Abstract
BACKGROUND
Antibodies specific for the neutrophil antigen HNA-3a cause severe, sometimes fatal transfusion-related acute lung disease (TRALI) when transfused, but it has not been possible to screen blood donors for anti-HNA-3a because using neutrophils as targets was impractical and molecular properties of the antigen were unknown. Recently it was shown that HNA-3a is carried on choline transporter–like protein-2 (CTL2) and that the HNA-3a/b phenotype is closely correlated with an R154Q amino acid polymorphism in CTL2. However, it has not been shown by direct experiment that R154 is essential for the HNA-3a epitope.
STUDY DESIGN AND METHODS
Preliminary attempts to express recombinant full-length CTL2 (R154) recognized by anti-HNA-3a were unsuccessful. We therefore tested HNA-3a–specific antibodies from donors implicated in TRALI reactions for reactivity against chemically synthesized linear and cyclic CTL2 peptides containing R154 or Q154.
RESULTS
Nine of 20 HNA-3a antibodies recognized the R154, but not the Q154 version of a cyclic 36-residue CTL2 peptide (D131-K166). However, 11 others failed to distinguish between the two versions of this peptide.
CONCLUSION
The findings provide direct evidence that R154 in the context of CTL2 D131-K166 is necessary to create the HNA-3a epitope but, in the context of cyclic CTL2 peptide D131-K166, is sufficient to detect only about one-half of the HNA-3a–specific antibodies implicated in TRALI. It is likely that fragments of CTL2 longer than can be made on a large scale with an automated synthesizer will be needed to produce a target capable of detecting all examples of anti-HNA-3a in donated blood.
Antibodies specific for the white blood cell antigen HNA-3a are particularly prone to cause severe, often fatal transfusion-related acute lung injury (TRALI),1–3 but it has not been possible to screen blood donors routinely for anti-HNA-3a because it is impractical to use neutrophils for antibody detection, and although the HNA-3a/b antigen system was described almost 50 years ago,4 its molecular properties were unknown. We recently showed that HNA-3a, previously thought to be neutrophil-specific, is also expressed on T and B lymphocytes and platelets (PLTs) and is carried on choline transporter–like protein-2 (CTL2) encoded by the gene SLC44A2.5 Genomewide association studies and sequencing of DNA from individuals typed serologically for HNA-3a/b demonstrated a tight association between a predicted amino acid polymorphism (R154Q) in CTL2 and HNA-3a antigen expression.5 Similar findings were reported independently by Greinacher and colleagues.6 However, neither of these studies provided direct evidence that arginine at Position 154 of CTL2 is essential to create the structure recognized by anti-HNA-3a. In initial studies, we found that recombinant full-length CTL2 is very difficult to express in a configuration recognized by alloantibodies, owing to its complex structure, which appears to have at least 10 transmembrane domains.7 As an alternative to using the full-length protein, we prepared synthetic CTL2 peptides containing arginine (R) or glutamine (Q) at Position 154 and characterized their reactions against a panel of anti-HNA-3a antibodies. Here, we show that a subset of 20 HNA-3a antibodies recognizes linear and cyclic versions of a 36-mer CTL2 peptide (D131-K166/R154) but is non-reactive with D131-K166/Q154. However, about half of the antibodies failed to make this distinction. The findings indicate that R154 in the context of CTL2 peptide D131-K166 is necessary to create the epitope recognized by HNA-3a antibodies, but that amino acid residues extending C-terminal and/or N-terminal from D131-K166 will be needed to create a target suitable for detecting all examples of anti-HNA-3a in blood donors.
MATERIALS AND METHODS
Antibodies
Nine serum samples containing HNA-3a–specific antibodies were identified by the Platelet and Neutrophil Immunology Laboratory of the BloodCenter of Wisconsin in persons whose donated blood was transfused to patients experiencing TRALI reactions. Antibody specificity was determined by the pattern of reactions against panels of typed neutrophils using flow cytometry and fluorescein isothiocyanate-labeled, anti-human immunoglobulin (Ig)G secondary antibodies8 and/or neutrophil agglutination9 and by showing that the donors were HNA-3a-negative. Other samples were gifts from the Neutrophil Antibody Reference Laboratory, American Red Cross (St Paul, MN; GTI Diagnostics, Waukesha, WI; and Dr Patricia Kopko, Sacramento, CA). Specificity of antibodies in the latter samples was determined by the referring laboratories from reactions against panels of typed neutrophils and was reconfirmed by our laboratory on the basis of flow cytometric and agglutination reactions against cell panels consisting of at least two HNA-3a–positive and two HNA-3a–negative neutrophils and T lymphocytes. Normal sera were obtained from normal healthy individuals and tested negative for neutrophil and lymphoycte-reactive antibodies by flow cytometry.
Peptides
Peptides comprising CTL2 Amino Acids N146-K166 and D131-K166 containing either R154 or Q154 (Fig. 1) and linked to biotin through a spacer group were chemically synthesized by the Protein Core Laboratory of the Blood Research Institute, high-performance liquid chromatography purified, and validated by mass spectrometry using standard methods.10 The design of the peptides selected was based on hydrophilic predictions (indicating likely antigenicity) using computer software (AbiePro3.0, Chang Bioscience, Castro Valley CA). Both a nonoxidized (linear peptide) and an oxidized cyclic version (cyclic peptide) of D131-K166 were produced. The cyclic peptide was produced by intramolecular linkage of cysteines (C) at Amino Acids 139 and 158 as described by Tam and colleagues.11 Oxidation of the purified cyclic peptide was validated by matrix-assisted laser desorption/ionization time-of-flight mass spectral analysis.
Fig. 1.
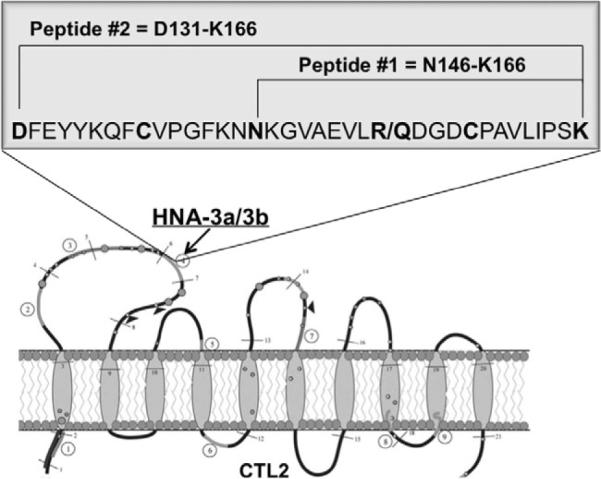
Predicted structure of CTL2 showing amino acid sequences of synthesized CTL2 peptides. Location of the R/Q154 polymorphism (arrow) associated with the HNA-3a/b antigens and sequences of peptides containing R154 or Q154 that were tested for reactivity against HNA-3a antibodies are shown. Large and small shaded circles indicate cysteine residues; large ones are predicted to have a high likelihood of disulfide bond formation. Open circles indicate asparagine residues; those with dark triangles are predicted to be glycosylated. Adapted from Nair et al.,7 where the basis for structural predictions is described.
Enzyme-linked immunosorbent assay
Biotin-labeled peptides (1.0 μg) were immobilized in wells of microtiter plates previously coated with 1.0 μg of streptavidin and blocked. Fifty microliters of HNA-3a antibody diluted 1:10 was added, incubated for 1 hour at room temperature, and washed four times with buffer. Bound immunoglobulin was detected with peroxidase-labeled goat anti-human IgG (Jackson ImmunoResearch Laboratories, West Grove, PA).
Statistical analysis
Log-transformed values were analyzed using mixed-effects modeling with random effect accounting for sample-to-sample variability and antibody type and peptide (R154 vs. Q154) as crossed fixed effects. The familywise Type I error rate was controlled over the pre-planned comparisons at 5% using Holm's method.12 All analyses were performed using computer software (2.10.1, R Foundation for Statistical Computing, Vienna, Austria; with the nlme 3.1–96 and multicomp 1.1–6 packages).
RESULTS
One of six HNA-3a antibodies recognized the 21-mer CTL2 peptide N146-K166 (R154)
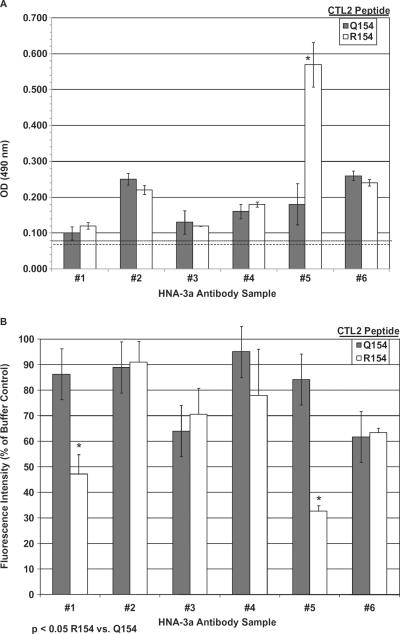
Initially, we studied reactions of six well-defined HNA-3a–specific antibodies (Antibodies 1–6) with 21-mer CTL2 peptides N146-K166 (R/Q154) (Fig. 1) using ELISA. Only Antibody 5 reacted preferentially with the R154 version (Fig. 2A). To determine whether antibodies testing negative might bind to N146-K166/R154 with low avidity, the six antibodies were diluted to a point at which they gave weak-positive reactions against HNA-3a–positive lymphocytes. The diluted antibodies were incubated with peptides N146-K166 (R154/Q154; 4 mg/mL) and then tested against HNA-3a–positive and –negative lymphocytes using flow cytometry. Reactions of Antibodies 1 and 5, but not those of Antibodies 2 through 4 and 6 against HNA-3a–positive lymphocytes, were inhibited significantly by the R154, but not the Q154 version of this peptide (p < 0.05; Fig. 2B). The findings indicate that Antibody 1 recognizes N146-K166/R154, but the interaction is not sufficiently strong to permit detection in an ELISA.
Fig. 2.
Reactions of serum samples containing HNA-3a–specific antibodies implicated in TRALI with linear 21-mer CTL2 peptides N144-K166 containing R or Q at Position 154. (A) Reactions of HNA-3a Antibodies 1 to 6 with CTL2 peptides N144-K166 (R/Q154) detected by ELISA. Results are mean ± 1 SD OD of triplicate measurements in ELISA. Asterisks indicate sera that gave significantly stronger reactions against N144-K166 (R154) than against N144-K166 (Q154; p < 0.05). Dotted line depicts mean OD obtained with plasma samples from 10 normal donors against the R154 peptide; dashed line depicts mean optical density obtained with normal plasma samples against the Q154 peptide. (B) Flow cytometric studies characterizing reactions of Antibodies 1 through 6 against HNA-3a–positive lymphocytes with and without prior incubation with 4 mg/mL CTL2 peptide N144-K166 (R154 or Q154). Reactions of Antibodies 1 and 5 were significantly inhibited by the R154 but not the Q154 peptide. Results are shown as median fluorescence intensities as a percentage of buffer control and are mean ± 1 SD of duplicate tests.
Nine of 20 HNA-3a–specific antibodies recognized both the cyclic and linear CTL2 peptides D131-K166 (R154)
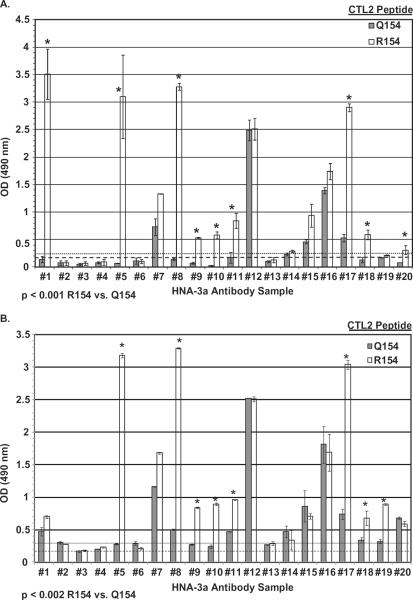
Failure of five of six HNA-3a antibodies to recognize the 21-mer peptide N146-K166 (R154) in ELISA (Fig. 2) made it desirable to study reactions of a larger panel of antibodies against a longer peptide. We therefore obtained 14 additional anti-HNA-3a sera from colleagues elsewhere and investigated reactions of all 20 antibodies against the CTL2 36-mer D131-K166 (R/Q154). Because D131-K166 contains cysteines at Positions 139 and 158 that might be disulfide linked,7 testing was done with a peptide in which the cysteines were S-S linked (cyclic peptide), and a peptide without linkage of the cysteines (linear peptide). As shown in Fig. 3, nine antibodies (1, 5, 8, 9, 10, 11, 17, 18, 20) reacted preferentially with the cyclic peptide D131-K166 (R154; p < 0.001, Fig. 3A), and nine antibodies (5, 8, 9, 10, 11, 17, 18, 19) reacted preferentially with the linear version of D131-K166 (R154; p = 0.002; Fig. 3B). Seven antibodies (2, 3, 4, 6, 13, 14, 19) failed to react with either peptide and four (7, 12, 15, 16) reacted with both. Interestingly, Antibodies 1 and 20 and Antibody 19 failed to distinguish between R154 and Q154 versions of the linear and cyclic peptides, respectively (Fig. 3).
Fig. 3.
Reactions of 20 serum samples containing HNA-3a–specific antibodies implicated in TRALI with oxidized (cyclic) and linear 36-mer CTL2 peptides D131-K166 containing R or Q at Position 154. (A) Antibodies 1, 5, 8, 9, 10, 11, 17, 18, and 20 reacted preferentially with the R154 form of the cyclic peptide. (B) Antibodies 5, 8, 9, 10, 11, 17, 18, and 19 reacted preferentially with the R154 form of the linear peptide. Results shown are mean ± 1 SD OD of triplicate measurements in ELISA. Asterisks indicate p < 0.001 (cyclic peptide) and p < 0.002 (linear peptide) for sera with preferential reaction against D131-K166 (R154). Dotted line is mean + 1 SD OD obtained with sera from 10 normal donors against the R154 peptide; dashed line indicates mean + 1 SD optical density obtained with normal sera against the Q154 peptide.
In general, antibodies that gave the strongest signals in ELISA had the highest titers against HNA-3a-positive lymphocytes in the flow cytometric assay. However, there were numerous exceptions. For example, Antibody 2 was negative in ELISA but reacted with HNA-3a-positive lymphocytes in flow cytometry at 1-in-80 dilution. Among the sera that reacted preferentially with cyclic D131-K166 (R154) in ELISA, Antibody 9 produced an optical density (OD) of only 0.50 to 0.80 in ELISA but could be detected by flow at 1-in-80 dilution, while Antibody 17 produced an OD about 3.0 in ELISA but could be detected by flow cytometry only at 1-in-20 dilution. Granulocyte agglutination test results are difficult to quantify, but as judged by visual inspection, all 20 HNA-3a-specific antibodies agglutinated HNA-3a-positive neutrophils equally well.
DISCUSSION
Ordinarily, confirmation that an amino acid polymorphism is responsible for a biallelic alloantigen system would be obtained by expressing both alleles of the carrier protein in an appropriate cell line and demonstrating that alloantibodies of known specificity produce the expected pattern of reactions against the two gene products. To date, we have attempted to express the two alleles of full-length CTL2 (R154, Q154) in insect cells, CHO cells, and two different human cell lines but have not yet succeeded in expressing proteins that can be consistently distinguished by anti-HNA-3a using flow cytometry to detect membrane-associated protein and immunoblotting to detect intracellular gene products. Difficulties encountered in expressing full-length CTL2 containing the epitope recognized by anti-HNA-3a appear to be related to its complex structure (Fig. 1). It is of interest that Greinacher and colleagues expressed CTL2 fragments containing R154 as GST fusion proteins (GST-CTL2) in Escherichia coli and found that GST-CTL2 55-231 (R154) was recognized in Western blot by two HNA-3a-specific antibodies and that a shorter peptide, GST 145-167 (R154), was recognized by a single antibody.6 However, the specificity of these reactions is uncertain because reactions of the antibodies with the Q154 (HNA-3a-negative) versions of the same peptides were not described. We have performed Western blotting studies of lysates from HNA-3a-positive and HNA-3a-negative T cells, but have been unable to distinguish between the two CTL2 alleles using various HNA-3a-specific antibodies (data not shown), suggesting that the HNA-3a epitope does not survive modifications of the protein resulting from detergent solubilization and sodium dodecyl sulfate electrophoresis. This behavior is similar to that of the red blood cell (RBC) D antigen carried on the 12-membrane-spanning RhD protein, which in general is not recognized by anti-D after solubilization by detergent.13
Because of problems encountered in expressing immunologically intact, full-length CTL2, we adopted the alternative approach of chemically synthesizing CTL2 peptides containing R154 or Q154 and studying their reactions with anti-HNA-3a to obtain direct evidence that R154 is critical for the HNA-3a epitope. Our finding that 9 of 20 HNA-3a antibodies recognized both cyclic and linear versions of peptide CTL2 D131-K166 (R154) but not the Q154 version of these peptides (Fig. 3) shows that R154 and adjacent peptides sequences are necessary to create the epitope recognized by many (and presumably all) HNA-3a-specific antibodies. However, failure of 11 antibodies to react preferentially with these D131-K166 (R154) peptides indicates that residues N- and/or C-terminal from D131-K166 and/or as yet undefined posttranslational modifications of the protein are required for about 50% of HNA-3a antibodies to bind with sufficient avidity to be detected by ELISA.
Reactions of Antibodies 7, 12, 15, and 16 with both the R154 and the Q154 versions of the cyclic and linear CTL2 peptides D131-K166 (Fig. 3) require comment. To characterize these reactions more fully, Antibodies 7 and 12 were absorbed with HNA-3a-positive and HNA-3a-negative lymphocytes. Reactions of the absorbed sera were comparable to those of unabsorbed sera (data not shown). At present, we have no satisfactory explanation for the reactions of these sera. Since all sera gave HNA-3a-specific reactions using intact lymphocytes and granulocytes as targets, it seems possible that the unexpected reactions of Sera 7, 12, 15, and 16 reflect an artifact introduced by use of the synthetic peptides as targets.
Our findings, limited information available about CTL2 structure, and prior studies of other alloantigens, enable some predictions to be made about the minimum CTL2 structure that may be needed to detect all examples of anti-HNA-3a. The first extracellular loop of CTL2 (Residues 55-231) where R154 is located contains eight cysteine residues, some of which are predicted to be disulfide linked.7 Our finding that nine of 20 HNA-3a-specific antibodies reacted preferentially with the R154 version of cyclic (S-S linked) peptide D131-K166 provides evidence that Cysteines 139 and 158 are probably disulfide linked naturally in the mature CTL2 protein, since if they were not, introducing an unnatural linkage between these residues would almost certainly distort the structure of the HNA-3a epitope in such a way that few, if any, of the antibodies would bind. It is of interest that the 20 HNA-3a antibodies studied reacted almost identically with the linear and cyclic versions of peptide D131-K166 (R154). The likely explanation for this is that, in solution, a structure that juxtaposes Cysteines 139 and 158 is energetically favored.14,15 Although reactions with the HNA-3a antibody panel indicate that the nonreduced peptide closely mimics the structure of the reduced peptide, the inherent instability of the former could explain its failure to provide a suitable target for Antibodies 1 and 20, which required the cyclic version of D131-K166 (R154) for binding. At present, we have no satisfactory explanation for the weak reaction of Antibody 19 with nonreduced D131-K166 (R154) only.
In addition to C139 and C158, the first extracellular loop of CTL2 contains cysteines at Positions 74, 95, 106, 112, 116, and 172. Valentin and colleagues16 showed that a long-range disulfide bond linking the GPIIIa PSI domain, where the HPA-1a epitope (determined by Leucine 33) is located, to C435 of the first EGF repeat of GPIIIa is required to stabilize the structure recognized by approximately one-half of HPA-1a-specific antibodies. By analogy, it is possible that linkage between two or more of the six unassigned cysteine residues in the CTL2 55-231 loop is required to produce a configuration suitable for binding of all HNA-3a antibodies. A second concern is that asparagine residues at Positions 187 and 200 of CTL2 may be glycosylated.7 Saccharide residues are important for recognition of the PLT antigen HPA-3a (Baka)17–19 and the RBC MN antigens20,21 by some alloantibodies. Therefore, the possible requirement for a glycan residue at Position 187 and/or 200 for binding of some HNA-3a antibodies deserves investigation. Finally, in addition to the R/Q mutation at Position 154 that is critical for expression of the HNA-3a and -3b antigens, mutations predicting amino acid substitutions at Positions 98 and 215 of the first extracellular loop of human CTL2, Position 413 of the third loop, and Positions 589 and 593 of the fifth loop are registered in the human genome SNP database.22 It is known that mutations in various extracellular loops of the 12-membrane-spanning D antigen influence expression of the D antigen.23 The possibility that binding of some HNA-3a antibodies to CTL2 is affected by mutations other than R/Q 154 therefore deserves consideration.
In summary, our findings show that, although the cyclic CTL2 peptide D131-K166 (R154) is suitable for detection of about one-half of HNA-3a-specific antibodies, the remainder require other structural elements of CTL2 to bind sufficiently well to be detected in an assay that requires washing of the target antigen. It is possible to synthesize peptides containing as many as 100 amino acid residues, but yield drops off significantly with increasing length. In the case of CTL2, a longer peptide centered on the polymorphic Amino Acid 154 could contain as many as eight cysteine residues, which would be unlikely to form the appropriate disulfide linkages upon oxidation. It is likely that fragments of CTL2 longer than can be made on a large scale with an automated synthesizer will be needed to produce a target capable of detecting all examples of anti-HNA-3a in donated blood.
ACKNOWLEDGMENTS
The authors wish to thank Randy Schuller, American Red Cross, St. Paul, MN, Dr Gian Visentin, GTI Diagnostics, Waukesha, WI, and Dr Patricia Kopko, Sacramento Blood Center, Sacramento, CA for their generosity in providing plasma samples containing HNA-3a antibodies.
This work was supported in part by Grant HL-13629 and HL-106286 from the National Heart, Lung, and Blood Institute and by Grant 1UL1RR031973 from the Clinical and Translational Science Award (CTSA) program of National Institutes of Health.
ABBREVIATION
- CTL2
choline transporter–like protein-2.
Footnotes
AUTHOR CONTRIBUTIONS BRC obtained reagents and samples, designed and performed experiments, analyzed data, and wrote and reviewed the manuscript. MJS designed and performed experiments and collected data. TH designed and produced CTL2 peptides and contributed to writing the manuscript. AZ performed statistical analysis of data and contributed to writing manuscript. DWB designed experiments and reviewed manuscript. RHA helped with design of experiments, analyzed data, and wrote and reviewed the manuscript.
CONFLICT OF INTEREST A patent application covering typing for the HNA-3a/b antigens and detection of HNA-3a-specific antibodies has been filed by BloodCenter of Wisconsin. GTI Diagnostics, Brookfield WI, has an agreement with BloodCenter of Wisconsin concerning technology for HNA-3 typing and detection of HNA-3 antibodies. These relationships involve only BRC and RHA. None of the other authors declare conflicts of interest.
REFERENCES
- 1.Kopko PM, Marshall CS, MacKenzie MR, Holland PV, Popovsky MA. Transfusion-related acute lung injury: report of a clinical look-back investigation. JAMA. 2002;287:1968–71. doi: 10.1001/jama.287.15.1968. [DOI] [PubMed] [Google Scholar]
- 2.Davoren A, Curtis BR, Shulman IA, Mohrbacher AF, Bux J, Kwiatkowska BJ, McFarland JG, Aster RH. TRALI due to granulocyte-agglutinating human neutrophil antigen-3a (5b) alloantibodies in donor plasma: a report of 2 fatalities. Transfusion. 2003;43:641–5. doi: 10.1046/j.1537-2995.2003.00374.x. [DOI] [PubMed] [Google Scholar]
- 3.Reil A, Keller-Stanislawski B, Gunay S, Bux J. Specificities of leucocyte alloantibodies in transfusion-related acute lung injury and results of leucocyte antibody screening of blood donors. Vox Sang. 2008;95:313–7. doi: 10.1111/j.1423-0410.2008.01092.x. [DOI] [PubMed] [Google Scholar]
- 4.Van Leeuwen A, Eernisse JG, Van Rood JJ. A new leucocyte group with two alleles: leucocyte group five. Vox Sang. 1964;9:431–46. doi: 10.1111/j.1423-0410.1964.tb03311.x. [DOI] [PubMed] [Google Scholar]
- 5.Curtis BR, Cox NJ, Sullivan MJ, Konkashbaev A, Bowens K, Hansen K, Aster RH. The neutrophil alloantigen HNA-3a (5b) is located on choline transporter-like protein 2 and appears to be encoded by an R>Q154 amino acid substitution. Blood. 2010;115:2073–6. doi: 10.1182/blood-2009-11-248336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greinacher A, Wesche J, Hammer E, Fürll B, Völker U, Reil A, Bux J. Characterization of the human neutrophil alloantigen-3a. Nat Med. 2010;16:45–8. doi: 10.1038/nm.2070. [DOI] [PubMed] [Google Scholar]
- 7.Nair TS, Kozma KE, Hoefling NL, Kommareddi PK, Ueda Y, Gong TW, Lomax MI, Lansford CD, Telian SA, Satar B, Arts HA, El-Kashlan HK, Berryhill WE, Raphael Y, Carey TE. Identification and characterization of choline transporter-like protein 2, an inner ear glycoprotein of 68 and 72 kDa that is the target of antibody-induced hearing loss. J Neurosci. 2004;24:1772–9. doi: 10.1523/JNEUROSCI.5063-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curtis BR, Reno C, Aster RH. Neonatal alloimmune neutropenia attributed to maternal immunoglobulin G antibodies against the neutrophil alloantigen HNA-1c (SH): a report of five cases. Transfusion. 2005;45:1308–13. doi: 10.1111/j.1537-2995.2005.00199.x. [DOI] [PubMed] [Google Scholar]
- 9.Lalezari P, Jiang A, Lee S. A microagglutination technique for the detection of leukocyte agglutinins. In: Ray JG, editor. NIAID manual of tissue typing techniques. Department of Health Education and Welfare; Bethesda (MD): 1979. pp. 20–2. NIH Publication; No. 80-545. [Google Scholar]
- 10.Albericio F, Kates SA. Solid-phase synthesis: a practical guide. 1st ed Marcel Dekker, Inc.; New York: 2000. [Google Scholar]
- 11.Tam JP, Wu CR, Liu W, Zhang JW. Disulfide bond formation in peptides by dimethyl sulfoxide. Scope and applications. J Am Chem Soc. 1991;113:6657–62. [Google Scholar]
- 12.Holm S. A simple sequentially rejective Bonferroni test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- 13.Avent ND, Reid ME. The Rh blood group system: a review. Blood. 2000;95:375–87. [PubMed] [Google Scholar]
- 14.Kyte J. Folding and assembly—sequences of polymers—posttranslational modifications. In: Kyte J, editor. Structure in protein chemistry. Garland Publishing; New York: 1995a. pp. 93–101. [Google Scholar]
- 15.Kyte J. Folding and assembly—thermodynamics of folding. In: Kyte J, editor. Structure in protein chemistry. Garland Publishing; New York: 1995b. pp. 445–69. [Google Scholar]
- 16.Valentin N, Visentin GP, Newman PJ. Involvement of the cysteine-rich domain of glycoprotein IIIa in the expression of the human platelet alloantigen, PlA1: evidence for heterogeneity in the humoral response. Blood. 1995;85:3028–33. [PubMed] [Google Scholar]
- 17.Take H, Tomiyama Y, Shibata Y, Furubayashi T, Honda S, Mizutani H, Nishiura T, Tsubakio T, Kurata Y, Yonezawa T, Tarui S. Demonstration of the heterogeneity of epitopes of the platelet-specific alloantigen, Baka. Br J Haematol. 1990;76:395–400. doi: 10.1111/j.1365-2141.1990.tb06374.x. [DOI] [PubMed] [Google Scholar]
- 18.Djaffar I, Vilette D, Pidard D, Wautier JL, Rosa JP. Human platelet antigen 3 (HPA-3): localization of the determinant of the alloantibody Lek(a) (HPA-3a) to the C-terminus of platelet glycoprotein IIb heavy chain and contribution of O-linked carbohydrates. Thromb Haemost. 1993;69:485–9. [PubMed] [Google Scholar]
- 19.Socher I, Zwingel C, Santoso S, Kroll H. Heterogeneity of HPA-3 alloantibodies: consequences for the diagnosis of alloimmune thrombocytopenic syndromes. Transfusion. 2008;48:463–72. doi: 10.1111/j.1537-2995.2007.01550.x. [DOI] [PubMed] [Google Scholar]
- 20.Issitt PD, Wilkinson SL. Further studies on the dependence of some examples of anti-M and anti-N on the presence of red-cell-borne sialic acid. Transfusion. 1983;23:117–9. doi: 10.1046/j.1537-2995.1983.23283172846.x. [DOI] [PubMed] [Google Scholar]
- 21.Springer GF, Desai PR. Human blood-group MN and precursor specificities: structural and biological aspects. Carbohydr Res. 1975;40:183–92. doi: 10.1016/s0008-6215(00)82680-4. [DOI] [PubMed] [Google Scholar]
- 22.National Center for Biotechnology Information “ENTREZ SNP” [monograph on the Internet] 2011 [cited 2011 Jan 20]. Available from: URL: http://www.ncbi.nlm.nih.gov/snp?term=Human%20CTL2.
- 23.Avent ND, Madgett TE, Lee ZE, Head DJ, Maddocks DG, Skinner LH. Molecular biology of Rh proteins and relevance to molecular medicine. Expert Rev Mol Med. 2006;8:1–20. doi: 10.1017/S1462399406010969. [DOI] [PubMed] [Google Scholar]