Abstract
Laboratory courses in neurophysiology fulfill a critical need for inquiry-based training in undergraduate programs in neuroscience and biology. These courses typically use classical electrophysiological preparations to explore the basic features of neuronal function. However, current neuroscience research also focuses on elucidating the molecular and genetic mechanisms of neuronal function, using model systems that include mutant and transgenic animals. To bridge laboratory training in neurophysiology with modern molecular genetics, we describe a teaching model based on electroretinography of the fruit fly Drosophila melanogaster, a long-established model system for basic neuroscience research. Drosophila are easily maintained, economical, and have hundreds of neurophysiologically relevant mutant strains and genetic tools readily available. The Drosophila electroretinogram (ERG) is a simple and accessible extracellular recording of a neural signal in the fly eye in response to flashes of light. The signal is multifaceted and the response is sensitive to stimulation parameters such as intensity, duration and wavelength, thus forming a rich source of analysis for students. Most importantly, different mutations affecting key components of intracellular signaling, synaptic transmission or neuronal function can affect the ERG waveform in characteristic ways. Recording wild type and mutant ERGs allows students to examine firsthand the connection between genetics, biochemical pathways, and electrophysiology. This neurophysiology laboratory course can facilitate and enhance an understanding of the cellular and molecular contributions to neurophysiological recordings.
Keywords: Drosophila, electroretinograms, visual system, phototransduction
With the rising popularity of neuroscience as a discipline, the number of colleges and universities offering specialized programs in neuroscience at the undergraduate level is rapidly increasing. Within the standard neuroscience curriculum, neurophysiology lab courses play a central role. There is no substitute for this type of hands-on experience, where students learn how to set up a preparation, record electrical signals from living organisms in real time, and analyze the generated data. Neurophysiology lab courses teach students how to think in terms of practical experiments, ask tractable questions about a biological process, and design experiments to probe these questions in a scientific way. This is where the information learned in the lecture room is synthesized into a deeper understanding of how nervous systems function, and how we know what we know about this process.
Traditionally, neurophysiology labs have used several standard preparations, chosen because they are robust model systems (Johnson et al., 2002). Crayfish, snails, and leeches are among the most common classical preps, because they are readily available, easy to maintain, and survive well on electrophysiology “rigs.” They provide excellent access to muscle cells, neuromuscular junction synapses, and/or large neuronal cell bodies. However, much of modern biology relies on genetically manipulable model systems. Molecular biological tools allow us to experimentally modify the underlying genetics of neurons, and thereby understand how nervous systems work at a more fundamental level. Students in neuroscience need practical, hands-on experience in applying these tools to questions in neurophysiology (Frantz et al., 2006). Therefore, incorporating organisms that are genetically accessible into our electrophysiology labs has many benefits. Ideally, the experimental animal will have available mutations in genes affecting neuronal function, such as ion channels, intracellular messenger molecules, or neuronal development. There should also be transgenic lines allowing expression of proteins to visualize or modify the function of nerves, muscle or glia. The fruit fly Drosophila melanogaster is an outstanding model system that combines genetic and physiological accessibility, a variety of model neurons, and a vast array of mutant and transgenic lines available to investigate genes and processes at the cellular and genetic level. This system has long been used in genetics courses, and has recently more been recognized as a valuable tool in undergraduate neuroscience education (Krans et al., 2005; Berni et al., 2006; Pulver et al., 2011a, b). Here, we highlight the utility of the Drosophila electroretinogram (ERG) as an appropriate undergraduate neurophysiological model system.
The Drosophila ERG has been used for more than 40 years (Hotta and Benzer, 1969; Heisenberg, 1971; Stark and Wasserman, 1972) and was instrumental in characterizing many of the key genes in phototransduction (reviewed in Pak, 1995; Montell, 1999; Hardie and Raghu, 2001). The ERG recording method uses an extracellular electrode to record a compound field potential from photoreceptors and downstream neurons within the fly eye in response to flashes of light (Dolph et al., 2011). Transient spikes at the onset and offset of a light flash correspond to postsynaptic potentials in visual system neurons, while a sustained potential during the light stimulus results from the depolarization of photoreceptor cells (Stark and Wasserman, 1972a; Wu and Wong, 1977; Montell, 1999; Hardie and Raghu, 2001). The ERG response can be triggered by light ranging from the visible spectrum to ultraviolet, which can be produced by a standard stereoscope light source or an ultra-bright white LED. The different components of the ERG signal reflect distinct events during visual signal transduction in the eye (Heisenberg, 1971). Mutations in genes coding for proteins involved in visual transduction result in characteristic changes in the ERG waveform. By recording from wild type (i.e., “normal”) animals, and comparing the resulting traces with those from specific mutant lines, students can correlate gene sequence, protein structure, and neuronal physiology (Stark and Wasserman, 1972a; Wu and Wong, 1977). ERG recording in a number of invertebrate species has already been successfully adapted for teaching purposes by several groups (Limulus, Wald et al., 1966; crayfish, Olivo, 2003, 2012; Musca and Neobellieria flies, Krans et al., 2006; other insects, Silver and Smith, 1999). This system provides a powerful demonstration of the structure-function relationship between subcellular protein machinery and nervous system function. In addition, by manipulating the intensity, wavelength, duration or other components of the stimulus, students can explore both the capabilities, and limitations, of the insect eye.
The visual system of Drosophila principally consists of two compound eyes, each with approximately 800 functional units called ommatidia (reviewed in Ranganathan et al., 1995; Montell, 1999). Each ommatidium is surrounded by pigment cells, and contains eight photoreceptors. Photoreceptors 1–6 (R1–6) express a rhodopsin most sensitive to blue light, while R7 detects UV and R8 either green or blue light. Light passes through the transparent cornea at the surface of the eye, and is focused on the highly convoluted membrane-rich rhabdomeres that house rhodopsin. Drosophila phototransduction is initiated with the isomerization of 11-cis 3-hydroxyretinal to all-trans 3-hydroxyretinal by a photon of light. This light-sensitive vitamin A derivative is housed within all of the Drosophila rhodopsins, and its isomerization results in the activation of rhodopsin (forming metarhodopsin). Metarhodopsin triggers the exchange of GDP for GTP on the alpha subunit of a heterotrimeric G-protein, activating the G-alpha subunit. This G-alpha subunit is a member of the G-alpha q family, and activates phospholipase C (PLC) that cleaves phosphatidyl inositol 4,5 bisphosphate into inositol trisphosphate and diacyl glycerol (DAG). DAG is thought to indirectly gate the cation selective channels Trp and Trpl, triggering the influx of Ca++ and Na+ into the photoreceptor. The resulting photoreceptor depolarization causes a release of histamine (His) at the synapse, which triggers a hyperpolarization of the downstream neurons (Reviewed in Montell, 1999; Hardie and Raghu, 2001).
The Drosophila ERG is a straightforward, robust preparation that can easily be learned during the course of one lab session by undergraduates. While the size of the fly itself is small, recording ERGs from the eye is no more difficult than using such standard extracellular preps as crayfish or insect sensory nerves. Under the dissection scope, with the use of micromanipulators, this setup proves to be easier than many common teaching labs. The recording is also robust, with a reasonable learning curve: after some practice, students can easily and reliably obtain recordings from several animals within one class session. This allows gathering of data in significant enough quantities to be useful for statistical analysis.
The ERG signal itself provides a rich resource for analysis in the teaching lab. ERGs are field potentials that show aggregate activity in a sensory organ, and therefore provide information different from extracellular recordings of individual spikes in a nerve. The signal demonstrates both sustained receptor potentials in primary sensory neurons (the photoreceptors), and the activity of downstream laminar neurons receiving synaptic input from the photoreceptors. These different components may be isolated experimentally, selectively modified by specific mutations or differences in light stimulus, and analyzed separately. In this way, the multifaceted ERG prep engages students, as each response potentially carries a wealth of information about the function of the eye.
The most significant advantage of Drosophila ERG recording is the ability to apply the vast resources of the Drosophila toolkit to the neurophysiology teaching lab. Many readily available mutant fly lines affect the electrophysiology of the fly eye (Table 1). The presence and amplitude of individual ERG components may be specifically affected by mutations, transgenic constructs, and genetic backgrounds. Students are, therefore, able to connect an electrophysiological phenotype with the underlying molecular mechanisms and, ultimately, with genes coding for components of neuronal function. This approach concretely demonstrates how genes shape the structure and function of neurons, nervous systems, and ultimately the behavior of the organism.
Table 1:
Relevant genes and proteins involved in phototransduction in Drosophila, and mutant Drosophila strains available for these genes at the Bloomington Stock Center (http://fly.bio.indiana.edu/).
| Gene | Protein encoded | Genotype | ERG Phenotype | Bloomington Stock # |
|---|---|---|---|---|
| Canton S | Wildtype strain | Canton S | Wildtype | 1 |
| white | ABC transporter for pigment deposition | W1118 | Distinct negative deflection following on-transient | 3605, 5905 (amorph) |
| ninaE | Rhodopsin 1 | w*; sr1, ninaE17, es ninaE5 | Loss of on- and off-transients, reduced receptor potential | 5701 (amorph) 3545 (hypomorph) |
| norpA | Phospholipase C beta | norpA36 | Ablation of ERG signal | 9048 (amorph) |
| Trp | Cation channel | Trp1 | Failure of receptor potential maintenance during light pulse | 5692 (amorph) |
| Trpl | Cation channel | cn1, trpl302, bw1 | Failure of receptor potential during prolonged stimulation | 31433 (amorph) |
| Ort | Histamine receptor | Ort1 | Loss of on- and off-transients, normal receptor potential amplitude and dynamics | 1133 (amorph) |
MATERIALS AND METHODS
Fly rearing and stock maintenance:
Labs without experience rearing Drosophila may at first be intimidated by the challenge of handling a small insect capable of escape. In reality, Drosophila are easier to maintain and manipulate than many animals commonly used in neurophysiology laboratory courses. Basic care of Drosophila and more advanced genetic manipulations using mutant lines are described in several useful references (Greenspan, 2004; Ashburner et al., 2005; Rissing and Cogan, 2009; Zhang et al., 2010). While many Drosophila research labs use incubators and CO2 anesthesia stations, neither of these is required for the basic fly handling described in this report. Fly vials can be kept in racks on open laboratory shelves. In our own teaching labs, fly vials are kept in large plastic storage bins, with air holes to maintain a reasonable humidity. We also use “fly pooters” (described below) to manipulate individual flies in place of CO2 anasthesia, greatly simplifying the collection procedure.
Most colleges and universities have at least one Drosophila lab, whether for research or teaching (usually genetics laboratory courses). These are often the best resources for on-site training in fly handling, and for obtaining prepared fly food vials. The requirements of neurophysiology teaching laboratories are typically a small fraction of the fly vials and bottles used by a research lab, and it is relatively easy to set up a collaboration where fly food can be provided by a common “fly kitchen.” If this is not possible, companies such as Ward’s or Carolina Biological sell Drosophila kits with vials and ready-made food that only needs to be mixed with water (Carolina Biological Formula 4–24). While this is more expensive on a per-unit basis, it is nevertheless very reasonable for the numbers of animals required for this laboratory.
Mounting flies:
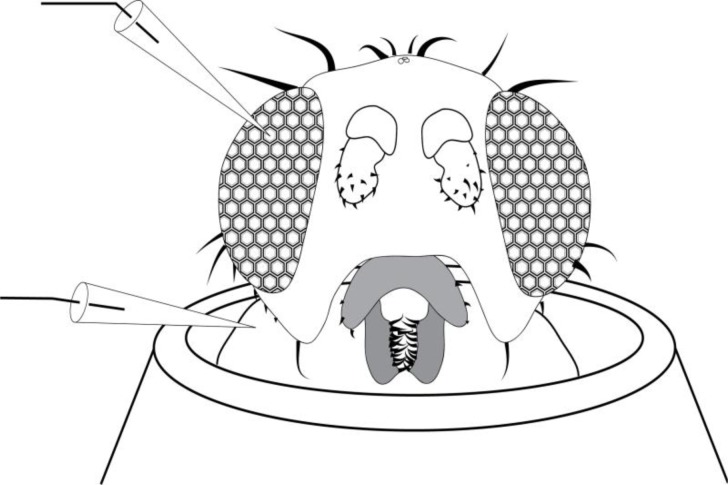
Drosophila were immobilized and mounted for electophysiological recordings using a simple technique that avoids the use of anesthetics. First, a “fly pooter” was made by cutting a 1-foot section of 3/8″ Tygon tubing. A Nitex nylon membrane (64μM; Genesee Scientific) was placed over the back of two P1000 tips, and these tips were inserted wide end first into the Tygon tubing. The tapered part of one P1000 tip, and a 1cm section of the other P1000 tip, was cut off with a razor blade. The narrower end of the pooter was used to collect a fly from a vial by mouth aspiration. With a fly inside the P1000 tip at the end of the pooter, the wide end of a P200 tip was placed over the cut P1000 tip. The fine 2mm taper was cut off the P200 tip, and the fly was lodged in the end of the P200 tip by blowing into the pooter. Once the fly was trapped at the narrow end of the P200 tip, the P200 tip was removed from the pooter, and a fine piece of cotton was pushed into the tip behind the fly. This cotton not only traps the fly at the end of the P200 tip, but is also used to push the fly’s head out the end of the P200 tip (Figure 1). With the fly mounted in the narrow end of a P200 tip, a piece of modeling clay was placed on the wide end of the tip. This mount was placed on a petri dish and taken to the electrophysiology rig.
Figure 1.
Diagram of a Drosophila positioned in the opening of a micropipette tip. The opening has been cut to allow the head to fit through, yet keep the thorax, wings and legs secure within the pipette. A sharp recording electrode is inserted into the eye while a reference electrode is inserted into the thorax. Alternatively, the recording electrode could consist of a thin cotton wick placed on to the eye.
Setting up the electrophysiology rig and recording ERGs:
There is a wide range of electrophysiology rigs that can accommodate ERG recordings in Drosophila. The rigs used in this experiment had two micromanipulators (Narishige M-3333, W. Nuhsbaum, Inc.), each positioning a standard electrode holder (Warner Instruments E series, straight configuration) into which the reference and recording electrodes were clamped. The specific choice of micromanipulator and electrode holder is not critical: this technique does not require very fine positioning resolution, and is not sensitive to the precise shape or resistance of the reference or recording glass electrodes. The recording electrode was connected to the input headstage of an amplifier (A-M Systems model 1600 Neuroprobe or A-M Systems AC/DC differential amplifier model 3000), while the reference electrode was connected to a common aluminum ground bar located inside a Faraday cage surrounding the electrophysiology rig. The ground input of the amplifier, as well as grounding connections from the Faraday cage and microscope, were connected to the same ground bar. Data from the amplifier were routed to an A/D converter (AD Instruments PowerLab 4/30) and thereafter to a PC. For ERG recording, any basic intracellular or DC-capable extracellular amplifier would work. Reliable vendors of such amplifiers include A-M Systems (www.a-msystems.com), World Precision Instruments (www.wpiinc.com) and Warner Instruments (www.warneronline.com).
Two glass electrodes (World Precision Instruments, model 1B150F-4) were pulled using a standard intracellular program (approximate tip size: 0.5μm) from the pre-programmed library on an electrode puller (Sutter Instruments P-97). The reference electrode, to be inserted into the fly’s thorax, was slightly broken back with forceps or on a laboratory wipe, while the recording electrode was left as is. The recording electrode was filled with 3M NaCl, while the reference electrode was filled with a 0.9% w/v NaCl solution. Both electrodes were fixed to the electrode holders, with chloride silver wire immersed in the saline. An alternative recording electrode configuration used in our lab course consists of a thread wick, either cut from a white cotton sewing thread, or hand-spun from a cotton ball. The wick is inserted into the barrel of an (un-pulled) glass capillary of the type used to pull our electrodes, then filled with 0.9% NaCl or standard phosphate-buffered saline. Using a dissecting stereomicroscope, the reference electrode was inserted into the thorax of the fly, and the recording electrode was placed either against the cornea of the eye, or was poked just through the cornea into the retina (Figure 1).
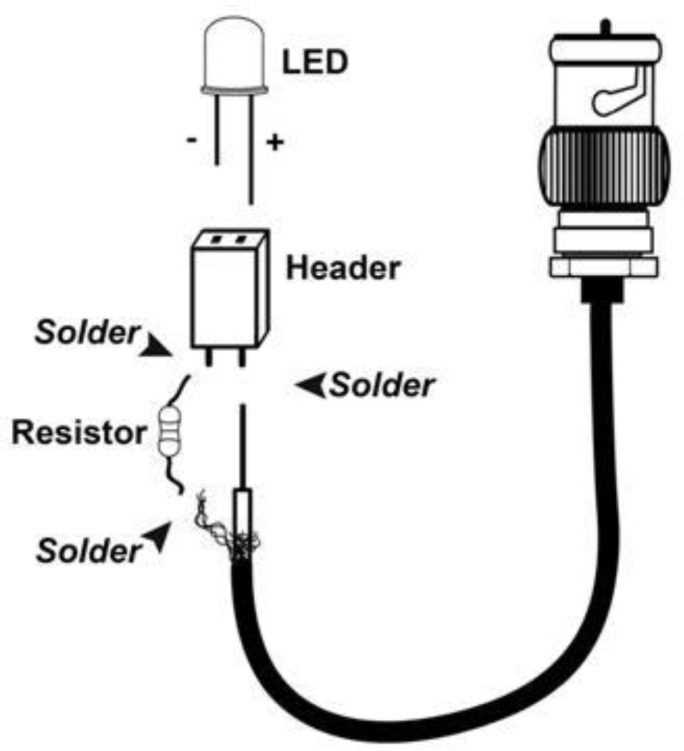
Light stimulation was achieved with the fiber light that illuminates specimens under the dissection scope. Some light assemblies may have an iris or dimmer that can be engaged quickly enough to use as a makeshift shutter. Alternatively, one can use a simple, modular light emitting diode (LED) assembly fitted with a high-brightness 5mm LED (Figure 2). A standard BNC cable was cut to a length of about 1m (3 ft.) and stripped at one end to separate the core conductor and the surrounding wire mesh shielding. The exposed mesh was twisted into a lead and soldered to a small 47 Ohm resistor (Digikey, part #P47BBTB-ND) as a minimal current limiting protection measure. The exact type and value of the resistor is not critical as long as the stimulator is able to supply enough current through the circuit to activate the LED. This step may be omitted if the students are careful not to exceed the current limit of the LEDs used. The other end of the resistor and the core conductor of the BNC cable are each soldered to one terminal of a two-position header (Digikey, part #S5438-ND). The assembly was then insulated with electrical tape. The BNC plug was connected directly to the analog output channel of an A/D board. During experiments the LED assembly was driven by software with pulses of 5V amplitude. This allows students to deliver light pulses of precisely controlled duration with a very sharp on and off. The data reported here were collected using a 30 degree angle, 18,000 mcd white LED (www.superbrightleds.com, part # RL5-W18030) as a light stimulus. The maximum available current (20 mA) of our analog output channel was below the 70 mA maximum current rating of the LEDs used.
Figure 2.
Schematic of the LED light stimulus assembly. A standard BNC and components are soldered together as indicated by arrows.
All data were acquired, analyzed and displayed by students using the program Chart 5 (AD Instruments). Images were generated by screen capture, imported into Microsoft Word, and cropped to generate figures used in student lab reports and in this report. Diagrams were drawn using Adobe Illustrator and processed in Adobe Photoshop (Adobe Systems, software version CS5).
RESULTS
The wild type ERG signal is comprised of several distinct components, as shown in Figure 3 (derived from student data). The observed extracellular signal is of opposite polarity to potentials within photoreceptors and laminar neurons. During the course of a light pulse stimulus, an initial corneal-positive “on” transient voltage spike reflects the hyperpolarization of laminar neurons, which are the synaptic targets of the photoreceptors R1–R6. In the fly retina, photoreceptors release His as a neurotransmitter, which opens chloride-permeable ionotropic His receptors (Gengs et al., 2002). This is followed by a sustained corneal-negative potential, corresponding to depolarization of the photoreceptors, and reaching a peak of 10–20 mV within approximately 100 msec of the onset of the light pulse. Termination of the light pulse elicits a corneal-negative “off” transient corresponding to the repolarization of the laminar neurons in response to the cessation of His release by the photoreceptors. The magnitudes of the on and off transients are dependent on the precise placement of the recording electrode, and as such are a challenging parameter to compare between fly strains. A poor connection of the electrode to the eye may also result in apparent loss of transients and a reduction in the observed receptor potential. The comparison to a model ERG trace gives students immediate feedback on the quality of their preparation and setup. We find that a practiced lab group can obtain consistent transient and receptor potential amplitudes, and are able to reliably and quantitatively compare wild type and mutant ERG recordings.
Figure 3.
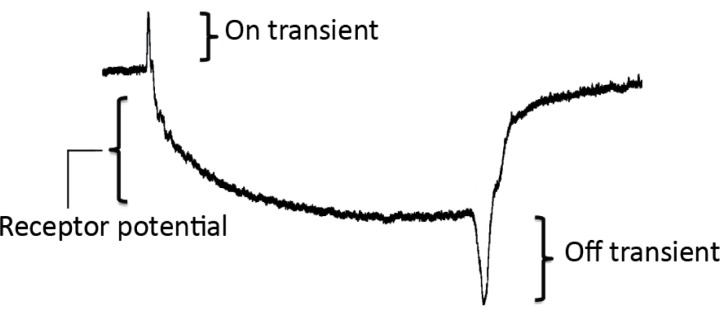
A typical ERG trace, recorded by students from a wild-type Drosophila. Brackets indicate the three major components of the signal. The positive on-transient appears within ten milliseconds of the initiation of the light pulse. The negative receptor potential reaches peak amplitude between 0.5–1 seconds after initiation of the light pulse. A negative off-transient peaks within ten milliseconds of the termination of the light pulse.
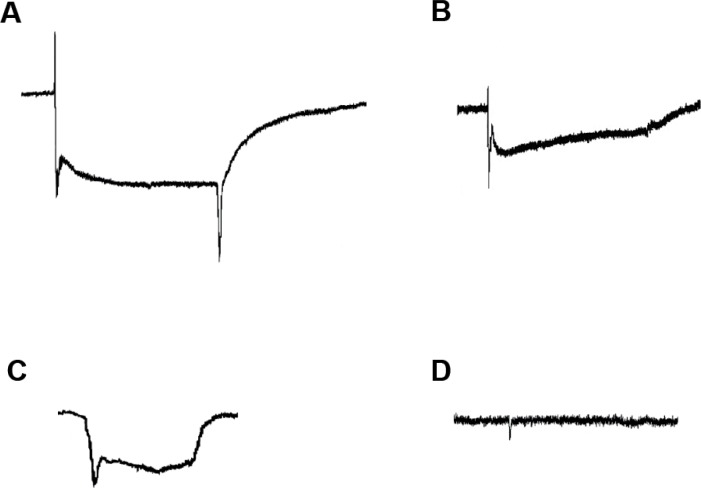
Representative ERG traces from some of the mutant strains used in our courses are shown in Figure 4. All recordings were generated by students in experiments that they themselves set up, conducted, analyzed and presented. Figure 4A is a trace recorded from a white mutant strain (w−), in which the eyes appear white rather than their normal bright red color, due to a lack of screening pigment that normally separates each ommatidium, resulting in an eye that can detect light but has compromised resolution (Kalmus, 1949; Wu and Wong, 1977). The main phenotype of this mutation is in the initial segment of the ERG trace, where a sharp negative voltage deflection immediately follows the on transient spike (Stark and Wasserman, 1972b). This is potentially caused by an increased initial photoreceptor response in the absence of screening pigment (W. Stark, personal communication). For the purposes of tracking phenotypes while designing genetic crosses, many mutant and transgenic strains of Drosophila are maintained on a w− background, including mutations that would themselves affect the ERG signal shape. The characteristic w− ERG waveform will often appear in conjunction with the effects of other mutations of interest, resulting in a compound phenotype. Therefore, it is important for students to know the characteristic electrophysiological signature of this mutant strain.
Figure 4.
Sample student data showing ERG traces from mutant strains, indicating selective effects on the electrophysiological signal. A. white mutant, showing the characteristic negative deflection following the on-transient. B. trp mutant, showing the decay in the receptor potential during the duration of the light pulse. C. ninaE mutant, showing the lack of on- and off-transients. D. norpA mutant, showing nearly complete absence of signal from the eye in response to light.
Figure 4B shows a trace recorded from a transient receptor potential (or trp mutant), which is a loss-of-function mutation in a cation channel, permeable to Na+ and Ca++, present in Drosophila photoreceptors (Hardie and Minke, 1992). This is the principal ion channel that opens in response to light, and drives much of the sustained potential of the ERG. trp mutants are unable to conduct enough Ca++ current to allow maintenance of the open state of the remaining photoresponsive channels (the trpl channels). The resulting signal thus shows an on-transient followed by a reduced receptor potential, which quickly decays to baseline. This mutant line is significant in that this was a key tool in the discovery and understanding of this critical ion channel family (Montell, 2011).
A recording from a w; ninaE double mutant, containing mutations in both the white gene and in the gene coding for the opsin expressed in the majority of Drosophila photoreceptors (R1–R6) is shown in Figure 4C. ninaE mutants have depleted rhodopsin levels, and thus have a defect in the first step in visual transduction, where energy from the stimulus (photons) is transformed into a chemical signal (Scavarda et al., 1983). The resulting ERG signal lacks on- and off-transients, though the corneal-negative spike characteristic of the w− phenotype is still present in w; ninaE double mutants. The remaining receptor potential is due to the presence of functional rhodopsin in a subset of photoreceptors (R7 and R8).
Figure 4D shows a trace from a norpA strain, which lacks functional PLC in photoreceptors (Bloomquist et al., 1988). PLC is a key biochemical link in the visual transduction cascade within photoreceptors. Metarhodopsin formed by absorption of photons activates a G-protein, which in turn activates PLC and drives DAG synthesis. norpA mutants cause complete failure of the visual response by ablating this key biochemical signal transduction component. Without functional PLC, Trp channels fail to open, and photoreceptors do not exhibit a membrane potential change in response to a light stimulus. PLC is a key component of many signal transduction pathways, critical to the function of neurons and other cell types. Once students are confident in their ERG recording ability, this dramatic phenotype provides an example of how complex cascades can critically depend on the integrity of individual components.
These mutant strains show clearly visible defects in their ERGs, and provide a basic set of mutants that are useful in a teaching laboratory setting. Many other mutant lines are available (Table 1), and in combination with manipulating the intensity, wavelength and duration of the light stimulus, these strains allow students to design independent research projects examining how both genes and stimulus parameters affect the visual response.
DISCUSSION
The Drosophila ERG is a technically simple, yet analytically rich, extracellular electrophysiological preparation. It is an elegant experimental system that allows students to investigate visual system function at the level of the receptor organ itself, and makes a great complement to other extracellular preps that record action potentials in mechanosensory or motor nerves. The light stimulus can be easily applied without having to directly contact the rig, and can be varied in intensity, wavelength and duration to produce a range of different stimulus qualities. The ERG signal is multifaceted, containing different components that correspond to distinct cells and processes within the Drosophila visual system, which can be analyzed individually. These qualities alone make the Drosophila ERG preparation an excellent teaching model. However, the real power of this system is that it links genetics and molecular biology to neurophysiology in a practical, hands-on laboratory that is readily accessible to undergraduates. Drosophila has long been used to probe the molecular basis of nervous system function (reviewed in Pak, 1995). As a result, hundreds of lines with specific mutations in neurophysiologically relevant genes have been generated, and are easily obtained from organizations such as the Bloomington Drosophila Stock Center.
In our own courses, we begin with a concise overview of Drosophila visual transduction, briefly explaining the signaling pathways that generate the photoreceptor response to light and the transmission of this signal to downstream neurons. In our experience, the lecture method of teaching biochemical pathways is less conducive to forming long-term memory and true understanding. Here, we can describe the signal transduction pathways in the context of the waveform of the ERG, explaining how each step in the process shapes the resulting signal that the students will record. Once students begin recording their own ERG traces, they can measure the time courses and amplitudes of components such as the on- and off-transients, the receptor potential, and the time to recovery to baseline. Thus they can correlate the quality of the light stimulus with the amplitudes of each component, and they can see traces that they have themselves acquired as direct manifestations of the underlying cellular and molecular biology. This puts the visual signal transduction pathways into a context of the students’ own experience, and in this way directly links material learned in lecture with data that they have personally produced.
At this point, students are given mutant lines with which to experiment. We often use the “mystery mutant” paradigm, where students record ERG traces from animals without being given the genotype. Without knowing what to expect students become acutely attuned to all details of the experiment, and begin to see the data that they collect as valuable information; clues to a mystery that they need to solve, rather than as an exercise that they need to replicate. They must explicitly explain any alterations in physiology that they observe in terms of the molecular mechanisms that they just learned. They begin to form hypotheses about the identity of the mutations. At this stage, they can be guided in exploring methods to test their hypotheses, or perhaps given a specific list of possible mutations that they should choose from. In this way, the Drosophila ERG allows students to “change” molecular machinery in real time, in living animals. A task that might have been a dry task of biochemical pathway memorization is thus adapted into an interesting and engaging process of discovery, hypothesis testing and data analysis.
The mutant lines that we present here (Table 1) are only a few of the array of different mutants available in Drosophila that affect visual signal transduction at several key points. Four mutant lines commonly used in our labs are described in Results and their traces are depicted in Figure 3. Another interesting and informative strain listed as a resource in Table 1, is the ort mutant line. ort ablates the His neurotransmitter receptor on laminar cells which receive synaptic input from photoreceptors (Gengs et al., 2002). Here, the receptor potential and amplitude are nearly identical to wild type, yet the on- and off-transients are gone. Laminar monopolar cells in the fly eye may thus be thought of as having a function analogous to the “on” and “off” bipolar neurons in the vertebrate retina (Sanes and Zipursky, 2010). While vision in flies is not directly homologous to vision in vertebrates (with the important exception of certain melanopsin-containing retinal ganglion cells, Hankins et al., 2008), each component of the visual transduction cascade in Drosophila is relevant to neuronal function in general.
We find that students are usually able to master the setup and recording within the first hour or two of a lab session. There are two main areas in which students have trouble, and where specific instructor attention is critical. The first is in stably mounting the fly into the pipette tip, with enough of the animal protruding to allow easy access to the eyes and thorax for electrode placement, while at the same time remaining restrained to minimize motion. The second is in obtaining a good connection between the recording electrode and the cornea (or retina) of the eye. Instructors not familiar with Drosophila husbandry may find fly manipulation challenging at first, but electrode placement should pose no special challenge within an electrophysiology lab. In our courses, all student groups have been successful in obtaining multiple recordings from wild type flies and at least one mutant. These techniques were taught to 71 2-person lab groups over the course of six years at Pomona College, and to 35 3-person lab groups over the course of three years at the University of Cincinnati, representing a total of 243 students. All groups in our courses were successful in recording ERGs from Drosophila by the end of the first day of class. Most groups are able to collect enough data to conduct a statistical analysis comparing ERGs between wild type and at least two different mutant strains or light stimulus conditions.
The mutant strains described here are a few of the many available lines that affect genes crucial to neuronal function, and that show distinctive electrophysiological phenotypes. In fact, many key intracellular signaling pathways were first discovered by studying Drosophila mutations affecting the visual system (Muqit and Feany, 2002). In our courses, we put fly visual transduction into a broader cell biology context. Our background reading assignments include a mix of classic research papers and current, broad reviews (see references). This provides students with context and an understanding of the relevance of their laboratory exercises, as well as instilling a sense of the history of research in neurogenetics and neurophysiology.
We have also used this experimental approach to allow students to design multi-week independent research projects to explore some aspect of Drosophila vision. Previous research topics have included: How do mutant lines differ in the dynamics of their response to light levels or light pulse length? How do Drosophila eyes recover from prolonged light exposure? Do different Drosophila mutants have different spectral sensitivities? What is the flicker fusion frequency in Drosophila? How quickly does the phosphatidylinositol bisphosphate (PIP2) pool recover in photoreceptors? Do different rearing conditions affect visual system function in adults? Once students have deduced the identity of the mutation in their experimental Drosophila lines, they can generate hypotheses and predictions around the roles of the gene of interest. Is the given mutation a loss-of-function allele (reducing or ablating the function of the protein product), a gain-of-function allele (increasing the rate or extent of the normal function of the protein), or a neomorph (resulting in a protein with a novel function)? What would they expect the ERG trace to look like in each case? Where in the visual transduction pathway could they imagine a change that would compensate for or reduce the severity of the mutant ERG phenotype?
Finally, while the Drosophila ERG is a relatively simple and readily accessible teaching prep, it is also commonly used in the research lab. For example, ERG recording is often a first-line technique used to quickly gauge the role of a gene in synaptic transmission (Stowers and Schwarz 1999) and intracellular signaling (reviewed in Pak 1995). For this reason, research labs using Drosophila as a model system, even if they do not normally use electrophysiology or are not focused on nervous system function, may benefit from expertise in this technique. Thus, students trained in ERG recording may continue using this experimental approach well beyond the laboratory exercises described here, perhaps to examine mutations affecting development, metabolism, or other neuronal processes. Just as importantly, neurophysiology lab courses teaching ERG recording could make use of novel mutations discovered locally and characterized in other ways. This technique could thus help integrate undergraduate neuroscience education with current research at their college or university.
Acknowledgments
This work was supported by the National Science Foundation DUE-1044643 to I.V. and RUI-0841551 to K.G.J.
I. V. would like to thank Elke Buschbeck for her role in helping to develop ERG teaching labs and many helpful discussions relating to this manuscript, Annette Stowasser for helping to refine the electrophysiological training methods described here, and Marisano James for comments and proofreading.
REFERENCES
- Ashburner M, Golic KG, Hawley RS. Drosophila: a laboratory handbook. 2nd Edition. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- Berni J, Muldal AM, Pulver SR. Using neurogenetics and the warmth-gated ion channel TRPA1 to study the neural basis of behavior in Drosophila. J Undergrad Neurosci Edu. 2006;9:A5–A14. [PMC free article] [PubMed] [Google Scholar]
- Bloomquist BT, Shortridge RD, Schneuwly S, Perdew M, Montell C, Steller H, Rubin G, Pak WL. Isolation of a putative phospholipase C gene of Drosophila, norpA, and its role in phototransduction. Cell. 1988;54:723–733. doi: 10.1016/s0092-8674(88)80017-5. [DOI] [PubMed] [Google Scholar]
- Dolph P, Nair A, Raghu P. Electroretinogram recordings of Drosophila. Cold Spring Harb Protoc Jan. 2011;1(1) doi: 10.1101/pdb.prot5549. [DOI] [PubMed] [Google Scholar]
- Frantz KJ, DeHaan RL, Demetrikopoulos MK, Carruth LL. Routes to research for novice undergraduate neuroscientists. CBE Life Sci Educ. 2006;5:175–187. doi: 10.1187/cbe.05-09-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengs C, Leung HT, Skingsley DR, Iovchev MI, Yin Z, Semenov EP, Burg MG, Hardie RC, Pak WL. The target of Drosophila photoreceptor synaptic transmission is a histamine-gated chloride channel encoded by ort (hclA) J Biol Chem. 2002;277:42113–42120. doi: 10.1074/jbc.M207133200. [DOI] [PubMed] [Google Scholar]
- Greenspan RJ. Fly pushing : the theory and practice of Drosophila genetics. 2nd Edition. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2004. [Google Scholar]
- Hankins MW, Peirson SN, Foster RG. Melanopsin: an exciting photopigment. Trends Neurosci. 2008;31:27–36. doi: 10.1016/j.tins.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Hardie RC, Minke B. The trp gene is essential for a light-activated Ca2+ channel in Drosophila photoreceptors. Neuron. 1992;8:643–651. doi: 10.1016/0896-6273(92)90086-s. [DOI] [PubMed] [Google Scholar]
- Hardie RC, Raghu P. Visual transduction in Drosophila. Nature. 2001;413:186–193. doi: 10.1038/35093002. [DOI] [PubMed] [Google Scholar]
- Heisenberg M. Separation of receptor and lamina potentials in the electroretinogram of normal and mutant Drosophila. J Exp Biol. 1971;55:85–100. doi: 10.1242/jeb.55.1.85. [DOI] [PubMed] [Google Scholar]
- Hotta Y, Benzer S. Abnormal electroretinograms in visual mutants of Drosophila. Nature. 1969;222:354–356. doi: 10.1038/222354a0. [DOI] [PubMed] [Google Scholar]
- Johnson BR, Wyttenbach RA, Hoy RR. The crawdad project: Crustaceans as model systems for teaching principles of neuroscience. Berlin: Springer Vehgtthhyirlag; 2002. [Google Scholar]
- Kalmus H. Optomotor responses in Drosophila and Musca. Physiol Comp Ocol Int J Comp Physiol Ecol. 1949;1:127–147. [PubMed] [Google Scholar]
- Krans JL, Rivlin PK, Hoy RR. Demonstrating the Temperature Sensitivity of Synaptic Transmission in a Drosophila Mutant. J Undergrad Neurosci Edu. 2005;4:1. [PMC free article] [PubMed] [Google Scholar]
- Krans JL, Gilbert C, Hoy R. Teaching insect retinal physiology with newly designed, inexpensive micromanipulators. Adv Physiol Edu. 2006;30:254–61. doi: 10.1152/advan.00029.2006. [DOI] [PubMed] [Google Scholar]
- Montell C. Visual transduction in Drosophila. Annu Rev Cell Dev Biol. 1999;15:231–268. doi: 10.1146/annurev.cellbio.15.1.231. [DOI] [PubMed] [Google Scholar]
- Montell C. The history of TRP channels, a commentary and reflection. Pflugers Arch. 2011;461:499–506. doi: 10.1007/s00424-010-0920-3. [DOI] [PubMed] [Google Scholar]
- Muqit MM, Feany MB. Modelling neurodegenerative diseases in Drosophila: a fruitful approach? Nat Rev Neurosci. 2002;3:237–243. doi: 10.1038/nrn751. [DOI] [PubMed] [Google Scholar]
- Olivo RF. An online lab manual for neurophysiology. J Undergrad Neurosci Edu. 2003;1:A16–A22. [PMC free article] [PubMed] [Google Scholar]
- Olivo RF. 2012. Biology 301 Laboratory webpage, Smith College, www.science.smith.edu/departments/NeuroSci/courses/bio330/labs.html.
- Pak WL. Drosophila in vision research. The Friedenwald Lecture. Invest Ophthalmol Vis Sci. 1995;36:2340–2357. [PubMed] [Google Scholar]
- Pulver SR, Hornstein NJ, Land BL, Johnson BR. Optogenetics in the teaching laboratory: using channelrhodopsin-2 to study the neural basis of behavior and synaptic physiology in Drosophila. Adv Physiol Educ. 2011;31:82–91. doi: 10.1152/advan.00125.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulver SR, Cognigni P, Denholm B, Fabre C, Gu WXW, Linneweber G, Prieto-Godino L, Urbancic V, Zwart M, Miguel-Aliaga I. Why flies? Inexpensive public engagement excersises to explain the value of basic biologicla research on Drosophila melanogaster. Adv Physiol Edu. 2011;35:384–392. doi: 10.1152/advan.00045.2011. [DOI] [PubMed] [Google Scholar]
- Ranganathan R, Malicki DM, Zucker CS. Signal transduction in Drosophila photoreceptors. Annu Rev Neurosci. 1995;18:283–317. doi: 10.1146/annurev.ne.18.030195.001435. [DOI] [PubMed] [Google Scholar]
- Rissing SW, Cogan JG. Can an inquiry approach improve college student learning in a teaching laboratory? CBE Life Sci Educ. 2009;8:55–61. doi: 10.1187/cbe.08-05-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, Zipursky L. Design principles of insect and vertebrate visual systems. Neuron. 2010;66:15–36. doi: 10.1016/j.neuron.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scavarda NJ, O'Tousa J, Pak WL. Drosophila locus with gene-dosage effects on rhodopsin. Proc Natl Acad Sci U S A. 1983;80:4441–4445. doi: 10.1073/pnas.80.14.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver WL, Smith JC. Sensory biology: examination of electroretinograms from photoreceptors in insects. Proceedings of the 20th Workshop/Conference of the Association for Biology Laboratory Education (ABLE); 1999. pp. 327–333. [Google Scholar]
- Stark WS, Wasserman GS. Transient and receptor potentials in the electroretinogram of Drosophila. Vision Res. 1972a;12:1771–1775. doi: 10.1016/0042-6989(72)90049-1. [DOI] [PubMed] [Google Scholar]
- Stowers RS, Schwarz TL. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics. 1999;152:1631–1639. doi: 10.1093/genetics/152.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald G, Abersheim P, Dowling J, Hopkins J, Lacks S. Twenty-Six afternoons of biology: an introductory laboratory manual. Reading, MA: Addison-Wesley Publishing Company Inc; 1962. [Google Scholar]
- Wu CF, Wong F. Frequency characteristics in the visual system of Drosophila: genetic dissection of electroretinogram components. J Gen Physiol. 1977;69:705–724. doi: 10.1085/jgp.69.6.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Freeman MR, Waddell S. Drosophila neurobiology: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2010. [Google Scholar]