Abstract
Three hydroxylated derivatives of PCBs, including 2′-hydroxy-4-chlorobiphenyl (2′-OH-4-CB), 3′-hydroxy-4-chlorobiphenyl (3′-OH-4-CB), and 4′-hydroxy-4-chlorobiphenyl (4′-OH-4-CB), were transformed by the PCB degrader, Burkholderia xenovorans LB400. When the bacterium was growing on biphenyl (biphenyl pathway-inducing conditions), all three hydroxylated isomers were significantly transformed. On the contrary, only 2′-OH-4-CB was transformed by the bacterium growing on succinate (conditions non-inductive of the biphenyl pathway). Gene expression analyses showed a strong induction of key genes of the biphenyl pathway (bph) when cells were grown on biphenyl, which is consistent with the transformation of the three isomers by biphenyl-grown cells. When cells were grown on succinate, only exposure to 2′-OH-4-CB resulted in expression of biphenyl pathway genes, which suggests that this isomer was capable of inducing the biphenyl pathway. These results provide the first evidence that bacteria are able to metabolize PCB derivatives hydroxylated on the non-chlorinated ring.
Keywords: Biodegradation, biphenyl dioxygenase, Burkholderia xenovorans, hydroxylated polychlorinated biphenyl
Introduction
Polychlorinated biphenyls (PCBs) are toxic environmental contaminants that are rather recalcitrant to biodegradation. Due to their high chemical and physical stability and high dielectric constant, PCBs have been used widely for a variety of industrial applications, including lubricants, dielectric fluids, and plasticizers. Because of their toxicity and recalcitrance to biodegradation, the production and usage of PCBs were banned in most countries by the late seventies (Field and Sierra-Alvarez 2008; Pieper and Seeger 2008). In the meanwhile, PCBs have been largely dispersed in the environment and they are today detected in every compartment of the ecosystem.
The first step of the PCB metabolism by higher organisms results in the formation of hydroxylated derivatives, which are increasingly suspected to be responsible for the toxicity of PCBs. Hydroxylated PCBs (OH-PCBs) are known to induce various deleterious effects, including disruption of transmembrane proton-gradient, generation of reactive oxygen species (ROS), oxidative damage to DNA, inhibition of proteins (e.g., transthyretin), and endocrine disruption (Camara et al., 2004; Kitamura et al. 2005). OH-PCBs have been detected in a variety of environmental samples, including animal tissues and feces, water, and sediments, and they are today increasingly considered as a new class of environmental contaminants (Kawano et al. 2005; Ueno et al. 2007).
Although PCBs are known to be susceptible to microbial degradation under both aerobic and anaerobic conditions, little information is available about the bacterial metabolism of OH-PCBs (Field and Sierra-Alvarez 2008; Pieper and Seeger 2008). Two publications reported the bacterial transformation of OH-PCBs bearing both hydroxyl and chlorine substituents on the same ring (Francova et al. 2004; Sondossi et al. 2004). On the other hand, bacterial degradation of PCB metabolites hydroxylated on the non-chlorinated ring has received little attention, even though hydroxylation of lower-chlorinated PCBs by higher organisms have been shown to occur on both chlorinated and non-chlorinated rings (Safe et al. 1975; Rezek et al. 2007).
The objective of this study was to characterize the biodegradation of three mono-hydroxylated isomers of 4-chlorobiphenyl (4-CB), with the hydroxyl group located on the non-chlorinated ring, by the PCB degrader, B. xenovorans LB400.
Materials and Methods
Chemicals
Hydroxylated derivatives of 4-CB, 2′-hydroxy-4-chlorobiphenyl (2′-OH-4-CB), 3′-hydroxy-4-chlorobiphenyl (3′-OH-4-CB), and 4′-hydroxy-4-chlorobiphenyl (4′-OH-4-CB), were custom-synthesized in purity higher than 99% by the Superfund Research Program Synthesis Core at the University of Iowa (Iowa City, IA).
Bacterium Strain and Culture Conditions
B. xenovorans LB400 (U.S. Department of Agriculture, Peoria, IL) was cultivated in 50-mL test vials closed with PTFE-lined stoppers and containing K1 mineral medium supplemented with 10 mM sodium succinate (1,180 mg L−1) or 5 mM biphenyl (770 mg L−1) as the carbon source. The vials were inoculated with 0.5% v/v late exponential phase bacteria previously acclimated on succinate or biphenyl and incubated on a rotary shaker at 185 rpm, 30°C (Denef et al. 2004). For growth inhibition tests, 4-CB and OH-4-CBs were dosed at concentrations ranging from 2 to 50 mg L−1 and bacterial growth was monitored by the optical density at 600 nm (OD600) for 96 hours. The OD600 was converted into cell dry weight concentration (mg L−1) using standard curves established separately for cells growing on succinate and on biphenyl. For biodegradation tests, 4-CB and OH-4-CBs were dosed at sub-inhibitory concentrations (i.e., 10 mg L−1 with succinate and 2 mg L−1 with biphenyl) and analyzed by GC-MS after 96 hours of exposure. Gene expression studies were conducted either by comparing cells growing on biphenyl (10 mM) with cells growing on succinate (5 mM) or by comparing cells exposed to 4-CB and OH-4-CBs (5 mg L−1) with non-exposed cells (growing on succinate). Cell suspensions were collected after 18 hours of growth/exposure, mixed with two volumes of RNAprotect® (Qiagen, Valencia, CA), and frozen at −80°C prior to RNA extraction. Experiments were conducted in triplicates. Positive controls were dosed with an equivalent volume of acetone and negative controls were not inoculated.
Extraction and Analysis of 4-CB, 2′-OH-, 3′-OH-, 4′-OH-4-CB, and 4-Chlorobenzoate (4-CBA)
4-CB was extracted with Triton X100 and n-hexane as described by Masse et al. (1989). OH-4-CBs and 4-CBA were extracted with ethyl acetate and derivatized using a mixture of N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA)/1-% trimethylchlorosilane (TMCS) as described by Maltseva et al. (1999). Analyses were conducted using a 7890A GC equipped with a 5975C MSD (Agilent, Santa Clara, CA) and a HP-5MS high-efficiency capillary column (30 m × 0.25 mm, 0.25 μm; Agilent) using published protocols (Masse et al. 1989).
RNA Extraction and Gene Expression Analyses
The expression of key genes of the biphenyl pathway, bphA, bphB, bphC, and bphD, was quantified using reverse-transcription real-time PCR (RT-qPCR). Total RNA was extracted from cell suspensions using RNeasy® Tissue Mini Kit (Qiagen) according to the modified protocol for bacterial cells. Residual DNA was digested in the silica gel column using RNase-free DNase (Qiagen). RNA was reverse-transcribed using SuperScriptTM Reverse Transcriptase III (Invitrogen, Carlsbad, CA). Negative controls were run without reverse-transcriptase. Quantitative PCRs were performed on an ABI StepOne™ System using SYBR® Green PCR Master Mix (Applied Biosystems, Foster City, CA) using cycling conditions prescribed by the manufacturer. Control reactions contained nucleic acid-free water or negative-control complementary DNA (cDNA) as a template. Sequences of B. xenovorans bphA, bphB, bphC, and bphD were retrieved from the National Center for Biotechnology Information (NCBI) database and used to design specific primers (Supplementary Information, Table S1). 16S rDNA was used as an internal standard and amplification levels were expressed by reference to non-exposed controls using the ΔΔCT method (StepOne™ Software, version 2.1, Applied Biosystems). The amplification efficiency was determined for all primer sets according to standard procedures.
Statistical Analyses
Analytical results are presented as means and standard deviations of experimental replicates. The statistical significance of the differences was assessed by performing paired t-tests using Microsoft Excel.
Results
The potential inhibitory effect of 4-CB and its three hydroxylated derivatives, 2′-OH-, 3′-OH-, and 4′-OH-4-CB, on B. xenovorans LB400 was determined by growing cells in the presence of increasing concentrations of the target compounds and monitoring the OD600. Although exposure to 4-CB did not result in significant growth inhibition at the highest concentration tested (50 mg L−1), the three OH-4-CBs exerted inhibitory effects above 10 mg L−1 when the cells were growing on succinate (i.e., conditions non-inductive of the biphenyl pathway) and above 2 mg L−1 when cells were growing on biphenyl (i.e., biphenyl pathway-inducing conditions) (Fig. 1 and Supplementary Information, Fig. S1).
Fig. 1.
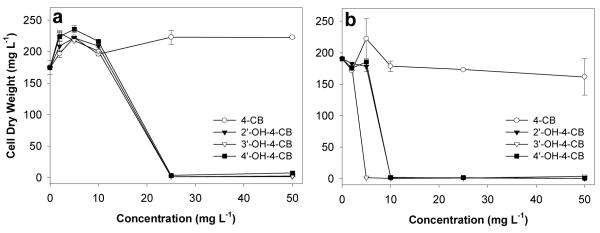
Dose-response curves showing the effect of increasing concentrations of 4-chlorobiphenyl (CB), 2′-hydroxy- (OH-), 3′-OH-, and 4′-OH-4-CB (0.0 to 50 mg L−1) on the growth of B. xenovorans LB400. Bacterial growth was expressed as the cell dry weight (mg L−1) that was reached in the stationary phase (48 h). Panel a: Cells were growing using succinate (1,180 mg L−1, 10 mM) as the carbon source. Panel b: Cells were growing using biphenyl (770 mg L−1, 5 mM) as the carbon source
Transformation of OH-PCBs by B. xenovorans LB400 was studied by growing the bacterium on K1 mineral medium containing sub-inhibitory concentrations of 4-CB, 2′-OH-, 3′-OH-, and 4′-OH-4-CB (i.e., 10 mg L−1 in the presence of succinate and 2 mg L−1 in the presence of biphenyl). Concentrations of 4-CB, OH-4-CBs, and the metabolite, 4-chlorobenzoic acid (4-CBA), were determined after 96 hours of incubation (Table 1).
Table 1.
Molar fraction (%) of 4-chlorobiphenyl (CB), 2′-hydroxy- (OH-), 3′-OH-, 4′-OH-4-CB, and the metabolite, 4-chlorobenzoate (4-CBA), recorded in cell suspensions of B. xenovorans LB400 growing on succinate (1,180 mg L−1, 10 mM) and biphenyl (770 mg L−1, 5 mM). Results are presented as mean and standard deviation of three replicates
| Succinate as Carbon Sourcea |
Biphenyl as Carbon Sourceb |
|||||
|---|---|---|---|---|---|---|
| Compounds | Initial (0 h) | Final (96 h) | 4-CBA (96 h) | Initial (0 h) | Final (96 h) | 4-CBA (96 h) |
| 4-CB | 100.0±10.3 | 0.0±0.0 | 25.6±5.9 | 100.0±10.1 | 0.7±0.4 | 0.0±0.0 |
| 2′-OH-4-CB | 100.0±7.7 | 0.0±0.0 | 99.4±4.7 | 100.0±2.2 | 0.0±0.0 | 0.0±0.0 |
| 3′-OH-4-CB | 100.0±11.9 | 103.3±6.9 | 0.0±0.0 | 100.0±1.1 | 19.0±1.4 | 0.0±0.0 |
| 4′-OH-4-CB | 100.0±0.6 | 96.9±1.8 | 0.0±0.0 | 100.0±8.4 | 45.7±5.7 | 0.0±0.0 |
100% corresponded to 10 mg L−1 of 4-CB, 2′-OH-, 3′-OH-, and 4′-OH-4-CB.
100% corresponded to 2 mg L−1 of 4-CB, 2′-OH-, 3′-OH-, and 4′-OH-4-CB.
In the presence of biphenyl, all congeners tested were significantly transformed after 4 days: ~100% of 4-CB and 2′-OH-4-CB, 81% of 3′-OH-4-CB, and 54% of 4′-OH-4-CB. In the presence of succinate, ~100% of 4-CB and 2′-OH-4-CB were transformed, but no significant transformation of 3′-OH-4-CB (0%) and 4′-OH-4-CB (3%) were recorded over the time of the experiment. The metabolite, 4-CBA, was only detected in cultures growing on succinate and exposed to 4-CB and 2′-OH-4-CB. No other metabolites were detected (relevant GC-MS chromatograms are presented as Supplementary Information, Figs. S2, S3 and S4). Abiotic losses recorded in the non-inoculated controls were negligible, suggesting that disappearance of 4-CB and OH-4-CBs observed in the presence of cells accounted for biotransformation.
The expression of key genes of the biphenyl pathway, bphA, bphB, bphC, and bphD, in response to 4-CB and OH-4-CBs was characterized using RT-qPCR (Table 2). We first compared the expression level of biphenyl pathway genes (bph) in cells growing in the presence of biphenyl (5 mM) and succinate (10 mM): biphenyl-grown cells showed higher expression of bph genes as compared to succinate-grown cells, with relative expression levels ranging from 6.5 to 8.8. On the other hand, when cells were grown on succinate as the carbon source, higher expression of bph genes was observed in the presence of biphenyl, 4-CB, and 2′-OH-4-CB (5 mg L−1) as compared to non-exposed controls, with relative expression levels ranging from 2.5 to 3.8, 2.0 to 2.8, and 1.6 to 2.4, respectively. On the contrary, downregulation of bph genes was observed in the presence of 3′-OH-4-CB and 4′-OH-4-CB.
Table 2.
Expression levels of genes of the biphenyl pathway, bphA, bphB, bphC, and bphD, in B. xenovorans LB400 after 18 hours of cultivation. Left Panel: Cells growing on biphenyl (BP) (770 mg L−1, 5 mM) relative to cells growing on succinate (1,180 mg L−1, 10 mM). Right Panel: Cells growing on succinate (1,180 mg L−1, 10 mM) exposed to 5 mg L−1 of BP, 4-chlorobiphenyl (CB), 2′-hydroxy- (OH-), 3′-OH-, and 4′-OH-4-CB relative to non-exposed cells
| Biphenyl |
Succinate |
|||||
|---|---|---|---|---|---|---|
| Genes | BP (770 mg L−1) |
BP (5 mg L−1) |
4-CB (5 mg L−1) |
2′-OH-4-CB (5 mg L−1) |
3′-OH-4-CB (5 mg L−1) |
4′-OH-4-CB (5 mg L−1) |
|
|
|
|||||
| bphA | 6.5 (5.9-7.1) |
2.5 (2.4-2.6) |
2.0 (1.9-2.2) |
1.6 (1.4-1.7) |
0.25 (0.23-0.28) |
0.29 (0.28-0.30) |
| bphB | 6.7 (5.1-8.7) |
3.1 (2.9-3.2) |
2.2 (1.9-2.6) |
2.2 (1.9-2.5) |
0.66 (0.51-0.85) |
0.46 (0.37-0.58) |
| bphC | 7.6 (6.9-8.3) |
3.8 (3.6-3.9) |
2.8 (2.4.-3.2) |
2.4 (1.8-3.2) |
0.46 (0.42-0.49) |
0.41 (0.33-0.50) |
| bphD | 8.8 (8.0-9.6) |
2.9 (1.8-4.7) |
2.3 (1.6-3.2) |
2.2 (1.8-2.5) |
0.21 (0.19-0.24) |
0.16 (0.14-0.17) |
Results are presented as mean of three replicates, the ranges of amplification levels were calculated based on the standard deviations of ΔΔCT values
Based on t-tests, expression of bph genes upon exposure to biphneyl, 4-CB, and 2′-OH-4-CB relative to non-exposed controls was statistically significant for all genes tested (p < 0.05), with the exception of bphC exposed to 2′-OH-4-CB (p = 0.09). The amplification efficiencies for all sets of primers ranged from 97 to 105% (regression line slopes between −3.4 and −3.2, with R2 higher than 0.98).
Discussion
We showed that exposure of B. xenovorans LB400 to hydroxylated derivatives of 4-CB resulted in a stronger inhibition when the cells were growing in the presence of biphenyl as compared to succinate, suggesting that induction of the biphenyl pathway produced toxic metabolites from OH-4-CBs. Although an increase of toxicity associated with the oxidative metabolism of OH-PCBs has not been reported, a few publications have showed that hydroxylation of PCBs by bacteria resulted in a higher toxicity (Camara et al. 2004; Sondossi et al. 2004; Parnell et al. 2006). Besides potential specific toxic mechanisms, hydroxylation results in an increase of solubility and bioavailability of the molecules susceptible to explain a higher toxic effect on cells (Camara et al. 2004).
We have also shown that under biphenyl pathway-inducing conditions (in the presence of biphenyl), B. xenovorans LB400 was able to transform 4-CB and all hydroxylated congeners tested, while only 4-CB and 2′-OH-4-CB were significantly transformed without biphenyl induction (in the presence of succinate). The enhanced transformation rates observed in the presence of biphenyl as compared to succinate, as well as the detection of the metabolite, 4-CBA, strongly suggest that the metabolism of OH-4-CBs occurred through the biphenyl pathway. The quantitative conversion of 2′-OH-4-CB into 4-CBA in the presence of succinate also suggests that only the hydroxylated ring was attacked by dioxygenases. The absence of 4-CBA detection in the presence of biphenyl seems to indicate that 4-CBA was further transformed under biphenyl pathway-inducing conditions. Although B. xenovorans LB400 is commonly believed to be unable to transform 4-CBA, a recent publication from Gilmartin et al. (2003) suggests that strain LB400 metabolizes 4-CBA by the action of a glutathione S-transferase (GST) encoded by a bphK gene located in the bph operon. The important question of the degradation of 4-CBA by PCB-degrading bacteria needs to be further investigated. Previous publications have shown that hydroxylated lesser-chlorinated PCBs were transformed by PCB-degrading bacteria. Sondossi et al. (2004) reported the metabolism of 2-, 3-, and 4-hydroxybiphenyl, 2-, 3-, and 4-CB, and 4-hydroxyl-2-chlorobiphenyl (OH-2-CB), 4-hydroxyl-3-chlorobiphenyl (4-OH-3-CB), and 2-hydroxyl-5-chlorobiphenyl (2-OH-5-CB) by Comamonas testosteroni B-356 and a recombinant Pseudomonas putida strain harboring the biphenyl pathway system. As it was observed with most PCBs, the metabolism of these compounds involved dihydroxylation of the unsubstituted ring in positions 2,3 followed by meta-cleavage in position 1,2. Francova et al. (2004) reported the transformation of a series of ortho-substituted hydroxylated PCBs, including 2-hydroxyl-3-chlorobiphenyl (2-OH-3-CB), 2-OH-5-CB, and 2-hydroxyl-3,5-dichlorobiphenyl (2-OH-3,5-DCB), by biphenyl dioxygenase enzymes of B. xenovorans LB400 and C. testosteroni B-356. However, these two studies have focused on PCB derivatives that were hydroxylated on the chlorinated ring. In contrast, our investigation showed that bacteria are able to metabolize PCBs hydroxylated on the non-chlorinated ring, which may have important implications for natural attenuation of PCBs. Indeed, published evidence shows that the metabolism of lesser-chlorinated PCBs by plants, fungi, and mammals yields derivatives hydroxylated on both chlorinated and non-chlorinated rings. For instance, the metabolism of PCBs by plants has been reported to generate derivatives hydroxylated on both chlorinated and non-chlorinated rings (Francova et al. 2004; Rezek et al. 2007). On the other hand, 4′-OH-4-CB and 3′,4′-dihydroxy-4-chlorobiphenyl were the major urinary metabolites identified in rabbits inoculated with 4-CB (Safe et al. 1975).
The upper biphenyl pathway that is responsible for the bacterial transformation of PCBs into chlorobenzoates and chlorinated aliphatic acids typically involves seven genes (bphA to bphG) grouped into one operon (bph). Our gene expression results showed induction of four key genes of the biphenyl pathway (bphA, bphB, bphC, and bphD) when cells were growing on biphenyl as the primary carbon source. Biphenyl, as the natural substrate of the biphenyl pathway, is a strong inducer of bph genes, resulting in significant transformation of 4-CB, 2′-OH-, 3′-OH-, and 4′-OH-4-CB (Denef et al. 2004; Parnell et al. 2006). On the other hand, when cells were growing on succinate as the carbon source, expression of bph genes was observed in the presence of 4-CB and 2′-OH-4-CB, which were the only compounds significantly degraded in the absence of biphenyl. The relatively low induction levels of bph genes in the presence of 4-CB and 2′-OH-4-CB (as compared with biphenyl) may account for the absence of significant transformation of their metabolite, 4-CBA, when cells were growing on succinate.
The toxicity and recalcitrance of selected hydroxylated PCBs to biodegradation may provide a partial explanation for the persistence of PCBs in the environment.
Supplementary Material
Acknowledgements
This work was supported by the National Institute of Health (NIH), Award Number 2P42 ES013661-05. We thank Hans-Joachim Lehmler (University of Iowa) for providing the hydroxylated PCB congeners used in this study.
Contributor Information
Rouzbeh Tehrani, Department of Civil and Environmental Engineering, Temple University, Philadelphia, Pennsylvania 19122.
Monica M. Lyv, Department of Civil and Environmental Engineering, Temple University, Philadelphia, Pennsylvania 19122
Rashid Kaveh, Department of Civil and Environmental Engineering, Temple University, Philadelphia, Pennsylvania 19122.
Jerald L. Schnoor, Department of Civil and Environmental Engineering, University of Iowa, Iowa City, Iowa 52242
Benoit Van Aken, Department of Civil and Environmental Engineering, Temple University, Philadelphia, Pennsylvania 19122.
References
- Camara B, Herrera C, Gonzalez M, Couve E, Hofer B, Seeger M. From PCBs to highly toxic metabolites by the biphenyl pathway. Environ Microbiol. 2004;6:842–850. doi: 10.1111/j.1462-2920.2004.00630.x. [DOI] [PubMed] [Google Scholar]
- Denef VJ, Park J, Tsoi TV, Rouillard JM, Zhang H, Wibbenmeyer JA, Verstraete W, Gulari E, Hashsham SA, Tiedje JM. Biphenyl and benzoate metabolism in a genomic context: Outlining genome-wide metabolic networks in Burkholderia xenovorans LB400. Appl Environ Microbiol. 2004;70:4961–4970. doi: 10.1128/AEM.70.8.4961-4970.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field JA, Sierra-Alvarez R. Microbial transformation and degradation of polychlorinated biphenyls. Environ Pollut. 2008;155:1–12. doi: 10.1016/j.envpol.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Francova K, Mackova M, Macek T, Sylvestre M. Ability of bacterial biphenyl dioxygenases from Burkholderia sp. LB400 and Comamonas testosteroni B-356 to catalyse oxygenation of ortho-hydroxychlorobiphenyls formed from PCBs by plants. Environ Pollut. 2004;127:41–48. doi: 10.1016/s0269-7491(03)00257-4. [DOI] [PubMed] [Google Scholar]
- Gilmartin N, Ryan D, Sherlock O, Dowling D. BphK shows dechlorination activity against 4-chlorobenzoate, an end product of bph-promoted degradation of PCBs. FEMS Microbiol Lett. 2003;222:251–255. doi: 10.1016/S0378-1097(03)00309-4. [DOI] [PubMed] [Google Scholar]
- Kawano M, Hasegawa J, Enomoto T, Onishi H, Nishio Y, Matsuda M, Wakimoto T. Hydroxylated polychlorinated biphenyls (OH-PCBs): Recent advances in wildlife contamination study. Environ Sci. 2005;12:315–324. [PubMed] [Google Scholar]
- Kitamura S, Jinno N, Suzuki T, Sugihara K, Ohta S, Kuroki H, Fujimotoc N. Thyroid hormone-like and estrogenic activity of hydroxylated PCBs in cell culture. Toxicology. 2005;208:377–387. doi: 10.1016/j.tox.2004.11.037. [DOI] [PubMed] [Google Scholar]
- Masse R, Lalanne D, Messier F, Sylvestre M. Characterization of new bacterial transformation products of 1,1,1-trichloro-2,2-bis-(4-chlorophenyl)ethane (DDT) by gas-chromatography mass-spectrometry. Biomed Environ Mass Spectrom. 1989;18:741–752. doi: 10.1002/bms.1200180917. [DOI] [PubMed] [Google Scholar]
- Maltseva O, Tsoi T, Quensen J, Fukuda M, Tiedje J. Degradation of anaerobic reductive dechlorination products of Aroclor 1242 by four aerobic bacteria. Biodegradation. 1999;10:363–371. doi: 10.1023/a:1008319306757. [DOI] [PubMed] [Google Scholar]
- Parnell JJ, Park J, Denef V, Tsoi T, Hashsham S, Quensen J, Tiedje JA. Coping with polychlorinated biphenyl (PCB) toxicity: Physiological and genome-wide responses of Burkholderia xenovorans LB400 to PCB-mediated stress. Appl Environ Microbiol. 2006;72:6607–6614. doi: 10.1128/AEM.01129-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper DH, Seeger M. Bacterial metabolism of polychlorinated biphenyls. J Mol Microbiol Biotechnol. 2008;15:121–138. doi: 10.1159/000121325. [DOI] [PubMed] [Google Scholar]
- Rezek J, Macek T, Mackova M, Triska J. Plant metabolites of polychlorinated biphenyls in hairy root culture of black nightshade Solanum nigrum SNC-9O. Chemosphere. 2007;69:1221–1227. doi: 10.1016/j.chemosphere.2007.05.090. [DOI] [PubMed] [Google Scholar]
- Safe S, Hutzinger O, Jones D. Mechanism of chlorobiphenyl metabolism. J Agric Food Chem. 1975;23:851–853. doi: 10.1021/jf60201a039. [DOI] [PubMed] [Google Scholar]
- Sondossi M, Barriault D, Sylvestre M. Metabolism of 2,2′- and 3,3′-dihydroxybiphenyl by the diphenyl catabolic pathway of Comamonas testosteroni B-356. Appl Environ Microbiol. 2004;70:174–181. doi: 10.1128/AEM.70.1.174-181.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno D, Darling C, Alaee M, Campbell L, Pacepavicius G, Teixeira C, Muir D. Detection of hydroxylated polychlorinated biphenyls (OH-PCBs) in the abiotic environment: Surface water and precipitation from Ontario, Canada. Environ Sci Technol. 2007;41:1841–1848. doi: 10.1021/es061539l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


