Abstract
Drosophila researchers have developed a powerful suite of genetic techniques for studying the neural basis of animal behavior. Many of these tools can be exported to neuroscience teaching laboratories (Berni et al., 2010; Pulver et al., 2011a,b), but often neuroscience educators lack the basic knowledge and resources to obtain, generate and rear transgenic fruit flies on their own. Fly researchers in turn may take for granted resources that are readily available in research laboratories, but out of reach for educators. Our goal is to provide a primer for neuroscience educators who want to incorporate Drosophila genetics into their teaching, but have limited knowledge of fruit fly genetics, and/or small budgets. First we review the available methods for manipulating gene expression in Drosophila. Then we provide educators with blueprints for obtaining transgenic animals tailored for specific types of teaching modules. We outline simple techniques for rearing transgenic Drosophila, performing genetic crosses, and preparing a teaching laboratory without the use of expensive animal-care facilities. Overall, we try to break down the practical barriers educators may face when integrating modern neurogenetic experiments into teaching laboratories.
Keywords: genetics; animal behavior; neuroethology, transgenic organisms; neuroscience education
INTRODUCTION
Drosophila researchers are continually developing new genetic methods for studying the neural basis of animal behavior (For review, see Venken et al., 2011). Many of these Drosophila research advances are now being specifically adapted for use in neuroscience teaching laboratories (Krans et al., 2005; Hornstein et al., 2009; Berni et al., 2010; Zhang et al., 2010; Pulver et al., 2011b), secondary schools (T. Marzullo, personal comm.), and even public outreach events for children (Pulver et al., 2011a). At the 2011 FUN meeting, we presented a workshop on Drosophila neurogenetics. We showed how to use the light-activated ion channel channelrhodopsin-2 (ChR2) and the warmth gated Drosophila TRPA1 (dTRPA1) cation channel to study the neural basis of behavior in Drosophila larvae. We remotely increased neuronal activity in motor and sensory neurons with ChR2 and recorded excitatory junctional potentials (EJPs) at the larval neuromuscular junction (NMJ). We then evaluated locomotor behaviors evoked by dTRPA1 activation of specific neuronal ensembles. The goal of these exercises was to illustrate the principle of sufficiency in neural circuits (i.e., that activation of specific groups of neurons is sufficient to evoke specific behaviors).
Undergraduate biology courses commonly use Drosophila to illustrate principles of genetics. As a result, most neuroscience faculty have worked with Drosophila at some point during their studies. But unless they have pursued research careers in neurogenetics, many neuroscience educators are out of touch with the day-to-day realities of obtaining, rearing, and generating the animals necessary to run current teaching modules.
Recent efforts to promote Drosophila in neuroscience education have been led by research scientists. This ensures that the very newest advances are exported to classrooms; however, fly researchers (current authors included) tend to take for granted the basic knowledge and core resources readily available in research laboratories. As a result, we do not always provide detailed information on genetic procedures and basic animal care for teachers. Ironically, this is exactly the kind of ‘nuts-and-bolts’ practical information that educators often need the most.
There are excellent resources detailing genetic experiments in Drosophila and how to employ this work in genetics courses (Sofer and Tompkins, 1994; Greenspan, 2004; Ashburner et al., 2005). There are also many articles highlighting the exponentially growing array of neurogenetic tools available in flies (Hamada et al., 2008; Parisky et al., 2008; Pulver et al., 2009; Pulver & Griffith, 2010; Depetris-Chauvin et al., 2011; Venken et al., 2011). In contrast, there are relatively few publications on Drosophila genetics that are tailored for neuroscience educators. The need for such resources became clear during discussions with participants at the 2011 Faculty for Neuroscience Education (FUN) workshop at Pomona College. Many participants expressed a desire to use fly genetics in their teaching, but were unsure where to turn for help learning the relevant methodologies underlying recent publications (e.g., Berni et al., 2010).
Our aim here is to provide basic need-to-know information on fruit fly genetics for neuroscience educators. First, we review the genetic techniques in fruit flies that we think are most relevant for neuroscience educators. Then we explain in detail how educators can obtain and rear transgenic flies with limited resources. Finally, we outline the preparation of neuroscience teaching laboratories for Drosophila exercises. Throughout, we highlight potential pitfalls and common mistakes made when incorporating genetic strategies in neuroscience teaching laboratories.
WHAT GENETIC METHODS IN DROSOPHILA ARE MOST RELEVANT FOR NEUROSCIENCE EDUCATORS?
Drosophila has become a chosen system for genetic studies due in large part to the simplicity and malleability of the fly genome. Fruit flies have one sexual and only three autosomal chromosomes. The third autosomal chromosome has very few genes and can, therefore, be mostly ignored in genetic experiments. There are also numerous genetic tools for changing the fly genome, tracking where those changes occur, and stabilizing them over multiple generations (for review see Greenspan, 2004). As a result, anyone with basic knowledge of Mendelian inheritance can create strains of flies with permanent genetic modifications. In addition, the fruit fly genome was among the first to be completely sequenced, and it is now extensively annotated (Adams et al., 2000; Rubin, 2000). Consequently, geneticists know exactly the chromosomal location of each fly gene and in many cases, exactly what information it encodes.
Neuroethologists have exploited the genetic tractability of Drosophila to study the genetic and neural basis of animal behavior. There are several excellent resources that detail the methodologies, history, and most recent developments in Drosophila neurogenetics research (Ganetzky and Wu, 1986; Hall, 1998; Sokolowski, 2001; Reaume and Sokolowski, 2006; Vosshall and Stocker, 2007; Villella and Hall, 2008; Harbison et al., 2009; Venken et al., 2011). The most important thing to know is that there are two basic types of flies available for use in educational circles: mutants and transgenics.
What is a mutant animal?
Mutants are animals that are either missing whole genes or have changes in endogenous genes that disrupt function. Mutations can be either dominant or recessive. Importantly, some mutations decrease gene expression (loss-of-function mutation), while other mutations actually increase gene expression (gain-of-function mutation). Most mutants have been generated by either chemical mutagenesis or insertion of transposable elements. Crucially, in classical mutants, one does not control when and where gene expression is altered. Any cell that expresses the mutated gene can be affected and subsequently contribute to observed phenotypes.
What Drosophila mutants work best in neuroscience teaching laboratories?
Traditionally behavior geneticists exposed flies to mutagens, then screened for defects in specific behaviors. They then went on to track down which genes were mutated and where in the nervous system they were expressed. This approach has generated a large number of dominant and recessive mutants with specific defects in behavior attributable to defects in single genes. Table 1A shows a list of selected mutants (along with their associated genotypes and phenotypes) that we think are particularly useful for neuroscience educators. These mutants may work best in teaching laboratories because 1) behavioral phenotypes are robust and easy to measure, 2) electrophysiological phenotypes can be reliably measured in neurophysiology teaching labs and 3) the underlying genetic etiology is well characterized. This combination allows educators to integrate principles of genetics, cellular physiology and animal behavior in their teaching.
Table 1A.
Behavior mutants with easily measureable electrophysiological phenotypes.
| Name | Behavior Phenotype | Electrophysiology phenotype | Gene product(s) | Bloomington Stock number | Reference |
|---|---|---|---|---|---|
| ether-a-go-go | Slow locomotion, shaking under ether anesthesia | Spontaneous Excitatory Junction Potentials at larval neuromuscular junction (NMJ) | transient voltage gated potassium channel subunits | 3561 | (Wu et al., 1983) |
| slowpoke | Slow locomotion | Prolonged action potentials in flight muscles | Calcium activated potassium channel | 4587 | (Elkins et al., 1986); http://www.sdbonline.org/fly/hjmuller/slowpk1.htm |
| no receptor potential | Blind / near blind | Reduced response in electroretinagram | phosphatidylinositol-specific phospholipase C (PLC) | 5685 | (Pak et al., 1970); http://www.sdbonline.org/fly/hjmuller/norpa1.htm |
| Or83b (null mutant) | Cannot smell | No response to odors in electroantenngrams | G-Protein coupled odorant receptor | 23129 | (Larsson et al., 2004); http://www.sdbonline.org/fly/hjmuller/or83b-1.htm |
| skywalker | Larvae rear off substrate | Reduced response in electroretinagram; increased neurotransmitter release at NMJ | GTPase activating protein | N/A | (Uytterhoeven et al., 2011) |
| Nanchung (null mutant) | deaf | Antennal sound evoked potentials absent | Vanilloid receptor subfamily of transient receptor potential cation channels | 24902 | (Kim et al., 2003; Gong et al., 2004); |
Many other mutants have clear behavior phenotypes and genetic etiology, but do not have easy-to-measure electrophysiology phenotypes. These genetic strains can also make powerful teaching tools, especially in teaching laboratories without access to electrophysiology equipment. Selected examples of this category of mutant are shown in Table 1B. Table 1 is not an exhaustive list, but it is meant to give educators a sense for what is possible in laboratory exercises.
Table 1B.
Behavior mutants without easily measureable electrophysiological phenotypes.
| Name | Behavior Phenotype | Electrophysiology phenotype | Gene product(s) | Bloomington Stock number | References |
|---|---|---|---|---|---|
| fruitless | Males court males | N/A | Transcription factor | 684 | (Gailey & Hall, 1989); http://www.sdbonline.org/fly/dbzhnsky/frutles1.htm |
| Period | circadian rhythms defective, long period or arrhythmic in darkness | N/A | Transcription factor | 4671 | (Konopka & Benzer, 1971); http://www.sdbonline.org/fly/neural/period.htm |
| Cheap date | Reduced ethanol tolerance | N/A | PACAP-like neuropeptide | N/A | (Moore et al., 1998); http://www.sdbonline.org/fly/neural/amnesiac.htm |
| Trpa1ins | Cannot perform thermotaxis | N/A | Heat-gated transient receptor potential cation channel | N/A | (Hamada et al., 2008) http://www.sdbonline.org/fly/sturtevant/anktm1-dtrpa1.htm |
In general, mutants like those listed in Table 1 give educators a chance to illustrate how altering the expression of single specific gene can strongly influence behavior and that those behavioral traits can be inherited from generation to generation. It is to be noted that often changes in complex behavioral traits are attributable to many genes (Mackay et al., 2009).
Potential pitfalls, common mistakes: mutants
Many genes have both a functional and developmental role in neural networks. This complicates interpretation of electrophysiological and behavioral phenotypes in mutants. A given mutant phenotype could be caused by acute disruption of gene expression within neural circuits; it could also be caused by disruption of gene expression during early development, which then indirectly affects circuit function. Very often, there is no way to tell for sure. This is an important point to stress to students when working with Drosophila behavior mutants.
Spontaneously occurring mutations that resulted in morphological phenotypes were exploited by early fly geneticists to track changes in the fly genome. Many of these strains then became the genetic ‘background’ for further work. It is important to be aware that some lines used as controls are actually mutants. Any fly that has a permanent morphological phenotype (e.g., curly wings, white eyes, stubby bristles) has at least one dominant mutation in the genome. ebony flies are good examples.
These animals have a dominant mutation which leads to darkened cuticle. By eye, ebony flies are just a bit darker than wild-type animals. Closer examination, however, reveals that they have profoundly abnormal behaviors (Newby and Jackson, 1991). In addition, white (w) animals carry a mutation in a gene that determines eye color.
These animals have white eyes and are commonly used as a background for transgenic lines (see below); they also show abnormal behaviors (Diegelmann et al., 2006). Table 2 provides further information on the two most commonly used wild type controls (Canton S and Oregon R), as well as ebony and white (w1118).
Table 2.
Wild type and other control lines.
| Name | Bloomington Stock number | Reference |
|---|---|---|
| Canton S | 1 | http://flystocks.bio.indiana.edu/Reports/1.html |
| Oregon-R | 5 | http://flystocks.bio.indiana.edu/Reports/5.html |
| ebony | 497 | (Newby and Jackson, 1991; Phillips et al., 2005) |
| w1118 | 5905 | (Kurkulos et al., 1991) |
In general, behavior phenotypes are very sensitive to any kind of genetic mutation, so it is important to consider the genetic background being used in animal behavior exercises. An easy way to dilute the effects of genetic background in control animals is to mate them to a known wild-type strain (e.g., Canton S, Oregon R) and use the progeny. For further details on mutants, mutant phenotypes and genetic background issues in flies we recommend reading Greenspan (2004).
What is a transgenic animal?
In recent years, transgenic animals have emerged as the tools of choice for studying the neural basis of behavior in Drosophila. Transgenics have foreign functional genes (transgenes) inserted into their genome in germline cells. This is in contrast to mutants, which have disrupted endogenous gene function. Mutants and transgenics are not the same, and it is important not to use the two words interchangeably. In general, transgenic approaches are powerful because they allow scientists to mix and match genes that encode useful proteins from diverse organisms.
What transgenics work best in neuroscience teaching laboratories?
There are many different potential uses for transgenic flies. From a neuroscience education point of view, the most immediately useful transgenics are those that allow spatial control of transgene expression. The most widely used method for doing this makes use of the yeast bipartite expression system, GAL4/UAS. The system relies on the affinity of the yeast transcription factor GAL4 for the DNA sequence UAS (Upstream Activating Sequence) (Brand and Perrimon, 1993). A “driver line” expressing GAL4 in a particular cellular pattern is crossed with the “reporter line” coupled to a transgene of interest. In the parents, the GAL4 and UAS transgenes alone do virtually nothing. But in the progeny of a cross, GAL4 activates UAS, which in turn triggers the expression of the coupled transgene in a specific cellular pattern.
At this point, researchers have created thousands of GAL4 lines specifically targeting cells with various neurotransmitters, neuropeptides, receptors, and transcription factors (reviewed in Meinertzhagen et al., 2009). In parallel, scientists have created hundreds of UAS lines that allow visualization of neural circuits with genetically encoded fluorescent proteins or enzymatic markers (reviewed in Venken et al., 2011). More recently, Drosophilists have also created a large number of UAS lines that allow manipulation of circuit activity targeted by GAL4 expression (reviewed in Venken et al., 2011).
Table 2 shows a selection of GAL4 and UAS lines that we feel are most immediately useful for neuroscience educators. The selected GAL4 lines (Table 3A) target a range of different transmitter systems and defined populations of neurons. In most cases, the lines have been used in published teaching exercises (Hornstein et al., 2009; Pulver et al., 2011b; Berni et al., 2010; Krans et al., 2005).
Table 3A.
Recommended GAL4 driver lines.
| Name | Expression pattern | Chromosome | Bloomington Stock number | Reference |
|---|---|---|---|---|
| Elav-Gal4 | All neurons | II | 8760 | (Robinow & White, 1991) |
| Elav-Gal4 | All neurons | X | 458 | (Robinow & White, 1991) |
| Cha7.4-GAL4 | Cholinergic neurons | II | 6793 | (Salvaterra & Kitamoto, 2001) |
| OK371-GAL4 | Glutamatergic neurons | II | 26160 | (Mahr & Aberle, 2006) |
| 540-GAL4 | All peripheral nervous system | X | N/A | (Hughes & Thomas, 2007) |
| ppk1.9-GAL4 | Nociceptive neurons (pickpocket class IV sensory neurons) | II | N/A | (Ainsley et al., 2003) |
The first UAS line in Table 3B provides a simple way to visualize GAL4 expression patterns using green fluorescent protein (UAS-CD8-GFP, Lee and Luo, 1999). Table 2B also shows three UAS lines that can be used to acutely control activity using light and heat pulses. The first UAS line uses channelrhodopsin-2 (UAS-H134R-ChR2), a blue light gated cation channel originally isolated from the blue green algae, Chlamydomonas reinhardtii. When expressed in neurons, ChR2 opens in response to blue light, and can remotely evoke action potentials in a variety of animals (Nagel et al., 2003; Boyden et al., 2005; Pulver et al., 2009). The second UAS line uses the Drosophila transient receptor potential ion channel (UAS-dTRPA1), a heat gated cation channel. When ambient temperatures rise above 25°C, neurons expressing dTRPA1 depolarize and fire action potentials (Hamada et al., 2008). A third UAS line (UAS-ShibireTS) uses a temperature sensitive mutation in the synaptic vesicle recycling protein, dynamin. At room temperature, the mutated dynamin protein functions normally. When temperatures reach 28–30°C, dynamin dependent trafficking of synaptic vesicles to presynaptic sites is compromised, and synaptic release is inhibited (Kitamoto, 2001).
Table 3B.
Recommended UAS reporter/effector lines.
| Name | Reporter line type | Chromosome | Bloomington Stock number | Reference |
|---|---|---|---|---|
| UAS-CD8-GFP | Green fluorescent protein | II | 5137 | (Lee & Luo, 1999) |
| UAS-H134R-ChR2 | Channelrhodopsin-2 (Blue light gated cation channel) | II | 28995 | (Pulver et al., 2009) |
| UAS-dTRPA1 | Drosophila Transient Receptor Potential ion channel (heat gated cation channel) | II | 26263 | (Hamada et al., 2008) |
| UAS-shibirets1 | Temperature sensitive dynamin protein (heat gated blockade of synaptic vesicle cycling) | III | N/A | (Kitamoto, 2001) |
ChR2, dTRPA1 and ShibireTS are used in research laboratories to acutely manipulate neuronal activity in behaving animals by simply shining blue light on animals or by raising the ambient temperature. Recent publications have detailed exactly how to employ these techniques in teaching laboratories in a cost effective manner (ChR2: Hornstein et al., 2009; Pulver et al., 2011b; dTRPA1:Berni et al., 2010; ShibireTS: Krans et al., 2005).
Table 3 is also not a comprehensive list of available transgenic tools in Drosophila. The GAL4 and UAS lines here are meant instead as reliable ‘go-to’ lines for educators and as solid foundations for future exploration of the research literature. For a comprehensive review of transgenic tools currently available in Drosophila, see Venken et al. (2011).
Potential pitfalls, common mistakes: transgenics
The idea of crossing two lines of flies and using the progeny seems quite simple. However, some of the transgene insertions necessary to generate the driver or reporter lines are lethal to flies if present on both copies of the same chromosome (i.e., homozygous). To solve this problem and maintain stable lines, balancer chromosomes (mutated chromosomes that prevent recombination) with dominant markers (mutations causing morphological mutations) have been developed (Hochman, 1971).
If a driver or reporter line is located in a second chromosome that is balanced, it means that the flies only have one copy of the transgene. In consequence, half of the progeny will inherit the transgene while the other half the balancer. To recognize the flies that have received the transgene of interest, it is necessary to select the animals against the phenotypic marker of the balancer or for a positive marker associated with the reporter line. If the reporter line is associated with a fluorescent marker (like GFP) it will be visible under a fluorescent scope both in larvae and adults.
One unfortunate fact is that Drosophila GAL4 lines published as being specific for certain types of cells may actually include other cells that were not evaluated in original publications. This can lead to unexpected phenotypes or lethality in crosses. In Table 3A, we list drivers that do not have this problem. To double check or to test the specificity of other lines, simply cross the driver lines with UAS-CD8-GFP (Table 3B). This experiment is very useful and also illustrative for students. It shows them the distribution and morphology of the neurons that they are working with. To visualize, simply immobilize GFP expressing larvae between a microscope slide and a coverslip in a drop of water and observe under a compound microscope equipped with epi-fluorescence.
One final thing to remember is that some GAL4 and UAS lines are strong, while others are weak, depending on where a given transgene is inserted in the genome. If one does not see a predicted phenotype, then it is advisable to assess the strength of any GAL4 and/or UAS line used. Researchers who are actively using the lines are good sources of advice on this topic.
FINDING THE RIGHT FLY
To find basic information about any genetic fly line or to find out if a certain mutant or transgenic strain exists, Flybase http://flybase.org/ is a good first stop. Flybase is a central hub for Drosophila genetic research. It contains extensive information on genes, gene products, mutants, transgenics, and tools for doing genetics and molecular biology in flies. A video-based tutorial on how to use Flybase can be found here: http://www.youtube.com/watch?v=_oL8h47fOuQ&feature=plcp. This makes Flybase a powerful resource, but the sheer amount of data on this website is sometimes overwhelming. We recommend that beginners first use it simply as a way to track down different fly lines. Flybase provides links to references for each gene, so educators can use it to explore the primary literature. Another attractive feature of Flybase is that it is fully integrated with multiple stock centers worldwide. So with one search, a person can determine which fly lines are publicly available and where to get them.
Most Drosophila mutants and transgenics are available from the Bloomington Stock Center http://flystocks.bio.indiana.edu/. To order flies, register and obtain a Bloomington User Number (BUN), this can be done online by following clearly posted instructions: http://flystocks.bio.indiana.edu/Distribution/Order/how-to-order.htm. To our knowledge, anyone can register free of charge. To search for specific mutants and transgenics, use the name of the gene that has been mutated or the name of the inserted transgene. It will typically take 1–4 weeks to get the flies, depending on the destination country. There is a small processing fee for orders (∼US$5–15 per stock) and charges for shipping. Bloomington’s is able to keep costs down because 50% of its operating costs are funded by grants from the National Science Foundation and the National Institutes of Health.
When a specific fly line is not listed on Flybase or available at Bloomington’s, then one should contact researchers who have published articles using the line. It is customary in the Drosophila community to freely distribute published genetic lines. Most researchers will send flies in exchange for the costs of shipping. Be aware that it may take a little longer to receive animals this way. Individual researchers are not as organized as stock centers and although well-intentioned, they sometimes forget to respond to fly requests (present authors definitely included). Gentle email reminders that highlight the necessity of getting a given fly line in time for a prescheduled course are generally effective at galvanizing forgetful researchers into action.
Potential pitfalls, common mistakes: finding flies
Many countries place restrictions on imports of live animals, including flies. Regulations differ from country to country. In addition, many fast couriers (e.g., FEDEX) will not transport live animals over international borders. Stock centers, but not always individual laboratories, have experience and workable solutions in place for shipping internationally. We recommend always trying to find a local source of flies first.
MAINTAINING STOCKS WITHOUT A FLY FACILITY
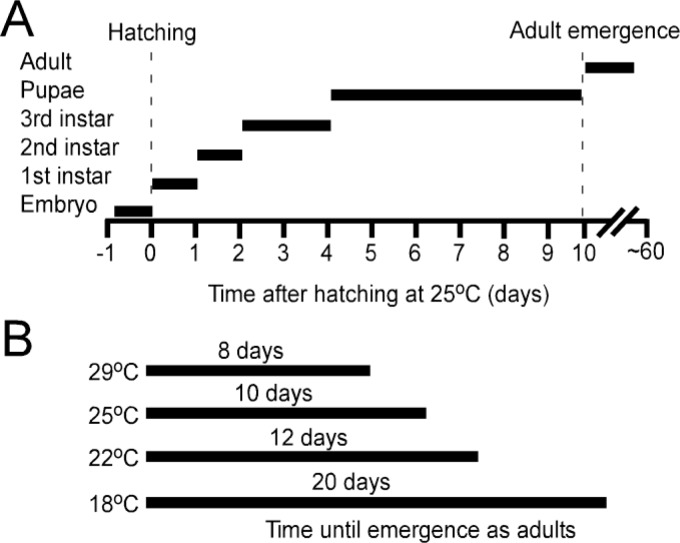
Unlike many model organisms, Drosophila can be reared with minimal infrastructure and expense. Fruit flies are quite easy to breed. Flies are holometabolous insects that go through embryonic, larval, pupal and adult life stages. Figure 1A shows time spent in different Drosophila developmental stages at 25°C. For most lines, all stages survive well at temperatures between 18°C and 29°C. However, development time varies considerably through this range (Fig. 1B). If possible, one should keep stocks at around 18°C for long-term storage, then amplify stocks and grow crosses at 25°C as needed. For simplicity, rearing and crossing can be done at room temperature without fine temperature control.
Figure 1.
Development times in Drosophila. A) Time spent in different life stages at 25°C. B) Time until adult emergence at various ambient temperatures.
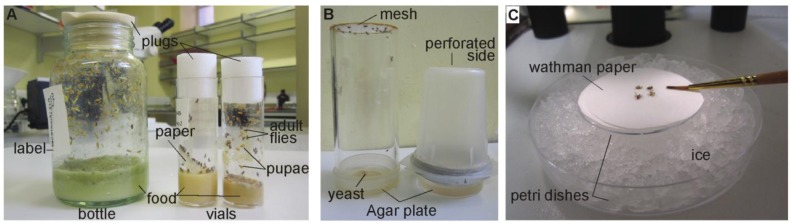
Flies can be kept in any glass or plastic receptacle containing corn-meal based food (Figure 2A). We generally keep stocks and crosses in vials of 95 mm height and 25 mm width. When we need a large population of flies, we transfer them into bottles (130 mm height). Foam or cotton plugs allow air circulation and prevent flies from escaping. After a few days of egg laying, larvae and adult flies live together in the same container. After approximately three weeks we transfer adult animals to new vials. To transfer flies, bang the bottle or vial a few times; the flies will drop to the bottom, then transfer them quickly into a new container and plug.
Figure 2.
Basic flying equipment. A) Example of bottle and vials for keeping stocks and making genetic crosses. B) Two types of laying pot. On left, a plastic cylinder that fits into a 5 cm agar lined petri dish. A plastic mesh has been glued to the top to allow air flow into the cylinder. On the right, a plastic beaker has been adapted for use as a laying pot. It has been perforated with a needle on top (5–10 holes). Holes must be made small enough to prevent adults from escaping. C) An example of the equipment necessary to collect virgins on a homemade cold block. One small petri dish is placed upside down in a larger dish filled with ice. It is essential to add a piece of paper over the petri dishes to avoid flies sticking to condensation on the dishes.
There are several recipes for fly food, including dry food that only has to be hydrated. A standard and simple recipe is shown below (One liter of this food will allow you to prepare 100 vials or 30 bottles):
| Fly Food Ingredients | Amounts (ml or g) |
|---|---|
| Water | 1000 ml |
| Yeast (dry) | 20 g |
| Agar | 10 g |
| Sucrose | 40 g |
| Corn Flour | 65 g |
| Propionic acid | 4.4 ml |
| 10 % Nipagin in 95% EtOH | 14 ml |
Cooking instructions:
Weigh all dry ingredients.
Put yeast, agar and sucrose in a large pot and add the water. Stir to remove most of the lumps.
Bring the mixture to a full boil stirring from time to time to avoid lumps.
When boiling, add the corn flour and cook while stirring for 10 minutes.
Turn off heat source.
Let cool until 70°C.
Stir in the propionic acid and Nipagin.
Dispense the medium.
Cool before plugging to avoid condensation on the walls of the tubes.
For teaching exercises with larvae, it may be convenient to make crosses in ‘laying pots’ instead of vials (Figure 2B). Laying pots allow flies to mate, then lay eggs on agar-lined petri dishes. To make them, prepare small petri dishes with the following agar recipe (1 liter for 100 5 cm-petri-dishes plates), then add a dab of hydrated bakers yeast on top of the plate where larvae feed:
| Agar ingredients | Amounts (ml or g) |
|---|---|
| Water | 1000 ml |
| Agar | 20 g |
| Sucrose | 30 g |
| 10 % Nipagin in 95% EtOH | 14 ml |
Cooking instructions:
Weigh and mix all the ingredients with water in a pan.
Heat the mixture until all the ingredients dissolve. Stir from time to time to avoid lumps. We recommend covering the pot during the cooking to avoid too much evaporation (this will affect the firmness of the agar).
Let cool until 70°C.
Stir in the Nipagin.
Dispense the medium and cover the plates.
Store at 4°C.
Inevitably, some flies will have to be killed. To prepare a ‘fly morgue’, fill a bottle with mineral oil or a mixture of mineral oil and ethanol. Put a funnel on top of the bottle and discard anaesthetized flies inside. Freeze old and unnecessary bottles and vials before discarding to avoid the release of transgenic flies into local ecologies.
Potential pitfalls, common mistakes: fly care
It is important to keep fly stocks healthy. The simplest way to do this is to transfer flies to new vials every three weeks. This will promote breeding and inhibit growth of parasites. Secondly, keep careful track of humidity. Fruit flies prefer humid environments (Drosophila translates as ‘dew loving’ in Latin), but food in vials tends to dry out and flies can slowly die of thirst. If fly food has lost its “creamy” consistency and/or pulled away from the vial itself, rehydrate immediately with a few drops of water.
Some fly lines are very unhealthy simply due to their mutations. Some mutants frequently get stuck in moist fly food. Adding dried bakers yeast and providing flies with emergent structures (bits of paper, Figure 2A) to climb and pupate away from food helps maintain sick lines.
BUILDING THE RIGHT FLY
Unless using a traditional mutant, most teaching laboratory exercises start by crossing flies to unite driver and reporter/effector lines. To do this, males and females must be differentiated. Furthermore, since females possess spermathecae, where sperm of several males can be stored for up to two weeks (Gilbert et al., 1981), selecting virgin females is crucial. To collect virgins with certainty, females have to be separated very soon after eclosing.
Anesthetizing flies
The first step of any fly selection is anesthetization. In a dedicated fly facility, there are porous mats connected to a CO2 source. When adults are put on the mat, they fall unconscious immediately. Flies can also be exposed to ether fumes or a commercial anaesthetic such as ‘flynap’ (http://www.carolina.com/product/flynap+anesthetic+kit.do); however, the simplest and least toxic (for flies and humans) alternative to CO2 is cooling. This technique requires only ice, a freezer, petri dishes and paper. First, transfer flies to a new vial with or without food (cold-anesthetized flies may fall and stick to used, mushy food). Then, transfer the vial to a freezer at −20°C. Wait until all flies fall to the bottom of the vial, but flies will start to die if held at −20°C for longer than 10 minutes. Meanwhile, fill a petri dish with ice, then cover with a second upside down petri dish and a piece of paper. This provides a cold-bottomed platform free of condensation. Place anesthetized flies on top of the paper and use a brush to gently select for appropriate genotypes. Individual flies will remain viable on this type of cold platform for 10–15 minutes.
Determining sex
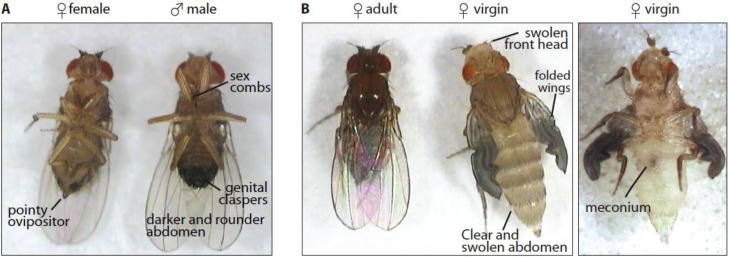
Male flies can be easily differentiated from females by examining the abdomen. The male abdomen is darker than that of females, and in males, genital claspers are clearly visible at the tip of the abdomen (Figure 3) (Greenspan, 2004).
Figure 3.
Determining gender in fruit flies. A) Males have darker and rounder abdomen than females. The females have a pointy ovipositor at the end of the abdomen. The sexual combs located in their first pair of legs are difficult to see but can be used. B) Selecting virgins by morphological markers. Newly eclosed flies will have their wings folded, their abdomen and head swollen and not yet colored. From the ventral view they will present a dark spot in their abdomen, the meconium. The most reliable markers are the wings and the meconium.
Selecting virgins
The easiest way to collect virgins is to remove all flies from a vial (check carefully that they are no flies on the food), wait five hours, then collect all newly eclosed females. Since newly eclosed females do not mate for approximately eight hours after eclosion, there is an extremely high probability that all females collected will be virgin. It is also possible to evaluate the youth and therefore virginity of anesthetized flies by morphological markers (Figure 3B).
Newly eclosed flies do not have fully inflated wings. In addition, their abdomens and heads are swollen and white. A dark spot is also visible on the ventral side of the abdomen, the meconium. By far, the most reliable markers are the wings and the meconium. If either of those can be observed, the flies are virgins.
As a side note, the cold anesthetization system mentioned above can also be used to make virgins from non-virgins. Most stored sperm in female flies is killed by 20°C for 10 minutes. This is a much less reliable method for selecting virgins (especially with females that have been mating for several days), but it can be used when large numbers of virgins are needed quickly. We advise leaving cold-virginized females in vials for 3–4 days, and then checking for the absence of larval progeny in the food before using in genetic crosses.
All sexual dimorphisms and virgin features described above are quite easy to observe under a dissecting stereomicroscope. A standard magnifying glass can also be used.
Crossing flies
Once appropriate virgins and males have been selected, we put them together in a fresh vial, bottle or laying pot with food. Crossing between 10–30 virgins and about 5–30 males is ideal for laying pots and bottles. Once the cross is started, transfer the flies to a new bottle every three days and replace the plates in the laying pot every day. To work with adult progeny grown in vials, we recommend setting up several crosses of three virgins and three males and then changing all vials every three days. This high transfer frequency prevents over-crowding and ensures a constant supply of animals.
Potential pitfalls, common mistakes: Building the right fly
One common problem doing genetic crosses in flies is the ‘stray-non-virgin.’ If just one female is not a virgin in a cross, then not all progeny will carry the driver and reporter line. To be sure of virginity, females can be selected and then housed in vials in groups of three or four for at least three days. At that point, lack of larval progeny in a vial confirms that the females in that vial are virgins.
The flies from the initial cross can be transferred to as many vial or bottles as needed, for as long as the flies are fertile (approximately one month). It is important to remember that crosses are designed to bring together two transgenes in just the first generation of progeny. Because the progeny are heterozygous for each one of the transgenes, it is not possible to keep these two transgenes stably in all the further progeny by simply breeding together the progeny of the initial cross. There are ways to use endogenous recombination events to create stable lines that have GAL4 and UAS permanently in all progeny. Details on strategies for doing this can be found in Greenspan (2004), but are beyond the scope of this article.
PREPARING FOR A TEACHING SESSION
We have published several detailed descriptions of laboratory exercises using specific genotypes. When preparing to do laboratory exercises with flies it is essential to first determine what experimental and control genotypes are needed. When examining phenotypes in mutants, the best controls are wild-type animals in the same genetic background as the mutants. For example, if a mutant was created in a Canton S background, then the appropriate controls are Canton S flies.
When examining phenotypes in flies with GAL4 and UAS combined, the most rigorous approach is to also examine the same GAL4 and UAS lines in heterozygosis with wild-type chromosomes. This way one can be sure that any given phenotype in flies with both GAL4 and UAS is not simply due to either of the constructs having an effect on their own. For simplicity, in teaching labs we usually use only one parent line as a control, but only after we have double-checked and are sure that both control lines give equivalent results. Next, estimate how many animals are needed and how long it will take to generate them. We regularly teach in the order of 15 students to do behavior and electrophysiology experiments with ChR2 expressing 3rd instar larvae in single afternoon sessions (Pulver et al., 2011b). For these sessions, we collect virgins intensively for one week, then start control lines and crosses in five vials (ideally each has 10 females and 5 males) one week ahead of the session. We maintain the flies at room temperature in a dark area because a co-factor (all-trans retinal) added to the food is photosensitive. Every day for the first four days, we transfer adults into new vials. All vials are retained, resulting in many animals at staggered larval stages. An even better solution is to set up two laying pots (experimental and control) for each group of two to three students. For this, we recommend following the same schedule as above: simply replace the agar lined petri dishes everyday instead of flipping adults into new vials.
We have also used adult flies in teaching exercises. When we need adults, we typically collect virgins for one week, then start crosses in vials three weeks ahead of teaching sessions. We maintain crosses at either room temperature or 25°C. For the first week, we flip the parent lines into new vials every other day. On the day of the lab session, we give each group of students their own vials containing both experimental animals and controls.
Potential pitfalls, common mistakes: Preparing for a teaching session
In our experience, researchers always underestimate how many animals are needed for teaching. We often run short of animals in exercises even when we explicitly try to generate large numbers of animals. We recommend always erring on the side of having extra animals.
One easy mistake to make when working with larvae is to forget to continually transfer parents to new vials during the first week of a cross. This results in a large number of third instar larvae after three to four days, and then very few animals afterwards. This is fine if precisely timed with a student session, but often it is wise to have a margin of error for unforeseen events. Staggering egg laying over several days in the week preceding session provides is a simple way to guarantee large numbers of larvae.
Often we find ourselves rushing to make crosses and generate appropriate genotypes at the last minute (despite all efforts to the contrary). We then make use of the fact that development times can be sped up at 29°C. In most cases, the increase in temperature poses no problems, and we encourage educators to maintain an incubator at 29°C just in case they run short on time. Just remember two things: 1) Temperatures above 30°C are unhealthly for flies and 2) Flies with dTRPA1 and/or shibireTS expression cannot be raised at temperatures above 24°C.
CONCLUSIONS
Drosophila as a model organism has many attributes that make it optimal for use in neuroscience teaching laboratories. Here we have outlined some of the advantages of using Drosophila in neuroscience education. We have also identified practical hurdles for those who want to incorporate Drosophila neurogenetics into their teaching, and provided simple, inexpensive strategies for overcoming common problems. Most importantly, we hope that the present work provides educators with a platform from which they can launch their own independent explorations into the field of Drosophila neurogenetics.
REFERENCES
- Adams MD, Celniker SE, Holt RA, Evans CA, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Ainsley JA, Pettus JM, Bosenko D, Gerstein CE, Zinkevich N, Anderson MG, Adams CM, Welsh MJ, Johnson WA. Enhanced locomotion caused by loss of the Drosophila DEG/ENaC protein Pickpocket1. Curr Biol. 2003;13:1557–1563. doi: 10.1016/s0960-9822(03)00596-7. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Golic K, Hawley R. Drosophila: a laboratory handbook. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2005. [Google Scholar]
- Berni J, Mudai A, Pulver SR. Using the warmth-gated ion channel TRPA1 to study the neural basis of behavior in Drosophila. J Undergrad Neurosci Ed. 2010;9:A5–A14. [PMC free article] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Depetris-Chauvin A, Berni J, Aranovich EJ, Muraro NI, Beckwith EJ, Ceriani MF. Adult-specific electrical silencing of pacemaker neurons uncouples molecular clock from circadian outputs. Curr Biol. 2011;21:1783–1793. doi: 10.1016/j.cub.2011.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diegelmann S, Zars M, Zars T. Genetic dissociation of acquisition and memory strength in the heat-box spatial learning paradigm in Drosophila. Learn Mem. 2006;13:72–83. doi: 10.1101/lm.45506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins T, Ganetzky B, Wu CF. A Drosophila mutation that eliminates a calcium-dependent potassium current in Proc Natl Acad Sci U S A. 1986;83:8415–8419. doi: 10.1073/pnas.83.21.8415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gailey DA, Hall JC. Behavior and cytogenetics of fruitless in Drosophila melanogaster: different courtship defects caused by separate, closely linked lesions. Genetics. 1989;121:773–785. doi: 10.1093/genetics/121.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganetzky B, Wu CF. Neurogenetics of membrane excitability in Drosophila. Annu Rev Genet. 1986;20:13–44. doi: 10.1146/annurev.ge.20.120186.000305. [DOI] [PubMed] [Google Scholar]
- Gilbert DG, Richmond RC, Sheehan KB. Studeis of esterase 6 in Drosophila melanogaster. VII Remating times of females inseminated by males having active or null alleles. Behav Gen. 1981;11(3):195–208. doi: 10.1007/BF01065458. [DOI] [PubMed] [Google Scholar]
- Gong Z, Son W, Chung YD, Kim J, Shin DW, McClung CA, Lee Y, Lee HW, Chang DJ, Kaang BK, Cho H, Oh U, Hirsh J, Kernan MJ, Kim C. Two interdependent TRPV channel subunits, inactive and Nanchung, mediate hearing in Drosophila. J Neurosci. 2004;24:9059–9066. doi: 10.1523/JNEUROSCI.1645-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan RJ. Fly pushing: the theory and practice of Drosophila genetics. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2004. [Google Scholar]
- Hall JC. Molecular neurogenetics of biological rhythms. J Neurogenet. 1998;12:115–181. doi: 10.3109/01677069809108556. [DOI] [PubMed] [Google Scholar]
- Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla TJ, Garrity PA. An internal thermal sensor controlling temperature preference in Drosophila. Nature. 2008;454:217–220. doi: 10.1038/nature07001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison ST, Mackay TF, Anholt RR. Understanding the neurogenetics of sleep: progress from Drosophila. Trends Genet. 2009;25:262–269. doi: 10.1016/j.tig.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochman B. Analysis of chromosome 4 in Drosophila melanogaster. II. Ethyl methanesulfonate induced lethals. Genetics. 1971;67:235–252. doi: 10.1093/genetics/67.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstein NJ, Pulver SR, Griffith LC. Channelrhodopsin2 mediated stimulation of synaptic potentials at Drosophila neuromuscular junctions. J Vis Exp. 2009;25 doi: 10.3791/1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CL, Thomas JB. A sensory feedback circuit coordinates muscle activity in Drosophila. Mol Cell Neurosci. 2007;35:383–396. doi: 10.1016/j.mcn.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chung YD, Park DY, Choi S, Shin DW, Soh H, Lee HW, Son W, Yim J, Park CS, Kernan MJ, Kim C. A TRPV family ion channel required for hearing in Drosophila. Nature. 2003;424:81–84. doi: 10.1038/nature01733. [DOI] [PubMed] [Google Scholar]
- Kitamoto T. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J Neurobiol. 2001;47:81–92. doi: 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krans JL, Rivlin PK, Hoy RR. Demonstrating the Temperature Sensitivity of Synaptic Transmission in a Drosophila Mutant. J Undergrad Neurosci Ed. 2005;4:A27–A33. [PMC free article] [PubMed] [Google Scholar]
- Kurkulos M, Weinberg JM, Pepling ME, Mount SM. Polyadenylylation in copia requires unusually distant upstream sequences. Proc Natl Acad Sci U S A. 1991;88:3038–3042. doi: 10.1073/pnas.88.8.3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Mackay TFC, Stone EA, Ayroles JF. The genetics of quantitative traits: challenges and prospect. Nat Rev Genet. 2009;10:565–577. doi: 10.1038/nrg2612. [DOI] [PubMed] [Google Scholar]
- Mahr A, Aberle H. The expression pattern of the Drosophila vesicular glutamate transporter: a marker protein for motoneurons and glutamatergic centers in the brain. Gene Expr Patterns. 2006;6:299–309. doi: 10.1016/j.modgep.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA, Takemura SY, Lu Z, Huang S, Gao S, Ting CY, Lee CH. From form to function: the ways to know a neuron. J Neurogenet. 2009;23:68–77. doi: 10.1080/01677060802610604. [DOI] [PubMed] [Google Scholar]
- Moore MS, DeZazzo J, Luk AY, Tully T, Singh CM, Heberlein U. Ethanol intoxication in Drosophila: genetic and pharmacological evidence for regulation by the cAMP signaling pathway. Cell. 1998;93:997–1007. doi: 10.1016/s0092-8674(00)81205-2. [DOI] [PubMed] [Google Scholar]
- Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby LM, Jackson FR. Drosophila ebony mutants have altered circadian activity rhythms but normal eclosion rhythms. J Neurogenet. 1991;7:85–101. doi: 10.3109/01677069109066213. [DOI] [PubMed] [Google Scholar]
- Pak WL, Grossfield J, Arnold KS. Mutants of the visual pathway of Drosophila melanogaster. Nature. 1970;227:518–520. doi: 10.1038/227518b0. [DOI] [PubMed] [Google Scholar]
- Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJ, Kang K, Liu X, Garrity PA, Rosbash M, Griffith LC. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron. 2008;60:672–682. doi: 10.1016/j.neuron.2008.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AM, Smart R, Strauss R, Brembs B, Kelly LE. The Drosophila black enigma: the molecular and behavioural characterization of the black1 mutant allele. Gene. 2005;351:131–142. doi: 10.1016/j.gene.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Pulver SR, Cognigni P, Denholm B, Fabre C, Gu WX, Linneweber G, Prieto-Godino L, Urbancic V, Zwart M, Miguel-Aliaga I. Why flies? Inexpensive public engagement exercises to explain the value of basic biomedical research on Drosophila melanogaster. Adv Physiol Educ. 2011a;35:384–392. doi: 10.1152/advan.00045.2011. [DOI] [PubMed] [Google Scholar]
- Pulver SR, Griffith LC. Spike integration and cellular memory in a rhythmic network from Na(+)/K(+) pump current dynamics. Nat Neurosci. 2010;13:53–59. doi: 10.1038/nn.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulver SR, Hornstein NJ, Land BL, Johnson BR. Optogenetics in the teaching laboratory: using channelrhodopsin-2 to study the neural basis of behavior and synaptic physiology in Drosophila. Adv Physiol Educ. 2011b;35:82–91. doi: 10.1152/advan.00125.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulver SR, Pashkovski SL, Hornstein NJ, Garrity PA, Griffith LC. Temporal dynamics of neuronal activation by Channelrhodopsin-2 and TRPA1 determine behavioral output in Drosophila larvae. J Neurophysiol. 2009;101:3075–3088. doi: 10.1152/jn.00071.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaume CJ, Sokolowski MB. The nature of Drosophila melanogaster. Curr Biol. 2006;16:R623–628. doi: 10.1016/j.cub.2006.07.042. [DOI] [PubMed] [Google Scholar]
- Robinow S, White K. Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J Neurobiol. 1991;22:443–461. doi: 10.1002/neu.480220503. [DOI] [PubMed] [Google Scholar]
- Rubin GM. Biological annotation of the Drosophila genome sequence. Novartis Found Symp. 2000;229:79–82. discussion 82–3. [PubMed] [Google Scholar]
- Salvaterra PM, Kitamoto T. Drosophila cholinergic neurons and processes visualized with Gal4/UAS-GFP. Brain Res Gene Expr Patterns. 2001;1:73–82. doi: 10.1016/s1567-133x(01)00011-4. [DOI] [PubMed] [Google Scholar]
- Sofer W, Tompkins L. Drosophila genetics in the classroom. Genetics. 1994;136:417–422. doi: 10.1093/genetics/136.1.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowski MB. Drosophila: genetics meets behaviour. Nat Rev Genet. 2001;2:879–890. doi: 10.1038/35098592. [DOI] [PubMed] [Google Scholar]
- Uytterhoeven V, Kuenen S, Kasprowicz J, Miskiewicz K, Verstreken P. Loss of skywalker reveals synaptic endosomes as sorting stations for synaptic vesicle proteins. Cell. 2011;145:117–132. doi: 10.1016/j.cell.2011.02.039. [DOI] [PubMed] [Google Scholar]
- Venken KJ, Simpson JH, Bellen HJ. Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron. 2011;72:202–230. doi: 10.1016/j.neuron.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villella A, Hall JC. Neurogenetics of courtship and mating in Drosophila. Adv Genet. 2008;62:67–184. doi: 10.1016/S0065-2660(08)00603-2. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Stocker RF. Molecular architecture of smell and taste in Drosophila. Annu Rev Neurosci. 2007;30:505–533. doi: 10.1146/annurev.neuro.30.051606.094306. [DOI] [PubMed] [Google Scholar]
- Wu CF, Ganetzky B, Haugland FN, Liu AX. Potassium currents in Drosophila: different components affected by mutations of two genes. Science. 1983;220:1076–1078. doi: 10.1126/science.6302847. [DOI] [PubMed] [Google Scholar]
- Zhang B, Freeman M, Waddell S. Drosophila neurobiology: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2010. [Google Scholar]