Abstract
Systematic studies of nonsense and sense suppression of the original and three derivative Methanosarcina mazei PylRS-tRNAPyl pairs and cross recognition between nonsense codons and various tRNAPyl anticodons in the Escherichia coli BL21(DE3) cell strain are reported. 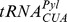 is orthogonal in E. coli and able to induce strong amber suppression when it is co-expressed with pyrrolysyl-tRNA synthetase (PylRS) and charged with a PylRS substrate, Nε-tert-butoxycarbonyl-l-lysine (BocK). Similar to
is orthogonal in E. coli and able to induce strong amber suppression when it is co-expressed with pyrrolysyl-tRNA synthetase (PylRS) and charged with a PylRS substrate, Nε-tert-butoxycarbonyl-l-lysine (BocK). Similar to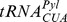 ,
, 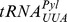 is also orthogonal in E. coli and can be coupled with PylRS to genetically incorporate BocK at an ochre mutation site. Although
is also orthogonal in E. coli and can be coupled with PylRS to genetically incorporate BocK at an ochre mutation site. Although 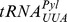 is expected to recognize a UAG codon based on the wobble hypothesis, the PylRS-
is expected to recognize a UAG codon based on the wobble hypothesis, the PylRS- pair does not give rise to amber suppression that surpasses the basal amber suppression level in E. coli. E. coli itself displays a relatively high opal suppression level and tryptophan (Trp) is incorporated at an opal mutation site. Although the PylRS-
pair does not give rise to amber suppression that surpasses the basal amber suppression level in E. coli. E. coli itself displays a relatively high opal suppression level and tryptophan (Trp) is incorporated at an opal mutation site. Although the PylRS- pair can be used to encode BocK at an opal codon, the pair fails to suppress the incorporation of Trp at the same site.
pair can be used to encode BocK at an opal codon, the pair fails to suppress the incorporation of Trp at the same site. 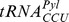 fails to deliver BocK at an AGG codon when co-expressed with PylRS in E. coli.
fails to deliver BocK at an AGG codon when co-expressed with PylRS in E. coli.
Introduction
Pyrrolysine (Pyl,
Figure 1
), the 22nd proteinogenic amino acid that was originally discovered in methanogenic methylamine methyltransferase, is genetically encoded by the RNA nucleotide triplet UAG, a stop codon that halts translation of mRNA during a regular protein translation process [1]. The delivery of Pyl to ribosome is mediated by a unique tRNA, 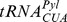 that is specifically aminoacylated by a unique aminoacyl-tRNA synthetase (aaRS), pyrrolysyl-tRNA synthetase (PylRS) [2].
that is specifically aminoacylated by a unique aminoacyl-tRNA synthetase (aaRS), pyrrolysyl-tRNA synthetase (PylRS) [2]. 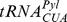 contains a special nucleotide triplet CUA, an anticodon that recognizes a UAG stop codon in mRNA. Unlike tRNASec that needs a special elongation factor (SelB in E. coli and EFsec in mammalian cells) and an mRNA secondary structure for its binding to the ribosome A site and recognition of a UGA stop codon for the delivery of selenocysteine,
contains a special nucleotide triplet CUA, an anticodon that recognizes a UAG stop codon in mRNA. Unlike tRNASec that needs a special elongation factor (SelB in E. coli and EFsec in mammalian cells) and an mRNA secondary structure for its binding to the ribosome A site and recognition of a UGA stop codon for the delivery of selenocysteine,  hijacks the regular translation elongation process to suppress a UAG codon for the incorporation of Pyl [3]–[5]. Previous studies also demonstrated that PylRS shows remarkably high substrate promiscuity and is able to charge
hijacks the regular translation elongation process to suppress a UAG codon for the incorporation of Pyl [3]–[5]. Previous studies also demonstrated that PylRS shows remarkably high substrate promiscuity and is able to charge 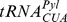 with a variety of noncanonical amino acids (NAAs). For these reasons and due to the naturally high orthogonality of the PylRS-
with a variety of noncanonical amino acids (NAAs). For these reasons and due to the naturally high orthogonality of the PylRS-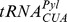 pair in bacteria, yeast, and mammalian cells, this pair has been directly transferred to E. coli, Saccharomyces cerevisiae, and human cells for the genetic incorporation of more than ten lysine derivatives including Nε-tert-butoxycarbonyl-l-lysine (BocK) into proteins at amber mutation sites[6]–[12]. Engineered PylRS-
pair in bacteria, yeast, and mammalian cells, this pair has been directly transferred to E. coli, Saccharomyces cerevisiae, and human cells for the genetic incorporation of more than ten lysine derivatives including Nε-tert-butoxycarbonyl-l-lysine (BocK) into proteins at amber mutation sites[6]–[12]. Engineered PylRS-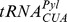 pairs have also been used to genetically encode other lysine derivatives and even phenylalanine derivatives that are structurally distinctive from Pyl[13]–[21]. The genetic incorporation of these NAAs into proteins and their following modifications allow a variety of biochemistry studies such as the functional investigation of protein posttranslational modifications, protein folding dynamic analysis, biosensor development, tracking signal transduction processes, and probing enzyme mechanisms [22]. Pioneered by Schultz and coworkers on the incorporation of two different NAAs [23], two methods were recently developed independently in the Chin Group and our group that couple the wild-type or a derivative PylRS-tRNAPyl pair with another suppressing aaRS-tRNA pair for the genetic incorporation of two different NAAs into one protein in E. coli
[24], [25]. The method developed in the Chin Group used one amber codon and one quadruple AGGA codon that were suppressed by the PylRS-
pairs have also been used to genetically encode other lysine derivatives and even phenylalanine derivatives that are structurally distinctive from Pyl[13]–[21]. The genetic incorporation of these NAAs into proteins and their following modifications allow a variety of biochemistry studies such as the functional investigation of protein posttranslational modifications, protein folding dynamic analysis, biosensor development, tracking signal transduction processes, and probing enzyme mechanisms [22]. Pioneered by Schultz and coworkers on the incorporation of two different NAAs [23], two methods were recently developed independently in the Chin Group and our group that couple the wild-type or a derivative PylRS-tRNAPyl pair with another suppressing aaRS-tRNA pair for the genetic incorporation of two different NAAs into one protein in E. coli
[24], [25]. The method developed in the Chin Group used one amber codon and one quadruple AGGA codon that were suppressed by the PylRS- pair and an evolved M. jannaschii tyrosyl-tRNA synthetase MjTyrRS-
pair and an evolved M. jannaschii tyrosyl-tRNA synthetase MjTyrRS- pair, respectively to code two different NAAs. A specially engineered ribosome, Ribo-Q1, was used to improve the AGGA suppression level. Our method relied on the suppression of two stop codons, namely one amber UAG codon and one ochre UAA codon, which was achieved by genetically encoding an evolved MjTyrRS-
pair, respectively to code two different NAAs. A specially engineered ribosome, Ribo-Q1, was used to improve the AGGA suppression level. Our method relied on the suppression of two stop codons, namely one amber UAG codon and one ochre UAA codon, which was achieved by genetically encoding an evolved MjTyrRS-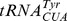 pair for amber suppression and a wild type or evolved PylRS-
pair for amber suppression and a wild type or evolved PylRS-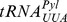 pair for ochre suppression in E. coli. The genetic incorporation of two different NAAs into one protein can be potentially applied to install a FRET pair in a protein for conformation and dynamic studies as we demonstrated in a separate publication [26], synthesize proteins with two different posttranslational modifications for their functional investigation, and build phage-displayed peptide libraries with the expanded chemical diversities.
pair for ochre suppression in E. coli. The genetic incorporation of two different NAAs into one protein can be potentially applied to install a FRET pair in a protein for conformation and dynamic studies as we demonstrated in a separate publication [26], synthesize proteins with two different posttranslational modifications for their functional investigation, and build phage-displayed peptide libraries with the expanded chemical diversities.
Figure 1. The structures of Pyl, BocK and AzF.
Although the PylRS- pair has been used extensively for the genetic incorporation of different NAAs in the past few years, two questions related to the pair have not been fully addressed. While our lab and other groups have showed that mutating the anticodon of
pair has been used extensively for the genetic incorporation of different NAAs in the past few years, two questions related to the pair have not been fully addressed. While our lab and other groups have showed that mutating the anticodon of  does not significantly affect its interaction with the catalytic domain of PylRS [27], further investigation need to be done to determine whether we can directly use mutant tRNAPyl forms for the genetic incorporation of NAAs at an opal or ochre codon or even a sense codon in E. coli. Another study is necessary to clarify whether an aminoacylated
does not significantly affect its interaction with the catalytic domain of PylRS [27], further investigation need to be done to determine whether we can directly use mutant tRNAPyl forms for the genetic incorporation of NAAs at an opal or ochre codon or even a sense codon in E. coli. Another study is necessary to clarify whether an aminoacylated  can lead to amber suppression since a UUA anticodon can recognize a UAG codon based on the wobble hypothesis [28]. In this study, we attempted to address these two questions and carried out all the experiments in the E. coli BL21(DE3) cell strain which has been broadly used for the genetic incorporation of NAAs.
can lead to amber suppression since a UUA anticodon can recognize a UAG codon based on the wobble hypothesis [28]. In this study, we attempted to address these two questions and carried out all the experiments in the E. coli BL21(DE3) cell strain which has been broadly used for the genetic incorporation of NAAs.
Materials and Methods
Materials
Phusion high-fidelity DNA polymerase, T4 DNA ligase, T4 polynucleotide kinase, and restriction enzymes were purchased from New England Biolabs. Oligonucleotide primers were ordered from Integrated DNA Technologies. Ni-NTA superflow resins were purchased from Qiagen. All polymerase chain reactions (PCRs) were performed using Phusion high-fidelity DNA polymerase. BocK was purchased from Chem Impex. p-Azido-l-phenylalanine (AzF) was synthesized according to a revised literature procedure [29].
Plasmids
Plasmid pETtrio-pylT(UUA)-PylRS-MCS was derived from pPylRS-pylT-GFP1TAG149TAA [25] and carries a 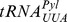 gene (a C34U mutant form of
gene (a C34U mutant form of  ) under control of the lpp promoter and the rrnC terminator, the wild type Methanosarcina mazei PylRS gene under control of the glnS promoter and terminator, and multiple cloning sites including NcoI, NotI, SalI and KpnI targeted sites under control of the T7 promoter and terminator. Plasmid pETtrio-pylT(UUA)-PylRS-sfGFP134TAG that carries an additional superfolder green fluorescent protein (sfGFP) gene with an amber mutation at N134 (sfGFP134TAG) was constructed by cloning the sfGFP134TAG gene to the NcoI and KpnI sites of pETtrio-pylT(UUA)-PylRS-MCS.
) under control of the lpp promoter and the rrnC terminator, the wild type Methanosarcina mazei PylRS gene under control of the glnS promoter and terminator, and multiple cloning sites including NcoI, NotI, SalI and KpnI targeted sites under control of the T7 promoter and terminator. Plasmid pETtrio-pylT(UUA)-PylRS-sfGFP134TAG that carries an additional superfolder green fluorescent protein (sfGFP) gene with an amber mutation at N134 (sfGFP134TAG) was constructed by cloning the sfGFP134TAG gene to the NcoI and KpnI sites of pETtrio-pylT(UUA)-PylRS-MCS.
Three plasmids pETtrio-pylT(CUA)-PylRS-sfGFP134TAG, pETtrio-pylT(UUA)-PylRS-sfGFP134TAA, and pETtrio-pylT(UCA)-PylRS-sfGFP134TGA that vary at the anticodon of tRNAPyl and have different nonsense mutations at N134 of the sfGFP gene were derived from pETtrio-pylT(UUA)-PylRS-sfGFP134TAG. Constructions of these plasmids were carried out using a site-directed mutagenesis protocol that was based on Phusion DNA polymerase. In brief, two oligonucleotide primers, one of which covers the mutation site were used to amplify the whole plasmid of pETtrio-pylT(UUA)-PylRS-sfGFP134TAG to give a blunt-end PCR product. This PCR product was phosphorylated by T4 polynucleotide kinase and then self-ligated using T4 DNA ligase. Plasmid pETtrio-pylT(CUA)-PylRS-sfGFP134TAG carries genes coding 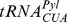 , PylRS, and sfGFP134TAG; plasmid pETtrio-pylT(UUA)-PylRS-sfGFP134TAA carries genes coding
, PylRS, and sfGFP134TAG; plasmid pETtrio-pylT(UUA)-PylRS-sfGFP134TAA carries genes coding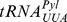 , PylRS, and sfGFP with an ochre mutation at N134 (sfGFP134TAA); and plasmid pETtrio-pylT(UCA)-PylRS-sfGFP134TGA carries genes coding
, PylRS, and sfGFP with an ochre mutation at N134 (sfGFP134TAA); and plasmid pETtrio-pylT(UCA)-PylRS-sfGFP134TGA carries genes coding , PylRS, and sfGFP with an opal mutation at N134 (sfGFP134TGA). Plasmid pETtrio-pylT(CUA)-PylRS-sfGFP134TAA that carries genes coding
, PylRS, and sfGFP with an opal mutation at N134 (sfGFP134TGA). Plasmid pETtrio-pylT(CUA)-PylRS-sfGFP134TAA that carries genes coding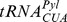 , PylRS, and sfGFP134TAA was derived from plasmid pETtrio-pylT(UUA)-PylRS-sfGFP134TAA.
, PylRS, and sfGFP134TAA was derived from plasmid pETtrio-pylT(UUA)-PylRS-sfGFP134TAA.
Plasmid pETtrio-sfGFP134TGA was derived from pETtrio-pylT(UCA)-PylRS-sfGFP134TGA by digesting it with SphI to remove the 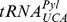 gene and parts of the PylRS gene and self-ligating the purified digested plasmid backbone. Plasmid pETtrio-pylT(UCA)-PylRS-sfGFP2TGA carries genes coding
gene and parts of the PylRS gene and self-ligating the purified digested plasmid backbone. Plasmid pETtrio-pylT(UCA)-PylRS-sfGFP2TGA carries genes coding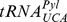 , PylRS, and sfGFP with an opal mutation at S2 (sfGFP2TGA). To construct this plasmid, the sfGFP2TGA gene was used to replace the sfGFP134TGA gene in pETtrio-pylT(UCA)-PylRS-sfGFP134TGA. Plasmid pETtrio-pylT(CCU)-PylRS-sfGFP2AGG was constructed from pETtrio-pylT(CUA)-PylRS-sfGFP2TGA using two consecutive Phusion DNA polymerase-based site directed mutagenesis steps.
, PylRS, and sfGFP with an opal mutation at S2 (sfGFP2TGA). To construct this plasmid, the sfGFP2TGA gene was used to replace the sfGFP134TGA gene in pETtrio-pylT(UCA)-PylRS-sfGFP134TGA. Plasmid pETtrio-pylT(CCU)-PylRS-sfGFP2AGG was constructed from pETtrio-pylT(CUA)-PylRS-sfGFP2TGA using two consecutive Phusion DNA polymerase-based site directed mutagenesis steps.
Three plasmids pETtrio-pylT(UCA)G73C-PylRS-sfGFP134TGA, pETtrio-pylT(UCA)G73U-PylRS-sfGFP134TGA, and pETtrio-pylT(UCA)G73A-PylRS-sfGFP134TGA carry mutations that change G73 of 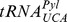 to C, U, and A, respectively. These plasmids were derived from pETtrio-pylT(UCA)-PylRS-sfGFP134TGA using Phusion DNA polymerase-based site-directed mutagenesis.
to C, U, and A, respectively. These plasmids were derived from pETtrio-pylT(UCA)-PylRS-sfGFP134TGA using Phusion DNA polymerase-based site-directed mutagenesis.
Amber, Opal, and Ochre Suppression
Plasmids pETtrio-pylT(CUA)-PylRS-sfGFP134TAG, pETtrio-pylT(UCA)-PylRS-sfGFP134TGA, and pETtrio-pylT(UUA)-PylRS-sfGFP134TAA were individually used to transform E. coli BL21(DE3) cells. For each plasmid, a single colony was selected and allowed to grow in 5 mL of LB medium with 100 µg/mL ampicillin at 37°C overnight. The overnight culture was inoculated into 200 mL of 2YT medium with 100 µg/mL ampicillin and allowed to grow at 37°C to OD600∼1.2. 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) and 5 mM BocK were then added to the medium to induce expression of sfGFP. Control experiments in which only 1 mM IPTG was added to the medium were also carried out. The induced cells were allowed to grow at 37°C overnight and then collected by centrifugation (4,200 rpm for 20 min). The collected cells were resuspended in 35 mL of lysis buffer (50 mM HEPES, 300 mM NaCl, 10 mM imidazole, pH 8.0) and lysed by sonication in an ice water bath. The lysed cells were clarified by centrifugation (10,000 rpm for 1 h). The supernatant was decanted and let bind to 5 mL of Ni-NTA superflow resins at 4°C for 1 h. The mixture of the supernatant and resins was then loaded to an empty Qiagen Ni-NTA superflow cartridge. The resins were washed with 5 volume times of lysis buffer and sfGFP was then eluted with buffer (50 mM HEPES, 300 mM NaCl, 250 mM imidazole, pH 8.0). To further purify the expressed sfGFP, the protein was equilibrated against buffer A (20 mM Bis-Tris, pH 6.1) and then loaded to a monoS column from GE Health Science. The protein was eluted out by running a gradient from buffer A to 100% of buffer B (20 mM Bis-Tris, 1 mM NaCl, pH 6.1). The finally purified protein was then concentrated to a desired volume and analyzed by SDS-PAGE. To analyze the purified protein by electrospray ionization mass spectrometry (ESI-MS) analysis, the buffer of the purified protein was changed to the phosphate buffer saline. Without further indication, protein purification and characterization in the following experiments were the same.
Suppression of an Opal Mutation at S2 of sfGFP
Plasmid pETtrio-pylT(UCA)-PylRS-sfGFP2TGA was used to transform E. coli BL21(DE3) cells. A single colony was then used to express sfGFP at two induction conditions: (1) 1 mM IPTG and (2) 1 mM IPTG and 5 mM BocK.
Basal Suppression at an Opal UGA Codon
Plasmid pETtrio-sfGFP134TGA was used to transform E. coli BL21(DE3) cells. A single colony was then selected to do protein expression that was induced by the addition of 1 mM IPTG.
Mutagenic Analysis of 
Plasmids pETtrio-pylT(UCA)G73C-PylRS-sfGFP134TGA, pETtrio-pylT(UCA)G73U-PylRS-sfGFP134TGA, and pETtrio-pylT(UCA)G73A-PylRS-sfGFP134TGA were used individually to transform E. coli BL21(DE3) cells. A single colony for each plasmid was then selected to do protein expression at two induction conditions: (1) 1 mM IPTG and (2) 1 mM IPTG and 5 mM BocK.
Anticodon-codon Cross Recognition
Plasmids pETtrio-pylT(CUA)-PylRS-sfGFP134TAG, pETtrio-pylT(CUA)-PylRS-sfGFP134TAA, pETtrio-pylT(UUA)-PylRS-sfGFP134TAG, and pETtrio-pylT(UUA)-PylRS-sfGFP134TAA were individually used to transform E. coli BL21(DE3) cells. A single colony for each plasmid was then selected and allowed to grow in 5 mL of LB medium with 100 µg/mL ampicillin at 37°C overnight. This overnight culture was then inoculated into 500 mL of 2YT medium with 100 µg/mL ampicillin and allowed to grow to OD600∼1.2. 1 mM IPTG and 5 mM BocK were then provided and cells were allowed to grow at 37°C overnight. For pETtrio-pylT(UUA)-PylRS-sfGFP134TAG, expression of sfGFP in the absence of BocK was also tested.
Competitive Recognition of the Third Nucleotide of an Amber Codon
Plasmid pEVOL-AzFRS was a gift from Dr. Peter Schultz at Scripps Research Institute [30]. It carries one  gene under control of a proK promoter and a proK terminator, an evolved AzF-specific MjTyrRS (AzFRS) gene under control of a glnS promoter and a glnS terminator, and an AzFRS gene under control of a pBAD promoter. This plasmid together with pETtrio-pylT(UUA)-PylRS-sfGFP134TAG was used to co-transform E. coli BL21(DE3) cells. One single colony was selected and allowed to grow in 5 mL of LB medium with 100 µg/mL ampicillin and 34 µg/mL chloramphenicol at 37°C overnight. This overnight culture was inoculated into 200 mL of 2YT medium with 100 µg/mL ampicillin and 34 µg/mL chloramphenicol and allowed to grow to OD600∼1.2. Expression of sfGFP was then induced. Four induction conditions were tested, including (1) 1 mM IPTG only, (2) 1 mM IPTG and 1 mM AzF, (3) 1 mM IPTG and 5 mM BocK, and (4) 1 mM IPTG, 1 mM AzF, and 5 mM BocK.
gene under control of a proK promoter and a proK terminator, an evolved AzF-specific MjTyrRS (AzFRS) gene under control of a glnS promoter and a glnS terminator, and an AzFRS gene under control of a pBAD promoter. This plasmid together with pETtrio-pylT(UUA)-PylRS-sfGFP134TAG was used to co-transform E. coli BL21(DE3) cells. One single colony was selected and allowed to grow in 5 mL of LB medium with 100 µg/mL ampicillin and 34 µg/mL chloramphenicol at 37°C overnight. This overnight culture was inoculated into 200 mL of 2YT medium with 100 µg/mL ampicillin and 34 µg/mL chloramphenicol and allowed to grow to OD600∼1.2. Expression of sfGFP was then induced. Four induction conditions were tested, including (1) 1 mM IPTG only, (2) 1 mM IPTG and 1 mM AzF, (3) 1 mM IPTG and 5 mM BocK, and (4) 1 mM IPTG, 1 mM AzF, and 5 mM BocK.
AGG Codon Suppression
Plasmid pETtrio-pylT(CCU)-PylRS-sfGFP2AGG was used to transform E. coli BL21(DE3) cells. A single colony was then selected and allowed to grow in 5 mL of LB medium with 100 µg/mL ampicillin at 37°C overnight. This overnight culture was then inoculated into 500 mL of 2YT medium with 100 µg/mL ampicillin and allowed to grow to OD600∼1.2. Two conditions were used to induce sfGFP expression. One is the addition of 1 mM IPTG and the other is the addition of 1 mM IPTG and 5 mM BocK.
Results
Amber, Opal, and Ochre Suppression Efficiencies of the PylRS-tRNAPyl Pairs
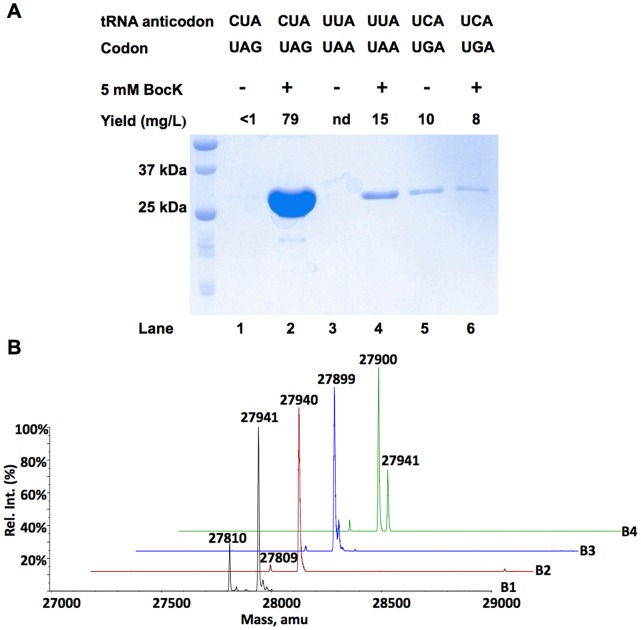
To demonstrate amber suppression efficiency of the PylRS-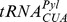 pair, E. coli BL21(DE3) cells transformed with pETtrio-pylT(CUA)-PylRS-sfGFP134TAG were used to express sfGFP both in the absence and in the presence of 5 mM BocK, a substrate of PylRS. Without BocK in the growth medium, low level of sfGFP expression was observed. On the contrary, the addition of BocK promoted sfGFP overexpression (
Figure 2A
). The ESI-MS analysis of the purified sfGFP displayed two major mass peaks at 27,810±1 Da and 27,941±1Da that agree well with the theoretical molecular weights of sfGFP with BocK incorporated at N134 (27,940 Da for the full-length protein; 27809 Da for the full-length protein without the first methionine (M1)) (
Figure 2B
1). E. coli BL21(DE3) cells transformed with pETtrio-pylT(UUA)-PylRS-sfGFP134TAA showed an undetectable expression level of sfGFP when BocK was absent in the growth medium. The addition of BocK induced sfGFP expression (
Figure 2A
). The ESI-MS analysis of the purified sfGFP showed two mass peaks (27,940±1 Da and 27,809±1 Da) that agree well with the theoretical molecular weights of sfGFP with BocK incorporated at N134 (
Figure 2B
2). In comparison to amber suppression, the expression level of sfGFP using ochre suppression is lower. E. coli BL21(DE3) cells transformed with pETtrio-pylT(UCA)-PylRS-sfGFP134TGA exhibited a high expression level of sfGFP both in the absence and in the presence of BocK in the growth medium (
Figure 2A
). The ESI-MS analysis of purified sfGFP expressed in the absence of BocK showed a mass peak at 27,899±1 Da that clearly matched a Trp residue at N134 of sfGFP (calculated mass: 27,898 Da) (
Figure 2B
3). The ESI-MS analysis of sfGFP expressed in the presence of BocK displayed a very interesting spectrum. Mass peaks for both Trp residue at N134 of sfGFP (27,900±1 Da) and BocK residue at N134 of sfGFP (27,941±1 Da) were observed. The mass peak for the Trp isoform was much more intensive than the BocK isoform (
Figure 2B
4).
pair, E. coli BL21(DE3) cells transformed with pETtrio-pylT(CUA)-PylRS-sfGFP134TAG were used to express sfGFP both in the absence and in the presence of 5 mM BocK, a substrate of PylRS. Without BocK in the growth medium, low level of sfGFP expression was observed. On the contrary, the addition of BocK promoted sfGFP overexpression (
Figure 2A
). The ESI-MS analysis of the purified sfGFP displayed two major mass peaks at 27,810±1 Da and 27,941±1Da that agree well with the theoretical molecular weights of sfGFP with BocK incorporated at N134 (27,940 Da for the full-length protein; 27809 Da for the full-length protein without the first methionine (M1)) (
Figure 2B
1). E. coli BL21(DE3) cells transformed with pETtrio-pylT(UUA)-PylRS-sfGFP134TAA showed an undetectable expression level of sfGFP when BocK was absent in the growth medium. The addition of BocK induced sfGFP expression (
Figure 2A
). The ESI-MS analysis of the purified sfGFP showed two mass peaks (27,940±1 Da and 27,809±1 Da) that agree well with the theoretical molecular weights of sfGFP with BocK incorporated at N134 (
Figure 2B
2). In comparison to amber suppression, the expression level of sfGFP using ochre suppression is lower. E. coli BL21(DE3) cells transformed with pETtrio-pylT(UCA)-PylRS-sfGFP134TGA exhibited a high expression level of sfGFP both in the absence and in the presence of BocK in the growth medium (
Figure 2A
). The ESI-MS analysis of purified sfGFP expressed in the absence of BocK showed a mass peak at 27,899±1 Da that clearly matched a Trp residue at N134 of sfGFP (calculated mass: 27,898 Da) (
Figure 2B
3). The ESI-MS analysis of sfGFP expressed in the presence of BocK displayed a very interesting spectrum. Mass peaks for both Trp residue at N134 of sfGFP (27,900±1 Da) and BocK residue at N134 of sfGFP (27,941±1 Da) were observed. The mass peak for the Trp isoform was much more intensive than the BocK isoform (
Figure 2B
4).
Figure 2. Suppression of amber, opal, and ochre mutations at N134 of sfGFP by their corresponding PylRS-tRNAPyl pairs in the absence and presence of BocK.
(A) Proteins shown in the gel represent their real relative sfGFP expression levels. Lanes 1 and 2 were transformed with pETtrio-PylT(CUA)-PylRS-sfGFP134TAG; lanes 3 and 4 were transformed with pETtrio-PylT(UUA)-PylRS-sfGFP134TAA; lanes 5 and 6 were transformed with pETtrio-PylT(UCA)-PylRS-sfGFP134TGA. ESI-MS spectra of sfGFP expressed in cells (B1) transformed with pETtrio-PylT(CUA)-PylRS-sfGFP134TAG and grown in the presence of 5 mM BocK, (B2) transformed with pETtrio-PylT(UUA)-PylRS-sfGFP134TAA and grown in the presence of 5 mM BocK, (B3) transformed with pETtrio-PylT(UCA)-PylRS-sfGFP134TGA and grown in the absence of BocK, and (B4) transformed with pETtrio-PylT(UCA)-PylRS-sfGFP134TGA and grown in the presence of 5 mM BocK.
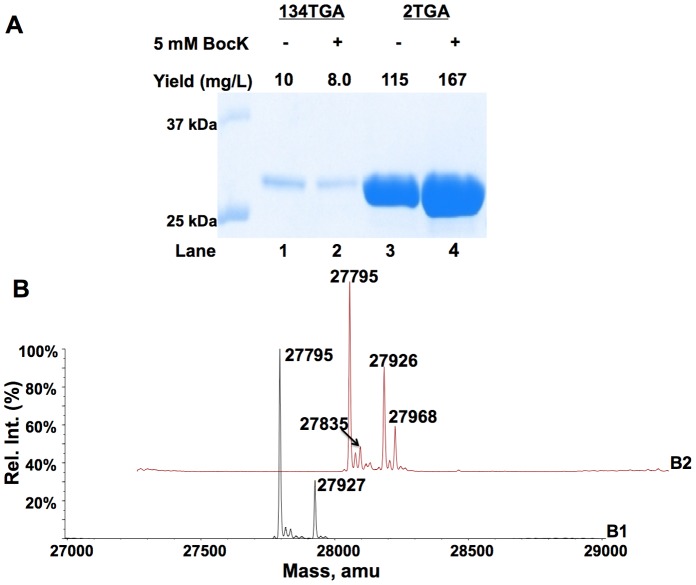
To test whether the incorporation of Trp at the 134 position is related to the nucleotide contents of mRNA around the UGA codon, plasmid pETtrio-pylT(UCA)-PylRS-sfGFP2TGA was constructed to test the opal suppression. E. coli BL21(DE3) cells transformed with pETtrio-pylT(UCA)-PylRS-sfGFP2TGA displayed high sfGFP expression levels both in the absence and in the presence of BocK that were much higher than in cells transformed with plasmid pETtrio-pylT(UCA)-PylRS-sfGFP134TGA and grown in the same conditions (
Figure 3A
). The ESI-MS analyses of purified sfGFP from cells transformed with pETtrio-pylT(UCA)-PylRS-sfGFP2TGA displayed similar patterns as sfGFP from cells transformed with pETtrio-pylT(UCA)-PylRS-sfGFP134TGA. Two mass peaks for sfGFP expressed both in the absence and in the presence of BocK (27,795±1 Da and 27,927±1 Da in
Figure 3B
1 and 27,795±1 Da and 27,926±1 Da in
Figure 3B
2) match the molecular weights of sfGFP with Trp incorporated at S2 (calculated mass: 27,926 Da for the full-length protein; 27,794 Da for the full-length protein without M1). For sfGFP expressed in the presence of 5 mM BocK, there were two small peaks at 27,968±1 Da and 27,835±1 Da that agree well with the molecular weights of sfGFP with BocK incorporated at S2 (calculated mass: 27,967 Da for the full-length protein; 27,836 Da for the full-length protein without M1). To further understand opal suppression in the E. coli BL21(DE3) cell strain, plasmid pETtrio-sfGFP134TGA that carries no opal suppressing 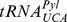 was used to transform E. coli BL21(DE3) cells. The transformed cells displayed a relatively high detectable level of sfGFP expression with a yield of 4 mg/L (
Figure 4A
).The ESI-MS analysis of sfGFP resulted from this basal opal suppression showed major mass peaks at 27,899±1 Da and 27,767±1 Da (
Figure 4B
). They match the molecular weights of sfGFP with Trp incorporated at N134.
was used to transform E. coli BL21(DE3) cells. The transformed cells displayed a relatively high detectable level of sfGFP expression with a yield of 4 mg/L (
Figure 4A
).The ESI-MS analysis of sfGFP resulted from this basal opal suppression showed major mass peaks at 27,899±1 Da and 27,767±1 Da (
Figure 4B
). They match the molecular weights of sfGFP with Trp incorporated at N134.
Figure 3. Suppression of an opal mutation at S2 of sfGFP by the PylRS-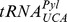 pair.
pair.
(A) Expression of sfGFP with an opal mutation. Lanes 1 and 2 were transformed with pETtrio-pylT(UCA)-sfGFP134TGA and grown in the absence or presence of 5 mM BocK; lanes 3 and 4 were transformed with pETtrio-pylT(UCA)-sfGFP2TGA and grown in the absence or presence of 5 mM BocK. Each protein shown in the gel represents their real relative expression levels. ESI-MS of sfGFP expressed in cells transformed with pETtrio-pylT(UCA)-sfGFP2TGA and grown in the (B1) absence or (B2) presence of 5 mM BocK.
Figure 4. Suppression of an opal mutation at N134 of sfGFP at different conditions.
(A) Proteins shown in the gel represent their real relative expression levels. Lane 1 was transformed with pET-sfGFP134TGA; lanes 2 and 3 were transformed with pET-pylT(UCA)-sfGFP134TGA and grown in the absence or presence of 5 mM BocK. (B) ESI-MS of sfGFP expressed in cells transformed with pETtrio-sfGFP134TGA.
The suppression efficiencies of three mutated forms of 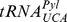 with mutations as G73A, G73C, and G73U, respectively were also examined. Cells transformed with either pETtrio-pylT(UCA)G73A-PylRS-sfGFP134TGA or pETtrio-pylT(UCA)G73C-PylRS-sfGFP134TGA displayed similar expression levels of sfGFP both in the presence and in the absence of BocK(
Figure 5A
). However, cells transformed with pETtrio-pylT(UCA)G73U-PylRS-sfGFP134TGA showed significantly different expression levels of sfGFP when grown in the absence and in the presence of BocK. The ESI-MS analysis of sfGFP expressed in the absence of BocK showed a major mass peak at 27,895±1 Da that matches sfGFP with Trp incorporated at N134 (
Figure 5B
1). The addition of 5 mM BocK promoted the sfGFP expression level to increase. The ESI-MS analysis of the purified protein confirmed sfGFP with BocK incorporated at N134 became dominant (
Figure 5B
2). The intensity of the mass peak at 27,940±1 Da that matches sfGFP with BocK incorporated at N134 is roughly twice of the mass peak at 27,898±1 Da that matches sfGFP with Trp incorporated at N134.
with mutations as G73A, G73C, and G73U, respectively were also examined. Cells transformed with either pETtrio-pylT(UCA)G73A-PylRS-sfGFP134TGA or pETtrio-pylT(UCA)G73C-PylRS-sfGFP134TGA displayed similar expression levels of sfGFP both in the presence and in the absence of BocK(
Figure 5A
). However, cells transformed with pETtrio-pylT(UCA)G73U-PylRS-sfGFP134TGA showed significantly different expression levels of sfGFP when grown in the absence and in the presence of BocK. The ESI-MS analysis of sfGFP expressed in the absence of BocK showed a major mass peak at 27,895±1 Da that matches sfGFP with Trp incorporated at N134 (
Figure 5B
1). The addition of 5 mM BocK promoted the sfGFP expression level to increase. The ESI-MS analysis of the purified protein confirmed sfGFP with BocK incorporated at N134 became dominant (
Figure 5B
2). The intensity of the mass peak at 27,940±1 Da that matches sfGFP with BocK incorporated at N134 is roughly twice of the mass peak at 27,898±1 Da that matches sfGFP with Trp incorporated at N134.
Figure 5. Suppression of an opal mutation at N134 of sfGFP by different 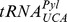 variants.
variants.
(A) Proteins shown in the gel represent their real relative expression levels. Lanes 1 and 2 were transformed with pETtrio-pylT(UCA)G73C-sfGFP134TGA and grown in the absence or presence of 5 mM BocK; lanes 3 and 4 were transformed with pETtrio-pylT(UCA)G73A-sfGFP134TGA and grown in the absence or presence of 5 mM BocK; lanes 5 and 6 were transformed with pETtrio-pylT(UCA)G73U-sfGFP134TGA and grown in the absence or presence of 5 mM BocK; lanes 7 and 8 were transformed with pETtrio-pylT(UCA)-sfGFP134TGA and grown in the absence or presence of 5 mM BocK. The ESI-MS analysis of sfGFP expressed in cells transformed with pETtrio-pylT(UCA)G73U-sfGFP134TGA and grown in the (B1) absence or (B2) presence of 5 mM BocK.
Anticodon-codon Cross Recognition
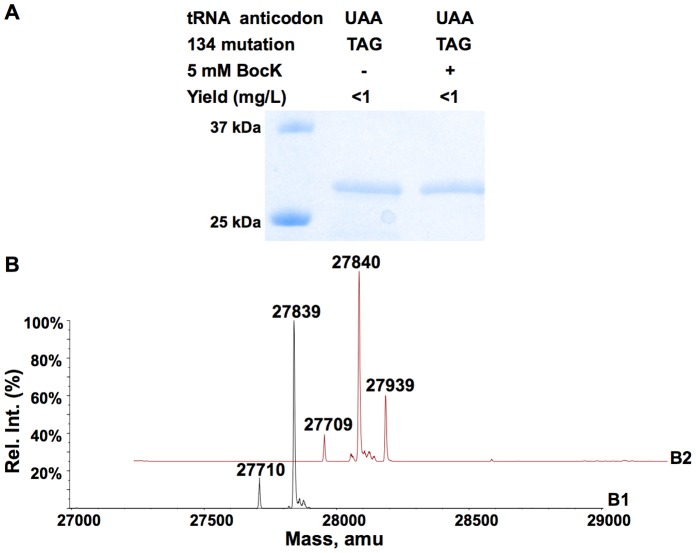
Transforming E. coli BL21(DE3) cells with pETtrio-pylT(CUA)-PylRS-sfGFP134TAA followed by growing cells in the presence of 5 mM BocK did not lead to detectable expression of sfGFP. E. coli BL21(DE3) cells transformed with pETtrio-pylT(UUA)-PylRS-sfGFP134TAG showed a detectable but low level of sfGFP expression (less than 1 mg/L) ( Figure 6 ). To see whether BocK promoted suppression at the amber mutation site, E. coli BL21(DE3) cells transformed with pETtrio-pylT(UUA)-PylRS-sfGFP134TAG was also grown in the absence of BocK. As shown in Figure 7A , the sfGFP expression levels both in the absence and in the presence of BocK were very similar. Addition of BocK did not lead to significant increase of amber suppression. As shown in Figures 7B 1&B2, sfGFP expressed in both conditions displayed mass peaks (27,839±1 Da and 27,710±1 Da in Figure 7B 1 and 27,840±1 Da and 27,709±1 Da in Figure 7B 2) that match sfGFP with glutamic acid (Glu), lysine (Lys), or glutamine(Gln) incorporated at N134. Figure 7B 2 did show a mass peak at 27,939±1 Da that matches the molecular weight of sfGFP with BocK incorporated at N134. However, its intensity was much lower than the mass peak of sfGFP at ∼27,840±1 Da.
Figure 6. Cross recognitions between different anticodons of tRNAPyl and nonsense mutations at N134 of sfGFP.

Cells were transformed with pETtrio-PylT(NNN)-PylRS-sfGFP134N’N’N’ and grown in the presence of 5 mM BocK (NNN and N’N’N’ denote anticodons and codons specified in the figure). Proteins shown in the gel represent their real relative expression levels.
Figure 7. Expression of sfGFP in cells transformed with pETtrio-PylT(UUA)-PylRS-sfGFP134TAG.
(A) Cells grown in 2YT medium supplemented without or with BocK. ESI-MS of sfGFP expressed in the (B1) absence or (B2) presence of 5 mM BocK.
Experiments were also carried out to demonstrate that the PylRS- pair does not interfere with suppression of an amber mutation mediated by an evolved MjTyrRS-
pair does not interfere with suppression of an amber mutation mediated by an evolved MjTyrRS- pair. Two plasmids, pETtrio-pylT(UUA)-pylRS-sfGFP134TAG and pEVOL-AzFRS were used to transform E. coli BL21(DE3)cells. As shown in
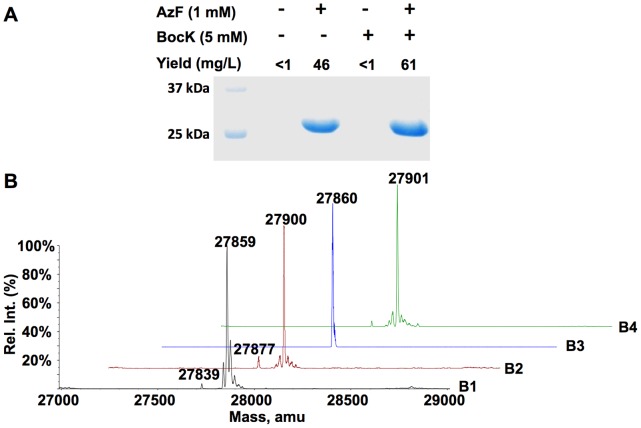
Figure 8A
, growing the transformed cells in four conditions led to different expression levels of sfGFP. When no NAA or only 5 mM BocK was provided in the medium, only a detectable but very low level of sfGFP expression (less than 1 mg/L) was detected. However, addition of 1 mM AzF to the medium supplemented with or without BocK promoted sfGFP overexpression. The ESI-MS analysis of purified sfGFP in all four conditions displayed very interesting spectra (
Figure 8B
). Although a mass peak for sfGFP expressed in two conditions with the supplement of AzF (27,900±1 Da in
Figure 8B
2and 27,901±1 Da in
Figure 8B
4) matches the calculated molecular weight (27,901 Da) of sfGFP with AzF incorporated at N134, a mass peak for sfGFP expressed in two conditions without the supplement of AzF (27,859±1 Da in
Figure 8B
1 and 27,860±1 Da in
Figure 8B
3) does not match the molecular weight of sfGFP with either Lys/Glu/Gln or BocK incorporated at N134. Instead, this mass peak agrees well with the molecular weight of sfGFP with Phe incorporated at N134 (calculated mass: 27,859 Da).
pair. Two plasmids, pETtrio-pylT(UUA)-pylRS-sfGFP134TAG and pEVOL-AzFRS were used to transform E. coli BL21(DE3)cells. As shown in
Figure 8A
, growing the transformed cells in four conditions led to different expression levels of sfGFP. When no NAA or only 5 mM BocK was provided in the medium, only a detectable but very low level of sfGFP expression (less than 1 mg/L) was detected. However, addition of 1 mM AzF to the medium supplemented with or without BocK promoted sfGFP overexpression. The ESI-MS analysis of purified sfGFP in all four conditions displayed very interesting spectra (
Figure 8B
). Although a mass peak for sfGFP expressed in two conditions with the supplement of AzF (27,900±1 Da in
Figure 8B
2and 27,901±1 Da in
Figure 8B
4) matches the calculated molecular weight (27,901 Da) of sfGFP with AzF incorporated at N134, a mass peak for sfGFP expressed in two conditions without the supplement of AzF (27,859±1 Da in
Figure 8B
1 and 27,860±1 Da in
Figure 8B
3) does not match the molecular weight of sfGFP with either Lys/Glu/Gln or BocK incorporated at N134. Instead, this mass peak agrees well with the molecular weight of sfGFP with Phe incorporated at N134 (calculated mass: 27,859 Da).
Figure 8. Expression of sfGFP in cells transformed with pETtrio-PylT(UUA)-PylRS-sfGFP134TAG and pEVOL-AzFRS.
(A)Cells were grown in 2YT medium supplemented with different combinations of NAAs. ESI-MS of sfGFP expressed in the(B1) absence of both AzF and BocK; (B2) presence of 1 mM AzF; (B3) presence of 5 mM BocK; and (B4) presence of both 1 mM AzF and 5 mM BocK.
AGG Codon Suppression
E. coli BL21(DE3) cells transformed with pETtrio-pylT(CCU)-PylRS-sfGFP2AGG showed similar sfGFP expression levels both in the absence and in the presence of BocK ( Figure 9A ). sfGFP proteins expressed in both conditions displayed one major mass peak at 27,895 Da that agrees well with the theoretical molecular weight of sfGFP with arginine (Arg) incorporated at N134 (calculated mass: 27,896 Da for the full-length protein) ( Figure 9B ).
Figure 9. Suppression of an AGG mutation at S2 of sfGFP by .
.

(A) Expression of sfGFP in cells transformed with pETtrio-pylT(CCU)-sfGFP2AGG and grown in the absence or presence of 5 mM BocK. (B) The ESI-MS analysis of sfGFP expressed in the presence of 5 mM BocK.
Discussion
Basal Nonsense Suppression in the E. coli BL21(DE3) Cell Strain
E. coli BL21(DE3) cells transformed with pETtrio-pylT(CUA)-PylRS-sfGFP134TAG and grown in the absence of BocK yielded a sfGFP expression level close to 1 mg/L. The ESI-MS spectrum of the purified sfGFP clearly indicated a Lys, Glu, or Gln residue at the amber mutation site. Since PylRS does not recognize Lys, Glu, and Gln and  itself does not mediate detectable amber suppression in the E. coli Top10 cell strain (data not shown), this low but detectable sfGFP expression level was due to the basal amber suppression in the E. coli BL21(DE3) cell strain [6], [27]. This basal amber suppression that was also demonstrated in one separate study from us [27] arises possibly from the recognitions of the UAG codon by its near-cognate tRNAs including
itself does not mediate detectable amber suppression in the E. coli Top10 cell strain (data not shown), this low but detectable sfGFP expression level was due to the basal amber suppression in the E. coli BL21(DE3) cell strain [6], [27]. This basal amber suppression that was also demonstrated in one separate study from us [27] arises possibly from the recognitions of the UAG codon by its near-cognate tRNAs including 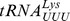 /
/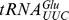 /
/ [31], [32].
[31], [32].
A similar test with E. coli BL21(DE3) cells transformed with pETtrio-pylT(UCA)-PylRS-sfGFP134TGA and grown in the absence of BocK yielded a sfGFP expression level of 8 mg/L. The ESI-MS spectrum of the purified protein showed a Trp residue at N134 of sfGFP. To rule out the possibility that  was charged by E. coli tryptophanyl-tRNA synthetase (TrpRS) for the delivery of Trp at the designated opal mutation site, we constructed plasmid pETtrio-sfGFP134TGA that did not contain the
was charged by E. coli tryptophanyl-tRNA synthetase (TrpRS) for the delivery of Trp at the designated opal mutation site, we constructed plasmid pETtrio-sfGFP134TGA that did not contain the  gene for further tests. E. coli BL21(DE3) cells transformed with this plasmid yielded a sfGFP expression level of 4 mg/L. The ESI-MS spectrum of the purified protein also showed the incorporation of Trp at N134 of sfGFP. Since we recently demonstrated that
gene for further tests. E. coli BL21(DE3) cells transformed with this plasmid yielded a sfGFP expression level of 4 mg/L. The ESI-MS spectrum of the purified protein also showed the incorporation of Trp at N134 of sfGFP. Since we recently demonstrated that 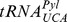 is not recognized by E. coli TrpRS, [27] we think sfGFP expressed in the above two conditions resulted from the basal opal suppression in the E. coli BL21(DE3) cell strain. The observed different sfGFP expression levels might be caused by different copy numbers of two plasmids in the transformed cells. This basal opal suppression is the consequence of the recognition of the opal codon by tRNATrp. E. coli tRNATrp has a CCA anticodon which is a near-cognate tRNA of UGA. It forms two Watson-Crick base pairs with the 5′ and middle nucleotides of UGA and a wobble base pair with the 3′ nucleotide of UGA, explaining its ability to recognize UGA. We have also excluded the possibility that nucleotide contents around the opal mutation at N134 of sfGFP facilitated the binding of tRNATrp and induced the Trp incorporation at this site. The E. coli BL21(DE3) cells transformed with pETtrio-pylT(UCA)-PylRS-sfGFP2TGA also expressed sfGFP in the absence of BocK and the expressed protein had Trp at the opal mutation site. The sfGFP expression level in E. coli BL21(DE3) cells transformed with pylT(UCA)-PylRS-sfGFP2TGA reached to 115 mg/L. This is roughly 20% of the expression level of wild-type sfGFP in E. coli BL21(DE3) cells (∼500 mg/L, unpublished data). This high efficiency to read across an opal codon with the binding of a near-cognate tRNATrp may correlate with the short distance from the opal codon to the start codon.
is not recognized by E. coli TrpRS, [27] we think sfGFP expressed in the above two conditions resulted from the basal opal suppression in the E. coli BL21(DE3) cell strain. The observed different sfGFP expression levels might be caused by different copy numbers of two plasmids in the transformed cells. This basal opal suppression is the consequence of the recognition of the opal codon by tRNATrp. E. coli tRNATrp has a CCA anticodon which is a near-cognate tRNA of UGA. It forms two Watson-Crick base pairs with the 5′ and middle nucleotides of UGA and a wobble base pair with the 3′ nucleotide of UGA, explaining its ability to recognize UGA. We have also excluded the possibility that nucleotide contents around the opal mutation at N134 of sfGFP facilitated the binding of tRNATrp and induced the Trp incorporation at this site. The E. coli BL21(DE3) cells transformed with pETtrio-pylT(UCA)-PylRS-sfGFP2TGA also expressed sfGFP in the absence of BocK and the expressed protein had Trp at the opal mutation site. The sfGFP expression level in E. coli BL21(DE3) cells transformed with pylT(UCA)-PylRS-sfGFP2TGA reached to 115 mg/L. This is roughly 20% of the expression level of wild-type sfGFP in E. coli BL21(DE3) cells (∼500 mg/L, unpublished data). This high efficiency to read across an opal codon with the binding of a near-cognate tRNATrp may correlate with the short distance from the opal codon to the start codon.
In contrary to UAG and UGA codons, the UAA codon displays high translation termination stringency in the E. coli BL21(DE3) cell strain. Cells transformed with pETtrio-pylT(CUA)-PylRS-sfGFP134TAA showed an undetectable basal ochre suppression level. This can be explained from several aspects. UAG and UGA are recognized by release factor 1 and release factor 2, respectively, whereas UAA is recognized by both release factor 1 and release factor 2. Its recognition by both release factor proteins, in theory, makes the translation termination at UAA more stringent than the other two stop codons. Another reason lies at the nucleotide contents of UAA. Unlike UAG and UGA that could involve a GC base pair interaction, UAA could only form AU pairs or wobble pairs. Its interactions with tRNAs are relatively weak, making its misrecognition less possible than UAG and UGA.
Amber, Opal, and Ochre Suppression Efficiencies of the PylRS-tRNAPyl Pairs
When co-expressed with PylRS, all three tRNAPyl isoforms  ,
,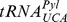 , and
, and  are capable to deliver BocK at their corresponding codon sites.
are capable to deliver BocK at their corresponding codon sites. 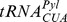 is orthogonal in E. coli and displays the highest efficiency in all three isoforms. Given that E. coli BL21(DE3) cells transformed with pETtrio-pylT(UUA)-PylRS-sfGFP134TAA did not show a detectable expression level of sfGFP in the absence of BocK, we could conclude that
is orthogonal in E. coli and displays the highest efficiency in all three isoforms. Given that E. coli BL21(DE3) cells transformed with pETtrio-pylT(UUA)-PylRS-sfGFP134TAA did not show a detectable expression level of sfGFP in the absence of BocK, we could conclude that 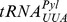 is fully orthogonal in E. coli. In comparison to sfGFP expressed in cells transformed with pETtrio-pylT(CUA)-PylRS-sfGFP134TAG and grown in the presence of BocK, the sfGFP expression level in cells transformed with pETtrio-pylT(UUA)-PylRS-sfGFP134TAA and grown in the presence of BocK is five times lower. The low ability of
is fully orthogonal in E. coli. In comparison to sfGFP expressed in cells transformed with pETtrio-pylT(CUA)-PylRS-sfGFP134TAG and grown in the presence of BocK, the sfGFP expression level in cells transformed with pETtrio-pylT(UUA)-PylRS-sfGFP134TAA and grown in the presence of BocK is five times lower. The low ability of 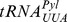 to deliver BocK is possibly due to the relative weak base pair interactions between its UUA anticodon, its UAA stop codon and the availability of both release factor 1 and release factor 2 to stop the translation at a UAA stop codon. In any case, the ochre suppression level achieved by the PylRS-
to deliver BocK is possibly due to the relative weak base pair interactions between its UUA anticodon, its UAA stop codon and the availability of both release factor 1 and release factor 2 to stop the translation at a UAA stop codon. In any case, the ochre suppression level achieved by the PylRS-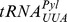 pair is sufficient to promote overexpression of a protein with an ochre mutation. Although a BocK-aminoacylated
pair is sufficient to promote overexpression of a protein with an ochre mutation. Although a BocK-aminoacylated  is able to suppress an opal codon for the incorporation of BocK, it does not inhibit the high incorporation of Trp at the same site. As an exogenous tRNA, the sequence and structure of
is able to suppress an opal codon for the incorporation of BocK, it does not inhibit the high incorporation of Trp at the same site. As an exogenous tRNA, the sequence and structure of 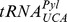 may not be optimal for the protein translation process in E. coli. When facing a competition from E. coli tRNATrp, the recognition of
may not be optimal for the protein translation process in E. coli. When facing a competition from E. coli tRNATrp, the recognition of  by the E. coli translation machinery may be inhibited. Since the 73rd nucleotide serves as a strong recognition element for most tRNAs [33], it was mutated in
by the E. coli translation machinery may be inhibited. Since the 73rd nucleotide serves as a strong recognition element for most tRNAs [33], it was mutated in 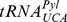 to U, A, and C and searched for a mutant that show a higher opal suppression efficiency. When co-expressed with PylRS in the presence of BocK,
to U, A, and C and searched for a mutant that show a higher opal suppression efficiency. When co-expressed with PylRS in the presence of BocK,  led to higher incorporation level of BocK compared to Trp at the same opal mutation site. However, further mutagenesis with
led to higher incorporation level of BocK compared to Trp at the same opal mutation site. However, further mutagenesis with  is necessary to fully inhibit the incorporation of Trp at an opal mutation site.
is necessary to fully inhibit the incorporation of Trp at an opal mutation site.
Anticodon-codon Cross Recognition
The wobble hypothesis was first introduced by Francis Crick in 1966 to explain the observation that a single tRNA is able to efficiently recognize multiple codons [28]. Based on this hypothesis, an ochre suppressor tRNAUUA is also capable of recognizing an amber UAG codon. This is a concern when both UAG and UAA codons are used to code two different NAAs. However, cells transformed with pETtrio-pylT(UUA)-PylRS-sfGFP134TAG and grown in the presence of 5 mM BocK showed a sfGFP expression level close to that from the basal amber suppression. This suggests very weak recognition of UAG by . Weak base pairing interactions involved with the UUA anticodon may contribute to the weak recognition of UAG. However, this is certainly not the determining factor since other tRNAs such as
. Weak base pairing interactions involved with the UUA anticodon may contribute to the weak recognition of UAG. However, this is certainly not the determining factor since other tRNAs such as 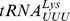 is also involve weak base pairing interactions to recognize multiple codons. One possible explanation for this weak recognition of UAG by
is also involve weak base pairing interactions to recognize multiple codons. One possible explanation for this weak recognition of UAG by  is the tRNA modifications. All cognate tRNAs are known to exhibit similar affinities for the ribosome A site when they bind to corresponding codons [34], [35]. This uniform binding is unexpected as certain codon-anticodon interactions are expected to be more stable than others due to factors such as the GC base pair content. It has been proposed that the specific sequence and post-transcriptional modification status of the tRNA in the region near the anticodon is tuned to ensure nearly indistinguishable binding of tRNAs to the ribosome A site [36]–[38]. This has been the case for tRNAs such as
is the tRNA modifications. All cognate tRNAs are known to exhibit similar affinities for the ribosome A site when they bind to corresponding codons [34], [35]. This uniform binding is unexpected as certain codon-anticodon interactions are expected to be more stable than others due to factors such as the GC base pair content. It has been proposed that the specific sequence and post-transcriptional modification status of the tRNA in the region near the anticodon is tuned to ensure nearly indistinguishable binding of tRNAs to the ribosome A site [36]–[38]. This has been the case for tRNAs such as  in which both nucleotides at 34 and 37 are post-transcriptionally modified to achieve similar recognitions of AAA and AAG codons [38]. Unlike endogenous tRNAs that have corresponding modification enzymes,
in which both nucleotides at 34 and 37 are post-transcriptionally modified to achieve similar recognitions of AAA and AAG codons [38]. Unlike endogenous tRNAs that have corresponding modification enzymes, 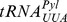 is exogenous and may not be targeted by tRNA modification enzymes in E. coli.
is exogenous and may not be targeted by tRNA modification enzymes in E. coli. 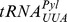 without modifications at its anticodon loop likely has a very weak binding affinity to the ribosome A site to associate UAG. Our finding points out that wobble base pairing at the 3′ nucleotide of a codon is not sufficient for recruiting a tRNA to the ribosome A-site. Additional interactions are required. This aspect needs to be further investigated. Since
without modifications at its anticodon loop likely has a very weak binding affinity to the ribosome A site to associate UAG. Our finding points out that wobble base pairing at the 3′ nucleotide of a codon is not sufficient for recruiting a tRNA to the ribosome A-site. Additional interactions are required. This aspect needs to be further investigated. Since  has a low ability to recognize UAG, it is feasible to use an amber suppressor aaRS-tRNA pair and a wild type or evolved PylRS-
has a low ability to recognize UAG, it is feasible to use an amber suppressor aaRS-tRNA pair and a wild type or evolved PylRS- pair to code two different NAAs at amber and ochre mutation sites, respectively, in E. coli.
pair to code two different NAAs at amber and ochre mutation sites, respectively, in E. coli.
During the anticodon-codon cross recognition analysis to examine whether 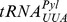 can compete against
can compete against  to bind UAG in the ribosome A site, we noticed that Phe was incorporated at N134 of sfGFP which was expressed in cells transformed with pEVOL-AzFRS and pETtrio-pylT(UUA)-PylRS-sfGFP134TAG and grown either in the absence or in the presence of BocK. We think AzFRS can recognize Phe, leading to the misincorporation of Phe. Since AzF is a hydrophobic amino acid and structurally very similar to Phe, one would expect that AzFRS that was originally evolved from MjTyrRS would probably recognize Phe at a relatively low level and charge
to bind UAG in the ribosome A site, we noticed that Phe was incorporated at N134 of sfGFP which was expressed in cells transformed with pEVOL-AzFRS and pETtrio-pylT(UUA)-PylRS-sfGFP134TAG and grown either in the absence or in the presence of BocK. We think AzFRS can recognize Phe, leading to the misincorporation of Phe. Since AzF is a hydrophobic amino acid and structurally very similar to Phe, one would expect that AzFRS that was originally evolved from MjTyrRS would probably recognize Phe at a relatively low level and charge  with Phe when AzF is absent in the medium. Although not clearly addressed in existing literature, most evolved MjTyrRS-
with Phe when AzF is absent in the medium. Although not clearly addressed in existing literature, most evolved MjTyrRS-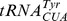 pairs did show significant background amber suppression even in minimal media [39], [40]. Since most evolved MjTyrRS variants are for Phe derivatives, it is highly possible that background amber suppression caused by these evolved MjTyrRS-
pairs did show significant background amber suppression even in minimal media [39], [40]. Since most evolved MjTyrRS variants are for Phe derivatives, it is highly possible that background amber suppression caused by these evolved MjTyrRS-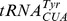 pairs was due to their recognition of either Phe or Tyr or both. In this study, the background amber suppression induced by the AzFRS-
pairs was due to their recognition of either Phe or Tyr or both. In this study, the background amber suppression induced by the AzFRS- pair inhibited both the basal amber suppression level and amber suppression induced by the PylRS-
pair inhibited both the basal amber suppression level and amber suppression induced by the PylRS- pair. This test also provided evidence that amber suppression mediated by the PylRS-
pair. This test also provided evidence that amber suppression mediated by the PylRS-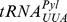 pair is too low to be a concern. It also points out that substrate specificities of evolved NAA-specific aaRSs need to be further characterized.
pair is too low to be a concern. It also points out that substrate specificities of evolved NAA-specific aaRSs need to be further characterized.
AGG Codon Suppression
Our lab and other groups have shown that mutating the anticodon of tRNAPyl does not significantly affect its interactions with PylRS. Three tRNAPyl isoforms that are specific for three stop codons are capable to deliver BocK at their corresponding codon sites when co-expressed with PylRS. We were also curious about the ability a tRNAPyl isoform to suppress a sensing codon. We chose to test on the suppression of the AGG codon since it is a rarely used codon and 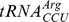 is limitedly expressed in the E. coli BL21(DE3) cell strain [41]. However, E. coli transformed with pETtrio-pylT(CCU)-PylRS-sfGFP2AGG only expressed sfGFP with Arg at its S2 position in the presence of 5 mM BocK. Increasing the BocK concentration to 10 mM did drive the expression of sfGFP with BocK incorporated at S2 of sfGFP, which was confirmed by the ESI-MS analysis of the purified proteins (data not shown). However, in comparison to the Arg-containing sfGFP isoform that showed a high intensity in the ESI-MS spectrum of the purified sfGFP, the intensity for the BocK-containing sfGFP isoform was very low. This indicates
is limitedly expressed in the E. coli BL21(DE3) cell strain [41]. However, E. coli transformed with pETtrio-pylT(CCU)-PylRS-sfGFP2AGG only expressed sfGFP with Arg at its S2 position in the presence of 5 mM BocK. Increasing the BocK concentration to 10 mM did drive the expression of sfGFP with BocK incorporated at S2 of sfGFP, which was confirmed by the ESI-MS analysis of the purified proteins (data not shown). However, in comparison to the Arg-containing sfGFP isoform that showed a high intensity in the ESI-MS spectrum of the purified sfGFP, the intensity for the BocK-containing sfGFP isoform was very low. This indicates 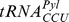 is charged by PylRS with BocK in E. coli and is able to deliver BocK at an AGG codon site. However, the PylRS-
is charged by PylRS with BocK in E. coli and is able to deliver BocK at an AGG codon site. However, the PylRS- 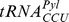 pair cannot compete efficiently against the endogenous Arg incorporation system at the AGG codon. Similar to
pair cannot compete efficiently against the endogenous Arg incorporation system at the AGG codon. Similar to 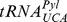 , the sequence and structure of
, the sequence and structure of  may not be optimized for the protein translation machinery in E. coli.
may not be optimized for the protein translation machinery in E. coli.
In summary, the suppression efficiencies of the original and three tRNAPyl variant, the cross recognition between nonsense codons and tRNAPyl anticodons in the E. coli BL21(DE3) cell strain have been investigated. Among all tRNAPyl isoforms, 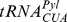 has the highest suppression efficiency for the delivery of BocK at its corresponding codon. Besides its orthogonal nature in E. coli,
has the highest suppression efficiency for the delivery of BocK at its corresponding codon. Besides its orthogonal nature in E. coli, 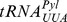 does not induce a significant level of suppression at an amber codon. This is contrary to the wobble hypothesis and makes it feasible to use amber suppressing aaRS-tRNA pair and the PylRS-
does not induce a significant level of suppression at an amber codon. This is contrary to the wobble hypothesis and makes it feasible to use amber suppressing aaRS-tRNA pair and the PylRS-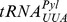 pair to code two different NAAs at amber and ochre codons respectively in E. coli. Our study also demonstrates the PylRS-
pair to code two different NAAs at amber and ochre codons respectively in E. coli. Our study also demonstrates the PylRS- pair cannot efficiently deliver BocK at an AGG codon site. Further work to optimize the sequence and structure of
pair cannot efficiently deliver BocK at an AGG codon site. Further work to optimize the sequence and structure of 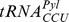 for the E. coli translation machinery may be necessary to increase the BocK incorporation efficiency and suppress the Arg incorporation at the AGG codon.
for the E. coli translation machinery may be necessary to increase the BocK incorporation efficiency and suppress the Arg incorporation at the AGG codon.
Acknowledgments
We thank Dr. Ryan Mehl at Franklin & Marshall College for providing us plasmids pBAD-sfGFP and pBAD-sfGFP134TAG and Dr. Peter G. Schultz at the Scripps Research Institute for providing us the plasmid pEVOL-AzFRS.
Funding Statement
This work was supported by the National Institute of Health (grant 1R01CA161158) and the Welch Foundation (grant A-1715). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hao B, Gong W, Ferguson TK, James CM, Krzycki JA, et al. (2002) A New UAG-Encoded Residue in the Structure of a Methanogen Methyltransferase. Science 296: 1462–1466. [DOI] [PubMed] [Google Scholar]
- 2. Srinivasan G, James CM, Krzycki JA (2002) Pyrrolysine Encoded by UAG in Archaea: Charging of a UAG-Decoding Specialized tRNA. Science 296: 1459–1462. [DOI] [PubMed] [Google Scholar]
- 3. Baron C, Böck A (1991) The length of the aminoacyl-acceptor stem of the selenocysteine-specific tRNA(Sec) of Escherichia coli is the determinant for binding to elongation factors SELB or Tu. J Biol Chem 266: 20375–20379. [PubMed] [Google Scholar]
- 4. Tujebajeva RM, Copeland PR, Xu XM, Carlson BA, Harney JW, et al. (2000) Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Reports 1: 158–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yuan J, O’Donoghue P, Ambrogelly A, Gundllapalli S, Sherrer RL, et al. (2010) Distinct genetic code expansion strategies for selenocysteine and pyrrolysine are reflected in different aminoacyl-tRNA formation systems. FEBS Lett 584: 342–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Polycarpo CR, Herring S, Berube A, Wood JL, Soll D, et al. (2006) Pyrrolysine analogues as substrates for pyrrolysyl-tRNA synthetase. FEBS Lett 580: 6695–6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mukai T, Kobayashi T, Hino N, Yanagisawa T, Sakamoto K, et al. (2008) Adding l-lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl-tRNA synthetases. Biochem Biophys Res Commun 371: 818–822. [DOI] [PubMed] [Google Scholar]
- 8. Yanagisawa T, Ishii R, Fukunaga R, Kobayashi T, Sakamoto K, et al. (2008) Multistep engineering of pyrrolysyl-tRNA synthetase to genetically encode N(epsilon)-(o-azidobenzyloxycarbonyl) lysine for site-specific protein modification. Chem Biol 15: 1187–1197. [DOI] [PubMed] [Google Scholar]
- 9. Nguyen DP, Lusic H, Neumann H, Kapadnis PB, Deiters A, et al. (2009) Genetic encoding and labeling of aliphatic azides and alkynes in recombinant proteins via a pyrrolysyl-tRNA Synthetase/tRNA(CUA) pair and click chemistry. J Am Chem Soc 131: 8720–8721. [DOI] [PubMed] [Google Scholar]
- 10. Ai HW, Lee JW, Schultz PG (2010) A method to site-specifically introduce methyllysine into proteins in E. coli. Chem Commun (Camb) 46: 5506–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fekner T, Li X, Lee MM, Chan MK (2009) A pyrrolysine analogue for protein click chemistry. Angew Chem Int Ed Engl 48: 1633–1635. [DOI] [PubMed] [Google Scholar]
- 12. Li X, Fekner T, Ottesen JJ, Chan MK (2009) A pyrrolysine analogue for site-specific protein ubiquitination. Angew Chem Int Ed Engl 48: 9184–9187. [DOI] [PubMed] [Google Scholar]
- 13. Neumann H, Peak-Chew SY, Chin JW (2008) Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat Chem Biol 4: 232–234. [DOI] [PubMed] [Google Scholar]
- 14. Huang Y, Wan W, Russell WK, Pai PJ, Wang Z, et al. (2010) Genetic incorporation of an aliphatic keto-containing amino acid into proteins for their site-specific modifications. Bioorg Med Chem Lett 20: 878–880. [DOI] [PubMed] [Google Scholar]
- 15. Wang YS, Wu B, Wang Z, Huang Y, Wan W, et al. (2010) A genetically encoded photocaged Nepsilon-methyl-L-lysine. Mol Biosyst 6: 1557–1560. [DOI] [PubMed] [Google Scholar]
- 16. Wang YS, Russell WK, Wang Z, Wan W, Dodd LE, et al. (2011) The de novo engineering of pyrrolysyl-tRNA synthetase for genetic incorporation of L-phenylalanine and its derivatives. Mol Biosyst 7: 714–717. [DOI] [PubMed] [Google Scholar]
- 17. Chen PR, Groff D, Guo J, Ou W, Cellitti S, et al. (2009) A facile system for encoding unnatural amino acids in mammalian cells. Angew Chem Int Ed Engl 48: 4052–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang M, Lin S, Song X, Liu J, Fu Y, et al. (2011) A genetically incorporated crosslinker reveals chaperone cooperation in acid resistance. Nat Chem Biol 7: 671–677. [DOI] [PubMed] [Google Scholar]
- 19. Takimoto JK, Dellas N, Noel JP, Wang L (2011) Stereochemical basis for engineered pyrrolysyl-tRNA synthetase and the efficient in vivo incorporation of structurally divergent non-native amino acids. ACS Chem Biol 6: 733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang YS, Fang X, Wallace AL, Wu B, Liu WR (2012) A rationally designed pyrrolysyl-tRNA synthetase mutant with a broad substrate spectrum. J Am Chem Soc 134: 2950–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gautier A, Nguyen DP, Lusic H, An W, Deiters A, et al. (2010) Genetically encoded photocontrol of protein localization in mammalian cells. J Am Chem Soc 132: 4086–4088. [DOI] [PubMed] [Google Scholar]
- 22. Liu WR, Wang YS, Wan W (2011) Synthesis of proteins with defined posttranslational modifications using the genetic noncanonical amino acid incorporation approach. Mol Biosyst 7: 38–47. [DOI] [PubMed] [Google Scholar]
- 23. Anderson JC, Wu N, Santoro SW, Lakshman V, King DS, et al. (2004) An expanded genetic code with a functional quadruplet codon. Proc Natl Acad Sci U S A 101: 7566–7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW (2010) Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 464: 441–444. [DOI] [PubMed] [Google Scholar]
- 25. Wan W, Huang Y, Wang Z, Russell WK, Pai PJ, et al. (2010) A facile system for genetic incorporation of two different noncanonical amino acids into one protein in Escherichia coli. Angew Chem Int Ed Engl 49: 3211–3214. [DOI] [PubMed] [Google Scholar]
- 26. Wu B, Wang Z, Huang Y, Liu WR (2012) Catalyst-free and site-specific one-pot dual-labeling of a protein directed by two genetically incorporated noncanonical amino acids. Chembiochem 13: 1405–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. O’Donoghue P, Prat L, Heinemann IU, Ling J, Odoi K, et al. (2012) Near-cognate suppression of amber, opal and quadruplet codons competes with aminoacyl-tRNA(Pyl) for genetic code expansion. FEBS Lett 586: 3931–3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Crick FH (1966) Codon–anticodon pairing: the wobble hypothesis. J Mol Biol 19: 548–555. [DOI] [PubMed] [Google Scholar]
- 29. Schwyzer R, Caviezel M (1971) p-Azido-L-phenylalanine: A photo-affinity ‘probe’ related to tyrosine. Helvetica Chimica Acta 54: 1395–1400. [DOI] [PubMed] [Google Scholar]
- 30. Young TS, Ahmad I, Yin JA, Schultz PG (2010) An enhanced system for unnatural amino acid mutagenesis in E. coli. J Mol Biol 395: 361–374. [DOI] [PubMed] [Google Scholar]
- 31. Swanson R, Hoben P, Sumner-Smith M, Uemura H, Watson L, et al. (1988) Accuracy of in vivo aminoacylation requires proper balance of tRNA and aminoacyl-tRNA synthetase. Science 242: 1548–1551. [DOI] [PubMed] [Google Scholar]
- 32. Fukunaga J-i, Ohno S, Nishikawa K, Yokogawa T (2007) A base pair at the bottom of the anticodon stem is reciprocally preferred for discrimination of cognate tRNAs by Escherichia coli lysyl- and glutaminyl-tRNA synthetases. Nucleic Acids Res 34: 3181–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Giege R, Sissler M, Florentz C (1998) Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res 26: 5017–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ogle JM, Ramakrishnan V (2005) Structural insights into translational fidelity. Annu Rev Biochem 74: 129–177. [DOI] [PubMed] [Google Scholar]
- 35. Zaher HS, Green R (2009) Fidelity at the Molecular Level: Lessons from Protein Synthesis. Cell 136: 746–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Agris PF, Vendeix FA, Graham WD (2007) tRNA’s wobble decoding of the genome: 40 years of modification. J Mol Biol 366: 1–13. [DOI] [PubMed] [Google Scholar]
- 37. Allner O, Nilsson L (2011) Nucleotide modifications and tRNA anticodon-mRNA codon interactions on the ribosome. RNA 17: 2177–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vold BS, Keith DE Jr, Buck M, McCloskey JA, Pang H (1982) Lysine tRNAs from Bacillus subtilis 168: structural analysis. Nucleic Acids Res 10: 3125–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang L, Brock A, Herberich B, Schultz PG (2001) Expanding the genetic code of Escherichia coli. Science 292: 498–500. [DOI] [PubMed] [Google Scholar]
- 40. Xie J, Wang L, Wu N, Brock A, Spraggon G, et al. (2004) The site-specific incorporation of p-iodo-L-phenylalanine into proteins for structure determination. Nat Biotechnol 22: 1297–1301. [DOI] [PubMed] [Google Scholar]
- 41. Gribskov M, Devereux J, Burgess RR (1984) The codon preference plot: graphic analysis of protein coding sequences and prediction of gene expression. Nucleic Acids Res 12: 539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]