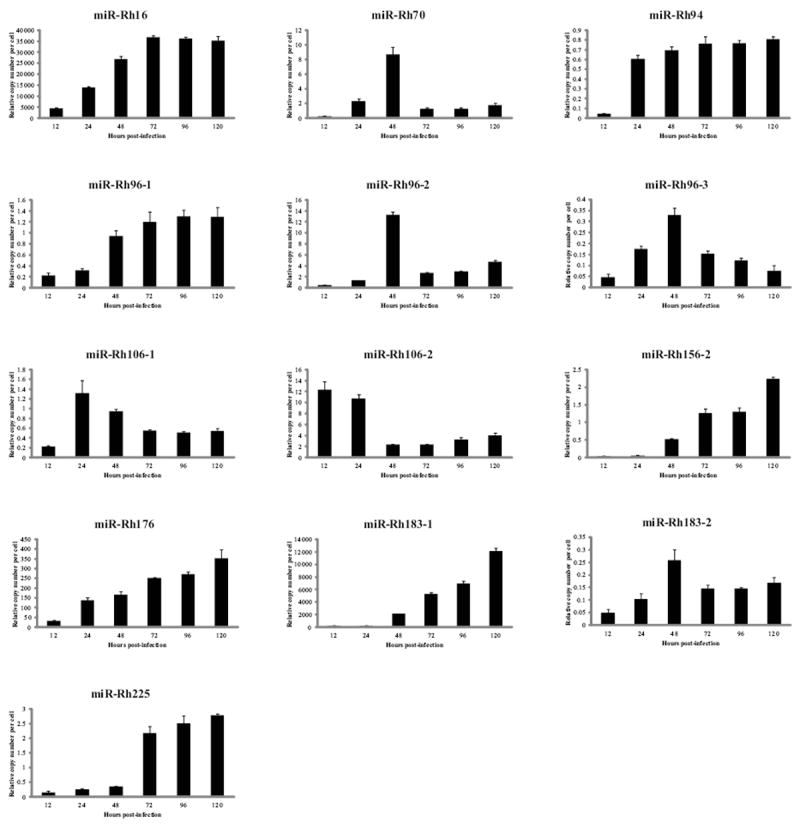
Immortalized rhesus fibroblasts were mock-infected or infected with 2 PFU/cell of RhCMV and RNA was harvested at the indicated timepoints. Stem-loop RT-PCR was performed using miRNA-specific primers. Viral miRNA copy numbers were determined using serial dilutions of oligo standards representing the miRNA sequence. Data was normalized to total RNA and used to determine the relative miRNA copy number per cell. 3 independent experiments are represented as mean copy number +/- standard deviation.
RhCMV miRNA expression was also examined in endothelial cells (EC) to determine if the pattern of miRNA expression was similar to infected fibroblasts. The replication of RhCMV in EC is delayed by approximately 24-48 hrs in comparison to fibroblasts as determined by viral DNA copy number as well as expression of immediate early 2 (IE2) protein (Figure 4A and data not shown). In addition to the delay in RhCMV expression, the total quantity of RhCMV DNA was approximately 2.5-fold lower in EC compared to fibroblasts (Figure 4A). Paralleling the decrease in viral DNA, miRNA expression was also lower in endothelial cells. Unlike the different kinetic patterns of miRNA expression observed in fibroblasts, all RhCMV miRNAs exhibited an increase in abundance throughout the time-course of infection in endothelial cells (Figure 4B).
Figure 4. RhCMV miRNA expression in endothelial cells.
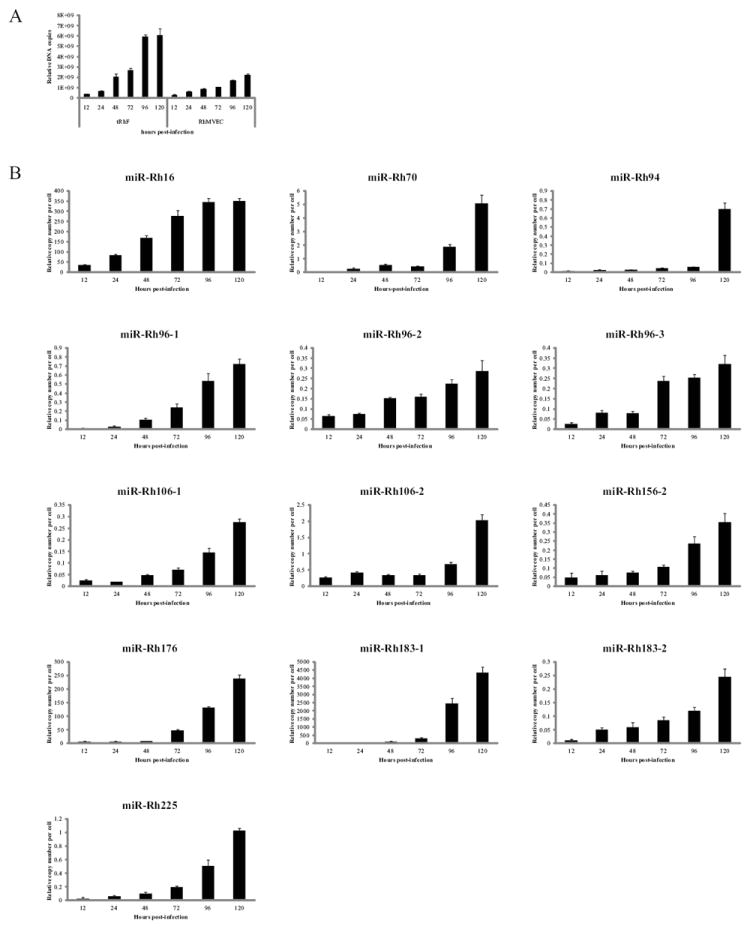
Rhesus microvascular endothelial cell (RhMVEC) or telomerized rhesus fibroblasts (tRhF) were mock-infected or infected with 2 PFU/cell of RhCMV and DNA and RNA was harvested at the indicated timepoints. (A) 100ng of DNA was used to perform Real-time PCR for Rh87. Data is presented as relative copy number as determined using serial dilution of RhCMV BAC DNA. Data was normalized to cellular β-actin. (B) Experiments were performed as in the legend of Figure 3.
RhCMV miRNAs are associated with Argonaute 2
Association of a small RNA with the RISC complex is one of the defining characteristics of a miRNA. Co-immunoprecipitation (Co-IP) of a viral miRNA with the RISC component Argonaute (Ago) suggests the formation of a functional miRNA complex during infection. To further elucidate the importance of the RhCMV miRNAs during infection of fibroblasts we determined whether a subset of RhCMV miRNAs are enriched in the RISC complex using RISC-IP (Grey et al., 2010). Cells were infected for 48 hours, followed by lysis and immunoprecipitation using pre-immune sera or an anti-Ago2 rabbit polyclonal antibody developed in our lab which cross-reacts with rhesus Ago2 (data not shown). RNA isolated from total RNA and IP samples was analyzed by stem loop RT-PCR for miR-Rh16 and miR-Rh183-1 as well as the low abundance miRNAs miR-Rh70 and miR-Rh225. Additionally, association of the cellular snRNA U6, which is not associated with the RISC complex (Riley, Rabinowitz, and Steitz, 2010), was examined. As demonstrated in Figure 5, each of the RhCMV miRNAs was enriched in the RISC complex from 190-fold to 1700-fold, while the cellular snRNA U6 showed little association with Ago2. Therefore, RhCMV miRNAs associate with the RISC component Ago2, suggesting the formation of RISC complexes containing both high and low abundance viral miRNAs during infection of fibroblasts.
Figure 5. RhCMV miRNAs are enriched in RISC.
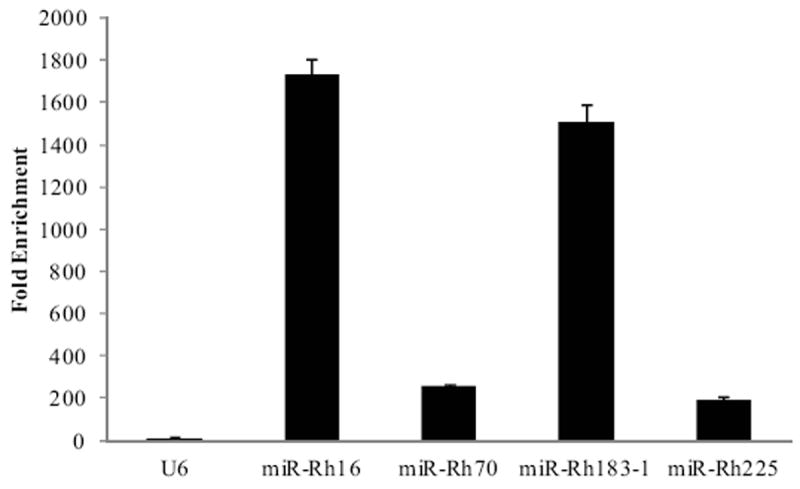
Rhesus fibroblasts were mock-infected or infected with 2 PFU/cell of RhCMV for 48 hours, after which cells were lysed and immunoprecipitations were performed using pre-immune sera or an anti-Ago2 antibody. Total and immunoprecipitated RNA was subjected to stem-loop RT-PCR for RhCMV miRNAs and the snRNA U6. Fold enrichment of a miRNA in Ago2 complexes was determined by comparing the IP/Total RNA values from samples immunoprecipitated with the Ago2 antibody to the IP/Total RNA values from samples immunoprecipitated with the pre-immune sera. Experiments were performed in duplicate.
RhCMV encodes a homologue of HCMV miR-US5-2
Despite the extensive sequence homology between HCMV and RhCMV, only one of the RhCMV miRNAs has a potential homologue in HCMV. Comparison of miR-Rh183-1 and miR-US5-2 sequences indicates that the seed sequence of miR-Rh183-1 is shifted by one nucleotide, leaving six of the core seed nucleotides identical (Figure 6A). In addition, eleven other nucleotides outside the seed region are also identical. miR-US5-2 is encoded antisense to the 3’ UTR of HCMV US7, while miR-Rh183-1 is encoded antisense to the 3’ UTR of Rh186 (Figure 2). Rh186 is a positional homologue of HCMV US7 that is located between Rh185 (US6) and Rh187 (US8) (Pande et al., 2005). Recently our group has reported that miR-US5-2 down-regulates expression of HCMV US7 through its 3’ UTR (Tirabassi et al., 2011). To determine whether miR-Rh183-1 functions similar to miR-US5-2, the putative 3’ UTRs of both Rh186 and US7 were cloned into a dual luciferase reporter vector. Transfection of 293 cells with miR-Rh183-1 and the Rh186 reporter vector or miR-US5-2 and the US7 reporter vector resulted in down-regulation of luciferase expression (Figure 6B). Mutation of the target sequences within the Rh186 and US7 3’ UTRs resulted in wild-type luciferase activity. Neither miR-Rh183-1 nor miR-US5-2 were able to downregulate US7 or Rh186 respectively, indicating that the 6 nucleotide match of the seed sequence is insufficient to mediate effective down-regulation of the reciprocal viral genes. Since miR-US5-2 and miR-Rh183-1 are encoded antisense to the 3’UTR of their target genes, the strong down-regulation of gene expression is likely mediated by the full 23 nucleotide match.
Figure 6. RhCMV miR-Rh183-1 is homologous to HCMV miR-US5-2.
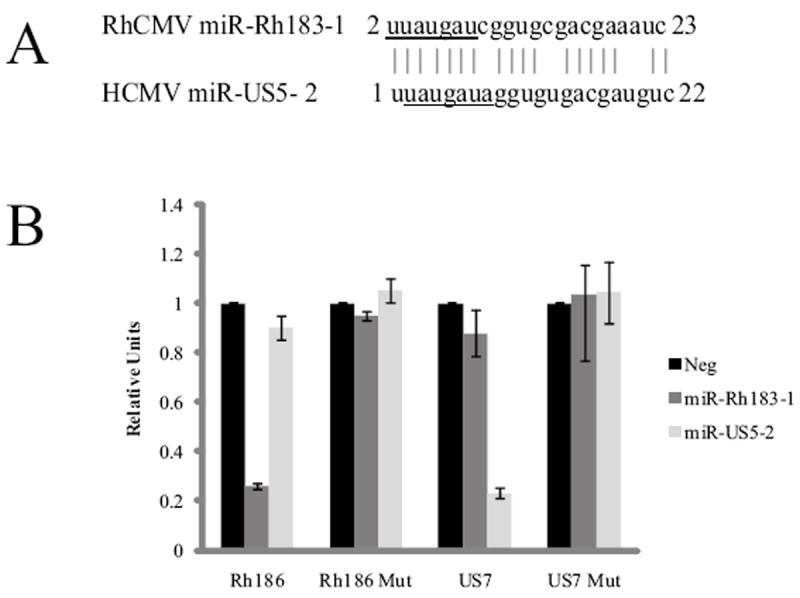
(A) Sequence alignment of miR-Rh183-1 and miR-US5-2. The seed sequence of each miRNA is underlined. (B) 293 cells were co-transfected with dual luciferase reporters encoding the WT Rh186 and US7 3’UTRs or reporters with mutations in the miRNA target sites (Rh186mut and US7mut) as well as miRNA mimics of miR-Rh183-1, miR-US5-2 or a negative control miRNA. 24 hours after transfection, cells were harvested and luciferase activity was monitored. Experiments were performed in triplicate and presented as mean +/- standard deviation with the negative control set to 1.
Expression of RhCMV miRNAs in rhesus macaque tissues
To examine viral miRNAs expression in naturally-infected rhesus macaques shedding RhCMV, RNA was extracted from persistently infected salivary gland tissue of four animals and examined for the presence of viral miRNAs. The amount of RhCMV in each salivary gland sample was determined by qPCR and viral DNA was detected in each animal at copy numbers ranging from approximately 2 × 104 to 2 × 105 (Figure 7A). RhCMV miRNA-specific stem-loop RT-PCR assays were used to assess the copy number of each miRNA in 100ng of total RNA isolated from homogenized tissue. Using oligonucleotide standards, this assay can reliably detect as little as 100 copies of miRNA. Five RhCMV miRNAs were detected in all animals, while a sixth miRNA was detected in one of four animals (Figure 7B). RhCMV miR-Rh16, the most abundant miRNA identified by NGS, was detected in all animals, as were miR-Rh96-1 and miR-Rh96-3 (Figure 7B). The other member of this cluster, miR-Rh96-2 was not detected above background levels. miR-Rh70 and miR-Rh225 were also detected in all the infected animals, while miR-Rh106-1 was detected in only one animal above background levels (Figure 7B). Interestingly, two of the most abundant RhCMV miRNAs detected in fibroblasts, miR-Rh176 and miR-Rh183-1, were not detected in the salivary gland tissue. These results suggest that there may be important roles for specific RhCMV miRNAs in the salivary gland during persistent infection.
Figure 7. RhCMV expression in salivary gland tissue from infected rhesus macaques.
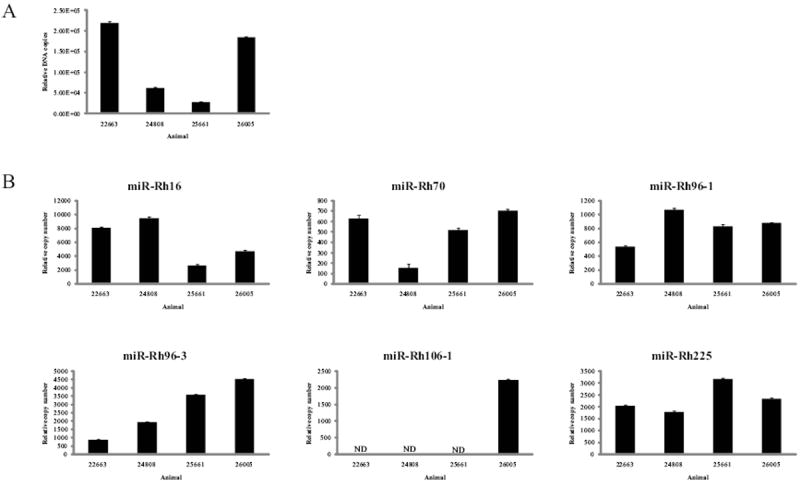
(A) 100ng of DNA was isolated from homogenized salivary gland tissue from four animals persistently infected with RhCMV and one RhCMV-negative animal and subjected to Real-time PCR using primers for Rh87. Relative DNA copy number was determined by serial dilution of RhCMV BAC DNA. Data was normalized to cellular β-actin. (B) 100ng of RNA was isolated from homogenized salivary gland tissue from four RhCMV-infected animals and one RhCMV-negative animal and subjected to stem-loop RT-PCR. Data was normalized to cellular miR-16 levels. ND – Not detected.
Discussion
In this study, seventeen RhCMV miRNAs were identified by NGS, thirteen of which were confirmed during infection of rhesus fibroblasts and endothelial cells by miRNA-specific RT-PCR. A subset of the RhCMV miRNAs were detected in the salivary glands of persistently infected rhesus macaques. Unlike HCMV, RhCMV exhibits distinct kinetic classes of miRNA expression in infected fibroblasts. Additionally, one of the RhCMV miRNAs was a positional and functional homologue of HCMV miR-US5-2.
Expression patterns for HCMV, MCMV and RCMV miRNAs in infected fibroblasts all follow a similar trend, with miRNAs increasing in abundance over the course of infection (Dolken et al., 2007; Grey et al., 2005; Meyer et al., 2011). In contrast, RhCMV miRNAs are expressed in three distinct patterns in infected fibroblasts. Five of the RhCMV miRNAs had peak expression at 24-48 hours post-infection and declined after this time. In addition, miR-Rh106-2 was expressed at the highest levels at the earliest times sampled, and declined considerably by 48 hours post-infection. Finally, seven of the RhCMV miRNAs increase in abundance throughout 120 hours of infection. Interestingly, Herpes B virus, a rhesus macaque homolog of herpes simplex virus (HSV), also displays several different kinetic patterns of miRNA expression during lytic infection of Vero cells (Amen and Griffiths, 2011), while HSV type 1 and 2 miRNAs accumulate throughout infection (Cui et al., 2006; Jurak et al., 2010). The different patterns of expression in rhesus fibroblasts may represent the expression patterns of the viral transcripts from which these miRNAs are derived. In support of this hypothesis, RhCMV miRNAs have decreased abundance and altered expression patterns in ECs, which could be the result of reduced expression from the viral genome in this cell type (Figure 4).
Calculating the approximate copy number of each miRNA during infection of fibroblasts identified three RhCMV miRNAs (miR-Rh16, miR-Rh176 and miR-Rh183-1) that are detected at hundreds to thousands of copies per cell while the remaining RhCMV miRNAs are much less abundant (Figure 3). Nevertheless, the RhCMV miRNAs identified in this report were detected by both NGS and RT-PCR, map to hairpin structures within the RhCMV genome and associate with the RISC component Ago2 (Figure 5). A recent study using NGS also reported low copy numbers for some HCMV miRNAs (Stark et al.). Factors that contribute to miRNA abundance include the rate of transcript accumulation, the efficiency of miRNA processing and the rate of miRNA decay. miRNA stability can be influenced by cellular modifications, Argonaute protein levels, exposure of the miRNAs to nucleases and target abundance (Kai and Pasquinelli, 2010). The relative contribution of each of these factors to miRNA abundance during CMV infection has not yet been assessed. The identification of low abundance RhCMV miRNAs enriched in the RISC complex strongly suggests that these miRNAs have an important role during viral infection. Additionally, a subset of the low-abundance miRNAs were specifically detected in persistently infected salivary gland tissue, suggesting a significant role for these miRNAs during infection in vivo (Figure 7). In contrast, miR-Rh176 and miR-Rh183-1, which both accumulate in high abundance in fibroblasts and ECs, could not be detected in the infected salivary gland. Thus, it is possible that some of the RhCMV miRNAs may be more highly expressed in cell types relevant to viral persistence and/or latency.
The detection of only six of the seventeen RhCMV miRNAs in salivary gland tissue from rhesus macaques suggests a dynamic situation in which only some of the transcripts from the RhCMV genome are being expressed and/or processed to form mature miRNAs during persistence. Correspondingly, RT-PCR analysis of salivary gland tissue from RCMV-infected animals demonstrated differential expression of miR-R87-1 and miR-r111.1-2 during the acute and persistent phases of infection (Meyer et al., 2011). The specific miRNAs detected in the macaque salivary gland may play important roles in maintaining the viral genome in this tissue, as has been suggested for MCMV miR-M23-2 and miR-m21-1. Infection of C57BL/6 mice with a miR-M23-2/m21-1 knockout virus resulted in a 100-fold decrease in virus production in the salivary gland (Dolken et al., 2010). This observation depended upon the mouse strain and viral load used, and was reversed by the combined depletion of NK and CD4+ T cells, suggesting a complex relationship in vivo between host genetic factors and the viral miRNAs which are required to maintain the viral genome.
Despite the significant sequence homology between HCMV and RhCMV, only one miRNA of RhCMV has an HCMV homologue (Figure 6). Homologous miRNAs have only rarely been identified in the herpesvirus family. Herpes B virus shares no miRNAs in common with HSV-1 or -2 (Amen and Griffiths, 2011). Rhesus rhadinovirus (RRV) has only one miRNA in common with its human counterpart, Kaposi’s sarcoma-associated herpesvirus (KSHV) (Umbach et al., 2010), while only 7 of the 68 miRNAs identified in rhesus lymphocryptovirus (rLCV) are closely related to Epstein Barr virus (EBV) miRNAs (Cai et al., 2006). The similarity between miR-Rh183-1 and miR-US5-2 suggests significant pressure to maintain the seed sequence during evolution in their respective hosts, indicating that regulating their viral and cellular targets is of significant importance for viral replication. Our laboratory has recently demonstrated that genetically removing miR-US5-2 from the HCMV genome results in an increase in US7 protein expression during infection of fibroblasts (Tirabassi et al., 2011). US7 is found within a cluster of genes involved in MHC down-regulation (Tortorella et al., 2000), however the role of this protein during viral infection is currently unclear. HCMV down-regulates expression of US7 using three target sites for two viral miRNAs (miR-US5-1 and miR-US5-2), while the Rh186 3’ UTR contains potential target sites for six RhCMV miRNAs (data not shown), underscoring the importance of regulating expression of this protein during the viral lifecycle. The full panel of viral and cellular targets for miR-US5-2 and miR-Rh183-1 has yet to be defined; however the conservation of miRNA sequence strongly implies that these miRNAs target similar genes that are important for viral replication and/or persistence. The rhesus macaque model provides a means to test the importance of the miR-US5-2 homologue in a relevant non-human primate system.
The species specificity of CMVs has hampered the identification of miRNAs important in HCMV pathogenesis and persistence. RhCMV infection of rhesus macaques is a valuable model system for CMV pathogenesis due to the genetic similarity of the viruses. Here we both identify the miRNAs encoded by RhCMV and provide a comprehensive analysis of their expression kinetics during lytic infection. This knowledge constitutes the basis for identifying cell-type and tissue-specific targets of the viral miRNAs. Given that at least one miRNA has a partially conserved seed sequence between HCMV and RhCMV, relevant questions regarding the importance of identified targets can be addressed in an in vivo model system of CMV infection. In addition, RhCMV and HCMV miRNAs may target similar genes or pathways using different miRNAs to achieve downregulation of target expression. This is a readily testable hypothesis in our model system now that the sequences of the RhCMV miRNAs have been identified.
Materials and Methods
Cells, tissue and virus
Rhesus fibroblasts that were life-extended with telomerase were a gift from Dr Scott Wong and were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Hyclone), 100U/mL of penicillin and 100ug/mL of streptomycin (Invitrogen). Primary rhesus brain microvascular endothelial cells were a gift from Dr Ashlee Moses and were maintained in endothelial cell basal medium supplemented with EGM-MV SingleQuots (Lonza). 293 cells were obtained from the American Type Culture Collection and maintained in DMEM as above. Cells were infected with RhCMV strain 68-1 at a multiplicity of 2 plaque forming units (PFU)/cell for two hours at 37°C. After this time, the inoculum was removed and replaced with fresh medium. Cells were harvested at various times post-infection using DNAzol or Trizol (Invitrogen) as appropriate. Samples were stored at 4°C (DNAzol) or -80°C (Trizol) prior to analysis. Rhesus macaque tissues were obtained from the Oregon National Primate Research Center and stored at -80°C immediately after necropsy. ~0.5 grams of tissue were suspended in Trizol or DNAzol and homogenized using glass beads.
miRNA Deep Sequencing
Next generation sequencing was performed by LC Sciences (Houston, TX). Briefly, total RNA was size-fractionated on a 15% tris-borate-EDTA-Urea (TBE) polyacrylamide gel. RNA fragments 15-50 nucleotides were eluted from the gel and ethanol precipitated. 5’ and 3’ adapters (Illumina) were ligated using T4 RNA ligase (Promega) and size-fractionated on 15% TBE polyacrylamide gels, followed by gel elution and ethanol precipitation. Next, cDNA was reverse transcribed using M-MLV (Invitrogen) and Illumina RT primers and amplified for 20 cycles with PFX DNA polymerase and the Illumina small RNA primer sets. These cDNAs were then purified on a 12% TBE polyacrylamide gel and a slice ~80-115bp was excised. cDNAs were eluted, ethanol precipitated and resuspended for deep sequencing using Illumina’s GAIIx sequencer following the vendor’s instructions. Raw sequencing data was filtered for composition, adapter dimers, length, sequence composition and copy number. The filtered data was mapped to miRBase, the Macaca mulatta genome and the RhCMV genome and stable hairpin structures were determined.
qPCR detection of RhCMV miRNAs
RT-PCR was used to quantitate the amount of viral DNA or miRNA in infected rhesus fibroblasts, endothelial cells and salivary gland tissue. Total RNA was isolated from infected cells or tissue using Trizol. cDNA was prepared using 100ng of total RNA and miRNA-specific stem-loop RT primers designed to the most abundant species detected by NGS for each miRNA (see Table 1). Samples were incubated at 16°C for 30 minutes, 42°C for 30 minutes and 85°C for 5 minutes. Real-time PCR (Taqman) was used to quantify the miRNA copy number from 10ng of the cDNA reaction. An ABI StepOnePlus Real Time PCR machine was used with the following program for 40 cycles: 95°C for 15 sec and 60°C for one minute. Forward primers and probes are outlined in Table 1. The same reverse primer (GTGCAGGGTCCGAGGT) was used in all assays. Standards were designed as oligonucleotides with the same sequence as the miRNA of interest and data is presented as relative copy number per cell. To determine relative copy number of each miRNA per cell, miRNA copy number was first determined in 10ng of total RNA using a known concentration of oligo standards. This value was multiplied by the total amount of RNA isolated from the infected cells and then divided by the total number of cells as determined prior to infection. Each experiment was performed in triplicate. This assay can detect as few as 100 copies of miRNA in 10ng of total RNA as determined using the oligonucleotide standards. For miRNA detection in persistently infected salivary gland tissue, miRNA copy numbers were normalized to the expression of cellular miR-16, detected using the miR-16 Taqman miRNA assay (Applied Biosystems). Expression of miR-16 was not affected by RhCMV during infection of fibroblasts (data not shown).100ng of viral DNA isolated using DNAzol was used in the Real-time PCR reactions. Rh87 Forward primer: GCGCCACAAACGTGGAATAACAGA Reverse primer: TACAGTGATCCCGTGGTGGTGTTT Probe: ATTGACGGATACAGGCTGGCAAAGTC Relative copy number was calculated using a serial dilution of BAC DNA and normalized to cellular β-actin. β-actin Forward primer: TCACCCACACTGTGCCCATCTACGA Reverse primer: CAGCGGAACCGCTCGTTGCCAATGG
Table 1.
miRNA RT-PCR primer and probe sets
| Target | RT primera | Taqman Forward Primer | Probe |
|---|---|---|---|
| miR-Rh04 | TCACCA | GCGGTCCTTTGTATA | TGGATACGACTCACCA |
| miR-Rh16 | CTAGAG | GCGCTGGGACCGAGTAGCT | TGGATACGACCTAGAG |
| miR-Rh70 | CACTGC | GCGGGACGCATCTGGGAGCT | TGGATACGACCACTGC |
| miR-Rh94 | CACGGC | GCGCTGGCGACCTGGGCAT | TGGATACGACCACGGC |
| miR-Rh96-1 | CGCCTG | GCGGGCAGTGCTGGATCGGG | TGGATACGACCGCCTG |
| miR-Rh96-2 | TGGCAT | GCGGAAGGCCGGCTTCTCGG | TGGATACGACTGGCAT |
| miR-Rh96-3 | AAGAGG | GCGGCGGGCCAACCTAGC | TGGATACGACAAGAGG |
| miR-Rh106-1 | ACGACC | GCGGAACTCTCGGATGAGGT | TGGATACGACACGACC |
| miR-Rh106-2 | ATGCTG | GCGGACTTGTGGTTTCTGG | TGGATACGACATGCTG |
| miR-Rh156-1 | CATGAG | GCGGATATGGCGTCTATGG | TGGATACGACCATGAG |
| miR-Rh156-2 | GGCCAC | GCGGCATTGACGTCAATGG | TGGATACGACGGCCAC |
| miR-Rh157 | AGTCCA | GCGGTTCAATATGGCGTTT | TGGATACGACAGTCCA |
| miR-Rh159 | ACAACC | GCGGTGCCTTATGGTCGTT | TGGATACGACACAACC |
| miR-Rh176 | ACGCGG | GCGGTAGGCCCTCTCTACC | TGGATACGACACGCGG |
| miR-Rh183-1 | GATTTC | GCGCTTTATGATCGGTGCGA | TGGATACGACGATTTC |
| miR-Rh183-2 | CACACC | GCGGGACGGTCAACGGTA | TGGATACGACCACACC |
| miR-Rh225 | TCTCCA | GCGGTCCGGAACGGGAGAC | TGGATACGACTCTCCA |
All RT primers share the sequence GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGAC, with the last six unique 3’ nucleotides listed in the table
RISC immunoprecipitation
RISC IPs were carried out as previously described (Grey et al., 2010). Rhesus fibroblasts were mock infected or infected with 2 PFU/cell of RhCMV for 48 hours. Cells were lysed and samples were taken for total RNA levels. miRNP complexes were immunoprecipitated using rabbit anti-Ago2 antibody or pre-immune sera and protein A sepharose beads (Sigma). RNA was isolated using Trizol and stem loop RT-PCR was performed as described above. Fold enrichment was calculated as IP/Total RNA from samples immunoprecipitated with the Ago2 antibody compared to IP/Total RNA from samples immunoprecipitated using the pre-immune sera. The U6 RT-PCR assay was obtained from Applied Biosystems.
Dual luciferase assay
The putative 3’ UTRs of RhCMV Rh186 and HCMV US7 were cloned into the dual luciferase reporter pSiCheck2 (Clontech). Site-directed mutagenesis was performed to mutate the single miR-Rh183-1 binding site in the Rh186 3’UTR and both miR-US5-2 binding sites in US7 3’UTR to produce Rh186mut and US7mut. 293 cells seeded into 96-well plates were co-transfected in triplicate with 100ng of plasmid and 100fmol of miRNA mimic (custom designed; IDT) using Lipofectamine 2000. Cells were incubated overnight, and then harvested for luciferase assay using the Dual-Glo Reporter Assay Kit (Promega) according to the manufacturer’s protocol. Luminescence was detected using a Veritas Microplate Luminometer (Turner Biosystems). All experiments were performed in triplicate and presented as mean +/- standard deviation.
Highlights.
RhCMV encodes 13 miRNAs as assessed by Next Generation sequencing and qPCR
RhCMV miRNAs display unique patterns of expression in rhesus fibroblasts
RhCMV miR-Rh183-1 is a homologue of HCMV miR-US5-2
A subset of RhCMV miRNAs are detected in salivary glands of persistently infected rhesus macaques
Acknowledgments
We would like to thank Dr. Scott Wong and Dr. Ashlee Moses for providing virus and cells. Additionally, we would like to thank Andrew Townsend for assistance with graphics and members of the Nelson and Streblow labs for critical discussions and review of the manuscript. This work was supported by NIH grant AI 21640 (J.A.N.) from the National Institute of Allergy and Infectious Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amen MA, Griffiths A. Identification and Expression Analysis of Herpes B Virus-Encoded Small RNAs. J Virol. 2011;85(14):7296–311. doi: 10.1128/JVI.00505-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin GB. Disseminated cytomegalovirus infection in immunodeficient rhesus monkeys. Am J Pathol. 1987;129(2):345–52. [PMC free article] [PubMed] [Google Scholar]
- Buck AH, Santoyo-Lopez J, Robertson KA, Kumar DS, Reczko M, Ghazal P. Discrete clusters of virus-encoded micrornas are associated with complementary strands of the genome and the 7.2-kilobase stable intron in murine cytomegalovirus. J Virol. 2007;81(24):13761–70. doi: 10.1128/JVI.01290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10(12):1957–66. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2(3):e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke LM, Duerr A, Feldman J, Sierra MF, Daidone BJ, Landesman SH. Factors associated with cytomegalovirus infection among human immunodeficiency virus type 1-seronegative and -seropositive women from an urban minority community. J Infect Dis. 1996;173(1):77–82. doi: 10.1093/infdis/173.1.77. [DOI] [PubMed] [Google Scholar]
- Cui C, Griffiths A, Li G, Silva LM, Kramer MF, Gaasterland T, Wang XJ, Coen DM. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J Virol. 2006;80(11):5499–508. doi: 10.1128/JVI.00200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolken L, Krmpotic A, Kothe S, Tuddenham L, Tanguy M, Marcinowski L, Ruzsics Z, Elefant N, Altuvia Y, Margalit H, Koszinowski UH, Jonjic S, Pfeffer S. Cytomegalovirus microRNAs facilitate persistent virus infection in salivary glands. PLoS Pathog. 2010;6(10):e1001150. doi: 10.1371/journal.ppat.1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolken L, Perot J, Cognat V, Alioua A, John M, Soutschek J, Ruzsics Z, Koszinowski U, Voinnet O, Pfeffer S. Mouse cytomegalovirus microRNAs dominate the cellular small RNA profile during lytic infection and show features of posttranscriptional regulation. J Virol. 2007;81(24):13771–82. doi: 10.1128/JVI.01313-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2010;223(2):102–15. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey F, Antoniewicz A, Allen E, Saugstad J, McShea A, Carrington JC, Nelson J. Identification and characterization of human cytomegalovirus-encoded microRNAs. J Virol. 2005;79(18):12095–9. doi: 10.1128/JVI.79.18.12095-12099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey F, Meyers H, White EA, Spector DH, Nelson J. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 2007;3(11):e163. doi: 10.1371/journal.ppat.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey F, Tirabassi R, Meyers H, Wu G, McWeeney S, Hook L, Nelson JA. A viral microRNA down-regulates multiple cell cycle genes through mRNA 5’UTRs. PLoS Pathog. 2010;6(6):e1000967. doi: 10.1371/journal.ppat.1000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundhoff A, Sullivan CS. Virus-encoded microRNAs. Virology. 2011;411(2):325–43. doi: 10.1016/j.virol.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404(6775):293–6. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Hansen SG, Strelow LI, Franchi DC, Anders DG, Wong SW. Complete sequence and genomic analysis of rhesus cytomegalovirus. J Virol. 2003;77(12):6620–36. doi: 10.1128/JVI.77.12.6620-6636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(5531):834–8. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Jurak I, Kramer MF, Mellor JC, van Lint AL, Roth FP, Knipe DM, Coen DM. Numerous conserved and divergent microRNAs expressed by herpes simplex viruses 1 and 2. J Virol. 2010;84(9):4659–72. doi: 10.1128/JVI.02725-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai ZS, Pasquinelli AE. MicroRNA assassins: factors that regulate the disappearance of miRNAs. Nat Struct Mol Biol. 2010;17(1):5–10. doi: 10.1038/nsmb.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur A, Kassis N, Hale CL, Simon M, Elliott M, Gomez-Yafal A, Lifson JD, Desrosiers RC, Wang F, Barry P, Mach M, Johnson RP. Direct relationship between suppression of virus-specific immunity and emergence of cytomegalovirus disease in simian AIDS. J Virol. 2003;77(10):5749–58. doi: 10.1128/JVI.77.10.5749-5758.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn EM, Stolte N, Matz-Rensing K, Mach M, Stahl-Henning C, Hunsmann G, Kaup FJ. Immunohistochemical studies of productive rhesus cytomegalovirus infection in rhesus monkeys (Macaca mulatta) infected with simian immunodeficiency virus. Vet Pathol. 1999;36(1):51–6. doi: 10.1354/vp.36-1-51. [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21(17):4663–70. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lima RT, Busacca S, Almeida GM, Gaudino G, Fennell DA, Vasconcelos MH. MicroRNA regulation of core apoptosis pathways in cancer. Eur J Cancer. 2011;47(2):163–74. doi: 10.1016/j.ejca.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Lockridge KM, Sequar G, Zhou SS, Yue Y, Mandell CP, Barry PA. Pathogenesis of experimental rhesus cytomegalovirus infection. J Virol. 1999;73(11):9576–83. doi: 10.1128/jvi.73.11.9576-9583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch DJ, Cook S, Dolan A, Jamieson FE, Telford EA. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J Mol Biol. 1995;247(3):443–58. doi: 10.1006/jmbi.1995.0152. [DOI] [PubMed] [Google Scholar]
- McGeoch DJ, Dolan A, Ralph AC. Toward a comprehensive phylogeny for mammalian and avian herpesviruses. J Virol. 2000;74(22):10401–6. doi: 10.1128/jvi.74.22.10401-10406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C, Grey F, Kreklywich CN, Andoh TF, Tirabassi RS, Orloff SL, Streblow DN. Cytomegalovirus microRNA expression is tissue specific and is associated with persistence. J Virol. 2011;85(1):378–89. doi: 10.1128/JVI.01900-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E, Vanicek J, Robins H, Shenk T, Levine AJ. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc Natl Acad Sci U S A. 2008;105(14):5453–8. doi: 10.1073/pnas.0711910105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande NT, Powers C, Ahn K, Fruh K. Rhesus cytomegalovirus contains functional homologues of US2, US3, US6, and US11. J Virol. 2005;79(9):5786–98. doi: 10.1128/JVI.79.9.5786-5798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L, Maidji E, McDonagh S, Tabata T. Insights into viral transmission at the uterine-placental interface. Trends Microbiol. 2005;13(4):164–74. doi: 10.1016/j.tim.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. Identification of virus-encoded microRNAs. Science. 2004;304(5671):734–6. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- Picker LJ, Hagen SI, Lum R, Reed-Inderbitzin EF, Daly LM, Sylwester AW, Walker JM, Siess DC, Piatak M, Jr, Wang C, Allison DB, Maino VC, Lifson JD, Kodama T, Axthelm MK. Insufficient production and tissue delivery of CD4+ memory T cells in rapidly progressive simian immunodeficiency virus infection. J Exp Med. 2004;200(10):1299–314. doi: 10.1084/jem.20041049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher CJ, Hagen SI, Walker JM, Lum R, Mitchell BL, Maino VC, Axthelm MK, Picker LJ. Development and homeostasis of T cell memory in rhesus macaque. J Immunol. 2002;168(1):29–43. doi: 10.4049/jimmunol.168.1.29. [DOI] [PubMed] [Google Scholar]
- Price DA, Bitmansour AD, Edgar JB, Walker JM, Axthelm MK, Douek DC, Picker LJ. Induction and evolution of cytomegalovirus-specific CD4+ T cell clonotypes in rhesus macaques. J Immunol. 2008;180(1):269–80. doi: 10.4049/jimmunol.180.1.269. [DOI] [PubMed] [Google Scholar]
- Rawlinson WD, Farrell HE, Barrell BG. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70(12):8833–49. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley KJ, Rabinowitz GS, Steitz JA. Comprehensive analysis of Rhesus lymphocryptovirus microRNA expression. J Virol. 2010;84(10):5148–57. doi: 10.1128/JVI.00110-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowshani AT, Bemelman FJ, van Leeuwen EM, van Lier RA, ten Berge IJ. Clinical and immunologic aspects of cytomegalovirus infection in solid organ transplant recipients. Transplantation. 2005;79(4):381–6. doi: 10.1097/01.tp.0000148239.00384.f0. [DOI] [PubMed] [Google Scholar]
- Stark TJ, Arnold JD, Spector DH, Yeo GW. High-resolution profiling and analysis of viral and host small RNAs during human cytomegalovirus infection. J Virol. doi: 10.1128/JVI.05903-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, Goldman-Wohl D, Greenfield C, Yagel S, Hengel H, Altuvia Y, Margalit H, Mandelboim O. Host immune system gene targeting by a viral miRNA. Science. 2007;317(5836):376–81. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N, Saleh N, Goldberg MD, Prichard M, Wolf DG, Mandelboim O. Analysis of human cytomegalovirus-encoded microRNA activity during infection. J Virol. 2009;83(20):10684–93. doi: 10.1128/JVI.01292-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirabassi R, Hook L, Landais I, Grey F, Meyers H, Hewitt H, Nelson J. Human Cytomegalovirus US7 is Regulated Synergistically by Two Virally-Encoded miRNAs and by Two Distinct Mechanisms. J Virol. 2011 doi: 10.1128/JVI.05443-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiscornia G, Izpisua Belmonte JC. MicroRNAs in embryonic stem cell function and fate. Genes Dev. 2011;24(24):2732–41. doi: 10.1101/gad.1982910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorella D, Gewurz B, Schust D, Furman M, Ploegh H. Down-regulation of MHC class I antigen presentation by HCMV; lessons for tumor immunology. Immunol Invest. 2000;29(2):97–100. doi: 10.3109/08820130009062289. [DOI] [PubMed] [Google Scholar]
- Tuddenham L, Pfeffer S. Roles and regulation of microRNAs in cytomegalovirus infection. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbagrm.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Umbach JL, Strelow LI, Wong SW, Cullen BR. Analysis of rhesus rhadinovirus microRNAs expressed in virus-induced tumors from infected rhesus macaques. Virology. 2010;405(2):592–9. doi: 10.1016/j.virol.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink C, Beuken E, Bruggeman CA. Complete DNA sequence of the rat cytomegalovirus genome. J Virol. 2000;74(16):7656–65. doi: 10.1128/jvi.74.16.7656-7665.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel P, Weigler BJ, Kerr H, Hendrickx AG, Barry PA. Seroepidemiologic studies of cytomegalovirus infection in a breeding population of rhesus macaques. Lab Anim Sci. 1994;44(1):25–30. [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JY, Strehle M, Frohn A, Kremmer E, Hofig KP, Meister G, Adler H. Identification and analysis of expression of novel microRNAs of murine gammaherpesvirus 68. J Virol. 2010;84(19):10266–75. doi: 10.1128/JVI.01119-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]


