Abstract
The heterotrimeric G-protein complex in Arabidopsis thaliana consists of one α, one ß and three γ subunits. While two of the γ subunits, AGG1 and AGG2 have been shown to provide functional selectivity to the Gßγ dimer in Arabidopsis, it is unclear if such selectivity is embedded in their molecular structures or conferred by the different expression patterns observed in both subunits. In order to study the molecular basis for such selectivity we tested genetic complementation of AGG1- and AGG2 driven by the respectively swapped gene promoters. When expressed in the same tissues as AGG1, AGG2 rescues some agg1 mutant phenotypes such as the hypersensitivity to Fusarium oxysporum and D-mannitol as well as the altered levels of lateral roots, but does not rescue the early flowering phenotype. Similarly, AGG1 when expressed in the same tissues as AGG2 rescues the osmotic stress and lateral-root phenotypes observed in agg2 mutants but failed to rescue the heat-stress induction of flowering. The fact that AGG1 and AGG2 are functionally interchangeable in some pathways implies that, at least for those pathways, signaling specificity resides in the distinctive spatiotemporal expression patterns exhibited by each γ subunit. On the other hand, the lack of complementation for some phenotypes indicates that there are pathways in which signaling specificity is provided by differences in the primary AGG1 and AGG2 amino acid sequences.
Introduction
Heterotrimeric G-proteins (G-proteins), consisting of three subunits Gα, Gß and Gγ, are involved in a diverse range of vital biological processes including hormone regulation, neurotransmission, light perception and cell proliferation [1], [2], [3]. In animal systems, G-proteins mediate signaling initiated by seven transmembrane (7 TM) spanning G-protein coupled receptors (GPCRs) after activation by an external stimulus. Activation of the GPCR promotes the exchange of GDP for GTP in the Gα subunit and as a result Gα-GTP dissociates from the Gßγ dimer, allowing Gα-GTP and Gßγ to activate their respective downstream effectors. Termination of signaling occurs when GTP is hydrolyzed to GDP by the intrinsic GTPase activity of Gα and the inactive heterotrimer re-associates back at the receptor [4]. In plants, the G protein complex is self-activating [5], [6], [7], [8] and therefore does not need a GPCR [9]. Instead, many plants utilize a 7 TM Regulator of G Signaling (RGS) protein, that regulates the GTP hydrolysis reaction of the Gα subunit (Urano. unpublished).
In humans, there are 23 Gα, 5 Gß and 12 Gγ subunits, allowing a large number of different heterotrimer combinations Gαxßyγz [10], [11], [12], [13], [14]. It is generally believed that the diversity of heterotrimer combinations provides the required coupling specificity to allow signaling by over 800 GPCRs [15]. It was initially thought Gα was the only subunit active in signaling relegating the role of the Gßγ dimer to Gα inactivation and escort to the receptor. It was subsequently established that different Gßγ dimers interact with specific effectors, proving that the Gßγ dimer contributes to both active signaling and heterotrimer specificity [16], [17]. Signaling specificity by the different Gßγ dimers can be a consequence of the dimer’s intrinsic structural properties but it can also be dictated by the tissue specificity of their expression patterns [18], [19], [20], [21], [22].
Plant G-proteins are also involved in numerous signaling processes including interactions with rhizobia [23], defense against pathogens [24], [25], [26], [27], [28], [29], [30], morphological development and growth [31], [32], [33], cell proliferation [34], [35], ion-channel regulation [36], stomatal control [37], [38], light perception [32], [39], [40], [41], [42], abiotic stress [43], [44], [45], [46] and hormonal responses including glucose, brassinosteroid, abscisic acid and jasmonate [47], [48], [49], [50], [51], [52], [53]. G-proteins have also been linked to yield related quantitative trait loci in important crops such as rice [54], [55]. Unlike animals, where G-proteins underwent extensive subunit duplication and divergence of function, the plant G-protein repertoire is much simpler. Only one Gα (GPA1), one Gß (AGB1) and three Gγ subunits (AGG1, AGG2 and AGG3) are encoded in the Arabidopsis thaliana genome [54], [56], [57], [58], [59].
Among the three Arabidopsis Gγ subunits, AGG1 (Gγ1) and AGG2 (Gγ2) strongly resemble the canonical mammalian Gγ [58], [59], [60]. AGG3 (Gγ3) on the other hand is quite different from AGG1 and AGG2 being more than twice the size (253 a.a.) and exhibiting a modular structure with a γ-like domain at its N terminus, followed possibly by a transmembrane domain and a long cysteine rich C-terminal region [29], [54], [58], [59]. Despite AGG1 and AGG2 sharing extensive sequence conservation (48% amino acid identity and 65% similarity considering conservative substitutions), Trusov et al. [61] reported that Arabidopsis agg1 and agg2 mutants exhibit distinct phenotypes, prompting the hypothesis that the different Gγ subunits confer specificity to the Gßγ dimer in plants. An important and still unanswered question is the molecular basis for such specificity. In normal circumstances it would be fair to assume that the basis for the specificity resides in the molecular structure of the two Gγ subunits (and ultimately in their amino acid composition). Nevertheless, promoter studies showed that the two closely related AGG1 and AGG2 subunits have tissue and developmental expression patterns that rarely overlap [61], [62], raising the possibility that the basis for the specificity could be either partially or totally provided by their mutually-exclusive expression patterns. In leaves, AGG1 expression was restricted to veins, while AGG2 expression was observed primarily in guard cells. In roots AGG1 expression was restricted to the stele while AGG2 expression was excluded from the stele yet found in the cortex and epidermis. This would provide a transcriptional means to control the level of the Gßγ subunit on the plasma membrane and therefore the capacity for signal output.
A number of hypothetical scenarios can be envisaged including (i) Gßγ1 and Gßγ2 may activate specific sets of effectors, therefore mediating different signaling processes; (ii) Gßγ1 and Gßγ2 may activate common sets of effectors, with their presence or absence in an individual tissue dictating their involvement in signaling and (iii) an intermediate case in which spatio-temporal separation of expression and a degree of effector specificity contribute to the final response.
In order to determine if the specificity observed for Gγ function resides in transcriptional control of the AGG1 and AGG2 genes we swapped gene promoters and tested for genetic complementation in the respective Arabidopsis agg1 and agg2 mutants. Phenotypic analyses revealed that AGG1 and AGG2 are able to complement some but not all mutant phenotypes, indicating the existence of both transcriptional spatial and temporal regulation of the Gßγ activity but also suggesting that some signaling specificity information resides in the primary amino acid composition of both subunits.
Materials and Methods
Plant Material
The agg1-1c mutant allele of AGG1 (At3g63420) and the agg2-1 mutant allele of AGG2 (At3g22942) in the Col-0 background, were described previously [61].
To generate the agg1-1c AGG1:AGG1 complementation lines, an AGG1 fragment from ∼2 kb 5′ of the start codon to ∼0.8 kb 3′ of the stop codon was amplified from wild-type genomic DNA using Elongase (Invitrogen). The primers used were: 5′-GAAAGAGAGGTCTGGTTAGCTATGC-3′ and 5′-GAAGGAGCTCTAATGAGGTCATCAAC-3′. The resulting 3.8 kb fragment was cloned into the pGEM-T Easy vector (Promega) and transferred using EcoRI sites into the binary vector pCAMBIA1380. Subsequently, the construct was transformed into Arabidopsis agg1-1c plants by Agrobacterium tumefaciens–mediated transformation [63]. Primary transformants were selected with hygromycin B. At least ten independent homozygous transgenic lines were obtained.
The agg1-1c AGG1:AGG2 transgenic Arabidopsis were generated as follows. Elongase (Invitrogen) was used to amplify sequences from wild-type Arabidopsis genomic DNA. The following primers were used: for the AGG1 promoter region, 5′-GGGGTACCGCGGCCGCTGATGAGACACACAATCAAAC-3′ and 5′-GGCTCGAGTCTCGCTAGCAGGTCGCA-3′; for the coding region and terminator of AGG2, 5′-GGCTCGAGTGATGGAAGCGGGTAGCTC-3′ and 5′-GCGGCCGCGTTTTGGTTCATGATGTTTCCT-3′. Restriction sites (underlined) were incorporated on the ends of fragment for cloning purposes. The PCR products were ligated into pGEM-T Easy vector (Promega). The AGG2 fragment was transferred into the pBluescript SK+ vector using XhoI and NotI restriction sites. AGG1 promoter fragment was inserted in front of AGG2 fragment in the pBluescript SK+ using KpnI and XhoI restriction sites. PCR was then performed on the AGG1 promoter-AGG2-pBluescript SK+ construct with the following primers: 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGTAAAACGACGGCCAG-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCAGGAAACAGCTATGAC-3′, in order to flank AGG1p::AGG2 fragment with attB1 and attB2 gateway recombination sequences. The attB1-AGG1p::AGG2-attB2 PCR product was recombined into pDONR™207 using Gateway® BP Clonase® (Invitrogen). A reaction was performed with Gateway® LR Clonase® system to clone AGG1p::AGG2 into the binary vector pMDC99. The construct was again transformed into agg1-1c by the floral dip method. Transformants were selected based on resistance to hygromycin B.
agg2-1 AGG2p::AGG2 transgenic lines were generated by transformation of a 4.7 kb AGG2 fragment, from1.6 kb 5′ of the start codon to 1.8 kb 3′ of the stop codon, into agg2-1. The primers used to amplify the 4.7 kb fragment were 5′-GGTACCGCGGCCGCATTGCCAGCCGATTTTTGCC-3′ and 5′-GCGGCCGCGTTTTGGTTCATGATGTTTCCT-3′. The resulting fragment was cloned into pGEM-T Easy (Promega),and transferred to the binary vector pUQC477 using terminal NotI restriction sites. The final construct was transformed into agg2-1 by floral dip, and transformants were selected using BASTA as described elsewhere [64].
The agg2-1 AGG2p::AGG1 mutant lines were generated as follows. The AGG2 promoter was amplified using the primers 5′-GGTACCGCGGCCGCATTGCCAGCCGATTTTTGCC-3′ and 5′-GGCTCGAGAAATTTCTCGAATTCAACCCTC-3′. The AGG1 coding region and terminator were amplified with 5′-GGCTCGAGGGATGCGAGAGGAAACTGT-3′ and 5′-GGGCGGCCGCTTTAACGGCTAACTTACTTATC-3′. The resulting two fragments were each ligated into pGEMT-Easy (Promega). The AGG1 coding region and terminator fragment was then transferred into pBluescript SK+ vector using XhoI and NotI restriction sites. The AGG2 promoter fragment was inserted in front of the AGG1 fragment using KpnI and XhoI restriction sites. The AGG2p::AGG1 fragment was then transferred into the pUQC227 vector using terminal NotI restriction sites. The final construct was transformed into agg2-1 by the floral dip method, and transformants were selected using BASTA.
Quantitative Real-time RT-PCR Analysis
Total RNA was extracted from two-week-old seedlings as described previously [65]. First strand cDNA synthesis was conducted using the SuperScript III RT kit (Invitrogen) according to the manufacturer’s instructions. qRT-PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems) and the 7900 HT Sequence Detection System (Applied Biosystems). The following primer pairs, designed using Primer Express software (Applied Biosystem), were used in the qRT-PCR: AGG1 5′UTR, forward, 5′-GAGAGAGACTTCGACGACAATTCA-3′, reverse, 5′-CTCGCTAGCAGGTCGCAGAT-3′; AGG1 exon2, forward, 5′- GGAGGTCGAGAACACAGATATTGTATC-3′, AGG1 exon3, reverse, 5′- CAACAGAGGATCGGGTCCTTT-3′; AGG1/AGG2, forward, 5′- TGCGACCTGCTAGCGAGACT-3′, reverse, 5′- CCTGTGTTTGCCTCTTGTATCAAC-3′; AGG2 5′ UTR, forward, 5′- CCCCAACTCATAACTTTGAATTTTCTA-3′, reverse, 5′- GGATTCAGAATCAAACAGATCTTGAGA-3′; AGG2 exon3, forward, 5′- GCATCAGCATCCTGCAAAGA-3′, reverse, 5′- GGACCTGTTGTTTCGGGAAGA-3′; AGG2/AGG1, forward, 5′- GTTTCGATTTTTATTTTGAGGGTTGA-3′, reverse, 5′- CCCGCCGTGAGAAACAGA-3′. The previously validated β-ACTIN2, β-ACTIN7 and β-ACTIN8 were used as reference genes to quantify relative expression [66]. Gene expression analysis was performed using SDS Version 2.2.2 software (Applied Biosystems). The results were average values from three independently prepared RNA samples.
Mutant Characterization
Plants were grown under a long-day conditions (16 h light/8 h dark) with cool white fluorescent bulbs at approximately 100 µmol m−2 s−1 and 22°C unless stated otherwise. All statistical analysis was performed with the GraphPad Prism version 5.04 for Windows (GraphPad Software, San Diego California USA, www.graphpad.com). All experiments were repeated at least three times with similar results.
Fusarium Culture Preparation and Inoculation
Fusarium oxysporum f. sp. conglutinans (BRIP 5176, Department of Primary Industries, http://www.dpi.vic.gov.au, Queensland, Australia) culture preparation and root inoculations were performed as previously described [67] with modifications. Briefly, F. oxysporum was grown for approximately 1 week on one-half-strength potato dextrose agar plates at 25°C. Two plugs were cut from these plates under sterile conditions and placed into a flask containing 250 mL of potato dextrose broth. The flask containing the inoculum was then grown for approximately 3 days at 28°C with shaking at 110 rpm. The culture was filtered through Miracloth (Calbiochem, San Diego) and quantified with a hemocytometer. The suspension was diluted with sterile distilled water to a concentration of 0.5 x 106 spores mL–1. Two-week-old plants which had been grown on steam sterilized soil were used for the assay. Before inoculation, the plants were carefully removed from the soil, the roots rinsed with water and dipped for at least 30 seconds into the fungal inoculum. The inoculated plants were replanted into fresh soil and grown at 27°C. Twenty plants from each of the wild-type and mutant lines were inoculated in two independent inoculation experiments. The degree of infection were scored as symptoms appeared and progressed in the window of days 7 to 12 post-inoculation, by counting the number of yellow and dead leaves as a percentage of the total number of leaves [66].
Flowering time Analysis
Seeds were sown on soil and stratified for 48 hours at 4°C in darkness. Thirty plants per line were grown at 22°C. Where flowering induction is required, seedlings were initially grown at 22°C for two weeks before being transferred to a 29°C growth room. Flowering time was determined by the age of the plant in days when the inflorescence reached approximately 1 cm in height from the rosette.
Plate Assays
All plates contained 0.5X MS basal salts (PhytoTechnology Laboratories), 0.8% phytagel (Sigma), and varying amount of sucrose [68]. No sucrose was added to plates used for germination assays. Seeds were dry sterilized by 4 hour incubation in a chamber filled with chlorine gas. After sowing onto solid media, all seeds were stratified for 48 h at 4°C in darkness. 6% w/v D-mannitol was added to the plates used for the osmotic stress germination assay. Germination was determined as an obvious protrusion of the radicle. For root assays, seedlings were grown at 26°C on vertical plates supplemented with 1% sucrose for 14 days, and the number of lateral roots per seedling was counted using a dissecting microscope. For adventitious root induction, media containing 3% sucrose was autoclaved. Once cooled to 55°C, NAA was added to a final concentration of 1 µM, from a stock solution of 10 mM. Hypocotyls from 5-day-old etiolated seedlings were aseptically excised and transferred onto the NAA supplemented media. Adventitious root development on the plate was photographed after 10 days incubation at 26°C.
Results
Complementation Constructs and Transgene Expression
To determine whether Gγ1 and Gγ2 are functionally interchangeable we designed a cross-complementation strategy. Since the expression profiles of AGG1 and AGG2 are non-overlapping we carefully designed the complementation constructs trying to reproduce as much as possible the genomic environment for each of the two genes. In order to complement the agg1 mutation, we fused 1 kb of the promoter region of AGG1 (including the 5′ UTR) to an AGG2 genomic fragment containing the entire gene, starting at the start ATG codon and ending 1.8 kb downstream of the 3′ UTR to include the terminator sequences (AGG1p::AGG2; Fig. 1A). To complement the agg2 mutation we fused 2 kb of the AGG2 promoter region, including the AGG2 5′ UTR, to a genomic fragment containing the entire AGG1 gene from the translational start codon and extending 1.8 kb downstream of the 3′ UTR (AGG2p::AGG1; Fig. 1C). As positive controls we prepared constructs containing the entire AGG1 and AGG2 genes, including promoter and terminator regions (AGG1p::AGG1 and AGG2p::AGG2 respectively in Fig. 1A and 1C).
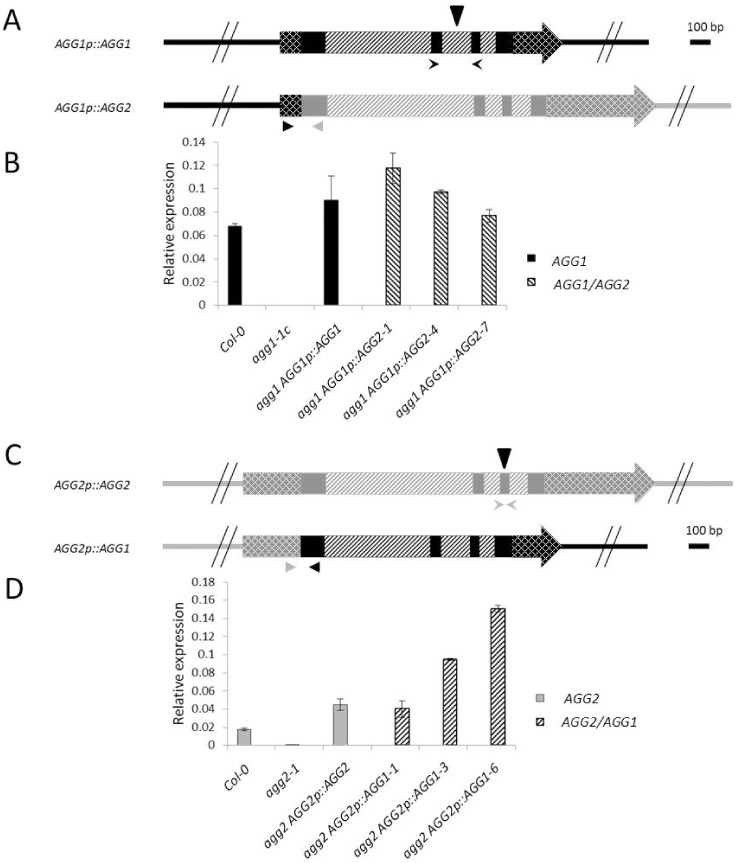
Figure 1. AGG1 and AGG2 complementation constructs and expression analysis in transgenic lines.
(A) AGG1 complementation constructs. The AGG1p::AGG1 construct contains the entire AGG1 gene starting 1 kb upstream of the 3′ untranslated region (UTR) and extending 1.8 kb downstream of the 3′ UTR region to include the native AGG1 termination sequence. The AGG1p::AGG2 construct contains the AGG1 promoter region as well as the AGG1 5′ UTR, fused to an AGG2 genomic fragment, starting at the ATG start codon and including 1.8 kb downstream of the 3′ UTR to include the native AGG2 termination sequence. Expression of the AGG1p::AGG2 construct will result in a ‘hybrid’ mRNA molecule containing the 5′UTR from AGG1 fused to the coding region from AGG2. (B) Relative mRNA expression levels. Solid black bars show the relative abundance of the AGG1 mRNA in WT (Col-0) and agg1-1c mutant plants. Self-complementation of the agg1-1c mutant with the AGG1p::AGG1 construct results in similar levels of AGG1 mRNA to those observed in WT plants. Cross-complementation of the agg1-1c mutant with the AGG1p::AGG2 construct results in similar levels of expression of the AGG2hybrid mRNA (dashed bars) to those observed for AGG1 mRNA in WT and self-complementation lines. (C) AGG2 complementation constructs. The AGG2p::AGG2 construct contains the entire AGG2 gene starting 2 kb upstream of the 3′ untranslated region (UTR) and extending 1.8 kb downstream of the 3′ UTR region to include the native AGG2 termination sequence. The AGG2p::AGG1 construct contains the AGG2 promoter region as well as the AGG2 5′ UTR, fused to an AGG1 genomic fragment, starting at the ATG start codon and including 1.8 kb downstream of the 3′ UTR to include the native AGG1 termination sequence. Expression of the AGG2p::AGG1 construct will result in a ‘hybrid’ mRNA molecule containing the 5′UTR from AGG2 fused to the coding region from AGG1. (D) Relative mRNA expression levels. Solid grey bars show the relative abundance of the AGG2 mRNA in WT (Col-0) and agg2-1 mutant plants. Self-complementation of the agg2-1 mutant with the AGG2p::AGG2 construct results in similar or higher levels of AGG2 mRNA to those observed in WT plants. Cross-complementation of the agg2-1 mutant with the AGG2p::AGG1 construct results in equal or higher levels of expression of the AGG1 hybrid mRNA (dashed bars) to those observed for AGG2 mRNA in WT and self-complementation lines. In (A) and (C), AGG1 genomic sequences are represented in black while AGG2 sequences are in grey. Regions upstream of the 5′UTRs (promoters) and downstream of the 3′ UTRs (terminators) are represented in solid lines; 5′ and 3′ UTRs are represented in dashed boxes, exons are represented in solid boxes and introns are represented in white boxes. Arrows represent the position of the primers used for real time quantitative PCR (RT-qPCR). The solid triangles show the position of the T-DNA insertions in the agg1-1c and agg2-1 mutants. In (B) and (D), transcript levels are shown as relative to ACTIN genes expression, mean ± SE of three replicas.
The AGG1p::AGG2 and AGG1p::AGG1 constructs were used to obtain transgenic Arabidopsis lines in the agg1-1c T-DNA mutant background (designated agg1 AGG1p::AGG2 and agg1 AGG1p::AGG1 respectively in this work) while the AGG2p::AGG1 and AGG2p::AGG2 were introduced into an agg2-1 background (designated agg2 AGG2p::AGG1 and agg2 AGG2p::AGG2 respectively, in this work) [61]. At least ten homozygous transgenic lines were generated from each of the constructs. Expression of the transgenes was analyzed in all transgenic lines and those with silencing or aberrant expression, as well as those with obvious insertional effects were discarded. Three lines for each of the complementation constructs and one line for each control were further characterized. The length of the promoter regions used in this study, especially in the case of AGG1, was chosen to maximize the length of the upstream region for each gene without including the full coding region of the neighboring genes which may lead to unwanted ectopic effects. The ultimate proof that we captured the entire promoter is the observation that the promoter segments chosen were able to drive genetic complementation of the cognate coding sequence. For example, the chosen AGG1 promoter segment driving expression of the AGG1 coding region genetically complemented the agg1 mutant (Figure 2).
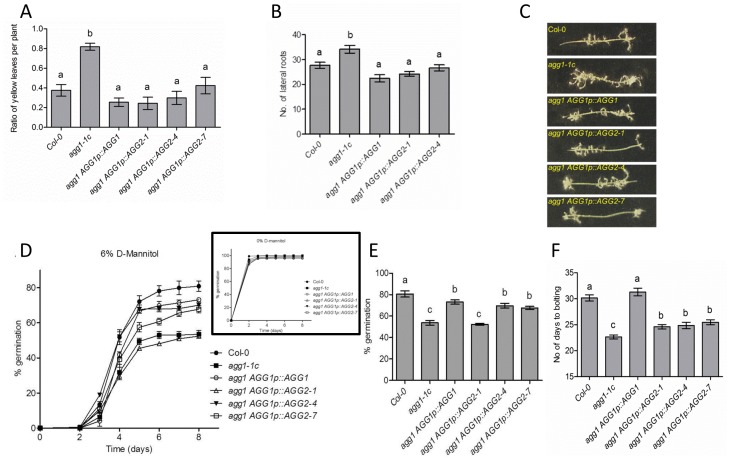
Figure 2. AGG2 complements some but not all agg1 mutant phenotypes.
(A) Sensitivity to F. oxysporum. Roots of two-week-old seedlings were inoculated with F. oxysporum spores and total number and number of chlorotic leaves were counted 9 days after inoculation for each plant. The ratio of chlorotic/total number of leaves was used to evaluate disease progression in infected plants. Bars on the graph represent average values estimated for 20 plants per each genotype. Error bars show standard errors. Letters indicate groups with statistically significant differences in disease progression (P<0.05, one-way ANOVA). (B) Total number of lateral roots was scored in two-week-old seedlings grown vertically on 0.5x MS supplemented with 1% sucrose. Bars represent average values ±SE of 15 plants per genotype. Letters indicate groups with statistically significant differences in number of lateral roots (P<0.05, one-way ANOVA). (C) Adventitious root development in excised hypocotyls was induced by supplementing media with 1µM NAA. Photos of representative hypocotyls from each tested genotype are shown. (D) Germination dynamics of wild-type, mutant and complementation lines grown on 0.5x MS supplemented with 6% D-mannitol during 8 days after stratification. Each genotype was analyzed in three replica plates with more than 100 seeds. Insert shows control germination without D-mannitol. (E) Percentage of germinated seeds at day 8 from panel (D) showing the highest difference between genotypes. Bars represent average value of three replicates (more than 100 seeds each). Error bars show standard errors. Letters indicate groups with statistically significant differences in seed germination (P<0.05, one-way ANOVA). (F) AGG2 partial rescue of the early flowering phenotype observed in agg1-1c mutants. Plants were grown under long day conditions (16 h light/8 h dark) at 23°C. Day of inflorescence appearance was recorded for at least 30 plants of each genotype. Bars represent average number of days from germination till inflorescence appearance ± SE. Letters indicate groups with statistically significant differences (P<0.05, one-way ANOVA).
To test the hypothesis that transcriptional regulation imparted at least part of the functional specificity of the Gγ subunits we expressed a hybrid messenger RNA containing the 5′UTR of AGG1 fused to the coding region and 3′UTR of AGG2. This was intentionally designed to include possible AGG1 regulatory elements present in this region, which could influence transcription or translation rates, and to account for any influence of this region in the stability of the mRNA. This strategy also provided a robust and reliable method to quantify the expression of the transgene using quantitative real time PCR with a forward primer located in the 5′UTR of AGG1 and a reverse primer located in the coding region of AGG2. This combination of primers detects only hybrid RNA molecules avoiding detection of the native AGG2 mRNA and any incomplete or aberrant AGG1 mRNA present in the agg1-1c T-DNA mutant.
A critical pre-requisite for the success of our approach is to obtain transgenic lines with transgene expression levels at least equal to those observed for the native gene in wild type plants. Fig. 1 shows the relative expression levels determined using Quantitative RT-PCR for all genotypes used in this study (wild-type, mutant and transgenic complementation lines). The agg1 AGG1p::AGG1 complementation line produced similar AGG1 transcript levels to wild-type plants while the levels of the AGG2 hybrid transcript in the AGG1p::AGG2 cross-complementation lines were also similar to wild-type AGG1 levels (Fig. 1B). As expected, no AGG1 transcript was detected in the agg1-1c mutant. Conversely, the AGG2p::AGG1 construct generates a hybrid messenger RNA containing the 5′UTR of AGG2 fused to the coding region and 3′UTR of AGG1. Analysis of the agg2 self- and cross-complementation lines showed transcript levels equal or greater to wild-type levels (Fig. 1D). In order for the cross-complementation strategy to work it was important to achieve at least the same levels of expression present in wild type plants, therefore higher expression levels in the transgenic lines, compared to wild type plants was still useful to determine if AGG1 complements agg2 mutants and vice versa.
AGG2 can Complement some but not all agg1 Mutant Phenotypes
The agg1 AGG1p::AGG1 self-complementation and agg1 AGG1p::AGG2 cross-complementation lines were characterized to determine their ability to revert several phenotypes observed in the agg1-1c mutant. In most assays, the control AGG1p::AGG1 construct was able to restore the agg1-1c mutant phenotype to wild-type, suggesting that the promoter region used in the constructs was sufficient to drive enough expression in the correct tissues to restore the function of the native AGG1 protein.
Fusarium oxysporum is a soil borne fungal pathogen which colonizes the vascular system of plants such as Arabidopsis. Symptoms of infection manifest in yellow chlorotic leaves hence disease progression can be quantified by counting the percentage of chlorotic rosette leaves per plant [69], [70]. As previously reported by Trusov et al. [61], agg1-1c mutants exhibit hypersensitivity to F. oxysporum as evidenced by the faster development of leaf chlorosis, being twice the ratio of wild-type nine days after inoculation (Fig. 2A). The hypersensitivity to F. oxysporum was restored to wild-type levels in the self-complementation line (agg1 AGG1p::AGG1) (Fig. 2A, P<0.05, one way ANOVA). Similarly, in all three cross-complementation lines assayed the ratios of chlorotic leaves were comparable to wild-type (Fig. 2A, P<0.05, one-way ANOVA), indicating that AGG2 fully rescued the agg1-1c F. oxysporum susceptibility phenotype.
It has been proven that Gβ attenuates auxin-induced cell division leading to lateral root proliferation, although it does not directly couple auxin signaling [32], [33]. Initial characterization revealed that the agg1-1c mutant also contains a larger number of lateral roots than wild-type. This fact, combined with the stele-specific expression pattern observed for AGG1 led to the hypothesis that AGG1 combines with AGB1 as a negative regulator of auxin-induced cell division with a possible role in the acropetal auxin stream [61]. Our assays confirmed that 14-days-old mutant agg1-1c seedlings produce on average ∼25% more lateral roots and root primordia than wild-type plants (Fig. 2B) (P<0.05, one way ANOVA). The self-complementation line restored the number of lateral roots to wild-type levels. Likewise, all cross-complementation lines rescued the phenotype with the numbers of lateral roots showing no significant differences with wild-type, indicating that AGG2 is able to rescue the lateral root phenotype of agg1-1c (Fig. 2B). In addition, agg1-1c hypocotyls incubated with exogenous 1-naphthaleneacetic acid (NAA) display increased adventitious root formation [61] (Fig. 2C). Our results confirm this observation and also show that either AGG1 or AGG2 can complement this phenotype returning the number of adventitious roots to wild-type levels. Therefore, for both auxin-related responses, AGG2 can successfully complement AGG1 in Arabidopsis.
A number of studies established the involvement of G proteins in germination [52], [71], [72], [73], [74], [75]. In particular, Gγ subunits have a role in the response to osmotic stress during germination with agg1-1c seeds being hypersensitive to mannitol [61]. To determine if AGG2 is able to rescue this agg1-1c mutant phenotype, relevant seed lines were sown on a single plate containing media supplemented with 6% D-mannitol. The seeds used in this assay were obtained from simultaneously grown plants to ensure synchronized germination and all the experiments were performed in triplicate. The germination percentage of each line was then scored on each plate and averaged between replicates. Our results confirmed the hypersensitivity to D-mannitol in the agg1-1c mutant (Fig. 2D and E). The difference with wild-type was most dramatic on day 8 when 80% of wild-type seeds had germinated, compared to 50% of the agg1-1c seeds. Interestingly the agg1 AGG1p::AGG1 self-complementation line did not completely restore germination levels, showing a small but statistically significant difference with wild-type plants (Fig. 2E). Two of the three cross-complementation lines showed similar germination dynamics to the self-complementation line while the third did not show any restoration of the germination levels, perhaps as a result of expression differences due to transgene positional effects (Fig. 2E).
Early flowering is another of the phenotypic characteristics shown by agg1-1c mutants [62]. When we determined the flowering times for wild-type, agg1-1c mutants, self- and cross-complementation lines, the early flowering phenotype was clearly observable in the agg1-1c mutants and was completely restored to wild-type levels in the self-complementation line (Fig. 2F). In open contrast, none of the cross-complementation lines rescued the early flowering phenotype. A small, but statistically significant increase was observed (P<0.05, one way ANOVA), but it was far from reaching the level observed in the wild-type or self-complementation line.
AGG1 can Complement some but not all agg2 Mutant Phenotypes
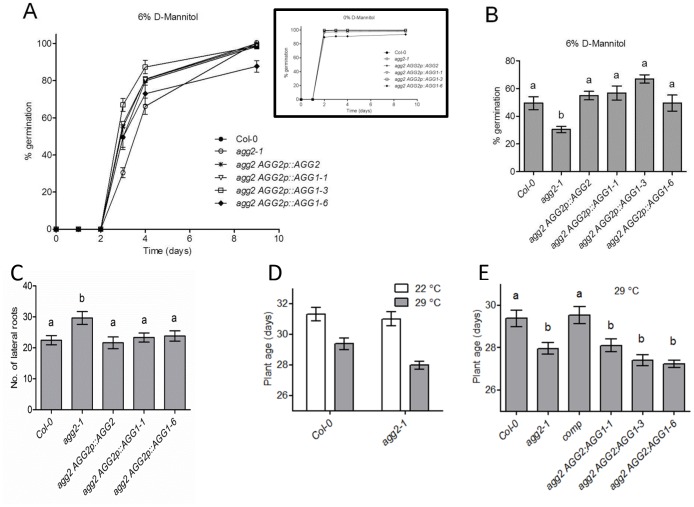
As observed for agg1-1c mutants, agg2-1 mutants are also hypersensitive to D-mannitol during germination [62]. When we tested sensitivity to D-mannitol in wild-type, agg1-1c mutants, agg2 AGG2p::AGG2 self-complementation line and three different agg2 AGG2p::AGG1 cross-complementation lines, the largest differences were observed in day 3 (Fig. 3A). On that time point, the germination percentage of agg2-1 seeds was significantly lower than wild-type (Fig. 3B; P<0.05, one way ANOVA). The self-complementation line restored germination to wild-type levels as did all three cross-complementation lines (Fig. 3B), suggesting that AGG1 can perform a similar function to AGG2 in the control of osmotic stress. This finding also supports the notion that Gγ1 and Gγ2 have a synergistic role in regulating the osmotic stress response component of germination.
Figure 3. AGG1 complements some but not all agg2 mutant phenotypes.
(A) Germination dynamics of wild-type, mutant and complementation lines on 0.5x MS supplemented with 6% D-mannitol during 9 days after stratification. Each genotype was analyzed in three replica plates with more than 100 seeds. Insert shows control germination without D-mannitol. (B) Percentage of germinated seeds at day 3 from panel (A) demonstrating highest difference between genotypes. Bars represent average value of three replicates (more than 100 seeds each). Error bars show standard errors. Letters indicate groups with statistically significant differences in seed germination (P<0.05, one-way ANOVA). (C) Total number of lateral roots was scored in two-week-old seedlings grown vertically on 0.5x MS supplemented with 1% sucrose. Bars represent average values ±SE of 15 plants per genotype. Letters indicate groups with statistically significant differences in number of lateral roots (P<0.05, one-way ANOVA). (D, E) AGG1 failed to complement the agg2-1 mutant on high temperature-induced flowering. (D) Effect of the agg2-1 mutation on high temperature-induced flowering. Col-0 and agg2-1 plants were initially grown at 22°C for two weeks and then divided into two groups: the first group was kept at 22°C, while the second group was transferred to 29°C. Day of inflorescence appearance was recorded for at least 30 plants of each genotype. Bars represent the average number of days from germination till inflorescence appearance ± SE. Letters indicate groups with statistically significant differences (P<0.05, one-way ANOVA). (E) Average number of days from germination till inflorescence appearance in at least 30 plants of each genotype induced at 29°C. Bars represent the average number of days from germination till inflorescence appearance ± SE. Letters indicate groups with statistically significant differences (P<0.05, one-way ANOVA).
Like agg1-1c mutants, agg2 mutants also produce more lateral roots than wild-type plants. Since the expression of AGG2 in roots is restricted to the cortex, it was hypothesized that this phenotype was due to defects in basipetal auxin transport or signaling [61]. We therefore investigated whether expressing AGG1 in the cortex region of agg2 mutants would restore the root phenotype. As expected, agg2-1 displayed significantly more lateral roots (including root primordia) than wild-type (P<0.05, one way ANOVA; Fig. 3C). The self- and cross-complementation lines restored the root numbers to wild-type levels indicating that AGG1 and AGG2 are functionally interchangeable in the control of lateral root formation (Fig. 3C).
The early flowering phenotype observed in agg1-1c mutants is not observed in agg2 mutants, at least when grown under long day conditions at 23°C. However, we observed early flowering in agg2-1 when grown at 29°C compared to wild-type plants (Fig. 3D). To confirm the initial observations, we simultaneously grew at least 30 plants on soil for each of the studied genotypes; i.e. wild-type, agg2-1 mutant, self-complementation line and three cross-complementation lines. Two trays were initially grown at 23°C under long day regime for each of the lines. After two weeks, one of the trays was moved to 29°C, with similar light conditions and plant age at bolting was recorded for both trays. While no differences in flowering time between wild-type and agg2-1 were apparent at 23°C, flowering time for agg2-1 mutants grown at 29°C was significantly shorter than wild-type (Fig. 3E; P<0.05, one way ANOVA). The self-complementation line restored flowering time at high temperature to wild-type levels but none of the three cross-complementation lines successfully rescued the agg2-1 mutation (Fig. 3E).
Discussion
Two proteins are functionally redundant when the absence of one can be compensated in vivo by the second one. Consequently, a null mutation in one of two redundant proteins will not result in phenotypic alteration (i.e. the mutant will have a wild-type phenotype). Two proteins are functionally interchangeable if they can perform the same biological functions, even if they do not belong to the same species. The Arabidopsis and rice Gγ subunits are obviously not redundant but can be functionally interchangeable if they can complement each other. The fact that agg1- and agg2-deficient mutants display distinct phenotypic alterations clearly established that AGG1 and AGG2 are not functionally redundant in planta and confer specificity to the Gßγ dimer in vivo [61]. The first and most obvious explanation for the observed specificity provided by AGG1 and AGG2 is that the Gßγ1 and Gßγ2 dimers activate different sets of effectors in response to diverse signals. However, with futher consideration, the non-overlapping expression patterns observed for AGG1 and AGG2 raise the possibility that the observed specificity could be due to the spatiotemporal separation observed for both proteins [61], [62]. It is therefore important to determine whether artificially expressing AGG2 in the same tissues and developmental stages in which AGG1 is normally present will rescue the agg1 mutant phenotypes. The same logic applies to the reverse. We hypothesized that, given the high degree of similarity between both proteins, AGG1 and AGG2 should be functionally interchangeable.
Our results summarized in Table 1 show that in most cases AGG1 and AGG2 are functionally interchangeable. Four out of five agg1-1c phenotypes tested were fully rescued by AGG2, demonstrating that AGG2 is able to functionally replace AGG1 in response to pathogen attack, auxin control of root development and osmotic stress during germination. Similarly, two out of three agg2-1 phenotypes were rescued by AGG1, proving that AGG1 is able to replace AGG2 in responses linked to auxin in root development and osmotic stress during germination. Collectively, the reciprocal complementation achieved by the two Gγ subunits suggests that both subunits are able to perform many of the same biochemical activities. However, both Gγ subunits are not always functionally interchangeable. AGG2 did not fully complement the agg1 early flowering phenotype, and AGG1 did not restore the thermosensitive flowering phenotype of agg2.
Table 1. Summary of Gγ1 and Gγ2 complementation studies.
| Mutant background | phenotype | Complementation |
| agg1-1c | Reduced resistance to Fusarium | Yes |
| Increased adventitious root growth under auxin induction | Yes | |
| Increased lateral root growth | Yes | |
| Increased sensitivity to osmotic stress during germination | Yes | |
| Early flowering time | Partial | |
| agg2-1 | Increased lateral root growth | Yes |
| Increased sensitivity to osmotic stress during germination | Yes | |
| Heat inducible early flowering time | No |
The fact that AGG1 and AGG2 complement each other in pathways responding to osmotic stress, auxin and defense suggests that the Gβγ1 and Gβγ2 dimers activate a number of common effectors, although the identity of those effectors is still unknown. While hypothetical, it is also possible that G-protein involvement could be somewhat indirect resulting from crosstalk among several pathways. Interestingly, there is accumulating evidence suggesting that osmotic stress, defense and auxin pathways modulate each other. Abiotic stress and wounding affect auxin responses [76], [77], [78]. While auxin is implicated in the regulation of plant defense [79], [80]. It is therefore tempting to speculate that Gβγ1 and Gβγ2 may modulate pathways responding to auxin, osmotic stress and pathogen attack at a point, or points of cross talk, using similar signaling mechanisms. This explains why a simplistic repertoire of G proteins functions in such divergent signaling processes.
Effector Activation by Gß
Alternatively, the fact that AGG1 and AGG2 are functionally interchangeable may indicate that the binding and activation of effectors resides on recognition sites predominantly or exclusively located on the surface of AGB1. Three recent studies revealed that several amino acid residues on AGB1 are essential for effector activation. The first identified acireductone dioxygenase 1 (ARD1) as an AGB1 interactor [81]. Physical interaction was proven in yeast 3-hybrid experiments, while genetic interaction was demonstrated by the rescue of the agb1-2 short hypocotyl and open apical hook phenotypes of etiolated two-day-old seedlings by ARD1 overexpression. ARD1 was shown to modulate cell division to control hypocotyl length and AGB1 was able to stimulate ARD1 enzymatic activity in vitro. The ability to stimulate ARD1 activity was abolished by several point mutations in AGB1, either single W109, double E248/R25 or triple Q120/T188/R235, suggesting that these residues are essential for ARD1 stimulation. This study proves that AGB1 contains key contact residues for some effectors, such as ARD1.
In the second study, site directed mutagenesis of Arabidopsis AGB1 and the ability of the different mutations to rescue agb1 phenotypes was tested [82]. Substitution of T65 for alanine rendered AGB1 unable to complement the hypersensitivity of the agb1 mutant to D-mannitol during germination. In addition, mutation at D250 failed to restore lateral root numbers in the agb1 mutant to wild-type levels. These observations highlight the importance of individual AGB1 residues in the activation of the effectors involved in osmotic response and lateral root formation. Our results showing that AGG1 and AGG2 are both able to restore D-mannitol sensitivity at germination and lateral root numbers are consistent with the above observations and may indicate that the effectors involved in these two responses form a direct contact with AGB1 residues for activation independently of the AGG subunit attached to AGB1 in the Gßγ dimer.
In a third study, a comparative approach was used to identify a set of residues on the AGB1 surface implicated in protein-protein interfaces [83]. The assumption was that these residues are critical in specific AGB1-effector contacts. Mutation of these residues in combination with genetic complementation assays enabled dissection of the AGB1 protein surface for a variety of ABG1-mediated physiologies (developmental, hormone responses, pathogen defense, and photosynthesis). Interestingly, residues R25 and E248 lie along the AGG-binding tract. Unfortunately, the AGG1-specific differences in flowering reported here were not tested in that study.
Gγ1 and Gγ2 are not Functionally Interchangeable in the Control of Flowering Time
Our results showed that AGG2 failed to complement the early flowering phenotype of agg1-1c, while AGG1 was unable to complement the thermo-sensitive flowering phenotype of agg2-1. Although both are flowering time phenotypes, our results suggest that Gßγ1 and Gßγ2 act in separate signaling pathways suggesting that they signal to different downstream effectors and are therefore not functionally interchangeable.
Flowering is a complex process whereby plants go through a transition between vegetative and reproductive phases and is influenced by many environmental factors including photoperiod, temperature, humidity and nutrient availability [84]. Endogenous factors such as carbohydrate reserves and genetic make-up also play a role during the transition phase [85], [86], [87]. Without a comprehensive study of G protein involvement in flowering induction, it is dangerous to speculate as to the specific roles of AGG1 and AGG2 with any confidence. Although speculative, it is interesting to note that G-proteins are implicated in modulating responses to gibberellins (GA) and brassinosteroids (BR), both of which promote flowering [88], [89], [90], [91]. It was recently suggested that G-proteins mediate the cross talk between auxin and BR signaling [91]. AGG1 is clearly implicated in auxin signaling as evidenced by the auxin sensitive traits of agg1-1c, opening the door to its involvement in cross talk with BR, therefore having an effect on flowering time through BR-mediated inhibition of FLC the potent flowering suppressor [89]. On the other hand, thermal induction of flowering is dependent on GA, suggesting that the thermosensitive flowering phenotype of agg2-1 may be due to a role of Gβγ2 in GA signaling [92].
Differential post-translational modification may contribute to the selective functions of AGG1 and AGG2 as has been proven in Cdc42, a GTPase with an important role in the regulation of cell polarity and the actin cytoskeleton [93]. Both AGG1 and AGG2 undergo prenylation, but AGG2 undergoes additional S-acylation, most probably by addition of a palmitoyl group. This second lipid modification was suggested to be the reason AGG2 is able to localize to the membrane more efficiently than AGG1 [94], [95]. Differential membrane affinity plays a major role in mammalian Gγ specialization [96]. It is possible palmitoylation of AGG2 could provide a defining functional difference between the two proteins due to altered membrane affinity. Additionally, lipid moieties are able to form direct contact with effectors and differential lipidation within a protein family can result in conformational variation, allowing different interaction surfaces to be available to different effector subsets [97].
Conclusions
There is ample proof that the Gβ plays a crucial role in the physical interaction with effectors in Gβγ dimer-mediated signaling in plants and animals [3], [29], [81], [98], [99]. It is also known that, in animal systems, interaction with effectors reside in different Gβ residues [100], [101]. This fact together with the existence of multiple Gβ subunits with divergent sequences can easily provide specificity for the multiple signaling pathways mediated by the Gβγ dimer. However, it is apparent that plants present a very different picture. The openly different phenotypes shown by the AGG1-, AGG2- and AGG3-defficient mutants together with the fact that there are single alpha and beta subunits clearly indicates that γ subunits provide functional selectivity in Gβγ dimer signaling in Arabidopsis. This study investigated the molecular basis for such selectivity in the two prototypical γ subunits, AGG1 and AGG2. AGG3 was not included in the study due to its atypical structural characteristics and strong differences with AGG1 and AGG2, making it highly unlikely to share effectors with the other two γ subunits. Our results show that for some pathways the selectivity is not embedded in their molecular structure, as proven by the ability of AGG2 and AGG1 to complement agg1 and agg2 mutants respectively. Effector contact points reside in Gβ and possibly in conserved residues between AGG1 and AGG2. In these cases, specificity is provided by the spatiotemporal differences in AGG1 and AGG2 expression patterns. Nevertheless this is not the case for all phenotypes, implicating that there are some pathways for which signaling specificity is at least partially provided by non-conserved amino acid residues in AGG1 and AGG2. In these cases, specificity is embedded in the molecular structure of AGG1 and AGG2, although differences in expression patterns could also contribute. Contact point/s between effectors and the Gβγ dimer are crucial for effector activation and there are a number of studies that have identified important amino acid residues in Arabidopsis Gβ [81], [82], [102]. The next obvious step is to perform similar mutagenesis studies in AGG1 and AGG2.
Funding Statement
This work was supported by the Australian Research Council [Grant number DP1094152]. This work was supported by grants from the National Institute of General Medical Sciences (R01GM065989 and National Science Foundation (MCB-0723515 and MCB-0718202) to AMJ. The Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the United States Department of Energy through the grant DE-FG02-05er15671 to AMJ funded preliminary studies on lateral root phenotypes. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Dupre DJ, Robitaille M, Rebois RV, Hebert TE (2009) The role of Gβγ subunits in the organization, assembly, and function of GPCR signaling complexes. Annual Review of Pharmacology and Toxicology 49: 31–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Milligan G, Kostenis E (2006) Heterotrimeric G-proteins: a short history. British Journal of Pharmacology 147: 46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Temple BRS, Jones AM (2007) The plant heterotrimeric G-protein complex. Annual Review of Plant Biology 58: 249–266. [DOI] [PubMed] [Google Scholar]
- 4. McCudden CR, Hains MD, Kimple RJ, Siderovski DP, Willard FS (2005) G-protein signaling: back to the future. Cellular and Molecular Life Sciences 62: 551–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johnston CA, Taylor JP, Gao Y, Kimple AJ, Grigston JC, et al. (2007) GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proceedings of the National Academy of Sciences of the United States of America 104: 17317–17322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones JC, Duffy JW, Machius M, Temple BRS, Dohlman HG, et al.. (2011) The crystal structure of a self-activating G protein α subunit reveals its distinct mechanism of signal initiation. Science Signaling 4. [DOI] [PMC free article] [PubMed]
- 7. Jones JC, Jones AM, Temple BRS, Dohlman HG (2012) Differences in intradomain and interdomain motion confer distinct activation properties to structurally similar Gα proteins. Proceedings of the National Academy of Sciences of the United States of America 109: 7275–7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jones JC, Temple BRS, Jones AM, Dohlman HG (2011) Functional reconstitution of an atypical G protein heterotrimer and regulator of G Protein Signaling Protein (RGS1) from Arabidopsis thaliana . Journal of Biological Chemistry 286: 13143–13150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Urano D, Jones JC, Wang H, Matthews M, Bradford W, et al. (2012) G protein activation without a GEF in the plant kingdom. PLoS Genet 8: e1002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Neer EJ (1995) Heterotrimeric G-proteins - Organizers of transmembrane signals. Cell 80: 249–257. [DOI] [PubMed] [Google Scholar]
- 11. Gautam N, Downes GB, Yan K, Kisselev O (1998) The G-protein βγ complex. Cellular Signalling 10: 447–455. [DOI] [PubMed] [Google Scholar]
- 12. Balcueva EA, Wang Q, Hughes H, Kunsch C, Yu ZH, et al. (2000) Human G protein γ11 and γ14 subtypes define a new functional subclass. Experimental Cell Research 257: 310–319. [DOI] [PubMed] [Google Scholar]
- 13. Moro S, Deflorian F, Spalluto G, Pastorin G, Cacciari B, et al. (2003) Demystifying the three dimensional structure of G protein-coupled receptors (GPCRs) with the aid of molecular modeling. Chemical communications (Cambridge, England) 24: 2949–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McIntire WE, Myung CS, MacCleery G, Wang Q, Garrison JC (2002) Reconstitution of G protein-coupled receptors with recombinant G protein α and βγ subunits. Methods in Enzymology 343: 372–393. [DOI] [PubMed] [Google Scholar]
- 15. Robishaw JD, Berlot CH (2004) Translating G protein subunit diversity into functional specificity. Current Opinion in Cell Biology 16: 206–209. [DOI] [PubMed] [Google Scholar]
- 16. Clapham DE, Neer EJ (1993) New roles for G-protein βγ dimers in transmembrane signaling. Nature 365: 403–406. [DOI] [PubMed] [Google Scholar]
- 17. Sternweis PC (1994) The active role of βγ in signal transduction. Current Opinion in Cell Biology 6: 198–203. [DOI] [PubMed] [Google Scholar]
- 18. Wolfe JT, Wang HG, Howard J, Garrison JC, Barrett PQ (2003) T-type calcium channel regulation by specific G-protein βγ subunits. Nature 424: 209–213. [DOI] [PubMed] [Google Scholar]
- 19. DePuy SD, Yao JL, Hu CL, McIntire W, Bidaudt I, et al. (2006) The molecular basis for T-type Ca2+ channel inhibition by G protein β2γ2 subunits. Proceedings of the National Academy of Sciences of the United States of America 103: 14590–14595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kisselev O, Gautam N (1993) Specific interaction with rhodopsin is dependent on the γ subunit type in a G-protein. Journal of Biological Chemistry 268: 24519–24522. [PubMed] [Google Scholar]
- 21. Ong OC, Yamane HK, Phan KB, Fong HKW, Bok D, et al. (1995) Molecular cloning and characterization of the G protein γ subunit of cone photoreceptors. Journal of Biological Chemistry 270: 8495–8500. [DOI] [PubMed] [Google Scholar]
- 22. Lee RH, Lieberman BS, Yamane HK, Bok D, Fung BKK (1992) A third form of the G protein β subunit. 1. Immunochemical identification and localization to cone photoreceptors. Journal of Biological Chemistry 267: 24776–24781. [PubMed] [Google Scholar]
- 23. Kelly MN, Irving HR (2003) Nod factors activate both heterotrimeric and monomeric G- proteins in Vigna unguiculata (L.) Walp. Planta 216: 674–685. [DOI] [PubMed] [Google Scholar]
- 24. Trusov Y, Rookes JE, Chakravorty D, Armour D, Schenk PM, et al. (2006) Heterotrimeric G proteins facilitate Arabidopsis resistance to necrotrophic pathogens and are involved in jasmonate signaling. Plant Physiology 140: 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhu HF, Li GJ, Ding L, Cui XQ, Berg H, et al. (2009) Arabidopsis Extra Large G-Protein 2 (XLG2) interacts with the Gβ subunit of heterotrimeric G protein and functions in disease resistance. Molecular Plant 2: 513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Llorente F, Alonso-Blanco C, Sanchez-Rodriguez C, Jorda L, Molina A (2005) ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina . Plant Journal 43: 165–180. [DOI] [PubMed] [Google Scholar]
- 27. Trusov Y, Sewelam N, Rookes JE, Kunkel M, Nowak E, et al. (2009) Heterotrimeric G proteins-mediated resistance to necrotrophic pathogens includes mechanisms independent of salicylic acid-, jasmonic acid/ethylene- and abscisic acid-mediated defense signaling. Plant Journal 58: 69–81. [DOI] [PubMed] [Google Scholar]
- 28.Trusov Y, Jorda L, Molina A, Botella JR (2010) G proteins and plant innate immunity. In: Yalovsky S, Baluska F, Jones A, editors. Integrated G Proteins Signaling in Plants: Springer. 221–250.
- 29.Thung L, Trusov Y, Chakravorty D, Botella JR (2012) Gγ1+Gγ2+Gγ3 = Gβ: The search for heterotrimeric G-protein γ subunits in Arabidopsis is over. Journal of Plant Physiology 169 542–545. [DOI] [PubMed]
- 30. Trusov Y, Botella JR (2012) New faces in plant innate immunity: heterotrimeric G proteins. Journal of Plant Biochemistry and Biotechnology 21: S40–S47. [Google Scholar]
- 31. Lease KA, Wen JQ, Li J, Doke JT, Liscum E, et al. (2001) A mutant Arabidopsis heterotrimeric G-protein β subunit affects leaf, flower, and fruit development. Plant Cell 13: 2631–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ullah H, Chen JG, Temple B, Boyes DC, Alonso JM, et al. (2003) The β subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell 15: 393–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen JG, Yajun G, Jones AM (2006) Differential roles of Arabidopsis heterotrimeric G-protein subunits in modulating cell division in roots. Plant Physiology 141: 887–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ma H (1994) GTP-binding proteins in plants - New members of an old family. Plant Molecular Biology 26: 1611–1636. [DOI] [PubMed] [Google Scholar]
- 35. Ullah H, Chen JG, Young JC, Im KH, Sussman MR, et al. (2001) Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science 292: 2066–2069. [DOI] [PubMed] [Google Scholar]
- 36. Armstrong F, Blatt MR (1995) Evidence for K+ channel control in Vicia guard-cells coupled by G-proteins to a 7TMS receptor mimetic. Plant Journal 8: 187–198. [Google Scholar]
- 37. Assmann SM (1996) Guard cell G proteins. Trends in Plant Science 1: 73–74. [Google Scholar]
- 38. Zhang W, He SY, Assmann SM (2008) The plant innate immunity response in stomatal guard cells invokes G-protein-dependent ion channel regulation. Plant Journal 56: 984–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Warpeha KM, Hamm HE, Rasenick MM, Kaufman LS (1991) A blue- light- activated GTP-binding protein in the plasma-membranes of etiolated peas. Proceedings of the National Academy of Sciences of the United States of America 88: 8925–8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Okamoto H, Matsui M, Deng XW (2001) Overexpression of the heterotrimeric G-protein α subunit enhances phytochrome-mediated inhibition of hypocotyl elongation in Arabidopsis. Plant Cell 13: 1639–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Botto JF, Ibarra S, Jones AM (2009) The heterotrimeric G-protein complex modulates light sensitivity in Arabidopsis thaliana seed germination. Photochemistry and Photobiology 85: 949–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jones AM, Ecker JR, Chen J-G (2003) A reevaluation of the role of the heterotrimeric G protein in coupling light responses in Arabidopsis. Plant Physiology 131: 1623–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Booker FL, Burkey KO, Overmyer K, Jones AM (2004) Differential responses of G-protein Arabidopsis thaliana mutants to ozone. New Phytologist 162: 633–641. [DOI] [PubMed] [Google Scholar]
- 44. Joo JH, Wang SY, Chen JG, Jones AM, Fedoroff NV (2005) Different signaling and cell death roles of heterotrimeric G protein α and β subunits in the Arabidopsis oxidative stress response to ozone. Plant Cell 17: 957–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bhardwaj D, Lakhanpaul S, Tuteja N (2012) Wide range of interacting partners of pea G beta subunit of G-proteins suggests its multiple functions in cell signalling. Plant Physiology and Biochemistry 58: 1–5. [DOI] [PubMed] [Google Scholar]
- 46. Misra S, Wu Y, Venkataraman G, Sopory SK, Tuteja N (2007) Heterotrimeric G-protein complex and G-protein-coupled receptor from a legume (Pisum sativum): role in salinity and heat stress and cross-talk with phospholipase C. Plant Journal. 51: 656–669. [DOI] [PubMed] [Google Scholar]
- 47. Wang L, Xu YY, Ma QB, Li D, Xu ZH, et al. (2006) Heterotrimeric G protein α subunit is involved in rice brassinosteroid response. Cell Research 16: 916–922. [DOI] [PubMed] [Google Scholar]
- 48. Pandey S, Assmann SM (2004) The Arabidopsis putative G protein-coupled receptor GCR1 interacts with the G protein α subunit GPA1 and regulates abscisic acid signalling. Plant Cell 16: 1616–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Okamoto H, Gobel C, Capper RG, Saunders N, Feussner I, et al. (2009) The α subunit of the heterotrimeric G-protein affects jasmonate responses in Arabidopsis thaliana. . Journal of Experimental Botany 60: 1991–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen JG, Willard FS, Huang J, Liang JS, Chasse SA, et al. (2003) A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 301: 1728–1731. [DOI] [PubMed] [Google Scholar]
- 51. Huang J, Taylor JP, Chen JG, Uhrig JF, Schnell DJ, et al. (2006) The plastid protein THYLAKOID FORMATION1 and the plasma membrane G-protein GPA1 interact in a novel sugar-signaling mechanism in Arabidopsis. Plant Cell 18: 1226–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ullah H, Chen JG, Wang SC, Jones AM (2002) Role of a heterotrimeric G protein in regulation of Arabidopsis seed germination. Plant Physiology 129: 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kushwah S, Jones AM, Laxmi A (2011) Cytokinin interplay with ethylene, auxin, and glucose signaling controls Arabidopsis seedling root directional growth. Plant Physiology 156: 1851–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chakravorty D, Trusov Y, Zhang W, Acharya BR, Sheahan MB, et al. (2011) An atypical heterotrimeric G-protein γ subunit is involved in guard cell K+ channel regulation and morphological development in Arabidopsis thaliana . Plant Journal 67: 840–851. [DOI] [PubMed] [Google Scholar]
- 55. Botella JR (2012) Can heterotrimeric G-proteins help to feed the world? Trends in Plant Science 17: 563–568. [DOI] [PubMed] [Google Scholar]
- 56. Ma H, Yanofsky MF, Meyerowitz EM (1990) Molecular-cloning and characterization of Gpa1, a G-protein α subunit gene from Arabidopsis thaliana . Proceedings of the National Academy of Sciences of the United States of America 87: 3821–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Weiss C, Garnaat C, Mukai K, Hu Y, Ma H (1994) Isolation of cDNAs encoding guanine nucleotide-binding protein β- subunit homologues from maize (ZGB1) and Arabidopsis (AGB1). Proceedings of the National Academy of Sciences of the United States of America 91: 9554–9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mason MG, Botella JR (2000) Completing the heterotrimer: Isolation and characterization of an Arabidopsis thaliana G protein γ subunit cDNA. Proceedings of the National Academy of Sciences of the United States of America 97: 14784–14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mason MG, Botella JR (2001) Isolation of a novel G-protein γ subunit from Arabidopsis thaliana and its interaction with Gβ. Biochimica Et Biophysica Acta 1520: 147–153. [DOI] [PubMed] [Google Scholar]
- 60. Trusov Y, Chakravorty D, Botella JR (2012) Diversity of heterotrimeric G-protein γ subunits in plants. BMC Research Notes 5: 608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Trusov Y, Rookes JE, Tilbrook K, Chakravorty D, Mason MG, et al. (2007) Heterotrimeric G protein γ subunits provide functional selectivity in Gβγ dimer signaling in Arabidopsis. Plant Cell 19: 1235–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Trusov Y, Zhang W, Assmann SM, Botella JR (2008) Gγ1+ Gγ2 ≠ Gβ: Heterotrimeric G protein Gγ-deficient mutants do not recapitulate all phenotypes of Gβ-deficient mutants. Plant Physiology 147: 636–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant Journal 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 64. Wever W, McCallum EJ, Chakravorty D, Cazzonelli CI, Botella JR (2010) The 5' untranslated region of the VR-ACS1 mRNA acts as a strong translational enhancer in plants. Transgenic Research 19: 667–674. [DOI] [PubMed] [Google Scholar]
- 65. Purnell MP, Botella JR (2007) Tobacco isoenzyme 1 of NAD(H)-dependent glutamate dehydrogenase catabolizes glutamate in vivo . Plant Physiology 143: 530–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, et al. (2004) Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16: 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Campbell EJ, Schenk PM, Kazan K, Penninckx I, Anderson JP, et al. (2003) Pathogen-responsive expression of a putative ATP-binding cassette transporter gene conferring resistance to the diterpenoid sclareol is regulated by multiple defense signaling pathways in Arabidopsis. Plant Physiology 133: 1272–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Anderson DJ, Botella JR (2007) Expression analysis and subcellular localization of the Arabidopsis thaliana G-protein beta-subunit AGB1. Plant Cell Reports 26: 1469–1480. [DOI] [PubMed] [Google Scholar]
- 69. Mauchmani B, Slusarenko AJ (1994) Systemic acquired-resistance in Arabidopsis thaliana induced by a predisposing infection with a pathogenic isolate of Fusarium oxysporum. . Molecular Plant-Microbe Interactions 7: 378–383. [Google Scholar]
- 70.Agrios GN (2005) Plant Pathology: New York: Elsevier Academic Press.
- 71. Lapik YR, Kaufman LS (2003) The Arabidopsis cupin domain protein AtPirin1 interacts with the G protein α subunit GPA1 and regulates seed germination and early seedling development. Plant Cell 15: 1578–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chen JG, Pandey S, Huang J, Alonso JM, Ecker JR, et al. (2004) GCR1 can act independently of heterotrimeric G-protein in response to brassinosteroids and gibberellins in Arabidopsis seed germination. Plant Physiology 135: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chen Y, Ji FF, Xie H, Liang JS, Zhang JH (2006) The regulator of G-protein signaling proteins involved in sugar and abscisic acid signaling in Arabidopsis seed germination. Plant Physiology 140: 302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Pandey S, Chen JG, Jones AM, Assmann SM (2006) G-protein complex mutants are hypersensitive to abscisic acid regulation of germination and postgermination development. Plant Physiology 141: 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 75. Warpeha KM, Upadhyay S, Yeh J, Adamiak J, Hawkins SI, et al. (2007) The GCR1, GPA1, PRN1, NF-Y signal chain mediates both blue light and abscisic acid responses in Arabidopsis. Plant Physiology 143: 1590–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kreps JA, Wu YJ, Chang HS, Zhu T, Wang X, et al. (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiology 130: 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Song Y, Wang L, Xiong L (2009) Comprehensive expression profiling analysis of OsIAA gene family in developmental processes and in response to phytohormone and stress treatments. Planta 229: 577–591. [DOI] [PubMed] [Google Scholar]
- 78. Cheong YH, Chang HS, Gupta R, Wang X, Zhu T, et al. (2002) Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiology 129: 661–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kidd BN, Kadoo NY, Dombrecht B, Tekeoglu M, Gardiner DM, et al. (2011) Auxin signaling and transport promote susceptibility to the root-infecting fungal pathogen Fusarium oxysporum in Arabidopsis. Molecular Plant-Microbe Interactions 24: 733–748. [DOI] [PubMed] [Google Scholar]
- 80. Kazan K, Manners JM (2009) Linking development to defense: auxin in plant-pathogen interactions. Trends in Plant Science 14: 373–382. [DOI] [PubMed] [Google Scholar]
- 81. Friedman EJ, Wang HX, Jiang K, Perovic I, Deshpande A, et al. (2011) Acireductone Dioxygenase 1 (ARD1) is an effector of the heterotrimeric G protein β subunit in Arabidopsis. Journal of Biological Chemistry 286: 30107–30118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chakravorty D, Trusov Y, Botella JR (2012) Site-directed mutagenesis of the Arabidopsis heterotrimeric G protein β subunit suggests divergent mechanisms of effector activation between plant and animal G proteins. Planta 235: 615–627. [DOI] [PubMed] [Google Scholar]
- 83. Jiang K, Frick-Cheng A, Trusov Y, Delgado-Cerezo M, Rosenthal DM, et al. (2012) Dissecting Arabidopsis Gβ signal transduction on the protein surface. Plant Physiol 159: 975–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hensel LL, Grbic V, Baumgarten DA, Bleecker AB (1993) Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in Arabidopsis. Plant Cell 5: 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Madueno F, RuizGarcia L, Salinas J, MartinezZapater JM (1996) Genetic interactions that promote the floral transition in Arabidopsis. Seminars in Cell & Developmental Biology 7: 401–407. [Google Scholar]
- 86. Eskins K (1992) Light-quality effects on Arabidopsis development- red, blue and far-red regulation of flowering and morphology. Physiologia Plantarum 86: 439–444. [Google Scholar]
- 87. Koornneef M, Leon-Kloosterziel KM, Schwartz SH, Zeevaart JAD (1998) The genetic and molecular dissection of abscisic acid biosynthesis and signal transduction in Arabidopsis. Plant Physiology and Biochemistry 36: 83–89. [Google Scholar]
- 88. Komeda Y (2004) Genetic regulation of time to flower in Arabidopsis thaliana . Annual Review of Plant Biology 55: 521–535. [DOI] [PubMed] [Google Scholar]
- 89. Domagalska MA, Schomburg FM, Amasino RM, Vierstra RD, Nagy F, et al. (2007) Attenuation of brassinosteroid signaling enhances FLC expression and delays flowering. Development 134: 2841–2850. [DOI] [PubMed] [Google Scholar]
- 90. Hooley R (1999) A role for G proteins in plant hormone signalling? Plant Physiology and Biochemistry 37: 393–402. [Google Scholar]
- 91.Wang L, Chong K (2010) Auxin, brassinosteroids, and G-protein signaling. In: Yalovsky S, Baluska F, Jones A, editors. Integrated G Proteins Signaling in Plants. Berlin: Springer-Verlag Berlin. 135–154.
- 92. Balasubramanian S, Sureshkumar S, Lempe J, Weigel D (2006) Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. Plos Genetics 2: 980–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kang R, Wan J, Arstikaitis P, Takahashi H, Huang K, et al. (2008) Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature 456: 904–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Adjobo-Hermans MJW, Goedhart J, Gadella TWJ (2006) Plant G protein heterotrimers require dual lipidation motifs of Gα and Gγ and do not dissociate upon activation. Journal of Cell Science 119: 5087–5097. [DOI] [PubMed] [Google Scholar]
- 95. Zeng Q, Wang XJ, Running MP (2007) Dual lipid modification of Arabidopsis Gγ-subunits is required for efficient plasma membrane targeting. Plant Physiology 143: 1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kassai H, Aiba A, Nakao K, Nakamura K, Katsuki M, et al. (2005) Farnesylation of retinal transducin underlies its translocation during light adaptation. Neuron 47: 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wedegaertner PB, Wilson PT, Bourne HR (1995) Lipid modification of trimeric G-proteins. Journal of Biological Chemistry 270: 503–506. [DOI] [PubMed] [Google Scholar]
- 98. Klopffleisch K, Phan N, Augustin K, Bayne RS, Booker KS, et al. (2011) Arabidopsis G-protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Molecular Systems Biology 7: 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ford CE, Skiba NP, Bae HS, Daaka YH, Reuveny E, et al. (1998) Molecular basis for interactions of G protein βγ subunits with effectors. Science 280: 1271–1274. [DOI] [PubMed] [Google Scholar]
- 100. Li Y, Sternweis PM, Charnecki S, Smith TF, Gilman AG, et al. (1998) Sites for Gα binding on the G protein β subunit overlap with sites for regulation of phospholipase C β and adenylyl cyclase. Journal of Biological Chemistry 273: 16265–16272. [DOI] [PubMed] [Google Scholar]
- 101. Liu MY, Yu B, Nabanishi O, Wieland T, Simon M (1997) The Ca2+-dependent binding of calmodulin to an N-terminal motif of the heterotrimeric G protein β subunit. Journal of Biological Chemistry 272: 18801–18807. [DOI] [PubMed] [Google Scholar]
- 102. Smrcka AV, Kichik N, Tarrago T, Burroughs M, Park MS, et al. (2010) NMR analysis of G-protein βγ subunit complexes reveals a dynamic Gα-Gβγ subunit interface and multiple protein recognition modes. Proceedings of the National Academy of Sciences of the United States of America 107: 639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]