Abstract
Deregulated expression of microRNAs (miRNAs) is common and biologically relevant in cervical carcinogenesis and appears only partly related to chromosomal changes. We recently identified 32 miRNAs showing decreased expression in high-grade cervical intraepithelial neoplasia (CIN) and carcinomas not associated with a chromosomal loss, 6 of which were located within a CpG island. This study aimed to investigate to what extent these miRNAs are subject to DNA methylation-mediated transcriptional repression in cervical carcinogenesis.
Methylation-specific PCR (MSP) analysis on a cell line panel representing different stages of human papillomavirus (HPV) induced transformation revealed an increase in methylation of hsa-miR-149, -203 and -375 with progression to malignancy, whereas expression of these miRNAs was restored upon treatment with a demethylating agent. All three miRNAs showed significantly increased levels of methylation in cervical carcinomas, whereas methylation levels of hsa-miR-203 and -375 were also significantly increased in high-grade CIN. A pilot analysis showed that increased hsa-miR-203 methylation was also detectable in HPV-positive cervical scrapes of women with high-grade CIN compared with controls. Similar to recent findings on hsa-miR-375, ectopic expression of hsa-miR-203 in cervical cancer cells decreased both the proliferation rate and anchorage independent growth. We found evidence for methylation-mediated transcriptional repression of hsa-miR-149, -203 and -375 in cervical cancer. Methylation of the latter two was already apparent in precancerous lesions and represent functionally relevant events in HPV-mediated transformation. Increased hsa-miR-203 methylation was detectable in scrapes of women with high-grade CIN, indicating that methylated miRNAs may provide putative markers to assess the presence of (pre)cancerous lesions.
Keywords: microRNA, squamous cell carcinoma, DNA methylation, CIN lesion, HPV, MSP
Introduction
Cervical cancer is initiated by a persistent infection with high-risk (hr) types of the human papillomavirus (HPV) and represents the third most common cancer in women worldwide.1,2 The development of cervical squamous cell carcinomas (SCC) is characterized by well-defined precursor lesions, called cervical intraepithelial neoplasia (CIN), graded 1–3.
HPV infects the basal cells of the cervical epithelium and, normally, expression of the viral genes is tightly linked to the differentiation of the epithelial cells. In a small number of cases, this productive infection can persist and change into a so called transforming infection. This aberrant infection pattern is found in virtually all high-grade CIN lesions and carcinomas and is characterized by uncontrolled expression of the viral oncogenes E6 and E7 in the basal proliferating cells of the epithelium. Deregulated expression of E6 and E7 results in uncontrolled cell cycling and, consequently, genetic instability, which may ultimately lead to malignant transformation of the cell. More insight in the crucial alterations occurring in the host cell during hrHPV-mediated transformation will increase our current understanding of (cervical) carcinogenesis and contribute to future diagnostic and therapeutic strategies.
Previous studies by others and us have shown that both chromosomal alterations and epigenetic changes contribute to the necessary changes in gene expression during hrHPV-mediated transformation (for a review see ref. 3). More recently, both mechanisms were also shown to be involved in the deregulation of microRNA (miRNA) expression in cervical cancer.4-6 miRNAs are small non-coding RNA molecules that regulate the expression of protein-coding genes via base pairing to the 3′ untranslated region (3′ UTR) and subsequent induction of degradation, destabilization or translation inhibition of their target mRNAs. Due to their ability to alter the expression of protein-coding oncogenes and tumor suppressor genes, miRNAs are now widely accepted as crucial players in cancer development.
We previously investigated the genome-wide miRNA expression patterns in normal cervical epithelium, high-grade CIN lesions and SCCs and were able to link part of the observed changes in expression to chromosomal alterations in the same samples.6 However, decreased expression in both high-grade CIN lesions and SCCs or only in SCCs compared with normal cervical epithelium was observed for 34 miRNAs, whereas only for 2 miRNAs an association with a chromosomal loss was found. According to current estimations around half of all miRNA loci is associated with CpG islands, suggesting that DNA methylation may play a prominent role in (ab)normal miRNA expression regulation as well.7 Indeed, the importance of methylation-mediated miRNA silencing in cancer is becoming more and more apparent (for a review see ref. 8).
Therefore, this study aimed to investigate the potential contribution of DNA methylation to the altered miRNA expression profiles we previously observed in cervical (pre)cancerous lesions.6 Out of the 32 downregulated miRNAs not associated with a chromosomal loss, 6 were located completely within a CpG island, namely hsa-miR-149, -203, -210, -375, -572 and -638. For these miRNAs, methylation was investigated using a longitudinal cell line panel consisting of primary human foreskin keratinocytes, hrHPV-immortalized keratinocytes, which are reminiscent of high-grade CIN lesions both (epi)genetically and morphologically, and cervical cancer cell lines.9-11 For miRNAs showing increased methylation in hrHPV-transformed cells compared with primary keratinocytes and for which methylation was shown to be directly associated with transcriptional repression of the miRNA, methylation levels were subsequently determined in clinical specimens. Finally, the functional relevance of methylation-mediated transcriptional repression of hsa-miR-203 was investigated in vitro.
Results
Increased hsa-miR-149, -203, -375 and -638 methylation in HPV-immortalized keratinocytes and cervical cancer cell lines
In a previous study we identified 32 miRNAs with significantly decreased expression in cervical (pre)malignant disease not associated with a chromosomal loss.6 Examination of the genomic context of these miRNAs identified 6 miRNAs, hsa-miR-149, -203, -210, -375, -572 and -638, which were located completely within a CpG island. To investigate whether the decrease in expression observed for these miRNAs during cervical carcinogenesis was associated with an increase in DNA methylation, we developed methylation-specific PCR (MSP) assays targeting the promoter regions close to the start of the miRNA gene. Our previous results demonstrate that methylation patterns observed in HPV-immortalized keratinocytes (FK16A, FK18A and FK18B; reminiscent of high-grade CIN) and cervical cancer cell lines SiHa, HeLa and CaSki in general reflect the methylation pattern found in cervical (pre)cancerous lesions.5,9,12,13 Therefore, methylation for all 6 miRNA promoter regions was first determined in this cell line panel including primary human foreskin keratinocytes (HFKs) from 5 individuals as normal controls.
For hsa-miR-149, methylation was observed in all HFKs and cell lines tested, though the level of methylation was increased in HPV-immortalized keratinocytes and cervical cancer cell lines compared with HFKs (Fig. 1A). hsa-miR-203 (Fig. 1B) showed methylation in 2 out of 5 HFKs as well as in 2 out of 3 HPV-immortalized keratinocyte cell lines and cervical cancer cell lines. hsa-miR-210 was not methylated in any of the cells tested (data not shown). Methylation of hsa-miR-375 was only observed in one HFK and all three cancer cell lines (Fig. 1C), whereas hsa-miR-572 was strongly methylated in all cells tested (Fig. 1D). Finally, hsa-miR-638 showed low levels of methylation in 2 out of 5 HFKs and 2 out of 3 HPV-immortalized keratinocyte cell lines and all cervical cancer cell lines (Fig. 1E).
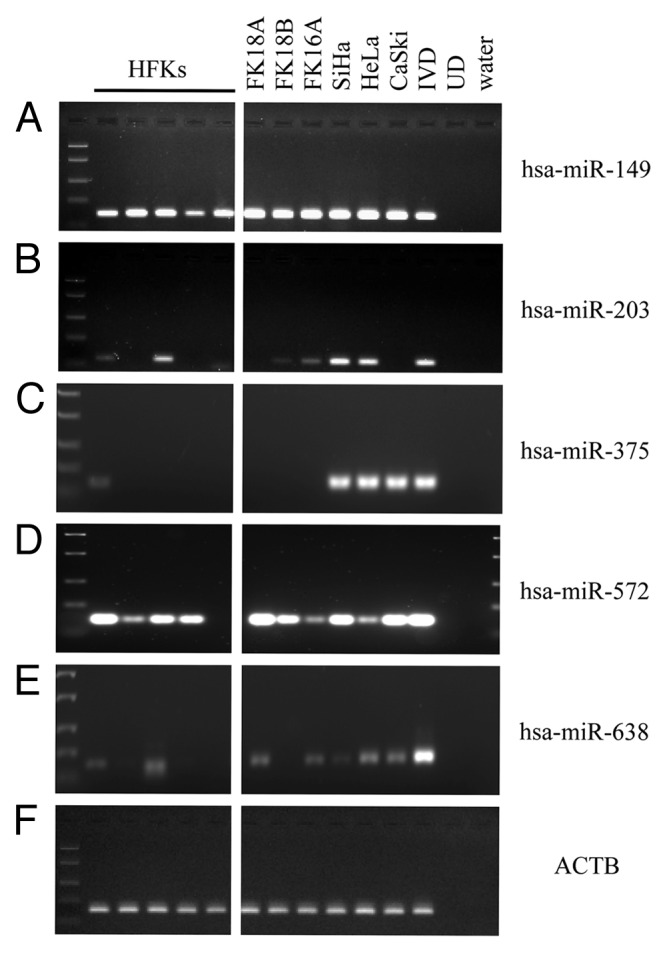
Figure 1. Methylation patterns of selected miRNAs in the cell line panel. Methylation as determined in human foreskin keratinocytes (HFKs), HPV-transformed keratinocyte cell lines FK16A, FK18A and FK18B (reminiscent of high-grade CIN), and cervical cancer cell lines SiHa, HeLa and CaSki is shown for (A) hsa-miR-149, (B) hsa-miR-203, (C) hsa-miR-375, (D) hsa-miR-572 and (E) hsa-miR-638. In (F) ACTB results are shown, indicating successful modification and comparable input for all samples. In vitro methylated DNA (IVD) and unmodified DNA (UD) were included as a positive and negative control, respectively.
In summary, methylation of hsa-miR-149, -203, -375 and -638 was found to increase in HPV-immortalized keratinocytes and/or cervical cancer cell lines compared with primary keratinocytes and, therefore, these miRNAs were selected for further examination.
Methylation-mediated transcriptional repression of hsa-miR-149, -203 and -375
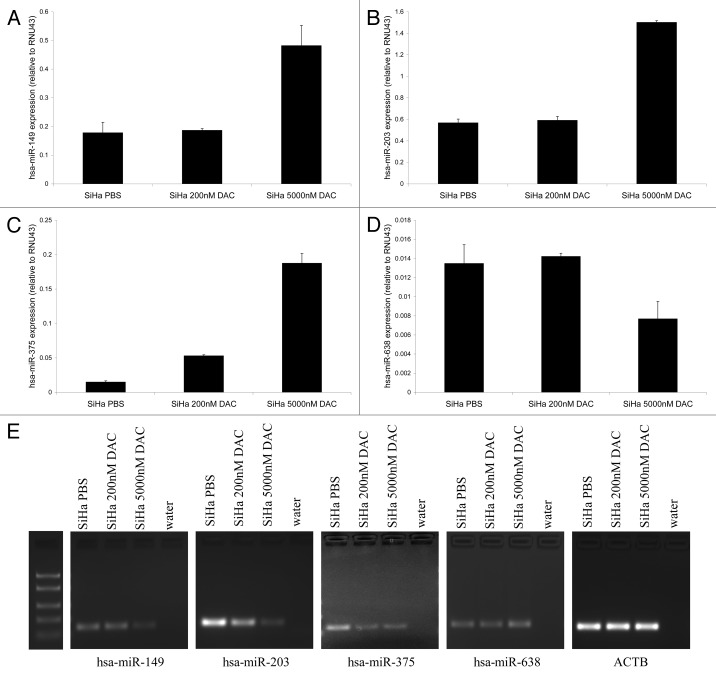
To investigate whether methylation of hsa-miR-149, -203, -375 and -638 was associated with transcriptional repression of these miRNAs, we treated SiHa cells with a demethylating agent (DAC). Expression of hsa-miR-149, -203, and -375 (Fig. 2A−C) increased after treatment with 5,000 nM DAC. However, no increase in hsa-miR-638 expression was observed following DAC treatment (Fig. 2D). Although some variation in expression was observed for hsa-miR-638 after DAC treatment, the overall low levels of hsa-miR-638 expression suggest these small differences may reflect normal biological variation. In concordance with these findings, decreased methylation was observed for hsa-miR-149, -203 and -375 in DAC-treated cells, but not for hsa-miR-638 (Fig. 2E). Potentially, higher concentrations of DAC are needed to reduce methylation at the hsa-miR-638 promoter region or more complex epigenetic mechanisms are involved at this locus. These results indicate that reduced hsa-miR-149, -203 and -375 expression in cervical cancer cells is associated with DNA methylation.
Figure 2. Re-expression of methylated miRNAs after demethylating treatment. Expression levels of (A) hsa-miR-149, (B) hsa-miR-203, (C) hsa-miR-375 and (D) hsa-miR-638 were determined in SiHa cells treated with PBS (mock), 200 nM and 5,000 nM DAC. (E) MSP results for hsa-miR-149, -203, -375 and -638 in SiHa cells treated with PBS (mock), 200 nM and 5,000 nM DAC are shown. The ACTB PCR results indicate successful modification and comparable input for all samples.
Increasing levels of hsa-miR-149, -203 and -375 methylation with progressive stages of cervical disease
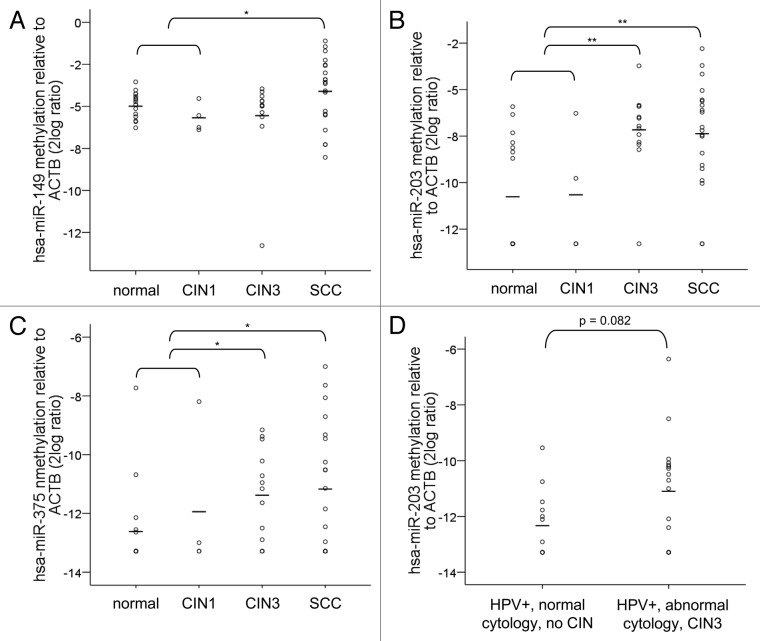
Next, methylation levels of hsa-miR-149, -203 and -375 were determined in paraffin-embedded specimens of normal cervix (n = 16), CIN1 (n = 4), CIN3 (n = 13) and SCCs (n = 20) by quantitative MSP (qMSP). For hsa-miR-149, an increase in methylation was only observed in SCCs and not in CIN3 lesions compared with normal and CIN1 together (≤ CIN1; p = 0.04) (Fig. 3A). Methylation levels of hsa-miR-203 and hsa-miR-375, on the other hand, were significantly increased in both CIN3 (p = 0.001 and p = 0.02, respectively) and SCCs (p = 0.002 and p = 0.03, respectively) compared with normal and CIN1 (Fig. 3B and C). Based on the fact that hsa-miR-203 showed the most significant increase in methylation level in CIN3 and SCC compared with normal and CIN1, we selected this miRNA for a pilot experiment using cervical scrapes. As is shown in Figure 3D, an increase in methylation of hsa-miR-203 was also detectable in a small set of hrHPV-positive cervical scrapes of women with abnormal cytology and underlying high-grade CIN disease compared with hrHPV-positive cervical scrapes of women with normal cytology and without underlying CIN disease (p = 0.082).
Figure 3. Methylation levels in cervical specimens. Methylation levels were determined in tissue specimens of normal cervix (n = 16), CIN1 (n = 4), CIN3 (n = 13) and SCCs (n = 20) for (A) hsa-miR-149, (B) hsa-miR-203 and (C) hsa-miR-375. In (D) methylation levels of hsa-miR-203 are shown in cervical scrapes of hrHPV-positive women with normal cytology and without any CIN disease during 5-y follow-up (n = 13) and in scrapes of hrHPV-positive women with abnormal cytology who presented with high-grade CIN disease within 18 mo (n = 17). hsa-miR-149, -203 and -375 methylation was undetectable in 0, 9 and 11 normals; 0, 2 and 2 CIN1; 1, 1 and 3 CIN3; and 0, 2, and 7 SCCs. Methylation of hsa-miR-203 was undetectable in 6 scrapes with normal cytology and 5 with abnormal cytology. Average methylation levels per sample group are indicated by the horizontal lines. * p < 0.05; ** p < 0.01.
Functional involvement of hsa-miR-203 in vitro
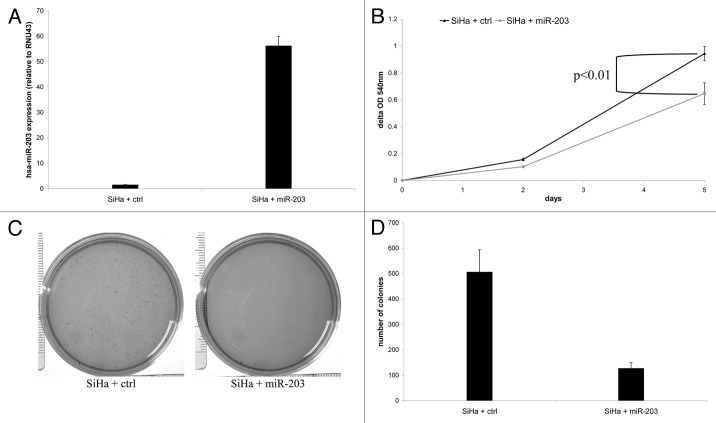
Others and we recently demonstrated that hsa-miR-375 is functionally involved in HPV-mediated transformation.14,15 Hence, we investigated whether hsa-miR-203 may also possess tumor suppressive traits in cervical cancer by transducing SiHa cells with either a hsa-miR-203 containing vector or an empty control vector. Ectopic hsa-miR-203 expression was verified by qRT-PCR (Fig. 4A). hsa-miR-203 overexpression significantly reduced the proliferation rate compared with empty vector control cells (Fig. 4B; p = 0.003). In addition, ectopic hsa-miR-203 expression resulted in a 4-fold reduction of the number of colonies formed in soft agar compared with empty vector control cells (Fig. 4C and D). Together, these results are supportive of a tumor suppressive role of hsa-miR-203 in hrHPV-mediated transformation.
Figure 4. The effect of ectopic expression of hsa-miR-203 in SiHa cells on cellular proliferation and anchorage independent growth. (A) Expression of hsa-miR-203 in SiHa cells transduced with either the control vector (SiHa + ctrl) or hsa-miR-203 (SiHa + miR-203). In (B) cell viability is shown in SiHa + ctrl cells and SiHa + miR-203 cells. In (C) representative pictures of colony formation in soft agar of SiHa + ctrl cells and SiHa + miR-203 cells and in (D) quantification of the number of colonies in soft agar formed by SiHa + ctrl cells and SiHa + miR-203 cells.
Discussion
In the present study we investigated the potential contribution of methylation-mediated transcriptional repression of miRNAs to the altered miRNA expression patterns we previously observed in cervical (pre)cancerous lesions.6 Using an in vitro model of hrHPV-mediated transformation, we found evidence for methylation-mediated transcriptional repression of 3 miRNAs, namely hsa-miR-149, -203 and -375, in HPV-mediated transformation.
Methylation-mediated transcriptional repression was not found for an additional 3 other candidates, hsa-miR-210, -572 and -638 that also showed reduced expression in cervical (pre)cancerous lesions and of which the entire gene body is located in a CpG island. Although low levels of hsa-miR-638 methylation were observed in HPV-immortalized keratinocytes and cervical cancer cell lines, treatment with a demethylating agent (DAC) did not restore its expression, indicating that expression of this miRNA may be subject to more complex epigenetic regulatory mechanisms.
We developed methylation specific PCR assays that were located close to the start of the miRNA gene, as in general this region is most closely linked to transcriptional silencing.16 Therefore, we cannot exclude that CpG-rich regions more distant from the miRNA gene may potentially be involved in regulation of these candidates. Moreover, as the current study only investigated miRNAs of which the whole gene body was located within a CpG island, it is possible that other miRNAs showing reduced expression in cervical (pre)cancerous lesions which are not entirely located in a CpG island are subject to DNA methylation-mediated transcriptional repression in cervical cancer as well. Future genome-wide methylation profiling studies will enable a more comprehensive analysis of methylation-mediated miRNA transcriptional repression in cervical cancer.
Since the percentage of hrHPV-positive samples was lower in our control group compared with high-grade CIN lesions and SCCs, differences in methylation levels between sample groups may also be associated with the presence of hrHPV. However, initial selection of the miRNAs investigated in this study was based on a decrease in expression in high-grade CIN and SCCs compared with hrHPV-positive normal cervical epithelium, suggesting that transcriptional repression of these miRNAs is not associated with hrHPV. This is further supported by the fact that we observed increased hsa-miR-203 methylation in hrHPV-positive scrapes with underlying disease compared with hrHPV-positive control scrapes.
So far, only few studies have investigated methylation of selected miRNAs, including hsa-miR-124, -203 and -34b, in cervical cancer.4,5 In this study, hsa-miR-149 methylation was identified as a novel methylation target in cervical cancers. To date, epigenetic transcriptional repression of hsa-miR-149 has only been described in colorectal cancer.17 The fact that elevated methylation levels were only detected in cervical carcinomas and not in CIN3 lesions, whereas expression levels were already decreased in high-grade CIN lesions, suggests that another mechanism is involved in the initial transcriptional repression of hsa-miR-149.6 Alternatively, since hsa-miR-149 methylation is detected in all samples, small differences in methylation level could already influence the expression of this miRNA. Since a significant increase in methylation of hsa-miR-149 is only detectable in invasive cancers, this miRNA may not provide a valuable tool for screening programs aiming at early detection of cervical precancerous lesions. Its potential prognostic or therapeutic value, however, remains to be determined.
In contrast to hsa-miR-149, methylation of hsa-miR-203 and -375 was already significantly increased in CIN3 lesions, which is concordant with the decrease in expression we previously observed in high-grade CIN lesions.6 The current study confirmed that methylation-mediated transcriptional repression of hsa-miR-203 is frequent in cervical (pre)cancer as described before.4 Here, we provide additional evidence for its functional relevance in cervical carcinogenesis. Ectopic expression of hsa-miR-203 in SiHa cells decreased both the proliferation rate and ability to grow anchorage independently, supporting a tumor suppressive role for this miRNA in cervical cancer. In breast cancer cells, decreased expression of hsa-miR-203 was associated with increased metastatic potential, which was (in part) regulated via direct targeting of SNAI2.18 hsa-miR-203 expression has also been described to be modulated by the HPV-encoded viral oncogenes. In particular, E7 was shown to decrease hsa-miR-203 expression upon differentiation of primary keratinocytes to retain cell cycle activity allowing viral amplification in the upper epithelial cell layers. Vice versa, increased hsa-miR-203 expression was shown to inhibit HPV genome amplification in differentiated cells.19 In addition, hsa-miR-203 expression was also shown to be dependent of the regulation of p53 levels by E6.20 However, as indicated before, the normal cervical epithelial samples used in our original expression profiling study, compared with which a strong decrease of hsa-miR-203 was observed in high-grade CIN lesions and SCCs, were already HPV-positive. This suggests that, next to an effect of HPV E6 and E7 on hsa-miR-203 expression in differentiating epithelium, additional transcriptional repression of this miRNA via DNA methylation in basal dividing cells is associated with a transforming hrHPV-infection. In addition to cervical cancer, methylation of hsa-miR-203 was recently described in oral and hepatocellular carcinomas.21,22
To the best of our knowledge, methylation-mediated transcriptional repression of hsa-miR-375 was not described in cervical cancer before, but has been demonstrated in esophageal cancer.23 In oral cancer, reduced hsa-miR-375 expression independent of methylation was observed.24 However, two separate CpG islands are present in the proximity of the hsa-miR-375 gene with opposite results on the expression of this miRNA, implicating that the exact region investigated for methylation will greatly influence the reported findings.25 We found evidence for methylation-mediated transcriptional repression of hsa-miR-375 in cervical (pre)cancer investigating the CpG island close to the start of the miRNA, which was shown to be associated with silencing before.25 Moreover, reduced expression of hsa-miR-375 was also shown to be associated with a focal chromosomal loss of its locus at 2q35 in high-grade cervical lesions and to be functionally relevant in cervical cancer development.14,15
In summary, we provide evidence for methylation-mediated transcriptional repression of hsa-miR-149 and -375 during cervical cancer development and validated previous findings by Botezatu et al. regarding hsa-miR-203 methylation in cervical (pre)cancerous lesions.4 Elevated methylation levels of hsa-miR-149 were only detectable in invasive cancers, whereas methylation levels of hsa-miR-203 and -375 already showed an increase in CIN3 lesions. Transcriptional repression of hsa-miR-203 was shown to be functionally relevant in cervical cancer cells in vitro, as was previously described for hsa-miR-375 as well. Previous studies by others and us have already highlighted the potential of aberrantly methylated gene detection for triage of hrHPV-positive women in cervical cancer screening by primary HPV testing.26,27 In the current study, we showed that increased methylation of hsa-miR-203 is also detectable in cervical scrapes of women with underlying high-grade cervical disease. Our earlier described results for hsa-miR-124, as well as current results for hsa-miR-203, warrant further investigation of the potential value of aberrant methylation-mediated transcriptional repression of miRNAs for improvement of HPV-based cervical cancer screening.5
Materials and Methods
Cell lines and clinical specimens
Establishment and culture of the HPV16 (FK16A) and HPV18 (FK18A and FK18B) immortalized keratinocyte cell lines has been described previously.28 The human cervical carcinoma cell lines SiHa, HeLa and CaSki were obtained from the American Type Culture Collection and cultured as described previously.29 Primary human foreskin keratinocytes (HFKs) were isolated and cultured as described previously.28 SiHa cells were treated with 200 nM and 5,000 nM 5-aza-2′-deoxycytidine (DAC; A3656; Sigma Chemical Co.) dissolved in PBS to analyze the effect of global methylation inhibition on expression of the selected miRNAs.
Formalin-fixed paraffin-embedded (FFPE) biopsies of 16 normal cervical squamous epithelial samples, 4 CIN1 lesions, 13 CIN3 lesions, and 20 SCCs were used. hrHPV testing of all biopsies was performed using the general primer GP5+/6+-mediated PCR-enzyme immunoassay method using a probe cocktail of 14 hrHPV types and hrHPV was detected in 0% of normals, 50% of CIN1, 92% of CIN3, and 100% of SCCs.30 All biopsies were collected during the course of routine clinical practice at the Department of Obstetrics and Gynaecology at the VU University Medical Center. This study followed the ethical guidelines of the Institutional Review Board of the VU University Medical Center.
Cervical scrapes were obtained from women participating in the population-based cervical screening trial POBASCAM.31,32 We used 13 hrHPV-positive scrapes from women with normal cytology and without any CIN disease during 5-y follow-up and 17 hrHPV-positive scrapes of women with abnormal cytology who presented with high-grade CIN disease within 18 mo.
Selection of miRNAs potentially silenced by methylation
The genomic locations of all miRNAs showing an early continuously (downregulated in both CIN2/3 and SCC compared with normal) or late (downregulated only in SCC compared with normal) decrease in expression as reported in Wilting et al. were examined for the presence of a CpG island using the UCSC Genome Browser (www.genome.ucsc.edu) (GRCh37/hg19 assembly).6 In this browser regions with a significantly higher density of CpG dinucleotides than found on average in the whole genome are considered a CpG island if the GC content is at least 50%, the region length is greater than 200 base pairs, and the ratio of the observed number of CpG dinucleotides to the expected number based on the total number of Gs and Cs in the region is greater than 0.6.33 In this way hsa-miR-149, -203, -210, -375, -572 and -638 were identified as potential methylation targets during cervical carcinogenesis.
RNA and DNA isolation, in vitro methylation of DNA, and DNA modification
Total RNA was isolated using TRIzol Reagent (15596-026; Life Technologies) according to the manufacturers’ instructions.
Total DNA was isolated by proteinase K digestion and purified using the High Pure PCR Template Preparation Kit (11796828001; Roche Diagnostics) following the manufacturer's recommendations or by standard phenol-chloroform extraction.34
In vitro methylated DNA (IVD) was generated as a positive control. In short, genomic DNA from SiHa was incubated with CpG methyltransferase (M.SssI; 1U/µg DNA; M0226S; New England Biolabs) in the presence of 1x NEBuffer 2 and 160 µM S-adenosylmethionine (E6021S and B9003S, respectively, New England Biolabs) at 37°C for 4 h. M.SssI was heat-inactivated at 65°C for 20 min.
Genomic DNA was modified using the EZ DNA Methylation kit (D5002, Zymo Research), which induces chemical conversion of unmethylated cytosines into uracils, whereas methylated cytosines are protected from this conversion.
(Quantitative) methylation-specific PCR [(q)MSP] analysis
MSP analysis for hsa-miR-149, -203, -210, -375, -572 and -638 was performed as described previously.35 Specific primers were designed to amplify the methylated DNA sequence of the promoter region (Table 1). In addition, the modified, unmethylated sequence of the housekeeping gene β-actin (ACTB) was amplified as a reference to verify sufficient DNA quality and successful DNA modification.36 All 6 MSP assays showed a positive signal with in vitro methylated DNA (IVD; positive control) and none showed signal with unmodified DNA from SiHa (UD; negative control).
Table 1. Primer and probe sequences used for (q)MSP analysis.
| Gene | Primer and probe sequences (5′−3′) |
Amplicon (bp) |
Annealing (°C) |
|---|---|---|---|
| hsa-miR-149 |
F: GTCGTGCGTTTAGTTTGTAGCG R: CGACCTAAAAACCCCCGAC P: CAAAACGAACGAAAACCTAAAAACCTCGAACA |
111 |
59 58 68 |
| hsa-miR-203 |
F: GTCGGTTTTCGGGATTCGTA R: CGATTCGCCGAACTACGAAA P: AACCGCCCGCGTC-Xsprobe |
78 |
59 60 70 |
| hsa-miR-210 |
F: GTCGGGTTGGGTAGGCG R: CACTACCGAATAAACGAACGAAAAAA |
90 |
59 61 |
| hsa-miR-375 |
F: GGGGCGTTGTGTAGTATTGAGTTC R: GAAACGAAAACGAAAAACCCG P: AAACCTAACGATCGCCCTCGATAATCTCCTAATC |
91 |
60 60 70 |
| hsa-miR-572 |
F: TCGGTTTATTTTTTAGTTTCGTAGTGC R: CACGACCTCGACACCTCCTAA |
111 |
59 59 |
| hsa-miR-638 |
F: TCGGTTAGAGTTGTTATTTGGATTTG R: CCGACCCACGAAATTCTCC |
96 |
59 59 |
| ACTB | F: TGGTGATGGAGGAGGTTTAGTAAGT R: AACCAATAAAACCTACTCCTCCCTTAA P: ACCACCACCCAACACACAATAACAAACACA |
133 | 58 59 69 |
F, forward; R, reverse; P, probe; Xsprobe, minor groove binder probe. Methylated CpG dinucleotides are in bold. Changed positions in the sequence due to the modification are indicated in italic.
For hsa-miR-149, -203 and -375 a methylation specific probe was also designed to allow the quantitative measurement of methylation levels for these miRNAs as described previously (Table 1).5 Only qMSP results for samples with a CT for ACTB below 32 were included in the analysis to exclude potential false negatives due to insufficient modified DNA quality and quantity.
Quantitative reverse-transcriptase PCR (qRT-PCR) analysis
Expression of hsa-miR-149, -203, -375 and -638 was measured using TaqMan microRNA assays following the manufacturer’s instructions (002255, 000507, 000564, 001588; Applied Biosystems) on the ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Nieuwerkerk a/d IJssel). The small nucleolar RNA transcript RNU43 was included as a reference gene (001095; Applied Biosystems). miRNA expression values were normalized to the reference using the comparative CT method (2-ΔCT).37
Retroviral transduction
Retroviral hsa-miR-203 and empty vector (ctrl) constructs previously described by Voorhoeve et al. were transfected into the Phoenix A retrovirus producer cell line and supernatants containing the replication-deficient hsa-miR-203-expressing retrovirus or empty vector retrovirus were harvested 48 h post-transfection.38 For the transduction experiments, SiHa cells were incubated for 16 h at 37°C with filtered viral supernatants supplemented with polybrene (15 µg/ml). SiHa + miR-203/SiHa + ctrl cells were selected by continuous culturing of the transduced cells in the presence of blasticidin (3 µg/ml) and ectopic expression was verified by qRT-PCR analysis.
Cell viability assay
Cell viability was measured using a colorimetric (MTT-tetrazolium) assay (02102227; MP Biomedicals, LLC.) as described before.12 In this assay the amount of dye conversion, as measured by the optical density at a wavelength of 540 nm, is directly related to the number of viable cells in each well. The proliferation rate was determined by subtracting the measurement of day 0 from all other time points.
Colony formation in soft agar
Anchorage-independent cell growth was analyzed as described before.13 In short, 5000 cells were suspended in medium containing 0.35% top agarose (Seaplague agarose; 50101; Lonza Group Ltd.) and plated on a surface of 0.6% bottom agarose in 6-cm dishes. Cells were incubated at 37°C for 3 weeks and were fed weekly by overlaying the agarose with fresh medium. Colonies were photographed and counted after 3 weeks of incubation.
Statistical analysis
Methylation levels were compared between the respective sample groups using the non-parametric Mann-Whitney U test. Proliferation rates between hsa-miR-203-expressing cells and control cells were compared using the Student's t test.
Acknowledgments
This research was funded by grants from the VU University Medical Center-Cancer Center Amsterdam (VUMC-CCA) and the Dutch Cancer Society (KWF2010-4668). The authors are grateful to Marlon Lindenbergh-Van der Plas, Tim Schutte, Annina van Splunter, Renée Overmeer, Suzanne Snellenberg, and Sylvia Duin for excellent technical assistance.
Glossary
Abbreviations:
- miRNA
microRNA
- MSP
methylation-specific polymerase chain reaction
- HPV
human papillomavirus
- hrHPV
high-risk human papillomavirus
- CIN
cervical intraepithelial neoplasia
- SCC
squamous cell carcinoma
- UTR
untranslated region
- HFK
human foreskin keratinocytes
- DAC
5-aza-2′-deoxycytidine
- qMSP
quantitative methylation-specific polymerase chain reaction
- qRT-PCR
quantitative reverse transcriptase polymerase chain reaction
- POBASCAM
population-based screening study Amsterdam
- IVD
in vitro methylated DNA
- UD
unmodified DNA
- MTT
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
Disclosure of Potential Conflicts of Interest
Dr RDM Steenbergen, Prof Dr PJF Snijders and Prof Dr CJLM Meijer are stockholders of Self-screen BV, The Netherlands. All other authors have no conflicts of interest to declare.
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/23605
References
- 1.Bosch FX, Lorincz A, Muñoz N, Meijer CJ, Shah KV. The causal relation between human papillomavirus and cervical cancer. J Clin Pathol. 2002;55:244–65. doi: 10.1136/jcp.55.4.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–9. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 3.Steenbergen RD, de Wilde J, Wilting SM, Brink AA, Snijders PJ, Meijer CJ. HPV-mediated transformation of the anogenital tract. J Clin Virol. 2005;32(Suppl 1):S25–33. doi: 10.1016/j.jcv.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Botezatu A, Goia-Rusanu CD, Iancu IV, Huica I, Plesa A, Socolov D, et al. Quantitative analysis of the relationship between microRNA‑124a, -34b and -203 gene methylation and cervical oncogenesis. Mol Med Report. 2011;4:121–8. doi: 10.3892/mmr.2010.394. [DOI] [PubMed] [Google Scholar]
- 5.Wilting SM, van Boerdonk RA, Henken FE, Meijer CJ, Diosdado B, Meijer GA, et al. Methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Mol Cancer. 2010;9:167–81. doi: 10.1186/1476-4598-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilting SM, Snijders PJ, Verlaat W, Jaspers A, van de Wiel MA, van Wieringen WN, et al. Altered microRNA expression associated with chromosomal changes contributes to cervical carcinogenesis. Oncogene. 2012 doi: 10.1038/onc.2012.20. In press. [DOI] [PubMed] [Google Scholar]
- 7.Weber B, Stresemann C, Brueckner B, Lyko F. Methylation of human microRNA genes in normal and neoplastic cells. Cell Cycle. 2007;6:1001–5. doi: 10.4161/cc.6.9.4209. [DOI] [PubMed] [Google Scholar]
- 8.Lopez-Serra P, Esteller M. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene. 2012;31:1609–22. doi: 10.1038/onc.2011.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henken FE, Wilting SM, Overmeer RM, van Rietschoten JG, Nygren AO, Errami A, et al. Sequential gene promoter methylation during HPV-induced cervical carcinogenesis. Br J Cancer. 2007;97:1457–64. doi: 10.1038/sj.bjc.6604055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steenbergen RD, Parker JN, Isern S, Snijders PJ, Walboomers JM, Meijer CJ, et al. Viral E6-E7 transcription in the basal layer of organotypic cultures without apparent p21cip1 protein precedes immortalization of human papillomavirus type 16- and 18-transfected human keratinocytes. J Virol. 1998;72:749–57. doi: 10.1128/jvi.72.1.749-757.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilting SM, Snijders PJ, Meijer GA, Ylstra B, van den Ijssel PR, Snijders AM, et al. Increased gene copy numbers at chromosome 20q are frequent in both squamous cell carcinomas and adenocarcinomas of the cervix. J Pathol. 2006;209:220–30. doi: 10.1002/path.1966. [DOI] [PubMed] [Google Scholar]
- 12.Overmeer RM, Henken FE, Bierkens M, Wilting SM, Timmerman I, Meijer CJ, et al. Repression of MAL tumor suppressor activity by promoter methylation during cervical carcinogenesis. J Pathol. 2009;9:327–36. doi: 10.1002/path.2598. [DOI] [PubMed] [Google Scholar]
- 13.Steenbergen RD, Kramer D, Braakhuis BJ, Stern PL, Verheijen RH, Meijer CJ, et al. TSLC1 gene silencing in cervical cancer cell lines and cervical neoplasia. J Natl Cancer Inst. 2004;96:294–305. doi: 10.1093/jnci/djh031. [DOI] [PubMed] [Google Scholar]
- 14.Bierkens M, Krijgsman O, Wilting SM, Bosch L, Jaspers A, Meijer GA, et al. Focal aberrations indicate EYA2 and hsa-miR-375 as oncogene and tumor suppressor in cervical carcinogenesis. Genes Chromosomes Cancer. 2012 doi: 10.1002/gcc.22006. In press. [DOI] [PubMed] [Google Scholar]
- 15.Wang F, Li Y, Zhou J, Xu J, Peng C, Ye F, et al. miR-375 is down-regulated in squamous cervical cancer and inhibits cell migration and invasion via targeting transcription factor SP1. Am J Pathol. 2011;179:2580–8. doi: 10.1016/j.ajpath.2011.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brenet F, Moh M, Funk P, Feierstein E, Viale AJ, Socci ND, et al. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One. 2011;6:e14524. doi: 10.1371/journal.pone.0014524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang F, Ma YL, Zhang P, Shen TY, Shi CZ, Yang YZ, et al. SP1 mediates the link between methylation of the tumour suppressor miR-149 and outcome in colorectal cancer. J Pathol. 2012 doi: 10.1002/path.4078. In press. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z, Zhang B, Li W, Fu L, Fu L, Zhu Z, et al. Epigenetic Silencing of miR-203 Upregulates SNAI2 and Contributes to the Invasiveness of Malignant Breast Cancer Cells. Genes Cancer. 2011;2:782–91. doi: 10.1177/1947601911429743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melar-New M, Laimins LA. Human papillomaviruses modulate expression of microRNA 203 upon epithelial differentiation to control levels of p63 proteins. J Virol. 2010;84:5212–21. doi: 10.1128/JVI.00078-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenna DJ, McDade SS, Patel D, McCance DJ. MicroRNA 203 expression in keratinocytes is dependent on regulation of p53 levels by E6. J Virol. 2010;84:10644–52. doi: 10.1128/JVI.00703-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–76. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 22.Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- 23.Li X, Lin R, Li J. Epigenetic silencing of microRNA-375 regulates PDK1 expression in esophageal cancer. Dig Dis Sci. 2011;56:2849–56. doi: 10.1007/s10620-011-1711-1. [DOI] [PubMed] [Google Scholar]
- 24.Wiklund ED, Gao S, Hulf T, Sibbritt T, Nair S, Costea DE, et al. MicroRNA alterations and associated aberrant DNA methylation patterns across multiple sample types in oral squamous cell carcinoma. PLoS One. 2011;6:e27840. doi: 10.1371/journal.pone.0027840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Souza Rocha Simonini P, Breiling A, Gupta N, Malekpour M, Youns M, Omranipour R, et al. Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor alpha in breast cancer cells. Cancer Res. 2010;70:9175–84. doi: 10.1158/0008-5472.CAN-10-1318. [DOI] [PubMed] [Google Scholar]
- 26.Eijsink JJ, Lendvai A, Deregowski V, Klip HG, Verpooten G, Dehaspe L, et al. A four-gene methylation marker panel as triage test in high-risk human papillomavirus positive patients. Int J Cancer. 2012;130:1861–9. doi: 10.1002/ijc.26326. [DOI] [PubMed] [Google Scholar]
- 27.Hesselink AT, Heideman DA, Steenbergen RD, Coupé VM, Overmeer RM, Rijkaart D, et al. Combined promoter methylation analysis of CADM1 and MAL: an objective triage tool for high-risk human papillomavirus DNA-positive women. Clin Cancer Res. 2011;17:2459–65. doi: 10.1158/1078-0432.CCR-10-2548. [DOI] [PubMed] [Google Scholar]
- 28.Steenbergen RD, Walboomers JM, Meijer CJ, van der Raaij-Helmer EM, Parker JN, Chow LT, et al. Transition of human papillomavirus type 16 and 18 transfected human foreskin keratinocytes towards immortality: activation of telomerase and allele losses at 3p, 10p, 11q and/or 18q. Oncogene. 1996;13:1249–57. [PubMed] [Google Scholar]
- 29.Steenbergen RD, Kramer D, Meijer CJ, Walboomers JM, Trott DA, Cuthbert AP, et al. Telomerase suppression by chromosome 6 in a human papillomavirus type 16-immortalized keratinocyte cell line and in a cervical cancer cell line. J Natl Cancer Inst. 2001;93:865–72. doi: 10.1093/jnci/93.11.865. [DOI] [PubMed] [Google Scholar]
- 30.Jacobs MV, Snijders PJ, van den Brule AJ, Helmerhorst TJ, Meijer CJ, Walboomers JM. A general primer GP5+/GP6(+)-mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J Clin Microbiol. 1997;35:791–5. doi: 10.1128/jcm.35.3.791-795.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bulkmans NW, Berkhof J, Rozendaal L, van Kemenade FJ, Boeke AJ, Bulk S, et al. Human papillomavirus DNA testing for the detection of cervical intraepithelial neoplasia grade 3 and cancer: 5-year follow-up of a randomised controlled implementation trial. Lancet. 2007;370:1764–72. doi: 10.1016/S0140-6736(07)61450-0. [DOI] [PubMed] [Google Scholar]
- 32.Rijkaart DC, Berkhof J, Rozendaal L, van Kemenade FJ, Bulkmans NW, Heideman DA, et al. Human papillomavirus testing for the detection of high-grade cervical intraepithelial neoplasia and cancer: final results of the POBASCAM randomised controlled trial. Lancet Oncol. 2012;13:78–88. doi: 10.1016/S1470-2045(11)70296-0. [DOI] [PubMed] [Google Scholar]
- 33.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–82. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 34.van Zeeburg HJ, Snijders PJ, Pals G, Hermsen MA, Rooimans MA, Bagby G, et al. Generation and molecular characterization of head and neck squamous cell lines of fanconi anemia patients. Cancer Res. 2005;65:1271–6. doi: 10.1158/0008-5472.CAN-04-3665. [DOI] [PubMed] [Google Scholar]
- 35.Overmeer RM, Henken FE, Snijders PJ, Claassen-Kramer D, Berkhof J, Helmerhorst TJ, et al. Association between dense CADM1 promoter methylation and reduced protein expression in high-grade CIN and cervical SCC. J Pathol. 2008;215:388–97. doi: 10.1002/path.2367. [DOI] [PubMed] [Google Scholar]
- 36.Harden SV, Guo Z, Epstein JI, Sidransky D. Quantitative GSTP1 methylation clearly distinguishes benign prostatic tissue and limited prostate adenocarcinoma. J Urol. 2003;169:1138–42. doi: 10.1097/01.ju.0000049627.90307.4d. [DOI] [PubMed] [Google Scholar]
- 37.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 38.Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–81. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]