SUMMARY
In eukaryotes, DNA is packaged into chromatin by canonical histone proteins. The specialized histone H3 variant CENP-A provides an epigenetic and structural basis for chromosome segregation by replacing H3 at centromeres. Unlike exclusively octameric canonical H3 nucleosomes, CENP-A nucleosomes have been shown to exist as octamers, hexamers, and tetramers. An intriguing possibility reconciling these observations is that CENP-A nucleosomes cycle between octamers and tetramers in vivo. We tested this hypothesis by tracking CENP-A nucleosomal components, structure, chromatin folding, and covalent modifications across the human cell cycle. We report that CENP-A nucleosomes alter from tetramers to octamers before replication and revert to tetramers after replication. These structural transitions are accompanied by reversible chaperone binding, chromatin fiber folding changes, and previously undescribed modifications within the histone fold domains of CENP-A and H4. Our results reveal a cyclical nature to CENP-A nucleosome structure and have implications for the maintenance of epigenetic memory after centromere replication.
INTRODUCTION
Every metaphase chromosome has a centromere, a unique chromatin structure to which kinetochore complexes and spindle microtubules attach during mitosis (Bloom and Joglekar, 2010). Centromeric chromatin is comprised of nucleosomes containing a centromere-specific histone H3 variant, CENP-A, which is required for establishing the kinetochore prior to every mitotic event over the replicative life span of eukaryotic cells. Thus, CENP-A is a key epigenetic determinant of centromere identity and function.
In contrast to canonical nucleosomes, which organize the bulk of eukaryotic genomes into octamers composed of H2A, H2B, H3, and H4, CENP-A nucleosomal structure remains controversial. Whereas yeast and human CENP-A can assemble into conventional octameric nucleosomes in vitro (Camahort et al., 2009; Tachiwana et al., 2011), human CENP-A also assembles into rigidified protein tetramers in vitro (Black et al., 2004; Sekulic et al., 2010). Furthermore, octameric (Camahort et al., 2009), hexameric (Mizuguchi et al., 2007), and right-handed (Furuyama and Henikoff, 2009) CENP-A nucleosomes have been documented in yeast, whereas tetrameric “hemisomes” containing CENP-A, H2A, H2B, and H4 have been identified in asynchronous Drosophila and human cells (Dalal et al., 2007; Dimitriadis et al., 2010). In contrast, a recent study using overexpressed CENP-A has reported the presence of unstable octamers in fly cells (Zhang et al., 2012). These studies point to an inexplicable variability in structure for a nucleosome whose function is both critical and conserved.
An unexplored possibility to explain such variability in structure might be that CENP-A nucleosomal organization is dynamic over the cell cycle, so that CENP-A forms octamers after completion of assembly at G1, but transits through stable tetrameric intermediates (Allshire and Karpen, 2008; Probst et al., 2009) that are generated after replication (Dalal and Bui, 2010; Henikoff and Furuyama, 2010; Black and Cleveland, 2011) or mitosis (Bloom and Joglekar, 2010). To investigate this hypothesis, we tracked CENP-A nucleosomes over the cell cycle in human cells by using a combination of chromatin biochemistry, atomic force microscopy (AFM), coimmunoprecipitation (co-IP) experiments, Förster resonance energy transfer (FRET), and liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS). We report that native CENP-A nucleosome are tetrameric at early G1, convert to octamers at the transition from G1 into S phase, and revert back to tetramers after replication, a state they assume for the rest of the cell cycle. These structural changes are accompanied by reversible binding of the CENP-A chaperone HJURP and changes in chromatin fiber folding. Furthermore, we uncover previously undescribed covalent modifications in both CENP-A and H4 histone fold domains, which occur at the key transition point from G1 into S phase. We discuss implications of our findings for the inheritance of centromeric domains after replication.
RESULTS
Heterotypic CENP-A Nucleosomes Bind the Chaperone HJURP at G1 and G2 Phases but Not at S Phase
We first examined whether histone or kinetochore components in the centromeric fiber change over the cell cycle. To address this, human cells were synchronized at early G1, G1/S, S, G2/M, and M phases (Experimental Procedures and Figure S1A available online). Chromatin arrays were released from these cells by mild nuclease digestion, followed by chromatin immunoprecipitation (ChIP) with an anti-CENP-A antibody to enrich for native CENP-A nucleosomes (Dimitriadis et al., 2010) (Figure S1B).
Components present within long- and short-length arrays of bulk chromatin (BC) and CENP-A chromatin were analyzed on high-sensitivity protein gels (Experimental Procedures). As expected, BC from these cells depicts the normal equivalence of canonical histones, within which CENP-A is detectable (Figure S2A, western blots [WB]). Our previous results demonstrated that CENP-A purified from asynchronous human cells associates with H2A, H2B, and H4 on long-, moderate-, and short-length chromatin arrays even when H3 is depleted, suggesting that CENP-A nucleosomes are heterotypic (Dimitriadis et al., 2010). We next examined whether CENP-A transits through a homotypic state (i.e., H2A/B free; Mizuguchi et al., 2007) during the human cell cycle. However, whether from G1, G1/S, S, and G2/M cells, long CENP-A chromatin arrays contain H2A, H2B, and H4 (Figure 1A). Such arrays are associated with key inner kinetochore proteins such as CENP-C and CENP-N (Figure 1A, WB) (Carroll et al., 2010; Screpanti et al., 2011) and contain H3 (Figure 1A, two-color WB), indicative of alternating domains typically found at centromeres (Sullivan and Karpen, 2004). Centromeric immunoprecipitates (IPs) are enriched in CENP-A and depleted in H3 (Figure S2B) and contain centromere-specific alpha satellite DNA (Figure S2C), supporting their centromeric origin. When the arrays are made shorter by prolonging nuclease treatment, until the input is almost exclusively mononucleosomes, CENP-A nucleosomes copurify with core histones H2A, H2B, and H4 in equimolar amounts, even when H3 is completely depleted within the IP (Figures 1B and 1C). Thus, these data indicate that H2A and H2B are intrinsic components of native CENP-A nucleosomes derived from active centromeres at all points of the cell cycle examined.
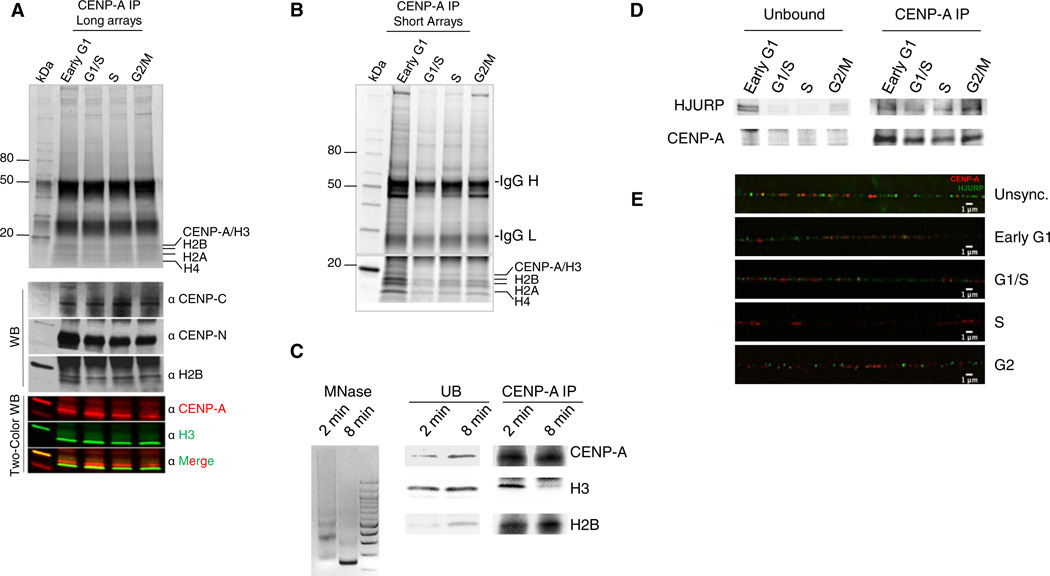
Figure 1. CENP-A Nucleosomes Are Heterotypic throughout the Cell Cycle and Bind the Chaperone HJURP at G1, G1/S, and G2 Phases but Not at S Phase.
Denaturing protein gel analysis of (A) native CENP-A IP from long chromatin arrays shows CENP-A contains H2A, H2B, and H4 at all points of the cell cycle and binds kinetochore proteins CENP-C and CENP-N (WB panel).
(B and C) Native CENP-A IP from short chromatin arrays (B) and mononucleosomal input (C) demonstrate that CENP-A is always associated with H2A, H2B, and H4, but not with H3.
(D) Western blot of HJURP within the CENP-A IP from (B) demonstrates loss of the HJURP at S phase, and return of HJURP at G2 phase.
(E) Extracted chromatin fibers stained for HJURP (green) and CENP-A as centromeric marker (red) demonstrate that HJURP is lost from centromeric fibers at S phase but enriched in G2 and G1 phases. Scale bars, 1 µm. See also Figure S2.
In parallel, we also analyzed the CENP-A histone chaperone HJURP in CENP-A chromatin arrays. At G1 phase, consistent with its role in depositing CENP-A (Dunleavy et al., 2009; Foltz et al., 2009; Shuaib et al., 2010), HJURP is bound to chromatin (Figure S2D, HJURP IP) and specifically to short CENP-A chromatin arrays (Figure 1D, CENP-A IP, HJURP WB). However, in contrast to its yeast homolog Scm3, which is bound to centromeres throughout the cell cycle (Xiao et al., 2011), we observed that HJURP is diminished in CENP-A chromatin from G1/S through S (Dunleavy et al., 2011) but reappears at G2 phase (Figure 1D, CENP-A IP, WB). A reciprocal HJURP IP likewise demonstrates that CENP-A copurifies with HJURP during G1, is depleted at S, but returns during G2/M phase (Figure S2D, HJURP IP, CENP-A WB).
To test this observation further, we performed HJURP immunofluorescence on centromeric chromatin fibers (Sullivan and Karpen, 2004). These data show that HJURP localizes onto centromeric fibers at G1 and G1/S, is absent at S phase, but returns at G2 phase (Figures 1E and S2E). Thus, HJURP association to CENP-A chromatin is cell-cycle-dependent (Figures 1D, 1E, S2D, and S2E).
CENP-A Nucleosomes Cycle between Tetramers and Octamers In Vivo
HJURP is known to be a CENP-A deposition chaperone (Hu et al., 2011, Foltz et al., 2009, Dunleavy et al., 2009, Shuaib et al., 2010). The loss of HJURP binding from CENP-A chromatin at the G1 into S transition might signal completion of assembly, heralding a structural shift in CENP-A nucleosomes. To test this idea, we turned to single-molecule analysis of native CENP-A chromatin. AFM is a powerful single-molecule imaging method (Zlatanova and van Holde, 2006) that has been used extensively to investigate chromatin dynamics (Shukla et al., 2010; Torigoe et al., 2011, Bintu et al., 2011). Using AFM, we have previously shown that native CENP-A nucleosomes contain H2A, H2B, and H4, but not H3, and are tetrameric rather than octameric in asynchronous fly and human cells (Dalal et al., 2007; Dimitriadis et al., 2010). To investigate whether CENP-A nucleosomes alter their structure over the cell cycle, we used AFM to measure dimensions of native CENP-A chromatin obtained from experiments described above (Figures 1A, 1B, and 1D).
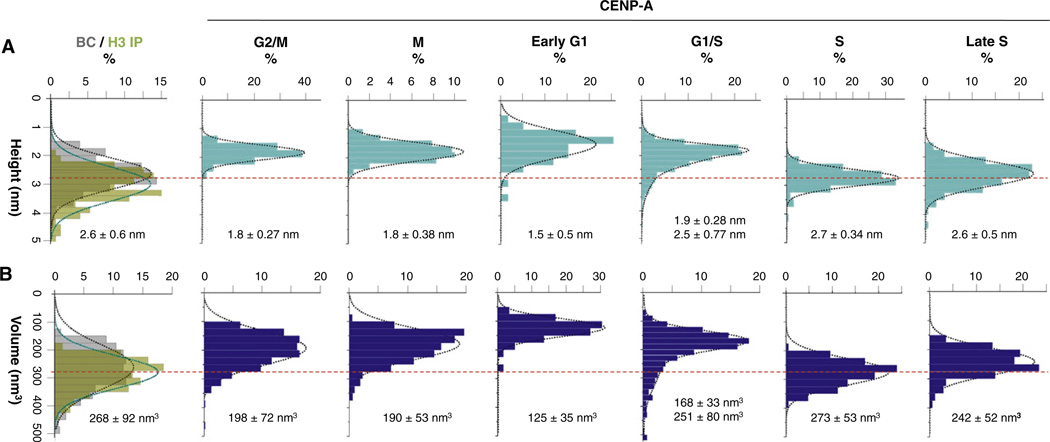
Consistent with previously published results (Dimitriadis et al., 2010), BC nucleosomes are exclusively octameric, averaging 2.6 nm in height and 268 nm3 in volume (Figures 2A, 2B, and S3A and Table 1). To ensure that IP or AFM conditions do not alter nucleosomal features, we immunopurified control H3 nucleosomes from G1/S and G2 phases of the cell cycle. Control H3 IP nucleosomes remain within the octameric range (Figures 2A and 2B overlay of BC and H3-IP, Table 1, and Figures S3B and S3C). In contrast, native CENP-A nucleosomes similarly purified present remarkably variable structures over the cell cycle. At early G1, the point at which new CENP-A assembles at centromeres (Hemmerich et al., 2008; Jansen et al., 2007; Schuh et al., 2007), CENP-A nucleosomes are tetrameric, averaging 1.5 nm high and 125 nm3 in volume (Figures 2A, 2B, and S3 and Table 1). At the G1/S transition, CENP-A nucleosomes have a broader distribution, with the majority averaging 1.9 nm high and 168 nm3 in volume, and an octameric subfraction, averaging 2.5 nm high and 251 nm3 in volume (Figures 2A, 2B, and S3 and Table 1). As cells enter S phase, the overwhelming majority of CENP-A nucleosomes are indistinguishable from H3 octameric nucleosomes, averaging 2.7 nm high and 273 nm3 in volume (Figures 2A, 2B,and S3 and Table 1). In late S phase, CENP-A nucleosomes persist as octamers, averaging 2.6 nm high and 242 nm3 in volume (Figures 2A, 2B, and S3 and Table 1). In contrast, postreplicative CENP-A nucleosomes from G2/M phase also form nucleosomal arrays (Figure S3) but are consistent with tetrameric dimensions, averaging 1.8 nm high and 198 nm3 in volume (Figures 2A and 2B and Table 1). CENP-A nucleosomes from early mitotic cells (Experimental Procedures) remain tetrameric, averaging 1.8 nm high and 190 nm3 in volume (Figures 2A, 2B, and S3 and Table 1). Thus, CENP-A nucleosomes cycle between tetrameric and octameric configurations in vivo.
Figure 2. CENP-A Nucleosomes Alter from Tetramers to Octamers at S Phase.
Single-molecule AFM measurements depicting (A) heights and (B) volumes of purified H3 and bulk chromatin (BC) nucleosomes show they have octameric dimensions. In contrast CENP-A nucleosomes are tetramers at G2/M, M, early G1, and G1/S but are octamers in early and late S phases. Red dotted line indicates octameric particle values. BC values are in gray, H3 values are in green, and CENP-A values are in blue. Average values for heights and volumes are indicated under each graph. Table 1 gives a detailed overview of the data. See also Figures S3, S4A, and S4B.
Table 1.
AFM Measurements of CENP-A Nucleosomal Dimensions Indicate a Change in Structure over the Cell Cycle
| Cell-Cycle Phase | Nucleosome | N | Heights (nm) | Volume (nm3) | Diameter (nm) | N | Inferred Organization |
|---|---|---|---|---|---|---|---|
| G2/M | CENP-A | 479 | 1.8 ± 0.27 | 198 ± 72 | 14.6 ± 2.20 | 584 | tetramer |
| M | CENP-A | 1177 | 1.8 ± 0.38 | 190 ± 53 | 14.2 ± 2.03 | 498 | tetramer |
| Early G1 | CENP-A | 169 | 1.5 ± 0.5 | 125 ± 35 | 13.7 ± 1.93 | 169 | tetramer |
| G1/S | CENP-A | 1372 | 1.9 ± 0.28 2.5 ± 0.77 |
168 ± 33 251 ± 80 |
12.7 ± 0.76 | 288 | transition to octamer |
| Early S | CENP-A | 188 | 2.7 ± 0.34 | 273 ± 53 | 13.3 ± 1.86 | 246 | octamer |
| Late S | CENP-A | 177 | 2.6 ± 0.5 | 242 ± 52 | 13.2 ± 1.36 | 124 | octamer |
| G1/S | BC | 281 | 2.9 ± 0.5 | 345 ± 56 | 14.4 ± 1.78 | 394 | octamer |
| G2/M | BC | 419 | 2.6 ± 0.6 | 268 ± 92 | 13.6 ± 1.25 | 497 | octamer |
| G1/S | H3 | 98 | 3.2 ± 0.4 | 300 ± 46 | 13.1 ± 1.32 | 98 | octamer |
| G2 | H3 | 126 | 3.3 ± 0.16 | 302 ± 65 | 12.8 ± 1.0 | 126 | octamer |
Bolded numbers denote values within the predicted range for canonical octameric nucleosomes, values rounded up to 1 decimal point. N = number of nucleosomes measured. BC = bulk chromatin; H3 = H3 IP; CENP-A = CENP-A IP.
Octameric volumes for S phase CENP-A nucleosomes could result from an increase not just in height but also in diameter, perhaps resulting from nonhistone binding. A priori, this possibility seems less likely because histone gels depict an equivalence of CENP-A, H2A, H2B, and H4, but no significant enrichment of other small proteins that might contribute to octameric dimensions at S phase (Figure 1B). Nevertheless, we also examined diameters of CENP-A nucleosomes. These data show that CENP-A nucleosomal diameters are similar over the cell cycle, and consistent with the observed lateral diameter of H3 nucleosomes (Figure S3). Therefore, volume increase in CENP-A nucleosomes from late G1 through late S reflects doubling of nucleosomal heights, supporting the interpretation that they form octamers.
To confirm that CENP-A nucleosomes transition to octamers at S phase and back to tetramers at G2/M, we examined their DNA content. G1/S, S, and G2/M CENP-A short chromatin arrays were treated with proteinase K to remove histones, and contour lengths of nucleosomal DNA released were measured by AFM (Experimental Procedures). At G1/S, CENP-A mononucleosomal DNA ranges from 100–150 bp (Figures S4A and S4B). In contrast, by S phase, CENP-A nucleosomes yield multiples of ~150–200 bp of DNA, consistent with classical octameric organization. By G2/M, CENP-A nucleosomal DNA decreases to ~115 bp in length, consistent with previous measurements of the CENP-A mononucleosomal footprint in vivo (Ando et al., 2002), and with diminutive heights and volumes observed in our analyses above (Figures 2A and 2B).
We sought an alternative approach to confirm that CENP-A:CENP-A intranucleosomal interactions are increased at S phase. In order to perform this experiment, we used a stable cell line expressing GFP-CENP-A alongside native (untagged) CENP-A (Dimitriadis et al., 2010) and collected cells at G1/S and S phase as before. Mononucleosomal input chromatin was extracted from these G1/S and S phase cells under conditions used previously (Figure 1C) and an anti-GFP antibody used to perform IP. Previous data have shown that EGFP-CENP-A in these cells is associated with the core histones H2A, H2B, and H4 (Dimitriadis et al., 2010). Here, the EGFP-tagged nucleosomes were analyzed for EGFP-CENP-A relative to native CENP-A content by using an anti-CENP-A antibody, followed by quantitative laser scanning. As can be seen, very little native CENP-A is present in the G1/S phase EGFP-CENP-A IP. In contrast, native CENP-A is enhanced 7-fold in the S phase EGFP-CENP-A IP (Figure S4C, CENP-A WB). These data support the interpretation that CENP-A:CENP-A intranucleosomal interactions are increased at S phase and are consistent with octameric dimensions uncovered by AFM experiments.
Thus, AFM analyses, DNA measurements, and co-IP analyses (Figure 2A, 2B, S4A–S4C, and Table 1) indicate that, whereas canonical H3 nucleosomes have octameric dimensions throughout the cell cycle, CENP-A nucleosomes are predominantly tetramers in early G1 phase, alter to octamers at the end of G1 through S phase, and revert to tetramers after replication.
The CENP-A Chromatin Fiber Is Highly Dynamic In Vivo
We were intrigued by the possibility that changes in CENP-A structure and components observed above might be reflected in chromatin fiber folding. To test whether chromatin fiber folding changes in vivo, we measured interactions between CENP-A molecules in living cells by using FRET. FRET occurs when two fluorescently tagged proteins are within 10 nm of each other in vivo, which is the average distance between nucleosomes in arrays, and thus has been used to study chromatin fiber interactions (Hellwig et al., 2008, 2011; Hemmerich et al., 2008). EGFP-CENP-A localizes accurately and can partially rescue CENP-A depletion within centromeres (Kalitsis et al., 2003). It associates with H2A, H2B, H4, and DNA in vivo (Dimitriadis et al., 2010), is able to interact with CENP-C (Trazzi et al., 2009), and has been widely used as a surrogate for tracking native CENP-A behavior (Wan et al., 2009).
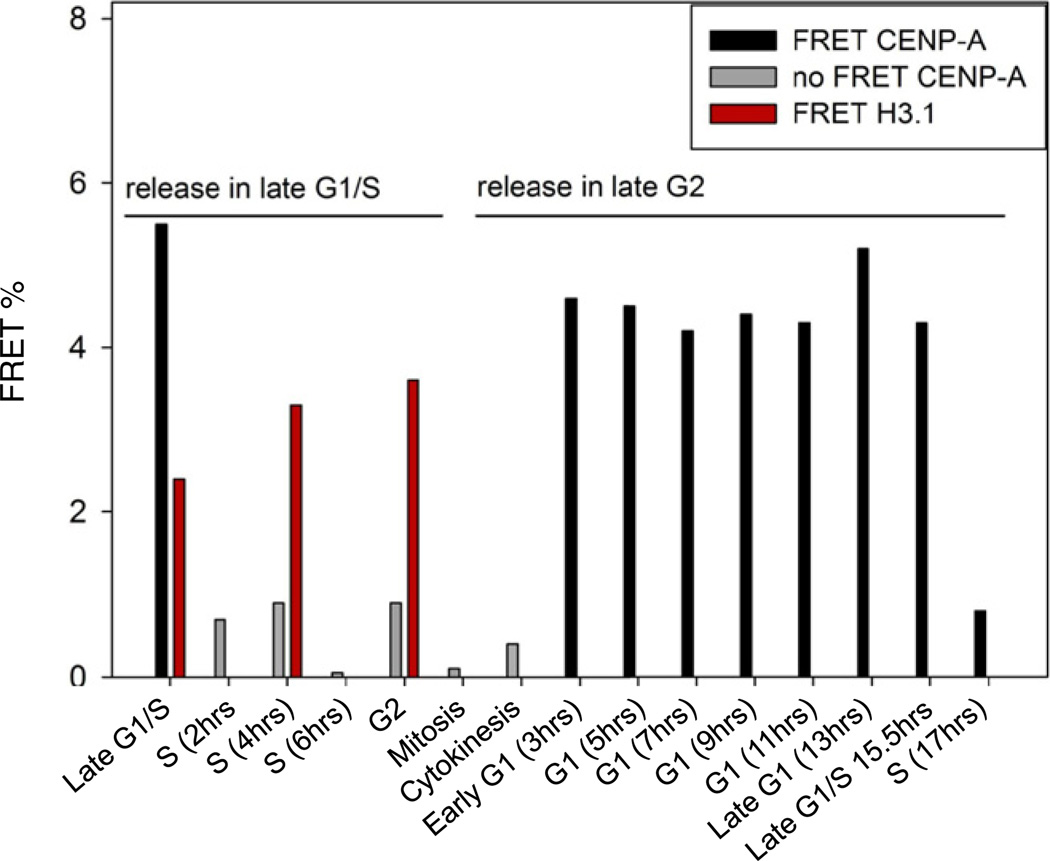
We assessed interactions between centromere-localized C-terminal and N-terminal EGFP- and mCherry- CENP-A pairs by using synchronized cell populations (Figures S1A and S5A). Equivalently tagged control H3.1 pairs show significant FRET between each other at all points of the cell cycle, including at mitosis, consistent with a globally compacted chromatin fiber (Figures 3, S5B and Table S1). In contrast, C terminally tagged CENP-A pairs yield remarkably different results. In early G1, we observed significant CENP-A-EGFP:CENP-A-mCherry FRET (p < 0.001; Figure 3 and Table S1). The FRET signal remains high over G1 and at G1/S (Figure 3 and Table S2). However, 2 hr into S phase, the FRET signal drops to background levels (Figure 3) and remains insignificant throughout the subsequent G2 and mitosis phases. Analysis of N terminally tagged CENP-As similarly yields high FRET during G1 and G1/S but no appreciable FRET during S, G2, and M phases (Figure S5C and Table S1).
Figure 3. FRET Measurements Reveals the CENP-A Chromatin Fiber Undergoes a Transition at G1/S Phase.
FRET measurements between CENP-A C termini and H3 C termini across the cell cycle. H3 FRET is constant across the cell cycle (red), whereas CENP-A FRET shows high FRET in G1 (Black bars) but no FRET from S through M (mitosis) or C (Cytokinesis) phase (Grey bars). Black or red bars, significant FRET (p < 0.001); gray bars, no FRET. See also Table S1, S2, and Figure S5.
FRET detection between CENP-As in G1 is consistent with CENP-A assembly, which peaks at G1 (Hemmerich et al., 2008; Jansen et al., 2007), increasing CENP-A occupancy on the centromeric chromatin fiber and possibly resulting in a highly compacted fiber (Marshall et al., 2008). Conversely, loss of FRET between N and C terminally tagged CENP-A pairs, from mid S to the next G1 is indicative of lack of physical proximity between CENP-A termini, possibly resulting from greater spatial distances between CENP-A nucleosomes, a more extended chromatin fiber (Dalal et al., 2007), or from inner kinetochore proteins binding to CENP-A termini (Hellwig et al., 2011; Carroll et al., 2010; Trazzi et al., 2009), thus preventing higher-order compaction.
In order to confirm our interpretation that the FRET data reflect chromatin fiber folding, we analyzed FRET before and after disrupting the chromatin fiber with mild nuclease treatment. We first confirmed FRET occurs between C terminally tagged CENP-A in untreated G1 phase nuclei (Figure S5D and Table S2), which recapitulated our in vivo results above (Figure 3). In contrast, upon treating G1 phase nuclei with mild nuclease (MNase), thus disrupting higher-order chromatin fiber folding in situ, FRET between C terminally tagged CENP-A in late G1 disappears (Figure S5D and Table S2). From these data, we infer that CENP-A FRET reflects interactions between CENP-A nucleosomes within a densely packed chromatin fiber during G1. Conversely, the loss of FRET between CENP-As in S phase suggests that the centromeric chromatin fiber is altered as cells transition from G1 into S phase.
We were curious to test whether disappearance of FRET between CENP-A arrays in early S phase could be due to disruption by of CENP-A chromatin fiber by DNA polymerases. We synchronized and released cells into G1/S in the presence of aphidocolin, an inhibitor of DNA polymerases α and δ, thus blocking replication. Because the cells are stalled at G1/S, we observe continued CENP-A/CENP-A FRET for 4 hr (Figure S5E). In contrast, after synchronizing and releasing cells at G1/S, but inhibiting DNA polymerase only one hour after replication has initiated, FRET between CENP-As is lost (Figure S5E, Table S2). This behavior is similar to that of wild-type cells, which lose FRET by mid S phase (Figure 3). Because centromeres are late replicating in human cells (Shelby et al., 2000), 1 hr of DNA polymerase activity is insufficient for the replication machinery to run through human centromeres. Thus, the loss of CENP-A FRET signal by mid S phase likely derives from a less folded centromeric chromatin fiber in advance of replication.
The Histone Fold Domains of CENP-A and H4 Have Modifications at G1/S
AFM and FRET experiments indicate that CENP-A structural and dynamic transitions peak during G1/S (Figures 2A, 2B, and 3), concurrent with the loss of HJURP binding in S phase (Figures 1C and 1D). We were curious to know whether covalent modifications within CENP-A/H4 could contribute to the changes noted above. We purified DNA-bound CENP-A/H4 complexes from G1/S, S, and G2/M cells (Figure S1C), excised them from gels (Figure S6A), and subjected them to LC-MS/MS (Experimental Procedures).
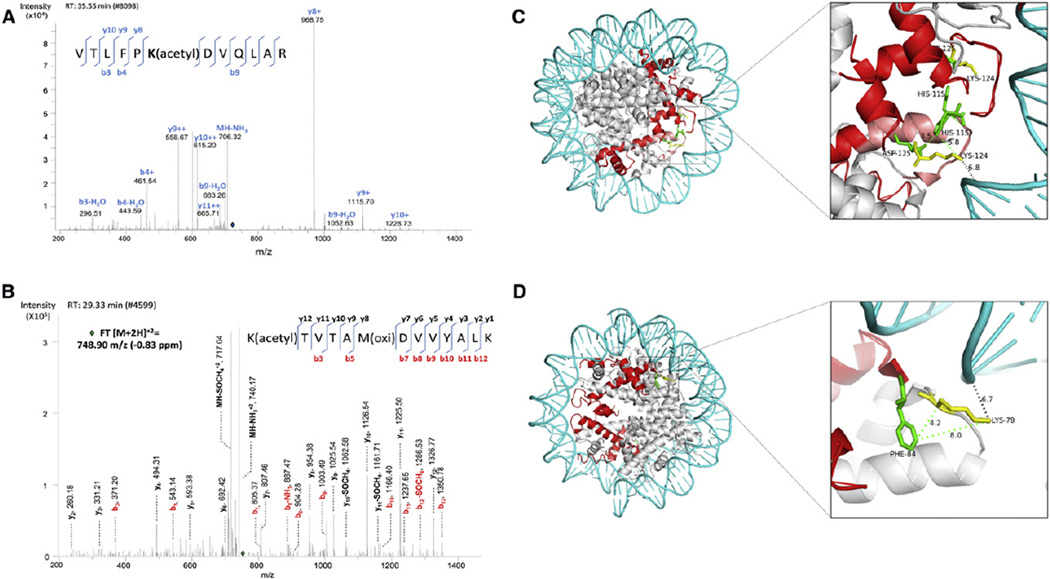
Six unique peptides from CENP-A were identified (Experimental Procedures). Remarkably, in G1/S phase-derived CENP-A, the mass spectra revealed enrichment of an ion that corresponds to a doubly charged tryptic peptide containing acetylated Lysine 124 (m/z 714.90, Figures 4A and S6B). Peptide fragmentation in MS/MS mode and manual validation allowed unequivocal confirmation of the CENP-A peptide as “119VTLFPK(acetyl)DVQLAR130” (Figures 4A and S6B). Similarly, LC-MS/MS analysis of CENP-A-bound H4 from G1/S phase cells also revealed an acetylation site on H4’s Lysine 79 residue in the tryptic peptide “79K(acetyl)TVTAMDVVYALK91,” showing the sequence of fragment ions that confirm the presence of the acetyl group on K79 (m/z 748.90, Figures 4B and S6C).
Figure 4. CENP-A Lys124 and H4 Lys79 Are Acetylated at G1/S Phase.
(A) MS/MS spectrum showing CENP-A K124 is acetylated in the peptide “VTLFPK(acetyl)DVQLAR”. Location of the parent peptide ion prior to fragmentation is indicated with a blue diamond. Peptide fragmentation ions identified are indicated in the spectra and on the peptide sequence. The masses of ions b9, y8, y9, y10 are diagnostic of K124 acetylation. See also Figure S6B.
(B) MS/MS spectrum showing fragmentation of H4 “K(acetyl)TVTAM(oxi)DVVYALK” as a doubly charged, monoisotopic ion, m/z 748.90 (−0.83 ppm difference from the theoretical m/z). Location of the parent ions prior to fragmentation is indicated with a green diamond. Peptide fragmentation ions identified are indicated in the spectra and on the peptide sequence. All fragment ions of the b series have masses diagnostic of K79 acetylation. See also Figure S6C.
(C) Model of the CENP-A octamer showing two CENP-As (red), DNA (blue), core histones (gray), CENP-A K124 (yellow), and CENP-A H115, R118, and D125 (green). Dotted lines represent distances (Å) between residues.
(D) Distance (Å) between H4K79 and DNA and H4K79 and CENP-A F84. Models were generated in PyMol by using published crystallographic coordinates (Tachiwana et al., 2011). See also Figure S6.
Whereas histone fold domain (HFD) modifications in histone H3 have been identified that impact the pseudo-dyad of the canonical octamer in vitro (Neumann et al., 2009; Simon et al., 2011), there have been no such reports for histone variants such as CENP-A. The structural consequence of CENP-A K124 and H4 K79 acetylation, buried in the interface of the HFD and DNA in octamers, is likely to be significant (Figures 4C and 4D). CENP-A K124 is located within the α3-helix domain and is within 7Å of the DNA double helix at the pseudo-dyad of the octamer (Figure 4C). Adding an acetyl group to K124 could have the structural consequence of neutralizing the positively charged lysine surface, which might loosen histone-DNA contacts. Similarly, H4 K79 is located within the Loop 2 domain juxtaposed near the DNA double helix (Figure 4D), and an acetylation on this residue could loosen the DNA-histone interface, thereby increasing accessibility of the CENP-A nucleosomal interior to nonhistone proteins or to chromatin remodelers.
DISCUSSION
Our data indicate that heterotypic CENP-A nucleosomes (Figures 1A–1C) undergo significant changes in binding of the chaperone HJURP (Figures 1D, 1E and S2), nucleosomal structure (Figures 2A, 2B, S3, and S4), chromatin fiber folding (Figures 3 and S5), and covalent modifications (Figures 4A, 4B, S6B, and S6C), during the transition from G1 into S phase (summarized in Figure S7). These findings are significant because they reconcile previous contradictory observations of CENP-A octamers and tetramers in diverse organisms, which likely reflect cycling of the CENP-A nucleosomes. Although we did not directly observe CENP-A hexameric or homotypic tetramer intermediates during the transition from G1 into S phase, it is plausible that such intermediates exist during interconversion between heterotypic tetramers and octamers.
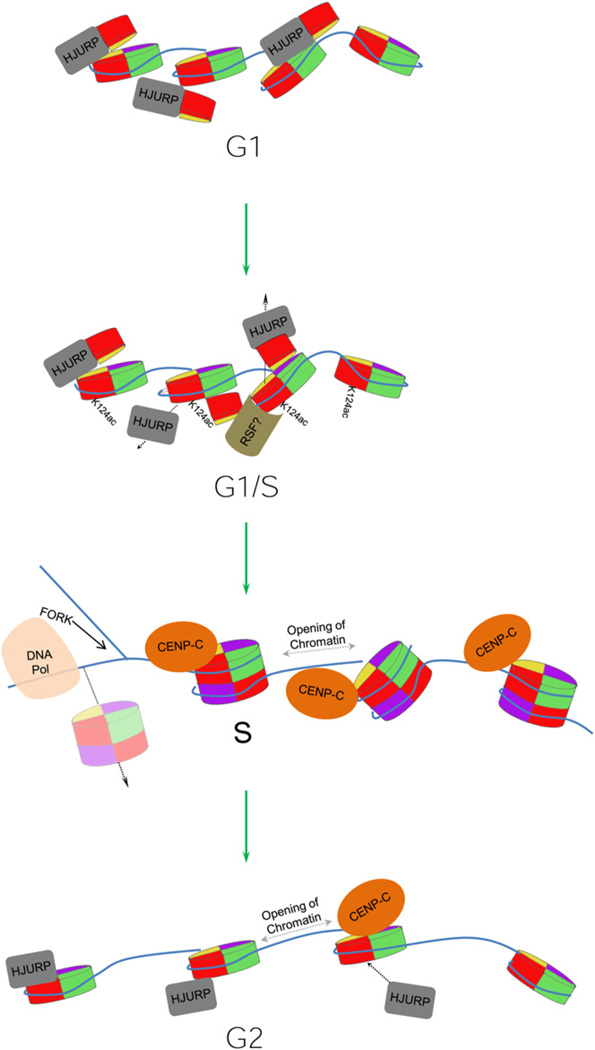
Our findings implicate novel mechanisms in the assembly-disassembly kinetics of the centromeric fiber. One speculative mechanism whereby CENP-A nucleosomes could be converted from one form to another in vivo is through the action of chromatin remodelers (Perpelescu et al., 2009; Torigoe et al., 2011). The chromatin remodeler RSF, which binds to centromeres at late G1 and is required for CENP-A inheritance (Perpelescu et al., 2009), could initiate reorganization of the CENP-A chromatin fiber and recruit histone acetyltransferases that catalyze acetylation of CENP-A and H4, delaying the formation of stable octamers (Figure 5). At the end of G1, loss of such modifications, accompanied by chromatin remodeling, could influence the release of the chaperone HJURP, thus resulting in the stabilization of the four helix bundle in the CENP-A octamer (Hu et al., 2011). A plausible alternative scenario is that changes in intrinsic stability (Conde e Silva et al., 2007), or processes such as transcription (Bintu et al., 2011), may alter CENP-A nucleosomal structure directly at various points of the cell cycle. CENP-A mononucleosomes have been demonstrated to be inherently unstable when subjected to unwinding stress in vitro (Dechassa et al., 2011), and, ectopically expressed CENP-A can be detected in the dimeric form in fly cells, suggesting that unstable CENP-A octameric nucleosomes might also exist in vivo (Zhang et al., 2012). Such unstable octameric CENP-A nucleosomes could disassemble during passage of DNA replication machinery. Consequently, our observation of the return of the chaperone HJURP concurrent with CENP-A tetrameric nucleosomes at G2 phase might reflect HJURP-dependent recycling of old CENP-A into tetrameric nucleosomal intermediates, after late replication of centromeric DNA in human cells (Dunleavy et al., 2011; Shelby et al., 2000). This state would persist until a burst of new CENP-A protein at the next G1 promotes completion of octamers prior to replication (Figure 5). New avenues that arise from this work include elucidating how HJURP is released and rebound over the cell cycle, identifying histone modifiers and chromatin remodelers that target CENP-A, and the mechanism by which CENP-A chromatin is reorganized throughout the cell cycle.
Figure 5. Cyclical Oscillations of HJURP and CENP-A Drive Centromere Dynamics.
Model depicting how cyclical oscillations in CENP-A nucleosomal structure could derive from changes in HJURP chaperone binding mediated by chromatin remodeling, histone modifications, and cell-cycle events such as replication.
In the absence of new CENP-A assembly during replication, random segregation of old CENP-A would be expected to result in unequal centromere domains at sister centromeres, unequal attachment to the mitotic spindle, and subsequently aneuploidy. It is plausible that an evolutionary conserved mechanism exists to ensure distribution of CENP-A nucleosomes after centromeric DNA replication. Thus, it is likely that cycling of CENP-A nucleosomal structure occurs in other organisms and that such cycling will track closely with assembly by CENP-A chaperones. Our observations that histone modifications, chromatin fiber folding, and chaperone binding changes accompany structural transitions in the centromere provide insight into a fundamental problem arising from replication-independent assembly of histone variants. These findings reveal a cyclical nature for CENP-A nucleosomes that is likely to play a critical role in centromere inheritance.
EXPERIMENTAL PROCEDURES
Chromatin Immunoprecipitation
Human cell lines HeLa and HEK293 were synchronized at various stages of the cell cycle by using a double-thymidine block (see Cell-Cycle Synchronization), harvested, washed twice with PBS + 0.1% Tween, and nuclei were extracted with TM2 Buffer (20 mM Tris [pH 8.0], 2 mM MgCl2) supplemented with 0.5% Nonidet P40 Substitute (Sigma Cat number 74385). For mitotic cells, early prophase cells were used which retain nuclear membranes, along with an antibody specific to phosphorylation of CENP-A Ser 7. To release chromatin, MNase (Sigma Cat number N3755) digestion was performed at 0.2–0.4 units/ml for 30 s, 2.5 min, and 4 min at 37°C to generate long, moderate, and short chromatin arrays, and the reaction stopped with 10 mM EGTA. An aliquot of nuclei was used for extracting and analyzing DNA to confirm bulk chromatin array length. Remaining nuclei were extracted overnight at 4°C in 10× volume of low-salt buffer (0.5× PBS, 5 mM EGTA, 0.5 mM PMSF). Chromatin IP was performed by using Dynabeads Protein G (Invitrogen catalog number 100-07D) or prehydrated sepharose-protein G, with primary antibodies as listed below. Three biological replicates were performed for each experiment.
Antibodies Used for ChIP and Western Blot
CENP-A, rabbit CENP-A (Santa Cruz catalog number sc-22787) and rabbit CENP-A (Millipore catalog number 07-574); Mitotic CENP-A, rabbit phospho-serine 7 CENP-A (Millipore catalog number 07-232) (early prophase cells still have nuclear membranes); HJURP, goat HJURP (Santa Cruz catalog number sc-168091); CENP-C, goat CENP-C (Santa Cruz catalog number sc-11285); and CENP-N, goat CENP-N (Santa Cruz catalog number sc-69152).
Chromatin Fiber Analysis by Immunofluorescence
PBS-washed HeLa cells were counted and diluted to 300,000 cells/ml in hypotonic buffer (75 mM KCl, 1 × PBS). After 10 min of incubation at room temperature, 200 µL of cells were cytospunned for 10 min at 4,000 rpm, lysed for 15 min in lysis buffer (2.5 mM Tris HCl [pH 7.5], 0.5 mM NaCl, 1% Triton X-100, 0.4 M urea), fixed for 10 min in fixation buffer (4% PFA [paraformaldehyde], 1× PBS), and permeabilized for 7 min in permeabilization buffer (0.1% Triton X-100, 1 × PBS). After blocking for 30 min in 1 × PBS supplemented with 0.5% BSA and 0.01% Triton X-100, extracted fibers were incubated overnight at 4°C with mouse anti-CENP-A (1:200, Abcam) or rabbit anti-HJURP (1:50, Santa Cruz) antibodies diluted in blocking buffer. After three washes with 1 × PBS supplemented with 0.05% Tween, slides were incubated for 3 hr at room temperature with Alexa Fluor 488 goat anti-rabbit IgG (1:300, Invitrogen) or Alexa Fluor 568 goat anti-mouse IgG (1:300, Invitrogen). DNA was counterstained with DAPI (50 ng/ml in 1 × PBS). Extracted fibers were observed with a DeltaVision RT system (Applied Precision) controlling an interline charge-coupled device camera (Coolsnap; Roper) mounted on an inverted microscope (IX-70; Olympus). Images were captured by using a 100× objective at 0.1 µm z sections, deconvolved, and projected by using softWoRx. Two biological replicates and three technical replicates each were performed.
AFM Imaging of Chromatin and DNA
Imaging of CENP-A, bulk chromatin and DNA was performed as described (Dimitriadis et al., 2010) with the following modifications. Imaging was performed by using standard AFM equipment (Multimode AFM and the Bioscope Catalyst, Bruker-Nano, Inc., Santa Barbara, CA) with silicon cantilevers (OTESPA and TESP-SS with nominal resonances of ~300 kHz, stiffness of ~42 N/m, and tip radii of 3–7 nm and FESP with ~75 kHz, 2.8 N/m and 7 nm, respectively, Bruker-Nano) in noncontact tapping mode. Usually, 5 µl stock solution of CENP-A chromatin or 1,000× diluted solution of bulk chromatin fraction was deposited on APS-mica pretreated with magnesium2+. APS-mica was prepared as previously described (Dimitriadis et al., 2010). The samples were incubated for 10 min, rinsed gently to remove salts, and dried in a stream of inert Argon before imaging. Images were acquired at high resolution and preprocessed on the NanoScope instrument software.
AFM Image Analysis
Automated image analysis was performed as described (Dimitriadis et al., 2010) by using NIH ImageJ software (NIH) and Nanoscope Software (Veeco/Bruker AFM). An algorithm was developed to first localize the nucleosomes under investigation and then perform automated statistical analysis of their height and volume distributions. To achieve this, images were thresh-holded to remove probe convolution that causes objects to appear dilated in AFM imaging. Using particle analysis routines, the base area (at half-height) and total height of all nucleosomes in each image were automatically measured. Filters were employed to reject particles that were not circular or elliptical in shape, thereby ensuring that measurements were made of nucleosomes attaching to the mica in the same orientation, and therefore projecting a uniform shape. For the particles segmented this way, the radii of the equivalent circles were calculated and their statistical distributions plotted and fitted with Gaussian functions. The histograms always displayed a peak of ~12–14 nm, slightly larger diameter than the known nucleosomal radius of ~11 nm, which represents residual dilation from the finite AFM tip size of 2–3 nm. Sub-population within one standard deviation of the peak of the Gaussian was chosen for further statistics of heights and volumes. Volumes were computed as the sum of pixel values within the segmented base of each particle. In addition to arrays ranging from 5–15 CENP-A nucleosomes, various large protein complexes (diameter > 25 nm, height > 10 nm) were observed associated with CENP-A tetrameric nucleosomes. These likely reflect kinetochore proteins, remodeling complexes or polymerases, and are the subject of ongoing analysis. Three biological replicates and 2 technical replicates were performed for each experiment. BC from the same preparation was imaged in parallel to get the baseline octameric range.
Contour Length Measurement of CENP-A Nucleosomal DNA by AFM
Aliquots of CENP-A and control BC nucleosomes were treated with proteinase K (0.2 mg/ml, Sigma) for 30 min, equilibrated with 1% SDS in 100 mM Tris pH 8.0, phenol:chloroform extracted (Sigma), ethanol precipitated and dissolved in 1 × PBS for imaging by AFM as above (see AFM imaging of chromatin). Images were collected and exported to Image J (NIH). DNA populations were thresholded at roughly half-height. The perimeter and area of each DNA chain were automatically measured by using particle analysis. Mean width and length were computed in OriginPro (OriginLab Corp., Nothampton, MA). For each molecule we measure both its projected area and its perimeter. These two quantities uniquely determine both the width and the length of the molecule by solving the two equations A = L*w+π*w2, p = 2*L+2*π*w for the length (L) and the width (w) of the molecule, where A is the projected area and P the perimeter. The second terms in the right hand sides of the two equations apply a small correction to the length at the two ends of the DNA chains for the AFM tip convolution. Particles were filtered to exclude contaminants by using low circularity criteria to restrict measurements to linear particles.
Cell-Cycle Synchronization of Hep-2 and HeLa Cells
For cell-cycle-dependent analysis in late G1, S phase, G2, and mitosis, cells were arrested by double-thymidine treatment in late G1, at the initiation of early S phase. Cells were blocked with a final concentration of 5 mM thymidine for 18 hr. Cells were released and grown in fresh media for 9 hr, followed by a second cell-cycle block with 5 mM thymidine for 18 hr. Synchronized cells were released into S phase by washing and cultivation in fresh media. For FRET analysis cells were harvested and fixed with 4% paraformaldehyde in PBS immediately before release and for 8 – 10 hr every 2 hr. Most cells were in late S phase 6 hr after release and were in G2 or mitosis 8 hr after release. For analysis of HEp-2 cells in G1 transfected cells were synchronized by a thymidine block followed by a release for six hours and a subsequent 12 hr treatment with the CDK1 inhibitor RO-3306 (Calbiochem) at a final concentration of 9 µM. RO-3306 reversibly arrests the cells in late G2 at the initiation of mitosis (Vassilev et al., 2006). Synchronized cells were washed and then released into mitosis in fresh media, to enter in G1, 1.5 – 2 hr after release. To identify the cell-cycle stages, cells in parallel experiments were transfected with EGFP-CENP-A and mCherry-PCNA and analyzed for the localization pattern of CENP-F and PCNA at each time point. G1 and G2 cells were identified by immunostaining of CENP-F. This protein is diffusely located in the nucleus during G2, forms distinct foci at the kinetochores during mitosis and is not detectable during G1 (Figure S6). Early, mid, and late S phase were identified via the localization pattern of PCNA (Figure S6). Mitotic phases were identified by DAPI staining of DNA, which was stimulated with 405 Diode laser at low intensity and detected via a 420 nm long path filter.
Imaging and Acceptor Bleaching FRET Measurements and Plasmids for FRET Study
Please refer to Supplemental Experimental Procedures for FRET analysis.
Large-Scale Purification of Endogenous CENP-A for LC-MS/MS
For each experiment, 10 large flasks of confluent HeLa and HEK cell lines were synchronized and released at G1/S, mid S, and G2/M phase, by performing double 5 mM thymidine blocks. Human CENP-A proteins were purified similarly to (Wang et al., 2008), with the following modifications. Cells were lysed and nuclear slurries prepared with 10% hydroxylapatite (HAP, Acros Organics Catalog number 37126-1000) in 1 × PBS supplemented with 0.35 M NaCl, 0.2 mM EDTA, 0.5 mM PMSF for several hours. The same buffer was used to gently remove nonhistone proteins away from the HAP-Chromatin matrix, 3 times for 15 min each. The HAP-chromatin matrix was then incubated with 1 × PBS supplemented with 2 M NaCl to release histones from the DNA-bound HAP matrix overnight. Under these conditions, histone H3/H4 dimers and H2A/H2B dimers are released from DNA (Dalal et al., 2005). The 2 M NaCl/PBS eluted histone preparation was concentrated ~10 fold by using Amicon centrifugation, dialyzed down to 0.35 M NaCl/PBS. This input was tested was sequentially immunoprecipitated with anti-H3 antibody (Santa Cruz Catalog number sc-8654) to preclear excess H3, followed by anti-CENP-A antibody (Santa Cruz Catalog number sc-22787). Eluted CENP-A IP complexes were analyzed on preparative SDS PAGE gels, an aliquot of which was confirmed by western blot analysis. The remaining was used for gel excision and LC-MS/MS analysis.
Mass Spectrometry Analysis
The components of histone IPs were processed for preparative SDS-PAGE (Invitrogen Novex Midi Gel System), and stained with Coomassie Brilliant Blue (Bio-Rad), bands around 11k and 13k were sliced for histone H4 and CENPA respectively, and each band was in-gel digested with sequencing grade modified trypsin (Promega, Madison, WI) (An et al., 2006). The resulting peptides were analyzed by LC-MS/MS at two different settings.
Liquid Chromatography-Tandem Mass Spectrometry
The first set of samples (HeLa and HEK293 cells) was run at the NIAID proteomics core facility. LC-MS/MS was performed on nano-HPLC system (Proxeon EASY; ThermoElectron, Bremen, Germany) connected to a hybrid mass spectrometer (Velos/Orbitrap; ThermoElectron, Bremen, Germany). Each sample was injected via an auto-sampler and directly loaded on to packed analytical column at the flow rate of 1.2ul/min and the sample was subsequently separated at the flow rate of 400 nl/min. The mobile phases consisted of water with 0.1% formic acid (A) and 100% acetonitrile with 0.1% formic acid (B). A linear gradient from 5 to 65% of solvent B was employed over a period of 60 min to separate peptide mixture. Eluted peptides were introduced into the mass spectrometer through a nano-electrospray source (ThermoElectron, Bremen, Germany). The spray voltage was set at 1.7 kV and the heated capillary at 250°C. Orbitrap was operated in the data-dependent mode in which each cycle consisted of one full-MS survey (m/z, 380–2,000) and subsequently 10th MS/MS scans. The targeted ions count in the mass spectrometer trap was 500,000 for full-MS scans and 10,000 for MS/MS scans. Peptides were fragmented in the linear ion trap by using collision-induced dissociation with helium and the normalized collision energy value set at 35%.
The rest of the samples were analyzed at the LSB Cellular Networks Proteomics Group, and nano-HPLC system (NanoLC 2D; Eksigent, Dublin, CA) connected to a hybrid mass spectrometer (LTQ Velos with ETD; Thermo-Electron, Bremen, Germany). Each sample was injected via an auto-sampler and loaded onto a C8 cap trap (0.2 2 mm, 2 µl bed volumn; Michrom bioresources, Auburn, CA) and the sample was subsequently separated by in house packed column with Magic C18 AQ beads (200 A, 5 µm, 50 µm × 30 cm, Michrom Bioresources, Auburn, CA) at a flow rate of 200 nl/min. The mobile phases consisted of water with 0.1% formic acid (A) and 100% acetonitrile with 0.1% formic acid (B). A linear gradient from 5 to 65% of solvent B was employed over a period of 60 min to separate peptides. Eluted peptides were introduced into the mass spectrometer through a nano-electrospray source (ThermoElectron, Bremen, Germany). The spray voltage was set at 2 kV and the heated capillary at 250°C. LTQ was operated in the data-dependent mode in which each cycle consisted of one full-MS survey and subsequently 10th order double play with CID and ETD decision tree. The targeted ions count was same as described previously. MS measurements were performed with the Orbitrap mass spectrometer at 60,000 resolution with accuracy better than 10 ppm. Peptides were fragmented in the linear ion trap by using CID set at 35% and electron transfer dissociation with reaction time 150 ms by using enabled supplemental activation.
Six different peptides (CENP-A amino acid residues: 17–28, 30–42, 57–63, 100–107, 119–124, and 119–130) of CENP-A were identified in this study by using trypsin (cleaving C-terminal to lysine, K or arginine, R). Sequence coverage was 37.14% (52 identified amino acids/140 total amino acids × 100).
Database Search
Protein identification was performed against the human UniProt database and database including CENPA, histone H4, and ubiquitin by using a software packages (Proteome Discoverer 1.2 equipped with the Sequest search engine; Thermo Fisher Scientific, San Jose, CA; and IP2 equipped with the Sequest and ProLuCID; Integrated Proteomics Application, San Diego, CA). For Proteome Discoverer, both databases were indexed with assumptions for fully enzymatic tryptic cleavage with two missed cleavages and the combination of following possible protein modifications: Met oxidation, Ser, Thr, Tyr phosphorylation, Lys acetylation, Lys ubiquitination, and Arg and Lys mono-, di-, and trymethylation.
The search result from Proteome Discoverer was filtered with high peptide confidence value and false discovery rate targeting 0.01. For IP2, MS spectra were extracted from raw data by using Raw Xtractor and the database containing CENPA, histone4, and ubiquitin was used for the search. Previously described PTMs were used and allow a maximum of three on each peptide. The search result from IP2 was filtered with DTASelect 2.0 (Tabb et al., 2002). The peptides passing the criteria were manually validated.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Shiv Grewal, Tom Misteli, Gordon Hager, Sam John, and anonymous reviewers for insightful comments and suggestions, and P. Donlin-Asp for preliminary FRET experiments. The NIH/NCI Intramural Research Programs (Y.D., A.N.-L., and E.K.D.), and the Deutsche Forschungsgemeinschaft (S.D.) supported this work. We thank Jennifer Gerton for sharing unpublished work complementary to our findings in this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, seven figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2012.05.035.
REFERENCES
- Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat. Rev. Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An E, Lu X, Flippin J, Devaney JM, Halligan B, Hoffman EP, Strunnikova N, Csaky K, Hathout Y. Secreted proteome profiling in human RPE cell cultures derived from donors with age related macular degeneration and age matched healthy donors. J. Proteome Res. 2006;5:2599–2610. doi: 10.1021/pr060121j. [DOI] [PubMed] [Google Scholar]
- Ando S, Yang H, Nozaki N, Okazaki T, Yoda K. CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol. Cell. Biol. 2002;22:2229–2241. doi: 10.1128/MCB.22.7.2229-2241.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu L, Kopaczynska M, Hodges C, Lubkowska L, Kashlev M, Bustamante C. The elongation rate of RNA polymerase determines the fate of transcribed nucleosomes. Nat. Struct. Mol. Biol. 2011;18:1394–1399. doi: 10.1038/nsmb.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- Bloom K, Joglekar A. Towards building a chromosome segregation machine. Nature. 2010;463:446–456. doi: 10.1038/nature08912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R, Shivaraju M, Mattingly M, Li B, Nakanishi S, Zhu D, Shilatifard A, Workman JL, Gerton JL. Cse4 is part of an octameric nucleosome in budding yeast. Mol. Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde e Silva N, Black BE, Sivolob A, Filipski J, Cleveland DW, Prunell A. CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J. Mol. Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- Dalal Y, Bui M. Down the rabbit hole of centromere assembly and dynamics. Curr. Opin. Cell Biol. 2010;22:392–402. doi: 10.1016/j.ceb.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal Y, Fleury TJ, Cioffi A, Stein A. Long-range oscillation in a periodic DNA sequence motif may influence nucleosome array formation. Nucleic Acids Res. 2005;33:934–945. doi: 10.1093/nar/gki224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechassa ML, Wyns K, Li M, Hall MA, Wang MD, Luger K. Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat Commun. 2011;2:313. doi: 10.1038/ncomms1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitriadis EK, Weber C, Gill RK, Diekmann S, Dalal Y. Tetrameric organization of vertebrate centromeric nucleosomes. Proc. Natl. Acad. Sci. USA. 2010;107:20317–20322. doi: 10.1073/pnas.1009563107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Dunleavy EM, Almouzni G, Karpen GH. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G1 phase. Nucleus. 2011;2:146–157. doi: 10.4161/nucl.2.2.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwig D, Emmerth S, Ulbricht T, Döring V, Hoischen C, Martin R, Samora CP, McAinsh AD, Carroll CW, Straight AF, et al. Dynamics of CENP-N kinetochore binding during the cell cycle. J. Cell Sci. 2011;124:3871–3883. doi: 10.1242/jcs.088625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwig D, Munch S, Orthaus S, Hoischen C, Hemmerich P, Diekmann S. Live-cell imaging reveals sustained centromere binding of CENP-T via CENP-A and CENP-B. J. Biophotonics. 2008;1:245–254. doi: 10.1002/jbio.200810014. [DOI] [PubMed] [Google Scholar]
- Hemmerich P, Weidtkamp-Peters S, Hoischen C, Schmiedeberg L, Erliandri I, Diekmann S. Dynamics of inner kinetochore assembly and maintenance in living cells. J. Cell Biol. 2008;180:1101–1114. doi: 10.1083/jcb.200710052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Furuyama T. Epigenetic inheritance of centromeres. Cold Spring Harb. Symp. Quant. Biol. 2010;75:51–60. doi: 10.1101/sqb.2010.75.001. [DOI] [PubMed] [Google Scholar]
- Hu H, Liu Y, Wang M, Fang J, Huang H, Yang N, Li Y, Wang J, Yao X, Shi Y, et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 2011;25:901–906. doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalitsis P, Fowler KJ, Earle E, Griffiths B, Howman E, Newson AJ, Choo KH. Partially functional Cenpa-GFP fusion protein causes increased chromosome missegregation and apoptosis during mouse embryogenesis. Chromosome Res. 2003;11:345–357. doi: 10.1023/a:1024044008009. [DOI] [PubMed] [Google Scholar]
- Marshall OJ, Marshall AT, Choo KH. Three-dimensional localization of CENP-A suggests a complex higher order structure of centromeric chromatin. J. Cell Biol. 2008;183:1193–1202. doi: 10.1083/jcb.200804078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol. Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perpelescu M, Nozaki N, Obuse C, Yang H, Yoda K. Active establishment of centromeric CENP-A chromatin by RSF complex. J. Cell Biol. 2009;185:397–407. doi: 10.1083/jcb.200903088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- Schuh M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr. Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- Screpanti E, De Antoni A, Alushin GM, Petrovic A, Melis T, Nogales E, Musacchio A. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr. Biol. 2011;21:391–398. doi: 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby RD, Monier K, Sullivan KF. Chromatin assembly at kinetochores is uncoupled from DNA replication. J. Cell Biol. 2000;151:1113–1118. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc. Natl. Acad. Sci. USA. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla MS, Syed SH, Montel F, Faivre-Moskalenko C, Bednar J, Travers A, Angelov D, Dimitrov S. Remosomes: RSC generated non-mobilized particles with approximately 180 bp DNA loosely associated with the histone octamer. Proc. Natl. Acad. Sci. USA. 2010;107:1936–1941. doi: 10.1073/pnas.0904497107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M, North JA, Shimko JC, Forties RA, Ferdinand MB, Manohar M, Zhang M, Fishel R, Ottesen JJ, Poirier MG. Histone fold modifications control nucleosome unwrapping and disassembly. Proc. Natl. Acad. Sci. USA. 2011;108:12711–12716. doi: 10.1073/pnas.1106264108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- Torigoe SE, Urwin DL, Ishii H, Smith DE, Kadonaga JT. Identification of a rapidly formed nonnucleosomal histone-DNA intermediate that is converted into chromatin by ACF. Mol. Cell. 2011;43:638–648. doi: 10.1016/j.molcel.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trazzi S, Perini G, Bernardoni R, Zoli M, Reese JC, Musacchio A, Della Valle G. The C-terminal domain of CENP-C displays multiple and critical functions for mammalian centromere formation. PLoS ONE. 2009;4:e5832. doi: 10.1371/journal.pone.0005832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, Heimbrook DC, Chen L. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl. Acad. Sci. USA. 2006;103:10660–10665. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X, O’Quinn RP, Pierce HL, Joglekar AP, Gall WE, DeLuca JG, Carroll CW, Liu ST, Yen TJ, McEwen BF, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Dalal Y, Henikoff S, Lindsay S. Single-epitope recognition imaging of native chromatin. Epigenetics Chromatin. 2008;1:10. doi: 10.1186/1756-8935-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Mizuguchi G, Wisniewski J, Huang Y, Wei D, Wu C. Nonhistone Scm3 binds to AT-rich DNA to organize atypical centromeric nucleosome of budding yeast. Mol. Cell. 2011;43:369–380. doi: 10.1016/j.molcel.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Colmenares SU, Karpen GH. Assembly of Drosophila centromeric nucleosomes requires CID dimerization. Mol. Cell. 2012;45:263–269. doi: 10.1016/j.molcel.2011.12.010. Published online Dec 28, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J, van Holde K. Single-molecule biology: what is it and how does it work? Mol. Cell. 2006;24:317–329. doi: 10.1016/j.molcel.2006.10.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.