Abstract
Chronic inflammation in the stomach induces metaplasia, the pre-cancerous lesion that precedes inflammation-driven neoplastic transformation. While Hedgehog signaling contributes to the initiation of some cancers, its role in gastric transformation remains poorly defined. We found that Helicobacter-infected C57BL/6 mice develop extensive mucous cell metaplasia at 6 month but not at 2 months post-infection. Gastric metaplasia coincided with the appearance of CD45+MHCII+CD11b+CD11c+ myeloid cells that were normally not present in the chronic gastritis at 2 months. The myeloid regulatory gene Schlafen-4 was identified in a microarray analysis comparing infected WT versus Gli1 null mice and was expressed in the CD11b+CD11c+ myeloid population. Moreover this same population expressed IL-1β and TNFα pro-inflammatory cytokines. By 6 months, the mucous neck cell metaplasia (SPEM) expressed IL-6, phosphorylated STAT3 and the proliferative marker Ki67. Expression was not observed in Gli1 mutant mice consistent with the requirement of Gli1 to induce this pre-neoplastic phenotype. Ectopic Shh ligand expression alone was not sufficient to induce SPEM, but with Helicobacter infection synergistically increased the histologic severity observed with the inflammation. Therefore Hedgehog signaling is required, but is not sufficient to generate pre-neoplastic changes during chronic gastritis. Gli1-dependent myeloid cell differentiation plays a pivotal role in the appearance of myeloid cell subtypes ostensibly required for SPEM development. Moreover, it suggests that therapies capable of targeting this phenotypic switch might prevent progression to metaplasia, the pre-neoplastic change that develops prior to dysplasia and gastric cancer, which also occurs in other epithelial-derived neoplasias initiated by chronic inflammation.
Introduction
Gastric metaplasia is the histologic change that precedes neoplastic transformation of the stomach in response to inflammation [1]. The gastric mucosa is primarily composed of acid-producing (parietal cells), pepsinogen-producing (chief cells), and mucus-producing (surface pit and neck) cells [2]. During Helicobacter pylori (H. pylori) infection, the gastric epithelium expands mucous cell compartments at the expense of parietal and chief cells, thereby reducing acid secretion [3]. Cytokine-secreting myeloid cells are a component of the bacteria-induced inflammation infiltrating the gastric stromal microenvironment [4]. Over time, these pro-inflammatory cytokines are sufficient to initiate pre-neoplastic changes in the epithelium [5].
Aberrant Hedgehog (Hh) signaling promotes the development of medulloblastoma, basal cell, pancreatic and colon carcinoma [6]. However, its role in gastric cancer development is less clear. We and others previously established that gastric parietal cells produce Sonic Hedgehog (Shh) ligand [7], [8], [9], [10], which activates one of several effector genes including the glioma-associated oncogene 1 (Gli1) transcription factor expressed in cells residing in the lamina propria such as immune cells and myofibroblasts [11]. A recent report showed that Hh signaling is necessary for myeloid cell recruitment to the stomach within two days of H. pylori infection [12], but the downstream effects of the Hh pathway leading to pre-neoplastic transformation were not examined. Therefore to test whether Hh signaling is required for gastric transformation, we infected wild type C57BL/6 (WT) and Gli1-deficient mice with Helicobacter felis (H. felis) to generate a robust inflammatory response (chronic gastritis) and gastric metaplasia.
Materials and Methods
Transgenic Mice
C57BL/6 and Gli1+/LacZ (Gli1+/−) mice were obtained from Jackson Labs (Bar Harbor, ME) and bred to generate Gli1LacZ/LacZ (Gli1−/−) mice. These mice contain a knock-in of the cDNA for nuclear β-galactosidase (nLacZ) inserted into exon 2 of the Gli1 gene abolishing endogenous gene expression. Transgenic founders were generated by micro-injecting CMV-ShhWT-HA, CMV-ShhF200H-HA, H+/K+-ATPase-β-ShhWT-HA and H+/K+-ATPase-β-ShhF200H-HA constructs into fertilized eggs obtained by mating (C57BL/6/J X SJL/J)F1 males and females of the same background (UM Transgenic Core, Ann Arbor, MI). The N-terminal Hemagglutinin (HA)-tagged mouse Shh constructs were generated by inserting four copies of the HA epitope (TACCCTTACGACGTTCCTGATTACGCT) downstream of nucleotide 96 (amino acid 32) in the Shh cDNA sequence. The F200H mutant Shh transgene was generated by substituting “TT” nucleotides for “CA” at position 597 of the mouse Shh cDNA using the QuickChange site-directed mutagenesis kit (Stratagene, Santa Clara, CA). These sequences were subcloned into the topoisomerase site of pcDNA3.1/V5-His-TOPO and into the BamHI restriction site of the mouse H+/K+-ATPase-β subunit expression plasmid (–1,035 to +24). Mice were housed under specific pathogen-free conditions and fasted overnight prior to use, with free access to water. All mouse studies were approved by the University of Michigan Institutional Animal Care and Use Committee.
Helicobacter felis culture and infection
H. felis (CS1 strain) stocks were stored in 50% glycerol solution at −80°C. Bacteria were cultured in sterile-filtered Brucella broth (BD, Franklin Lakes, NJ) plus 10% FBS (Atlanta Biologicals, Lawrenceville, GA) using the GasPak™ EZ Campy Container System (BD) at 37°C with 150 rpm shaking. The cultures were spun down at 2700 rpm at room temperature, and the pellets resuspended in Brucella broth plus 10% FBS (Thermo Fisher Scientific, Houston, TX). Cells were counted using a hemocytometer by diluting the cells 1∶100 in 9∶1 HBSS/Formalin solution. Mice were gavaged 3 times over 3 days with 108 H. felis cells in 100 μL of Brucella broth. Control mice were gavaged with Brucella broth alone.
H. felis DNA quantification
Gastric tissue from the corpus and fundus was snap frozen and stored at −80°C. Total DNA was extracted using the DNEasy Blood and Tissue Kit (Qiagen). Quantitative PCR was performed using the H. felis Fla-B primers-F: 5′TTCGATTGGTCCTACAGGCTCAGA, R: 5′TTCTTGTTGATGACATTGACCAACGCA 3′ on a CFX96 real-time PCR detection system (Bio-RAD).
Tissue Preparation
Mice were starved overnight then euthanized. The stomachs were removed, opened along the greater curvature, and cut into longitudinal strips for histology from the lesser and greater curvatures. Half of the strips were fixed in 4% formaldehyde (Fisher Scientific) and the other half directly embedded in OCT compound (Fisher Scientific) and snap-frozen. The remainder of the stomach, containing only fundus and corpus, was minced and processed for RNA extraction or digested for flow cytometric analysis.
Immunofluorescence
For frozen sections, 8 µm sections were fixed in 4% paraformaldehyde for 10 min, washed in PBS twice, and then blocked with 20% donkey serum (#017-000-121, Jackson ImmunoResearch, West Grove, PA) in PBS. Frozen sections were immunostained with the following antibodies: β-gal (gift from James Douglas Engel, Department of Cell and Developmental Biology, University of Michigan), TFF-2 (gift from Nicholas Wright, Barts and The London School of Medicine, London, UK), F4/80 (#MCA497GA, AbD Serotec, Raleigh, NC), CD11b (#ab6332-100, clone M1/70.15, Abcam, Cambridge, MA), CD11c-FITC (#553801, BD Pharmingen, BD Bioscience, Bedford, MA), α-SMA-Cy3 (#C6198, Sigma, St Louis, MO), CD19 (#MCA1439, AbD Serotec), MPO-FITC (#90812, Abcam), Slfn-4 (#sc-8903, Santa Cruz Biotechnology, Santa Cruz, CA), pSTAT-3 (#9131, Cell Signaling, Boston, MA), IL-1β (#AF-401-NA, R&D Systems, Minneapolis, MN), Ki-67 (#RM-9106-S1, Thermo Scientific, Fisher), Shh (#sc-1194, Santa Cruz, CA), E-cadherin (#3195, Cell Signaling), and GSII-FITC (#FL-1211, Vector Labs, Burlingame, CA). For unlabeled primary antibodies, the staining was developed using Alexa Fluor-conjugated secondary antibodies (Molecular Probes, Invitrogen, Carlsbad, CA).
For paraffin sections, 8 µm sections were deparaffinized in xylene and 100% ethanol. Sections were re-hydrated with distilled water and antigen retrieval was performed using 10 mM citric acid buffer (pH 6). Slides were washed in 0.01% Triton X-100 (Fisher) in PBS twice, incubated with 20% donkey serum (#017-000-121, Jackson ImmunoResearch) and immunostained with the following antibodies: H+/K+-ATPase-β (#D032-3, Medical and Biological Laboratories, Woburn, MA), HA-probe (#sc-7392, Santa Cruz), Shh (#sc-1194, Santa Cruz), intrinsic factor (gift from David Alpers, Washington University, St. Louis, MO), and GSII-FITC (#FL-1211, Vector Labs). For unlabeled primary antibodies, staining was developed using Alexa Fluor-conjugated secondary antibodies (Molecular Probes, Invitrogen). Concentration and species matched immunoglobulins were used as controls for each antibody. Staining was visualized using an Olympus Fluoview scanning confocal microscope (Olympus, Center Valley, PA).
Morphometric Analysis
Well-oriented gastric glands from the fundus and corpus were observed under a 20× objective lens (200× total magnification) and scored for neutrophilic infiltration (polymorphonuclear leukocytes [PMN]), gastritis, follicle formation, and metaplasia. Each microscopic field was scored only for the presence or absence of the lesion according to the following criteria: 1) neutrophilic inflammation was defined by the presence of several clusters of neutrophils within the mucosa; 2) “displacement of epithelial glands” is defined as the displacement of gastric glands by inflammatory cell infiltrates of any cell type; 3) metaplasia was defined by the presence of an expansion of the mucous cell lineage compartment. For each slide, the number of positive fields was divided by the total number of fields examined and was expressed as a percentage of the affected fields.
Real-time quantitative PCR
The forestomach was removed and tissue from the glandular stomach was minced and homogenized in TRIzol (Invitrogen). RNA clean-up was performed using the RNeasy Minikit (Qiagen, Valencia, CA), and cDNA synthesized from 1 µg of RNA using iScript reverse transcriptase (Bio-Rad, Hercules, CA). Real-time quantitative polymerase chain reaction (RT-qPCR) was performed using Platinum Taq DNA polymerase (Invitrogen) on a CFX96 real-time PCR detection system (Bio-RAD), using the following primer sequences (Tm = 65°C for all primers):
CD11b-F: 5′CTCCGGTAGCATCAACAACAT, R: 5′TGATCTTGGGCTAGGGTTTCT;
CD11c-F: 5′CTGGATAGCCTTTCTTCTGCTG, R: 5′GCACACTGTGTCCGAACTC;
F4/80-F: 5′CCCAGCTTCTGCCACCTGCA, R: 5′GGAGCCATTCAAGACAAAGCC;
Gli1-F: 5′TTGGGATGAAGAAGCAGTTG, R: 5′GGAGACAGCATGGCTCACTA;
HPRT-F: 5′AGGACCTCTCGAAGTGTTGGATAC, R: 5′AACTTGCGCTCATCTTAGGCTTTG;
IFN-γ-F: 5′TCAAGTGGCATAGATGTGGAAGAA, R: 5′TGGCTCTGCAGGATTTTCATG;
IL-12p35-F: 5′AAATGAAGCTCTGCATCCTGC, R: 5′TCACCCTGTTGATGGTCACG;
IL-12p40-F: 5′AAACCAGACCCGCCCAAGAAC, R 5′AAAAAGCCAACCAAGCAGAAGACAG;
IL-4 F: 5′ACAGGAGAAGGGACGCCAT, R: 5′GAAGCCCTACAGACGAGCTCA;
IL-5-F: 5′AGCACAGTGGTGAAAGAGACCTT, R: 5′TCCAATGCATAGCTGGTGATTT,
IL-6-F: 5′GAGGATACCACTCCCAACAGACC, R: 5′AAGTGCATCATCGTTGTTCATACA;
IL1-β-F: 5′CAACCAACAAGTGATATTCTCCATG, R: 5′GATCCACACTCTCCAGCTGCA;
IL17A-F: 5′GCTCCAGAAGGCCCTCAGA, R: 5′AGCTTTCCCTCCGCATTGA;
KC-F: 5′CTGCACCCAAACCGAAGTCAT, R: 5′TTGTCAGAAGCCAGCGTTCAC;
MPO-F: 5′CCAGCAGCCATGAAGTA, R: 5′CATAACGGAAAGCATTGGTG;
Ptch1-F: 5′CTGCTGTGGTGGTGGTATTC, R: 5′GGCTTGTGAAACAGCAGAAA;
SAA3-F: 5′TGCCATCATTCTTTGCATCTTGA, R: 5′CCGTGAACTTCTGAACAGCCT;
Shh-F: 5′ATGTTTTCTGGTGATCCTTGCT, R: 5′ATCGTTCGGAGTTTCTTGTGAT;
Slfn-4-F: 5′GCCCTCTGTTCAAGTCAAGTGTCC, R: 5′CCCAGATGAAATCCTTTCCACGA;
TNF-α-F: 5′CATCTTCTCAAAATTCGAGTGACAA, R: 5′TGGGAGTAGACAAGGTACAACCC;
CCL-19-F: 5′TGTGGCCTGCCTCAG ATTAT, R: 5′AGTCTTCCGCATCATTAGCAC;
CD3-F: 5′TGCTCTTGGTGTATATCTC ATTGC, R: 5′CAGAGTCTGCTTGTCTGAAGCTC.
Microarray analysis
Microarray analysis was performed by the Microarray Core Facility at the University of Michigan. Briefly, RNA was labeled by preparing biotinylated antisense RNA (aRNA) from 250 ng of total RNA according to the Affymetrix GeneChip 3′ IVT Express kit protocol (Affymetrix, Santa Clara, CA). Following fragmentation, 7.5 µg of aRNA were hybridized for 16 hr at 45°C to the Mouse Genome 430 2.0 Perfect Match Peg Arrays (Affymetrix) and scanned using the Affymetrix Gene Atlas system (software version 1.0.4.267, Affymetrix).
Laser Capture Microdissection
Frozen sections (10 µm) were mounted on Leica Pen-Membrane (2 µm) slides (Leica Microsystems Inc., Buffalo Grove, IL) and stored at −20°C until further use. Frozen sections were air-dried, fixed in RNase-free 70% ethanol (pre-chilled, −30°C) for 5 min at room temperature, air-dried, stained in 0.25% RNase-free toluidine blue for 10 sec, then washed in 70% ethanol (pre-chilled, −30°C). Sections were air-dried and the microdissectates collected using a Leica LMD 7000 microdissection microscope (Leica Microsystems Inc.).
Flow cytometry
Stomach tissue was minced, incubated in 10 ml HBSS supplemented with 1 mM DTT (Sigma) and 1 mM EDTA (#51201 Lonza, NJ, USA), and then shaken at 160 rpm for 1 hr at 37°C. The supernatant was collected by passing the 10 ml sample through a 40 µm cell strainer (#08-771-1 Fisher). The tissue collected in the cell strainer was further digested in 10 ml of RPMI (without L-glutamine) containing 5% BSA (#9048-46-8 Roche) and 1.5 mg/ml Dispase II (#04942078001 Roche) with 160 rpm shaking at 37°C for 90 min. The supernatant was collected by passing the digested tissue through a 40 µm cell strainer and both supernatants pooled. For bone marrow cells, the femur marrow was directly flushed with RPMI and treated with ACK lysis buffer (#10-548E Lonza) to lyse red blood cells. The cells were spun down at 1600 rpm, resuspended in 200 µl of PBS containing 5% BSA (Roche), blocked with purified rat anti-mouse DC16/CD32 mouse Fc block (#553141 BD Biosciences, San Diego, CA) at 4°C for 5 min, and incubated with F4/80-FITC (#MCA497FT AbD Serotec), CD11b-PE-Cy7 (#561098 BD Pharmingen, BD Biosciences), CD11c-APC (#561119 BD Biosciences), CD45-PE (#12-0451-81 eBioscience), pSTAT3 (pY705)-PerCP-Cy5.5 (#560114 BD Biosciences), EpCAM-APC (#17-5791-80 eBioscience), GSII-FITC (#FL-1211 Vector Labs), or IL-6-PE (#562050 BD Biosciences). The cells were fixed in 100 µl of Reagent A (#GAS001S5 Invitrogen), washed, and resuspended in 500 µl PBS containing 5% BSA in a BD Falcon tube (#352052 BD). For pSTAT3-PerCP-Cy5.5 an additional permeabilization step was added by incubating the cells in Reagent B (#GAS002S5 Invitrogen). The cells were analyzed using an Accuri C6 Flow Cytometer (Accuri Cytometers, Ann Arbor, MI).
To flow sort cells for subsequent RT-PCR analysis, gastric cells were prepared as described above and gated for CD45, F4/80 and CD11c-expressing cells using the CD45-PE (#12-0451-81 eBioscience), F4/80-FITC (#MCA497FT AbD Serotec) and CD11c-APC (#561119 BD Biosciences) antibodies. Cells were collected in buffer RLT containing β-mercaptoethanol and carrier RNA using a sy3200 (Synergy) sorter (iCyt, Champaign, IL) then extracted using the RNEasy Microkit (Qiagen, Valencia, CA) according to the manufacturer's instructions.
Cell Culture Experiments
NCI-N87 mucous cells (#CRL-5822 ATCC) were plated in 24 well plates, serum-starved overnight, and then treated with recombinant human IL-6 (100 ng/ml, 206-IL-010/CF, R&D Systems) or IL-1β (1 ng/ml, 201-LB-005, R&D Systems) overnight. IL-1RA (10 ng/ml) was added 2 hrs prior to IL-1β treatment. The cells were lysed in TRIzol for RNA extraction and RT-qPCR analysis.
Western Blot Analysis
The cell lysates of NIH-3T3 cells were homogenized in T-per extraction reagent (78510, Pierce, Rockford, IL). The protein lysate (100 µg) was resolved on a Novex 4–20% Tris-Glycine gel (#EC6025 Invitrogen) and then transferred to nitrocellulose using the iBlot Dry Blotting System (Invitrogen) according to the manufacturer's instructions. The membranes were incubated in KPL Detector Block (#71-83-00 Gaithersburg, MD) for 1 hour at room temperature and then incubated with 1∶100 Shh (#sc-1194; Santa Cruz) or 1∶1000 HA-tag (#sc-7392, Santa Cruz) antibodies at 4°C overnight. Membranes were washed with TBS containing 0.1% Tween, incubated with IRDye 680-conjugated antibodies (Licor, Lincoln, NB), and visualized with an Odyssey infrared imager (Licor).
Chromatin Immunoprecipitation (ChIP) assay
Gli1 binding sites in the Slfn-4, Slfn-2 and SAA3 promoters were identified using Genomatix (Munich, Germany). Primers were designed to amplify the nucleotide sequence between −3323 and −3101 of the Slfn4 promoter, which contains a GGCCACCCA Gli1 binding site (consensus sequence GACCACCCA). The primers were designed as follows: F 5′ AAGGCAGAAACATG GCAGCATCC 3′; R 5′ ACTTTGGCTTGTGTCAGGTTGGC 3′. A nucleotide sequence between −589 and −385 that lacks a Gli1 binding site was used as a negative control and was amplified using the following primers: F 5′ TGTGTTCCTGAGTGTGAGTGTTC 3′; R 5′ CTTCCCAGAGCAG CCTTAGC 3′. Flag-tagged pcDNA3.1-Gli1 (gift from Andrzej Dlugosz, Department of Dermatology, University of Michigan) or empty vector were transfected into RAW264.7 cells. ChIP was performed using the Flag-tag antibody (#2368 Cell Signaling) and the Chromatin Immunoprecipitation (ChIP) Assay Kit (#17–295 Millipore, Kankakee, IL).
Statistical Analysis
For in vivo qPCR analysis and pathological scoring of mice, each bar graph represented the mean +/− standard error from 5–10 mice. For cell culture experiments, error bars represented the means +/− standard error of the mean from 3 independent experiments. Data were tested for normality using the Shapiro-Wilk W test (Prism, GraphPad Software, La Jolla, CA). Data were compared using one-way analysis of variance (ANOVA) with Dunnet's (parametric) or Dunn's (non-parametric) multiple comparison tests (Prism). P values less than 0.05 were considered significant.
Microarray Accession Number
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (El-Zaatari et al., 2013) and are accessible through GEO Series accession number GSE43693 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE43693).
Results
Gli1 Deletion Prevents Gastric Metaplasia
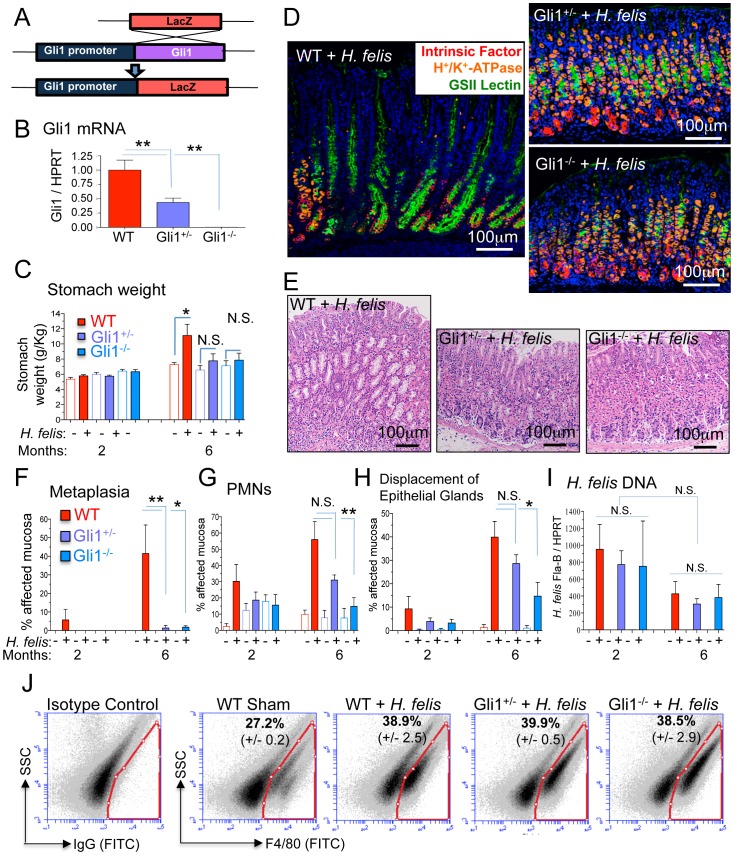
To understand the role of Hedgehog signaling in gastric metaplasia, WT, Gli1+/LacZ (Gli1+/−) and Gli1LacZ/LacZ (Gli1−/−) mice (Figure 1A) were infected with H. felis. Gli1+/− and Gli1−/− mice expressed significantly reduced or absent Gli1 mRNA in the stomach respectively (Figure 1B). Although inflammation was observed 2 months after inoculating the WT mice, by 6 months there was an increase in stomach weight (Figure 1C), loss of parietal and chief cells, and development of spasmolytic polypeptide-expressing metaplasia (SPEM) characterized by the expansion of the mucous neck cell compartment (Figure 1D and E) and co-labeling of mucous cells with the chief cell lineage marker intrinsic factor (Figure S1) [3]. The amount of SPEM (Figure 1F and Figure S1), polymorphic neutrophil (PMN) infiltration (Figure 1G) and epithelial gland displacement (Figure 1H) was greatest in the infected WT mice. In contrast, Gli1+/− and Gli1−/− mice were also inflamed but lacked any SPEM (Figure 1D-F). These differences were not due to differences in H. felis colonization between WT, Gli1+/− or Gli1−/− mice (Figure 1I). Despite the absence of metaplasia, there were similar amounts of F4/80+ myeloid cells infiltrating the infected WT, Gli1+/− and Gli1−/− stomachs (Figure 1J), demonstrating that Gli1 deletion prevented the transition from gastritis to SPEM.
Figure 1. Gli1 deletion prevents gastric metaplasia.
A) Schematic representation of LacZ substitution for the Gli1 gene to generate Gli1+/− and Gli1−/− mice. B) Gli1 mRNA in stomachs of WT, Gli1+/− and Gli1−/− mice. C) Stomach weight normalized to total body weight in infected and uninfected mice. D) Triple immunofluorescent staining of Griffonia simplicifolia II (GSII) lectin (green; mucous neck cells), intrinsic factor (red; chief cells), and H+/K+-ATPase (orange; parietal cells) in 6-month H. felis-infected WT, Gli1+/− and Gli1−/− stomachs. E) Hematoxylin and eosin (H&E) staining of the gastric mucosa in 6-month H. felis-infected WT, Gli1+/− and Gli1−/−. F–H) Histologic scoring of metaplasia, PMN infiltration, and displacement of epithelial glands by infiltrating inflammatory cells in 2- and 6-month H. felis-infected WT, Gli1+/− and Gli1−/− stomachs. I) qPCR quantification of H. felis flagellar filament B (Fla-B) DNA in the infected mucosa. J) Flow cytometric analysis of F4/80+ cells versus side-scatter in WT Sham and 6-month infected WT, Gli1+/− and Gli1−/− stomachs (N = 3 mice per group). Open and closed bars denote uninfected and infected mice respectively. For RT-qPCR experiments, N = 5–10 mice per group. Error bars represent the mean +/− SEM. ***p<0.001; **p<0.01; *p<0.05. N.S. = not significant.
Gli1 Deletion Prevents Th1, Th2, not Th17 inflammatory responses
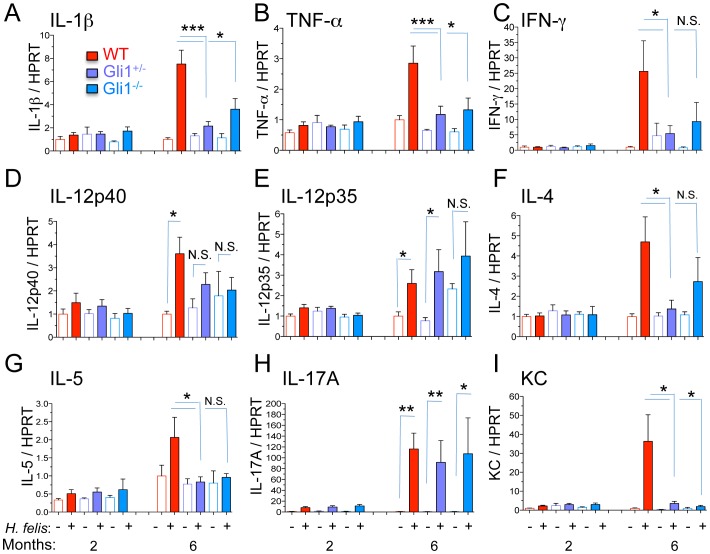
H. felis normally induces a complex inflammatory response in the stomach including increased Th1, Th2 and Th17 T helper lymphocyte subsets [13], [14], [15]. By 6-months, H. felis infection of WT mice induced the gene expression of Th1 (Figure 2 A–E), Th2 (Figure 2 F and G), and Th17 cytokines (Figure 2H) in addition to the chemokine KC (Figure 2I), the mouse IL-8 ortholog. In contrast, H. felis failed to induce several of these cytokines in the Gli1+/− and Gli1−/− stomachs except for the p35 subunit of IL-12 (Figure 2E) and IL-17A (Figure 2H), suggesting that these cytokines were not affected by loss of Gli1 and did not significantly contribute to the development of SPEM.
Figure 2. Gli1 deletion blocks increase in Th1 and Th2 cytokines.
A–E) RT-qPCR determination of mRNA for Th1-associated cytokines. F–G) RT-qPCR of mRNA for Th2-associated cytokines. H) RT-qPCR of mRNA for the Th17-associated cytokine IL-17A. I) RT-qPCR analysis of the mRNA for the mouse IL-8 ortholog, chemokine KC. Open and closed bars denote uninfected and infected mice respectively. N = 5–10 mice per group. Error bars represent the mean +/− SEM. **p<0.01; *p<0.05.
Gastric Myeloid Cells Express Gli1
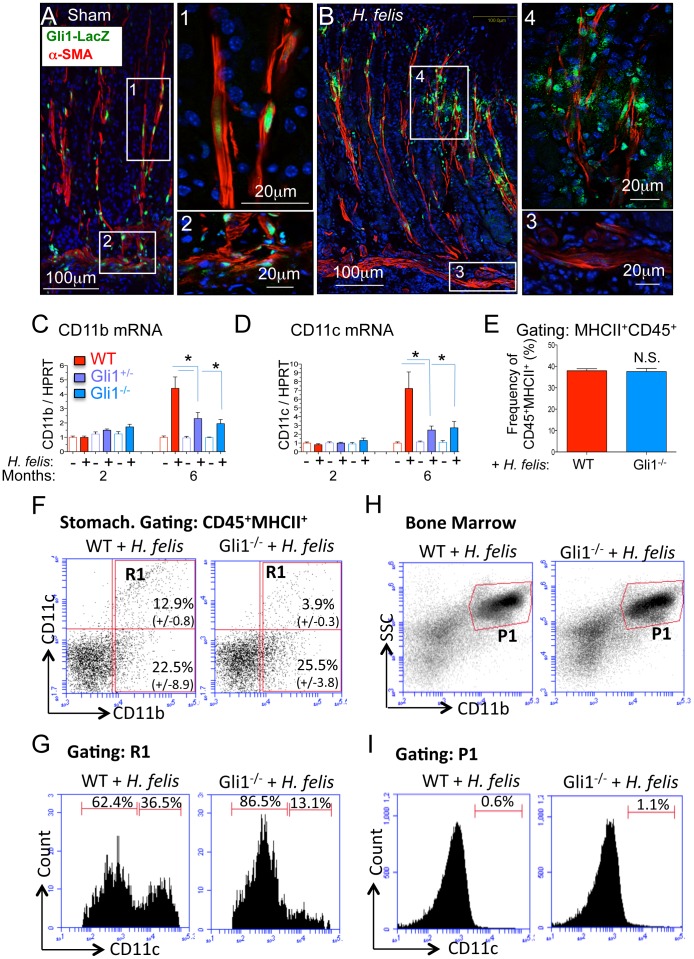
Gumucio and coworkers previously showed that intestinal myeloid cells and myofibroblasts express Gli1 [11]. Myeloid cells are a major source of IL-1β [16], one of the major cytokines sufficient to induce gastric metaplasia [5], [17]. In addition, the expected induction of IL-1β gene expression was reduced in the infected Gli1 mutant mice (Figure 2A), suggesting that Gli1 expression contributes to myeloid cell production of 1L-1β. To characterize the immunological defect in the Gli1 mutant mice, we identified myeloid cell subsets in the stomachs of infected WT and mutant mice. Using the LacZ reporter to identify Gli1-expressing cells, we found that the LacZ reporter was expressed primarily in α-SMA+ myofibroblasts and smooth muscle cells in the uninfected stomach as previously reported [11] (Figure 3A). However LacZ expression was significantly reduced in these resident mesenchymal cells 6 months after H. felis infection (Figure 3 A and B, insets 1–3), but was detected in a population of infiltrating cells that were F4/80+ (Figure 3B inset 4 and Figure S2A), CD11b+ (Figure S2B), and CD11c+ (Figure S2C). LacZ expression was not detected in CD19+ (B lymphocytes, except for a few transitional cells at the follicle border, Figure S3A, inset), CD3+ (T lymphocytes, Figure S3B) or myeloperoxidase+ cells (MPO, PMNs, Figure S3C). No LacZ expression was detected with the isotype control IgGs (Figure S3D) or in WT mice lacking the LacZ gene insert (Figure S3E). The immunohistochemical analysis was consistent with reduced CD11b and CD11c surface marker gene expression in the Gli1 mutant mice (Figure 3 C–D). However the total frequency of CD45+MHCII+ myeloid cells between 6-month infected WT and Gli1−/− mice was not affected (Figure 3E), indicating a shift in the composition of the myeloid population rather than a defect in total myeloid cell recruitment. Indeed, flow cytometry revealed no significant difference in CD11b+CD11c− cells but instead a 70% reduction in CD11b+CD11c+ myeloid cells in the infected Gli1−/− mice (Figure 3F). This corresponded to a shift in the frequency of CD11c+ cells within the CD11b+ population from 36.5% to 13.1% (Figure 3G). Consistent with this result, we detected by immunofluorescence a significant loss of CD11c+, but not CD11b+ cells in the infected Gli1+/− and Gli1−/− mouse stomachs (Figure S4). Analysis of the bone marrow revealed no detectable CD11b+CD11c+ cells (Figure 3H and I). Although we cannot exclude the presence of CD11b+CD11c+ cells in the bone marrow, which might be dwarfed by the abundance of single positive cells, we concluded that CD11b+CD11c− cells differentiate into a CD11b+CD11c+ population in the stomach in a Gli1-dependent manner. The acquisition of CD11b+CD11c+ cells in the Helicobacter-infected stomachs at 6 months mirrors the CD11b+ subset of tumor infiltrating myelomonocytoid cells described by Umemura et al. that acquire the CD11c+Gr1+ surface markers along with their T cell suppressor function (myeloid-derived suppressor function or MDSCs) [18]. Therefore we concluded that Gli1 is necessary for the differentiation of myeloid cells into CD11b+CD11c+ cells within the preneoplastic gastric microenvironment that correlates with the SPEM.
Figure 3. Gli1 deletion prevents expansion of CD11b+CD11c+ myeloid cell subsets.
A–B) Immunofluorescent detection of β-gal (green), α-SMA (red), and DAPI (blue) in sham- and H. felis-infected Gli1+/LacZ mice. C–D) RT-qPCR analysis of CD11b and CD11c in 2- and 6-month infected mice. E) Bar graphs representing the percentages of CD45+MHCII+ myeloid cells per total gastric cell number in 6-month H. felis-infected WT versus Gli1−/− mice. F) Flow cytometric analysis of CD11b and CD11c in 6-month infected WT and Gli1−/− mice. Values on dot plot represent the percentages of cells (relative to total CD45+MHCII+ myeloid cell population) +/− SEM from N = 3 mice per group. G) Flow cytometry of CD11c+ cells within the CD11b+ stomach population (R1 gate from Figure 3F) of 6-month H. felis-infected WT versus Gli1−/− mice. H) Flow cytometric analysis of CD11b+ bone marrow cells in 6-month H. felis-infected WT and Gli1−/− mice. I) Flow cytometry of CD11c+ cells within the CD11b+ bone marrow population (P1 gate from Figure 3H) of 6-month H. felis-infected WT and Gli1−/− mice. Open and closed bars denote uninfected and infected mice respectively. Error bars represent the mean +/− SEM. For RT-qPCR graphs, N = 5–10 mice per group. For flow sorting bar graph, N = 3 mice per group. Values on flow cytometric dot blots represent the mean +/− SEM from 3 mice per group. *p<0.05.
Schlafen-4 is a Gli1-target gene that marks the CD11b+CD11c+ population associated with gastric metaplasia
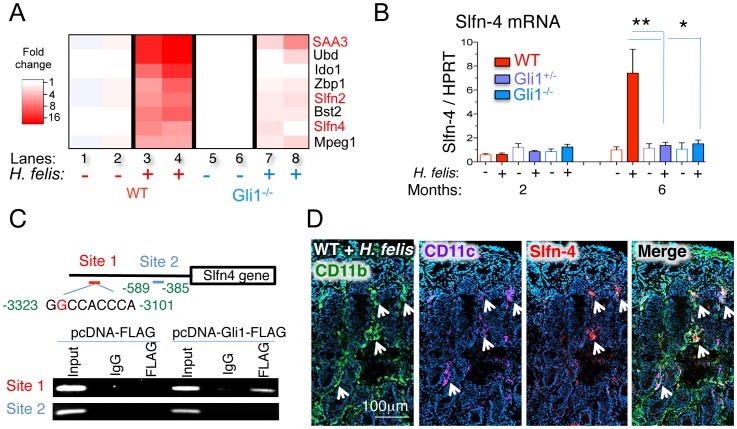
To identify Gli1-dependent myeloid regulatory factors, we performed a microarray analysis of gastric tissue from Helicobacter-infected WT and Gli1−/− mice. The genome-wide analysis revealed eight potential Gli1 targets associated with myeloid cell differentiation during H. felis infection (Figure 4A) in addition to several inflammatory pathways (Figure S5). Of particular interest were two Schlafen genes, Schlafen-4 (Slfn-4), Schlafen-2 (Slfn-2) and serum amyloid protein A3 (SAA3) (Figure 4A), whose gene products were previously implicated in macrophage differentiation [19], [20] and recruitment [21] respectively. RT-qPCR analysis confirmed that Slfn-4 and SAA3 were only induced in the H. felis-infected WT stomach (Figure 4B and Figure S6A). We performed in silico analysis to identify Gli binding sites and identified one site in the Slfn-4 (Figure 4C), one in Slfn-2 and several in the SAA-3 promoters (El-Zaatari and Merchant JL, unpublished data). Chromatin immunoprecipitation (ChIP) demonstrated that Gli1 is present at the Slfn-4 promoter near the Gli1 binding site GGCCACCCA (consensus site = GACCACCCA), but not at a different site that lacked a Gli1 binding sequence (Figure 4C). Since Slfn-4 is associated with aberrant myeloid cell differentiation [19], we further defined which myeloid subpopulations express Slfn-4+ in the stomach. Immunofluorescent staining (Figure 4D and Figure S6B) demonstrated that Slfn-4 was expressed in CD11b+CD11c+ myeloid cells. Flow-cytometric cell sorting coupled with RT-qPCR revealed that these CD11c+ myeloid cells produced significant amounts of IL-1β and TNF-α (Figure S6 C–E). Therefore, we concluded that Slfn-4 is a Gli1 gene target expressed in a specific IL-1β- and TNF-α-expressing subset of CD11b+CD11c+ myeloid population and could be used as an additional marker to identify these cells.
Figure 4. Schlafen-4 is a Gli1-target gene that marks the CD11b+CD11c+ gastric myeloid population.
A) Microarray heat map of sham- and H. felis-infected WT and Gli1−/− stomachs. Each column represents pooled stomach RNA samples from two mice. B) RT-qPCR analysis of Slfn-4 mRNA from sham- or 6 month H. felis-infected WT, Gli1+/−, or Gli1−/− stomachs. C) ChIP analysis of two Slfn-4 promoter sequences in mouse Gli1 promoter in RAW264.7 cells transfected with Gli1-FLAG-overexpressing or the empty vector plasmid. D) Immunofluorescence analysis of CD11b (green), CD11c (magenta), Slfn-4 (red) in 6-month H. felis infected WT mice. Error bars represent the mean +/− SEM. N = 5–10 mice per group. **p<0.01; *p<0.05.
Sonic Hedgehog Remains Epithelial in the Normal and Metaplastic Stomach
Although Shh ligand is secreted acutely from parietal cells within days of Helicobacter infection [12], the cellular source of Shh ligand responsible for Hh signaling has not been examined in the chronically infected stomach once parietal cell atrophy has occurred. We previously showed using a Shh-LacZ mouse that both surface pit and mucous neck cells express Shh despite parietal cell atrophy in 2 month H. felis infected mice [8]. Therefore to identify the source of Shh ligand in the 6-month infected mice, we immunostained WT and Gli1 mutant mice with Shh antibody. Indeed, the Shh ligand was exclusively expressed by E-cadherin+ but not F4/80+ or α-SMA+ cells (Figure S7 A and B) and then was primarily produced by the expanded mucous neck cell population in inflamed stomachs of H. felis-infected WT mice (Figure S7 A–C). Therefore Shh ligand expression remained epithelial and was produced by the expanded mucous neck cell population in the chronically inflamed stomach despite parietal cell atrophy.
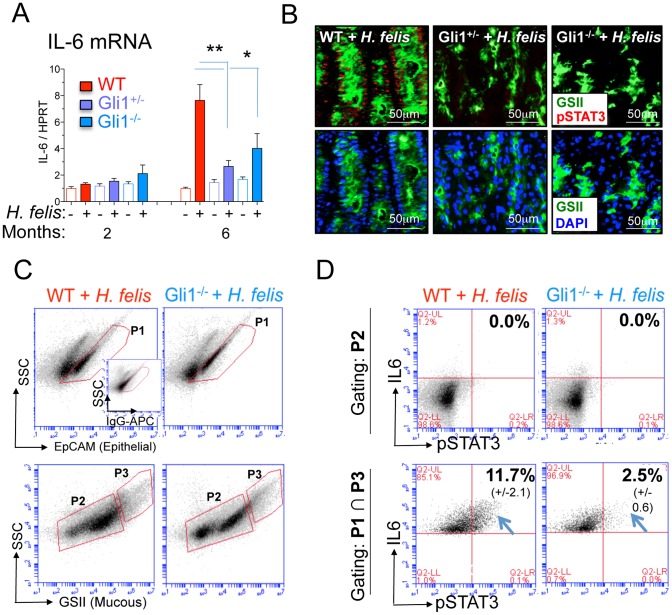
Epithelial Activation of IL-6/pSTAT3 pathway in Gastric Metaplasia
Given that IL-1β induces gastric metaplasia [5], [17] and stimulates expression of the proto-oncogene IL-6 in various cell types [22], [23], [24], [25], we hypothesized that IL-1β induces expansion of the SPEM phenotype by stimulating proliferation and IL-6 expression along with phosphorylation of STAT-3 (pSTAT-3). Surprisingly, we detected IL-6 protein expression primarily in epithelial cells, particularly in metaplastic mucous cells of the H. felis-infected WT mice (Figure S8A) and significantly lower levels of IL-6 in the mucosa of Gli1+/− and Gli1−/− mice (Figure S8A). Moreover, laser capture microdissection (LCM) confirmed that most of the IL-6 expression was produced by epithelial cells after 6 months of infection (Figure S8 B and C). RT-qPCR analysis confirmed the reduction of IL-6 in infected Gli1+/− and Gli1−/− stomach tissue compared to WT mice (Figure 5A). The expanded population of mucous cells from the infected WT stomachs also expressed higher levels of pSTAT3 than the gastric mucosa from the Gli1+/− and Gli1−/− (Figure 5B). Dissociated gastric cells were analyzed by flow cytometry using an epithelial (EpCAM) and mucous neck cell marker (GSII lectin) (Figure 5C). The GSII− [P2] population, which excludes mucous neck cells, was IL-6 and pSTAT3 negative (Figure 5 D, Upper Panel). However, a proportion of the EpCAM+GSII+ population (epithelial mucous neck cells, [P1] ∩ [P3], Figure 5C) co-expressed IL-6 and pSTAT3 (Figure 5D, lower panel). The proportion of epithelial mucous neck cells co-expressing IL-6 and pSTAT3 was significantly lower in infected Gli1−/− mice than WT mice (Figure 5D, lower panel). This corroborated the immunohistochemical observation that GSII+ mucous neck cells expressed higher levels of pSTAT-3 in 6-month infected WT than in Gli1-deficient mice (Figure 5B). Consistent with the induction of the proto-oncogene pSTAT3, mucous neck cells showed higher expression of the proliferation marker Ki67 in infected WT but not in Gli1-deficient mice (Figure S9A). To determine whether IL-1β induced IL-6 gene expression in mucous cells, we treated the NCI-N87 gastric mucous cell line with IL-1β. IL-1β induced IL-6 gene expression in these cells and the induction was inhibited by pre-incubating with the IL-1β receptor antagonist IL-1RA (Figure S9B). Therefore, we concluded that pro-inflammatory cytokines, such as IL-1β, stimulate epithelial IL-6 expression leading to increased pSTAT3 and proliferation of the mucous cell lineage.
Figure 5. Gli1 deletion prevents IL-6 and pSTAT3 expression in SPEM.
A) RT-qPCR analysis of IL-6 mRNA. Error bars represent the mean +/− SEM. N = 5–10 mice per group. Open and closed bars denote uninfected and infected mice respectively. B) Immunofluorescent analysis of pSTAT3 (red), GSII (green) and DAPI (blue) in stomachs of 6-month infected mice. C) Flow cytometric gating for EpCAM+ (epithelial cells) and GSII+ (mucous neck cells, SPEM) in 6-month infected WT and Gli1−/− mice. The inset shows the rat IgG-APC isotype control for the EpCAM-APC antibody. D) IL-6 and pSTAT3 expression in GSII− (non-mucous, gate P2), and EpCAM+GSII+ (mucous neck epithelial cells, gate P1 ∩ P3) in 6-month infected WT and Gli1−/− mice. Values on flow cytometric dot blots represent the mean +/− SEM from 3 mice per group. **p<0.01; *p<0.05.
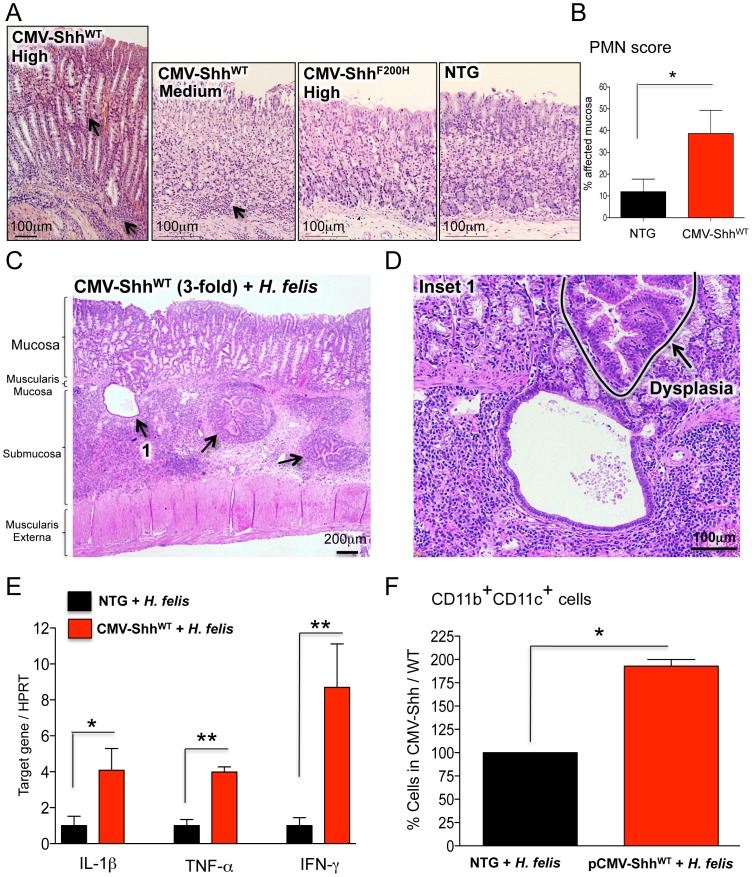
Sonic Hedgehog Ligand is Not Sufficient to Induce Gastric Metaplasia
Although Gli1 deletion demonstrated that a component of the Hh signaling pathway is necessary for inflammation and gastric metaplasia, we examined whether ectopic expression of the Shh ligand was sufficient to induce these lesions. We expressed N-terminally HA-tagged WT (ShhWT) and mutant (ShhF200H) Shh cDNA (F200H) from the CMV promoter. The ShhF200H mutant cannot be cleaved to the functional 19 kDa ShhN form (Figure S10A). As previously reported [9], WT-Shh is efficiently cleaved when expressed in NIH-3T3 cells while the F200H mutant Shh only generated the uncleaved, full-length form (Figure S10B). WT-Shh induced Gli1 and Patched-1 (Ptch1) mRNA indicating active signaling, whereas the F200H form was non-functional and did not induce these two target genes (Figure S10C and S10D). Transgenic mice overexpressing CMV-ShhWT and CMV-ShhF200H plasmids exhibited variable transgene expression in different founder lines (Table 1). Interestingly, the CMV promoter expressed the Shh transgene exclusively in gastric chief cells (Figure S10 E and F). Overexpression of ShhWT induced gastritis (3/3 medium expressing mice) at medium Shh levels and gastric metaplasia only in the highest-expressing founder (Figure 6 A and B and Table 1) characterized by the co-localization of TFF-2 and intrinsic factor (Figure S11A). By contrast, none of the CMV-ShhF200H mice developed gastritis despite high expression of the transgene in some of these mice (Figure 6A and Table 1). Overexpression of WT or the F200H Shh from the parietal cell-specific H+/K+-β-ATPase promoter produced a similar phenotype to the CMV-ShhWT mice marked by gastritis and infrequent metaplasia (Figure S10A, Figure S11 and Table 1). Therefore, overexpression of the Shh ligand was sufficient to induce gastric inflammation, but the presence of SPEM occurred infrequently in mice correlating with high expression of the transgene. Therefore, we concluded that Shh is sufficient to induce gastritis but not SPEM, although we cannot exclude the possibility that atrophy and metaplasia develop above a certain threshold level of Shh.
Table 1. Shh overexpression exacerbates Helicobacter gastritis.
| Founders | Average Fold Expression of Shh/NTG | Histology | ||
| Normal | Inflammation | Metaplasia | ||
| CMV-ShhWT (High Expressor) | 102.0 | 0/1 | 1/1 | 1/1 |
| CMV-ShhWT (Medium Expressor) | 6.8 (+/−1.9) | 0/3 | 3/3 | 0/3 |
| CMV-ShhWT (Low Expressor) | 0.78 (+/−0.3) | 5/5 | 0/5 | 0/5 |
| CMV-ShhF200H(High Expressor) | 82.5 | 1/1 | 0/1 | 0/1 |
| CMV-ShhF200H (Medium Expressor) | 19.5 (+/−9.3) | 7/7 | 0/7 | 0/7 |
| CMV-ShhF200H (Low Expressor) | 0.3 | 1/1 | 0/1 | 0/1 |
| H+/K+-β-ShhWT (Medium Expressor) | 5.23 (+/−0.6) | 1/3 | 2/3 | 2/3 |
| H+/K+-β-ShhWT (Low Expressor) | 1.62 (+/−0.3) | 6/6 | 0/6 | 0/6 |
| H+/K+-β-ShhF00H (Medium Expressor) | 13 | 1/1 | 0/1 | 0/1 |
| H+/K+-β-ShhF200H (Low Expressor) | 0.92 (+/−0.2) | 4/4 | 0/4 | 0/4 |
High expressor >70-fold; Medium expressor >3-fold; Low expressor <3-fold. Average transgenic expression for low-, medium- and high-expressing founders is indicated. The number of mice with normal gastric phenotype, inflammation or metaplasia is also indicated.
Figure 6. Shh overexpression exacerbates Helicobacter gastritis.
A) H&E staining of NTG, CMV-ShhWT and CMV-ShhF200H mouse stomachs. Arrows indicate regions of inflammatory cell infiltration and mucous neck cell expansion, SPEM. B) Polymorphonuclear (PMN) cell scoring in NTG and CMV-ShhWT mice. C) H&E staining of 6-month infected CMV-ShhWT stomach. D) High power H&E staining of herniated epithelial glands in the gastric submucosa in 6-month infected CMV-ShhWT stomach. A dysplastic area is indicated. E) RT-qPCR analysis of IL-1β, TNF-α and IFN-γ in 6 month-infected CMV-ShhWT versus NTG stomachs. F) Bar graphs representing flow cytometric analysis of CD11b+CD11c+ cells in 6-month infected NTG and CMV-ShhWT stomachs. Positive cell numbers in infected CMV-ShhWT mice are expressed as a percentage of cells present in infected NTG controls (set to a 100%). Error bars represent the mean +/− SEM. N = 3 mice per group. **p<0.01; *p<0.05.
Since the primary phenotype of the CMV-ShhWT mice was a mild gastritis, we tested whether infecting these mice with Helicobacter affected the severity of the inflammation and subsequent development of SPEM. CMV-ShhWT mice from founder line 314 (medium transgene expression levels) and non-transgenic littermates were infected for 6 months with H. felis. All three CMV-ShhWT mice displayed high-grade dysplasia (Figure 6 C and D), epithelial gland herniation into the submucosa (Figure 6 C and D), and elevated Th1 cytokines compared to infected non-transgenic mice (Figure 6E). Furthermore, these mice exhibited significantly higher CD11b+CD11c+ myeloid cells compared to infected non-transgenic controls (Figure 6F). Therefore, Shh overexpression exacerbated the histologic severity observed with H. felis infection and increased the amount of myeloid cells recruited to the inflamed stomach compared to non-transgenic mice.
Discussion
Cancer is a sequential process initiated by epithelial cell aberrations that can be triggered and sustained by immune and stromal cell types within the tumor microenvironment [26], [27]. In the stomach, infection by Helicobacter is correlated with the development of gastric cancer in the setting of chronic atrophic gastritis [28]. Since Hh signaling has been linked to a variety of cancers, some of which develop in the setting of inflammation [29], [30], [31], we examined the role of Gli1, a known component of the Hh signaling pathway, in the transition from chronic inflammation to mucous neck cell metaplasia or SPEM. Gli1 is expressed in both intestinal myeloid and myofibroblasts [11], consistent with prior studies showing Gli1 in hematopoietic stem cells [32], [33]. We can only speculate as to why Gli1 is lost only in α-SMA+ cells of the muscularis mucosa in response to H. felis, but not in the intraglandular mesenchyme. Nevertheless similar changes occurred in the H+/K+-ATPase-IFN-γ mice [34], and might pertain to the heterogeneity of the α-SMA+ cells responding to the inflammatory cytokines [5].
In the current study, we examined the role of Gli1 during chronic Helicobacter infection and found that Gli1 was required for SPEM to develop. In addition, the inflammation correlated with infiltration of a specific subpopulation of myeloid cells. SPEM arose in response to Helicobacter-induced expansion of the mucous neck cell compartment and acquired proliferative markers including Ki67, IL-6 and pSTAT3. Examining Gli1-dependent myeloid cell subpopulations that correlated with the metaplastic phenotype, we found that these populations were CD45+MHCII+CD11b+CD11c+. Interestingly, the myeloid subpopulation that expressed CD11c only appeared in the stomach and not in the bone marrow, suggesting that the acquisition of the CD11c surface marker occurs within the inflamed gastric environment. However, additional phenotypic and functional characterization of this CD11b+CD11c+ population requires their direct isolation. Since using Gli1 antibodies to sort for these myeloid subsets is not currently feasible, we used genome-wide analysis to identify Gli1 gene targets in myeloid cells, e.g., Slfn-4, which might be used to isolate these cells for further study. We demonstrated here that Slfn-4 contains a Gli1 binding site in its promoter. Moreover, Slfn4 expression has been linked to aberrant myelopoiesis [19], making it a relevant regulatory gene contributing to generation of the various Gli1-dependent myeloid populations during Helicobacter infection. Therefore, the current phenotype of the myeloid subset correlating with SPEM development is a CD45+MHCII+CD11b+CD11c+Slfn4+ cell that expresses IL-1β and TNFα. Although this phenotype is consistent with an “inflammatory monocyte” activated in the periphery by tissue damage and inflammation [18], [35], the actual function of these cells awaits their isolation and further characterization including chemokine profile and possible suppressor activity.
Although Shh ligand induces Gli1 expression, we previously reported that Shh is lost from parietal cells in the presence of inflammatory cytokines [8]. Indeed reduced Shh expression has been reported in H. pylori-infected human subjects [36]. Nevertheless we show here that metaplastic mucous cells express Shh ligand in the absence of parietal cells. In a study of myeloid cell progenitors using Gli1LacZ mice, Merchant et al. demonstrate loss of hematopoietic stem cell proliferation in Gli1 null mice and suggest that non-Hh ligand mechanisms regulate bone marrow myeloid cell populations [33]. Likewise, we also found no evidence for ligand-mediated induction of Gli1 in RAW 264.7 cells, a mouse myeloid cell line (El-Zaatari and Merchant JL, unpublished data). Indeed, ectopic Shh expression was generally not sufficient to induce metaplasia in the absence of the Helicobacter – mediated inflammation. This result was consistent with Gli1 regulation of myeloid subpopulations being required but not ligand-dependent. Thus while parietal cell secretion of Shh into the circulation is important for the initial recruitment of inflammatory cells to the stomach [12], the ligand does not appear to mediate further myeloid cell differentiation ostensibly required for the pre-neoplastic changes.
We also characterized the mechanism by which IL-1β induces mucous cell expansion and metaplasia. A recent study shows that IL-6 mediates IL-1β induction of Barrett's metaplasia of the esophagus [25]. IL-6 binds the gp130 receptor [37] and induces the proto-oncogene pSTAT3 [38], which stimulates proliferation. It is well documented that activation of the gp130 cytokine receptor is sufficient to mediate gastric cancer [39]. These studies also demonstrated that IL-11, rather than IL-6, mediates the effects of gp130 activation on gastric tumorigenesis [39]. However, transfer of bone marrow cells exhibiting overactive gp130 is not sufficient to induce gastric cancer indicating that gp130-induced gastric carcinogenesis is epithelial cell-mediated [39], and might represent a distinct mechanism from Helicobacter-induced carcinogenesis. Administration of IL-11 induces foveolar gastric hyperplasia but does not induce SPEM [40]. Therefore in contrast to gp130-mediated antral tumors driven by the IL-6 family member IL-11, we show here that Helicobacter infection drives IL-6 expression and SPEM. Surprisingly, the expanded mucous epithelial cells of the stomach corpus were a major source of IL-6. Correlating with increased IL-6 production by mucous cells, these cells expressed elevated levels of pSTAT3 and were highly proliferative. Indeed, IL-1β induced IL-6 expression in gastric mucous cells suggesting that myeloid cell-secreted cytokines target epithelial cells. This data demonstrated that stromal signals from myeloid cells activate the proliferation of gastric epithelial cells leading to SPEM.
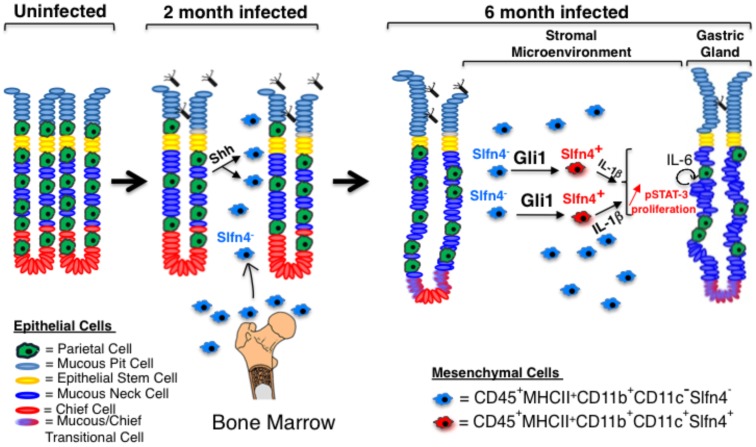
As suggested by Merchant et al., Gli1 is not required for myeloid cell development or maintenance but rather is required when stress-induced hematopoiesis increases the common progenitor pool of cells that differentiate into specific myeloid subsets [33]. We conclude that in the stomach, this pathway has two phases (Figure 7). First, bacterial infection induces the production and release of Shh ligand to stimulate the recruitment of Gli1+ myeloid cells to the stomach [12]. However during chronic infection, these same Gli1+ cells acquire new markers through Hh ligand-independent mechanisms that promote metaplasic, proliferative changes in the gastric epithelium, setting the stage for neoplastic transformation.
Figure 7. Proposed role of Hedgehog-dependent myeloid cells in Helicobacter-induced gastric metaplasia.
Upon Helicobacter infection, CD11b+CD11c−Slfn-4− myeloid cells migrate from the bone marrow to the stomach in response to Shh ligand secretion from parietal cells. Prolonged exposure of these cells to the inflamed gastric environment induced by H. felis (∼6 months) induces a phenotypic shift into CD11b+CD11c+ myeloid cells, which express the marker Slfn-4. Gli1 expression in myeloid cells is necessary for this shift. Myeloid cell-derived IL-1β triggers the IL-6/pSTAT-3 pathway in epithelial mucous cells leading to mucous neck cell proliferation at the expense of parietal and chief cells (gland atrophy) and metaplasia (SPEM).
Supporting Information
Infected wild-type mice develop gastric SPEM. Left Panel, Confocal imaging of trefoil factor 2 (TFF-2, green) and DAPI (blue) in the stomach of 6-month H. felis-infected wild-type mice. Insets (high power), co-immunofluorescent imaging of TFF-2 (green) and intrinsic factor (red), in 6-month H. felis-infected wild-type mice with and without DAPI (blue, middle and left panels respectively). Numbers on insets indicate magnified regions.
(TIF)
Infiltrating myeloid cells express Gli1. A) Immunofluorescent analysis of β-gal (red), F4/80 (green), and DAPI (blue) in H. felis-infected Gli1+/LacZ mice. B) Immunofluorescent detection of β-gal (red), CD11b (green), and DAPI (blue) in H. felis-infected Gli1+/LacZ mice. C) Immunofluorescent detection of β-gal (red), CD11c (green), and DAPI (blue) in H. felis-infected Gli1+/LacZ mice. Analysis was performed in 6-month infected mice. Numbers on insets indicate magnified regions.
(TIF)
B cells, T cells, and neutrophils do not express Gli1. A) Immunofluorescent analysis of β-gal (red), CD19 (green), and DAPI (blue). B) Immunofluorescence of β-gal (red), CD3 (green) and DAPI (blue). C) Immunofluorescence of β-gal (red), MPO (green), and DAPI (blue). D) Concentration-matched hamster, rat and rabbit isotype controls. E) Immunofluorescent analysis of β-gal expression in Gli1+/LacZ versus WT mice. Numbers on insets indicate magnified regions.
(TIF)
Gli1 deletion prevents CD11c+ cell expansion in 6-month H. felis -infected stomachs. Confocal imaging of CD11b (green, top panel), CD11c (green, lower panel) and DAPI (blue) in 6-month H. felis-infected WT, Gli1+/− and Gli1−/− stomachs.
(TIF)
Gli1 deletion prevents the induction of pro-inflammatory genes in H. felis -infected stomachs at after 6 months of infection. Microarray heat map of H. felis-induced and Gli1-dependent gene groups in the stomach. Each column represents pooled stomach RNA samples from two mice.
(TIF)
Gli1 is required for the appearance of a Slfn-4+ population that produces IL-1β and TNF-α. A) RT-qPCR analysis of SAA3 mRNA from sham- or 6 month H. felis-infected WT, Gli1+/−, or Gli1−/− stomachs. B) Immunofluorescent analysis of CD11c (green) and Slfn-4 (red) in 6-month infected WT mice. C) Flow cytometric analysis of F4/80 and CD11c in 6-month infected WT mouse stomachs. D) Flow cytometric analysis of F4/80 and CD11c subtypes in CD45+Slfn4+ myeloid cells from 6-month infected WT mouse stomachs. F) Semi-quantitative RT-PCR analysis of flow sorted F4/80−CD11c− (P9) versus F4/80+CD11c+ (P3) cells. Numbers on insets indicate magnified regions. Arrows indicate co-localization of Slfn4, CD11b and CD11c markers.
(TIF)
Epithelial cells express Shh ligand. A) Triple immunofluorescent staining of Shh (red), E-cadherin (epithelial, green) and F4/80 (mesenchymal, grey) in sham- and H. felis-infected WT, Gli1+/− and Gli1−/− mice. B) Double immunofluorescent staining of Shh (red) and α-SMA (green). C) Double staining of Shh (red) and GSII (green). White arrows indicate the expanded mucous neck cell lineage in H. felis-infected WT mice at 6 months.
(TIF)
Gli1 deletion prevents mucous neck cell compartment expansion and IL-6 production. A) Immunofluorescent analysis of GSII (green) and IL-6 (red) in 6-month infected mice. B) Laser Capture Microdissection of toluidine stained section. Shown are the micrographs showing the epithelial (top panel) versus stromal regions (lower panel) from a 6-month infected WT stomach with SPEM. C) Semi-quantitative RT-PCR analysis of IL-6 in epithelial versus stromal laser microdissectates.
(TIF)
Gli1 and IL-1β mediate mucous cell proliferation and IL-6 production by mucous cells respectively. A) Immunofluorescent staining for Ki67 (red) and GSII (green)-positive cells in 6-month infected stomachs. (B) RT-qPCR analysis of IL-6 in NCI-N87 cells treated with or without IL-6, IL-1β, and IL-1RA. Error bars represent the mean +/− SEM. *p<0.05.
(TIF)
Transgene expression of Shh ligand in mice. A) Diagrammatic representation of pCMV-ShhWT, pCMV-ShhF200H, H+/K+-ATPase-β-ShhWT, and H+/K+-ATPase-β-ShhF200H constructs. The HA tag is indicated. B) Western blotting analysis of NIH-3T3 conditioned media following transfection with pCMV-ShhWT and pCMV-ShhF200H. C–D) RT-qPCR of Gli1 and Ptch1 mRNA expression in NIH-3T3 cells transfected with pCMV-ShhWT or pCMV-ShhF200H plasmids. (E) Upper Panel: Immunofluorescent staining of GSII (green) and HA-tag (red) in NTG, pCMV-ShhWT-HA, and pCMV-ShhF200H-HA mouse stomachs. Middle Panel: Shh (green) and H+/K+-ATPase (red). Lower Panel: intrinsic factor (IF; green) and HA-tag (red). (F) Immunofluorescent staining of Shh (green) and HA-tag (red) in pCMV-ShhWT and pCMV-ShhF200H mouse stomachs. A short 15 min incubation with Shh antibody was used to detect transgenes overexpressing ShhWT and ShhF200H without cross-reacting with endogenous gastric Shh. Error bars represent the mean +/− SEM. *p<0.05.
(TIF)
SPEM in pCMV-ShhWT and H+/K+-ATPase-β-ShhWT mice. A) Left panel, Immunofluorescent staining of TFF-2 (green), intrinsic factor (IF, red), and DAPI (blue) in the highest expressing pCMV-ShhWT mouse stomach (102-fold expression of Shh, please refer to Table 1); right panel, high power magnification of the co-localization of IF (red) and TFF-2 (green). B) H&E staining of mouse stomach in two H+/K+-ATPase-β-ShhWT founder mice expressing medium levels of the transgene (please refer to Table 1). C) Left panel, immunofluorescent staining of TFF-2 (green), intrinsic factor (IF, red), and DAPI (blue) in an H+/K+-ATPase-β-ShhWT founder mouse expressing medium levels of the transgene (please refer to Table 1); right panel, high power magnification of the co-localization of IF (red) and TFF-2 (green). Arrows indicate regions of inflammatory cell infiltration and mucous neck cell expansion.
(TIF)
Acknowledgments
We thank Y. Zavros for providing the flow-cytometry protocol, D. Alpers for providing the intrinsic factor antibody, J. D. Engel for providing the β-gal antibody, N. Wright for providing the TFF-2 antibody, Stephanie Daignault-Newton for assistance with the statistical analysis of the data, A. Dlugosz for providing the pcDNA3.1-FLAG-Gli1 plasmid, and L. C. Samuelson and A. Dlugosz for providing critical feedback regarding the manuscript.
Funding Statement
This study was supported by Public Health Service Grants P01-DK62041 (JLM) and R01 DK087708-01 (JYK). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Correa P, Haenszel W, Cuello C, Tannenbaum S, Archer M (1975) A model for gastric cancer epidemiology. Lancet 2: 58–60. [DOI] [PubMed] [Google Scholar]
- 2. Goldenring JR, Nam KT, Mills JC (2011) The origin of pre-neoplastic metaplasia in the stomach: chief cells emerge from the Mist. Experimental cell research 317: 2759–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schmidt PH, Lee JR, Joshi V, Playford RJ, Poulsom R, et al. (1999) Identification of a metaplastic cell lineage associated with human gastric adenocarcinoma. Laboratory investigation; a journal of technical methods and pathology 79: 639–646. [PMC free article] [PubMed] [Google Scholar]
- 4. Houghton J, Stoicov C, Nomura S, Rogers AB, Carlson J, et al. (2004) Gastric cancer originating from bone marrow-derived cells. Science 306: 1568–1571. [DOI] [PubMed] [Google Scholar]
- 5. Tu S, Bhagat G, Cui G, Takaishi S, Kurt-Jones EA, et al. (2008) Overexpression of interleukin-1beta induces gastric inflammation and cancer and mobilizes myeloid-derived suppressor cells in mice. Cancer Cell 14: 408–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beachy PA, Karhadkar SS, Berman DM (2004) Tissue repair and stem cell renewal in carcinogenesis. Nature 432: 324–331. [DOI] [PubMed] [Google Scholar]
- 7. van den Brink GR, Hardwick JC, Tytgat GN, Brink MA, Ten Kate FJ, et al. (2001) Sonic hedgehog regulates gastric gland morphogenesis in man and mouse. Gastroenterology 121: 317–328. [DOI] [PubMed] [Google Scholar]
- 8.Waghray M, Zavros Y, Saqui-Salces M, El-Zaatari M, Alamelumangapuram CB, et al.. (2010) Interleukin-1beta promotes gastric atrophy through suppression of Sonic Hedgehog. Gastroenterology 138: 562–572, 572 e561–562. [DOI] [PMC free article] [PubMed]
- 9. Zavros Y, Waghray M, Tessier A, Bai L, Todisco A, et al. (2007) Reduced pepsin A processing of sonic hedgehog in parietal cells precedes gastric atrophy and transformation. The Journal of biological chemistry 282: 33265–33274. [DOI] [PubMed] [Google Scholar]
- 10.El-Zaatari M, Zavros Y, Tessier A, Waghray M, Lentz S, et al.. (2010) Intracellular calcium release and protein kinase C activation stimulate sonic hedgehog gene expression during gastric acid secretion. Gastroenterology 139: 2061–2071 e2062. [DOI] [PMC free article] [PubMed]
- 11. Kolterud A, Grosse AS, Zacharias WJ, Walton KD, Kretovich KE, et al. (2009) Paracrine Hedgehog signaling in stomach and intestine: new roles for hedgehog in gastrointestinal patterning. Gastroenterology 137: 618–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schumacher MA, Donnelly JM, Engevik AC, Xiao C, Yang L, et al.. (2012) Gastric Sonic Hedgehog acts as a macrophage chemoattractant during the immune response to Helicobacter pylori. Gastroenterology 142: 1150–1159 e1156. [DOI] [PMC free article] [PubMed]
- 13. Mohammadi M, Czinn S, Redline R, Nedrud J (1996) Helicobacter-specific cell-mediated immune responses display a predominant Th1 phenotype and promote a delayed-type hypersensitivity response in the stomachs of mice. Journal of immunology 156: 4729–4738. [PubMed] [Google Scholar]
- 14. Ihrig M, Whary MT, Dangler CA, Fox JG (2005) Gastric helicobacter infection induces a Th2 phenotype but does not elevate serum cholesterol in mice lacking inducible nitric oxide synthase. Infection and immunity 73: 1664–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Obonyo M, Rickman B, Guiney DG (2011) Effects of myeloid differentiation primary response gene 88 (MyD88) activation on Helicobacter infection in vivo and induction of a Th17 response. Helicobacter 16: 398–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Franchi L, Amer A, Body-Malapel M, Kanneganti TD, Ozoren N, et al. (2006) Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nature immunology 7: 576–582. [DOI] [PubMed] [Google Scholar]
- 17. El-Omar EM, Carrington M, Chow WH, McColl KE, Bream JH, et al. (2000) Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature 404: 398–402. [DOI] [PubMed] [Google Scholar]
- 18. Umemura N, Saio M, Suwa T, Kitoh Y, Bai J, et al. (2008) Tumor-infiltrating myeloid-derived suppressor cells are pleiotropic-inflamed monocytes/macrophages that bear M1- and M2-type characteristics. J Leukoc Biol 83: 1136–1144. [DOI] [PubMed] [Google Scholar]
- 19. van Zuylen WJ, Garceau V, Idris A, Schroder K, Irvine KM, et al. (2011) Macrophage activation and differentiation signals regulate schlafen-4 gene expression: evidence for Schlafen-4 as a modulator of myelopoiesis. PloS one 6: e15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berger M, Krebs P, Crozat K, Li X, Croker BA, et al. (2010) An Slfn2 mutation causes lymphoid and myeloid immunodeficiency due to loss of immune cell quiescence. Nat Immunol 11: 335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hiratsuka S, Watanabe A, Sakurai Y, Akashi-Takamura S, Ishibashi S, et al. (2008) The S100A8-serum amyloid A3-TLR4 paracrine cascade establishes a pre-metastatic phase. Nature cell biology 10: 1349–1355. [DOI] [PubMed] [Google Scholar]
- 22. Luo G, Hershko DD, Robb BW, Wray CJ, Hasselgren PO (2003) IL-1beta stimulates IL-6 production in cultured skeletal muscle cells through activation of MAP kinase signaling pathway and NF-kappa B. American journal of physiology Regulatory, integrative and comparative physiology. 284: R1249–1254. [DOI] [PubMed] [Google Scholar]
- 23. Tsukihara S, Harada T, Deura I, Mitsunari M, Yoshida S, et al. (2004) Interleukin-1beta-induced expression of IL-6 and production of human chorionic gonadotropin in human trophoblast cells via nuclear factor-kappaB activation. American journal of reproductive immunology 52: 218–223. [DOI] [PubMed] [Google Scholar]
- 24. Kauma SW, Turner TT, Harty JR (1994) Interleukin-1 beta stimulates interleukin-6 production in placental villous core mesenchymal cells. Endocrinology 134: 457–460. [DOI] [PubMed] [Google Scholar]
- 25. Quante M, Bhagat G, Abrams JA, Marache F, Good P, et al. (2012) Bile Acid and inflammation activate gastric cardia stem cells in a mouse model of barrett-like metaplasia. Cancer cell 21: 36–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hanahan D, Coussens LM (2012) Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21: 309–322. [DOI] [PubMed] [Google Scholar]
- 27. Mbeunkui F, Johann DJ Jr (2009) Cancer and the tumor microenvironment: a review of an essential relationship. Cancer Chemother Pharmacol 63: 571–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamb A, Chen LF (2012) Role of the Helicobacter pylori-induced inflammatory response in the development of gastric cancer. J Cell Biochem. [DOI] [PMC free article] [PubMed]
- 29. Wang J, Pham-Mitchell N, Schindler C, Campbell IL (2003) Dysregulated Sonic hedgehog signaling and medulloblastoma consequent to IFN-alpha-stimulated STAT2-independent production of IFN-gamma in the brain. J Clin Invest 112: 535–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lauth M, Toftgard R (2011) Hedgehog signaling and pancreatic tumor development. Adv Cancer Res 110: 1–17. [DOI] [PubMed] [Google Scholar]
- 31. Nakashima H, Nakamura M, Yamaguchi H, Yamanaka N, Akiyoshi T, et al. (2006) Nuclear factor-kappaB contributes to hedgehog signaling pathway activation through sonic hedgehog induction in pancreatic cancer. Cancer Res 66: 7041–7049. [DOI] [PubMed] [Google Scholar]
- 32. Bhardwaj G, Murdoch B, Wu D, Baker DP, Williams KP, et al. (2001) Sonic hedgehog induces the proliferation of primitive human hematopoietic cells via BMP regulation. Nat Immunol 2: 172–180. [DOI] [PubMed] [Google Scholar]
- 33. Merchant A, Joseph G, Wang Q, Brennan S, Matsui W (2010) Gli1 regulates the proliferation and differentiation of HSCs and myeloid progenitors. Blood 115: 2391–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Syu LJ, El-Zaatari M, Eaton KA, Liu Z, Tetarbe M, et al. (2012) Transgenic expression of interferon-gamma in mouse stomach leads to inflammation, metaplasia, and dysplasia. Am J Pathol 181: 2114–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Strauss-Ayali D, Conrad SM, Mosser DM (2007) Monocyte subpopulations and their differentiation patterns during infection. J Leukoc Biol 82: 244–252. [DOI] [PubMed] [Google Scholar]
- 36. Shiotani A, Iishi H, Uedo N, Ishihara R, Ishiguro S, et al. (2006) Helicobacter pylori-induced atrophic gastritis progressing to gastric cancer exhibits sonic hedgehog loss and aberrant CDX2 expression. Aliment Pharmacol Ther 24 Suppl 471–80. [DOI] [PubMed] [Google Scholar]
- 37. Taga T, Hibi M, Hirata Y, Yamasaki K, Yasukawa K, et al. (1989) Interleukin-6 triggers the association of its receptor with a possible signal transducer, gp130. Cell 58: 573–581. [DOI] [PubMed] [Google Scholar]
- 38. Gao SP, Mark KG, Leslie K, Pao W, Motoi N, et al. (2007) Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. The Journal of clinical investigation 117: 3846–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jenkins BJ, Grail D, Nheu T, Najdovska M, Wang B, et al. (2005) Hyperactivation of Stat3 in gp130 mutant mice promotes gastric hyperproliferation and desensitizes TGF-beta signaling. Nature medicine 11: 845–852. [DOI] [PubMed] [Google Scholar]
- 40. Howlett M, Chalinor HV, Buzzelli JN, Nguyen N, van Driel IR, et al. (2012) IL-11 is a parietal cell cytokine that induces atrophic gastritis. Gut 61: 1398–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Infected wild-type mice develop gastric SPEM. Left Panel, Confocal imaging of trefoil factor 2 (TFF-2, green) and DAPI (blue) in the stomach of 6-month H. felis-infected wild-type mice. Insets (high power), co-immunofluorescent imaging of TFF-2 (green) and intrinsic factor (red), in 6-month H. felis-infected wild-type mice with and without DAPI (blue, middle and left panels respectively). Numbers on insets indicate magnified regions.
(TIF)
Infiltrating myeloid cells express Gli1. A) Immunofluorescent analysis of β-gal (red), F4/80 (green), and DAPI (blue) in H. felis-infected Gli1+/LacZ mice. B) Immunofluorescent detection of β-gal (red), CD11b (green), and DAPI (blue) in H. felis-infected Gli1+/LacZ mice. C) Immunofluorescent detection of β-gal (red), CD11c (green), and DAPI (blue) in H. felis-infected Gli1+/LacZ mice. Analysis was performed in 6-month infected mice. Numbers on insets indicate magnified regions.
(TIF)
B cells, T cells, and neutrophils do not express Gli1. A) Immunofluorescent analysis of β-gal (red), CD19 (green), and DAPI (blue). B) Immunofluorescence of β-gal (red), CD3 (green) and DAPI (blue). C) Immunofluorescence of β-gal (red), MPO (green), and DAPI (blue). D) Concentration-matched hamster, rat and rabbit isotype controls. E) Immunofluorescent analysis of β-gal expression in Gli1+/LacZ versus WT mice. Numbers on insets indicate magnified regions.
(TIF)
Gli1 deletion prevents CD11c+ cell expansion in 6-month H. felis -infected stomachs. Confocal imaging of CD11b (green, top panel), CD11c (green, lower panel) and DAPI (blue) in 6-month H. felis-infected WT, Gli1+/− and Gli1−/− stomachs.
(TIF)
Gli1 deletion prevents the induction of pro-inflammatory genes in H. felis -infected stomachs at after 6 months of infection. Microarray heat map of H. felis-induced and Gli1-dependent gene groups in the stomach. Each column represents pooled stomach RNA samples from two mice.
(TIF)
Gli1 is required for the appearance of a Slfn-4+ population that produces IL-1β and TNF-α. A) RT-qPCR analysis of SAA3 mRNA from sham- or 6 month H. felis-infected WT, Gli1+/−, or Gli1−/− stomachs. B) Immunofluorescent analysis of CD11c (green) and Slfn-4 (red) in 6-month infected WT mice. C) Flow cytometric analysis of F4/80 and CD11c in 6-month infected WT mouse stomachs. D) Flow cytometric analysis of F4/80 and CD11c subtypes in CD45+Slfn4+ myeloid cells from 6-month infected WT mouse stomachs. F) Semi-quantitative RT-PCR analysis of flow sorted F4/80−CD11c− (P9) versus F4/80+CD11c+ (P3) cells. Numbers on insets indicate magnified regions. Arrows indicate co-localization of Slfn4, CD11b and CD11c markers.
(TIF)
Epithelial cells express Shh ligand. A) Triple immunofluorescent staining of Shh (red), E-cadherin (epithelial, green) and F4/80 (mesenchymal, grey) in sham- and H. felis-infected WT, Gli1+/− and Gli1−/− mice. B) Double immunofluorescent staining of Shh (red) and α-SMA (green). C) Double staining of Shh (red) and GSII (green). White arrows indicate the expanded mucous neck cell lineage in H. felis-infected WT mice at 6 months.
(TIF)
Gli1 deletion prevents mucous neck cell compartment expansion and IL-6 production. A) Immunofluorescent analysis of GSII (green) and IL-6 (red) in 6-month infected mice. B) Laser Capture Microdissection of toluidine stained section. Shown are the micrographs showing the epithelial (top panel) versus stromal regions (lower panel) from a 6-month infected WT stomach with SPEM. C) Semi-quantitative RT-PCR analysis of IL-6 in epithelial versus stromal laser microdissectates.
(TIF)
Gli1 and IL-1β mediate mucous cell proliferation and IL-6 production by mucous cells respectively. A) Immunofluorescent staining for Ki67 (red) and GSII (green)-positive cells in 6-month infected stomachs. (B) RT-qPCR analysis of IL-6 in NCI-N87 cells treated with or without IL-6, IL-1β, and IL-1RA. Error bars represent the mean +/− SEM. *p<0.05.
(TIF)
Transgene expression of Shh ligand in mice. A) Diagrammatic representation of pCMV-ShhWT, pCMV-ShhF200H, H+/K+-ATPase-β-ShhWT, and H+/K+-ATPase-β-ShhF200H constructs. The HA tag is indicated. B) Western blotting analysis of NIH-3T3 conditioned media following transfection with pCMV-ShhWT and pCMV-ShhF200H. C–D) RT-qPCR of Gli1 and Ptch1 mRNA expression in NIH-3T3 cells transfected with pCMV-ShhWT or pCMV-ShhF200H plasmids. (E) Upper Panel: Immunofluorescent staining of GSII (green) and HA-tag (red) in NTG, pCMV-ShhWT-HA, and pCMV-ShhF200H-HA mouse stomachs. Middle Panel: Shh (green) and H+/K+-ATPase (red). Lower Panel: intrinsic factor (IF; green) and HA-tag (red). (F) Immunofluorescent staining of Shh (green) and HA-tag (red) in pCMV-ShhWT and pCMV-ShhF200H mouse stomachs. A short 15 min incubation with Shh antibody was used to detect transgenes overexpressing ShhWT and ShhF200H without cross-reacting with endogenous gastric Shh. Error bars represent the mean +/− SEM. *p<0.05.
(TIF)
SPEM in pCMV-ShhWT and H+/K+-ATPase-β-ShhWT mice. A) Left panel, Immunofluorescent staining of TFF-2 (green), intrinsic factor (IF, red), and DAPI (blue) in the highest expressing pCMV-ShhWT mouse stomach (102-fold expression of Shh, please refer to Table 1); right panel, high power magnification of the co-localization of IF (red) and TFF-2 (green). B) H&E staining of mouse stomach in two H+/K+-ATPase-β-ShhWT founder mice expressing medium levels of the transgene (please refer to Table 1). C) Left panel, immunofluorescent staining of TFF-2 (green), intrinsic factor (IF, red), and DAPI (blue) in an H+/K+-ATPase-β-ShhWT founder mouse expressing medium levels of the transgene (please refer to Table 1); right panel, high power magnification of the co-localization of IF (red) and TFF-2 (green). Arrows indicate regions of inflammatory cell infiltration and mucous neck cell expansion.
(TIF)