Abstract
Apoptotic processes are important for physiologic renewal of an intact epithelial barrier and contribute some antimicrobial resistance for bacteria and viruses, as well as anti-inflammatory effects that benefits the mucosa. The oral cavity presents a model of host-bacterial interactions at mucosal surfaces, in which a panoply of microorganisms colonizes various niches in the oral cavity and creates complex multispecies biofilms that challenge the gingival tissues. This report details gene expression in apoptotic pathways that occur in oral mucosal tissues across the lifespan, using a nonhuman primate model. Macaca mulatta primates from 2 to 23 years of age (n = 23) were used in a cross-sectional study to obtain clinical healthy gingival tissues specimens. Further, mRNA was prepared and evaluated using the Affymetrix Rhesus GeneChip and 88 apoptotic pathway genes were evaluated. The results identified significant positive correlations with age in 12 genes and negative correlations with an additional five genes. The gene effects were predicted to alter apoptosis receptor levels, extrinsic apoptotic pathways through caspases, cytokine effects on apoptotic events, Ca+2-induced death signaling, cell cycle checkpoints, and potential effects of survival factors. Both the positively and negatively correlated genes within the apoptotic pathways provided evidence that healthy tissues in aging animals exhibit decreased apoptotic potential compared to younger animals. The results suggested that decreased physiologic apoptotic process in the dynamic septic environment of the oral mucosal tissues could increase the risk of aging tissues to undergo destructive disease processes through dysregulated inflammatory responses to the oral microbial burden.
Keywords: Aging, Apoptosis, Oral mucosa, Inflammation, Infection
Introduction
Mucosal surfaces of the body are under constant challenge from a wide array of microorganisms, the complexity of which has only recently been fully appreciated through the results of the Human Microbiome Project [1]. This lifelong challenge requires the development of a range of immune response pathways, cells and biomolecules that are required to maintain an effective homeostasis between the epithelial barrier and the autochthonous microbiota. These interlinked systems of defense are also required to discriminate between members of the commensal microbial ecology that contribute to protection of the host, and the acquisition or emergence of pathogens within these ecologies. Additionally, this constant “battle” between the mucosal bacteria and the host cells would be expected to take a toll on the survival and function of both non-immune and immune cells that inhabit mucosal tissues, such as those in the gingival tissues in the oral cavity. This facet of the cell biology can be deduced from the short lifespan of epithelial cells, and the importance of epithelial cell sloughing as a part of innate protection of the host. The mucosal tissues are also enriched in cells undergoing the process of programmed cell death, apoptosis [2]. This natural physiologic aspect of cell biology is critical in developmental biology [3, 4], renewal of tissues in wound healing, elimination of auto-reactive cells, and loss of mutated cells with neoplastic potential [5].Moreover, apoptotic processes appear important for physiologic renewal of an intact epithelial barrier and contribute some antimicrobial resistance, for both bacteria and viruses that benefits the mucosa [6]. Most recently, evidence dictates that apoptosis is an essential mechanism that regulates the immuno-inflammatory response against pathogens, through the generation of anti-inflammatory signals affecting phagocytes at the site of the infection, as well as contributing to the determination of the characteristics of the T helper response [2, 7, 8].
The oral cavity presents a model of host-bacterial interactions at mucosal surfaces in which a panoply of microorganisms colonize various niches in the oral cavity [9] and create complex multispecies biofilms that reflect environmental changes and disease processes in the gingival tissues [10–13]. Furthermore, this model system enables routine sampling of biological fluids and tissues that respond to these microbial changes, as well as likely contributing to selective pressures that alter the ecology. We have reported previously that examination of gene expression profiles in gingival tissues from young to aged nonhuman primates, Macaca mulatta, demonstrated higher levels of pro-apoptotic and lower levels of anti-apoptotic pathway genes in younger animals, with these patterns reversed in the aged animals [14]. This implied that the apoptotic events were a normal activity in the gingival tissues related to tissue renewal and health in this septic environment. An apparent decrease in this process with aging would suggest a loss of capacity of the aged gingival tissue to respond to the microbial challenge in a pathophysiologic fashion contributing to the increase in gingival disease and tissue destruction with aging [15–17]. Additional findings from our studies demonstrated unique patterns of apoptotic gene expression and resulting biological pathways with disease in adult compared to aged animals [14]. The interpretation being that alterations in apoptosis occur in gingival health with aging that is even further disrupted either resulting from the disease process, or contributing to the disease progression.
This report extends these observations, by specifically documenting a profile of genes in apoptotic pathways that are significantly correlated with aging in healthy gingival mucosal tissues. In addition, a number of these gene transcripts altered with aging interdigitate with other biologic pathways that would be predicted to enhance a homeostatic host-bacterial interaction, or contribute to a risk profile of the gingival tissues that could result in tissue destruction in response to the chronic infection in the older animals.
Materials and methods
Nonhuman primate model and oral clinical evaluation
Rhesus monkeys (M. mulatta) (n = 23; 11 females and 12 males) housed at the Caribbean Primate Research Center (CPRC) at Sabana Seca, Puerto Rico, were used in these studies. Animals were selected by age based on the following criteria: ≤3 years (young; n = 5), 3–7 years (adolescent; n = 5), 12–16 years (adult; n = 8) and 18–23 years (aged; n = 5). Nonhuman primates were fed a 20 %protein, 5 % fat, and 10 %fiber commercial monkey diet (diet 8773, Teklad NIB primate diet modified: Harlan Teklad). The diet was supplemented with fruits and vegetables, and water was provided ad libitum in an enclosed corral setting.
A protocol approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Puerto Rico, enabled anesthetized animals to be examined for clinical measures of periodontal including probing pocket depth (PD), and bleeding on probing (BOP) as we have described previously [18].
Tissue sampling and gene expression microarray analysis
A buccal gingival sample from a healthy site from the premolar/molar maxillary region of each animal was taken using a standard gingivectomy technique, and maintained frozen in RNAlater solution. Total RNA was isolated from each gingival tissue using a standard procedure as we have described, and tissue RNA samples submitted to the microarray core to assess RNA quality and analyze the transcriptome using the GeneChip® Rhesus Macaque Genome Array (Affymetrix) [14, 19]. Individual samples were used for gene expression analyses.
Data analysis
For each gene, a simple linear regression model was fit to the scatter plot of expression by age as a continuous variable. A p value ≤0.05 was used to evaluate the significance of the correlation. Equivalently, this p value tests if the slope of the regression line is zero or not. Genes whose expression showed significant correlation with age were mapped into the Kyoto Encyclopedia of Genes and Genomes (KEGG) apoptosis pathway (www.genome.jp) to develop an ontology analysis.
Results
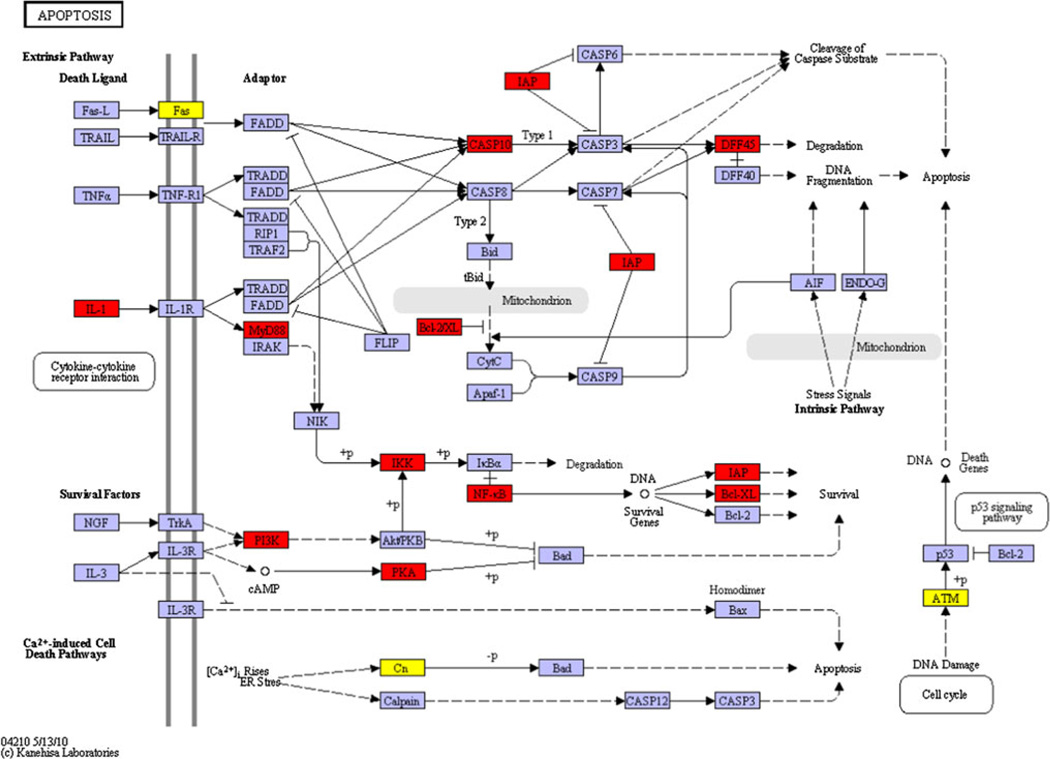
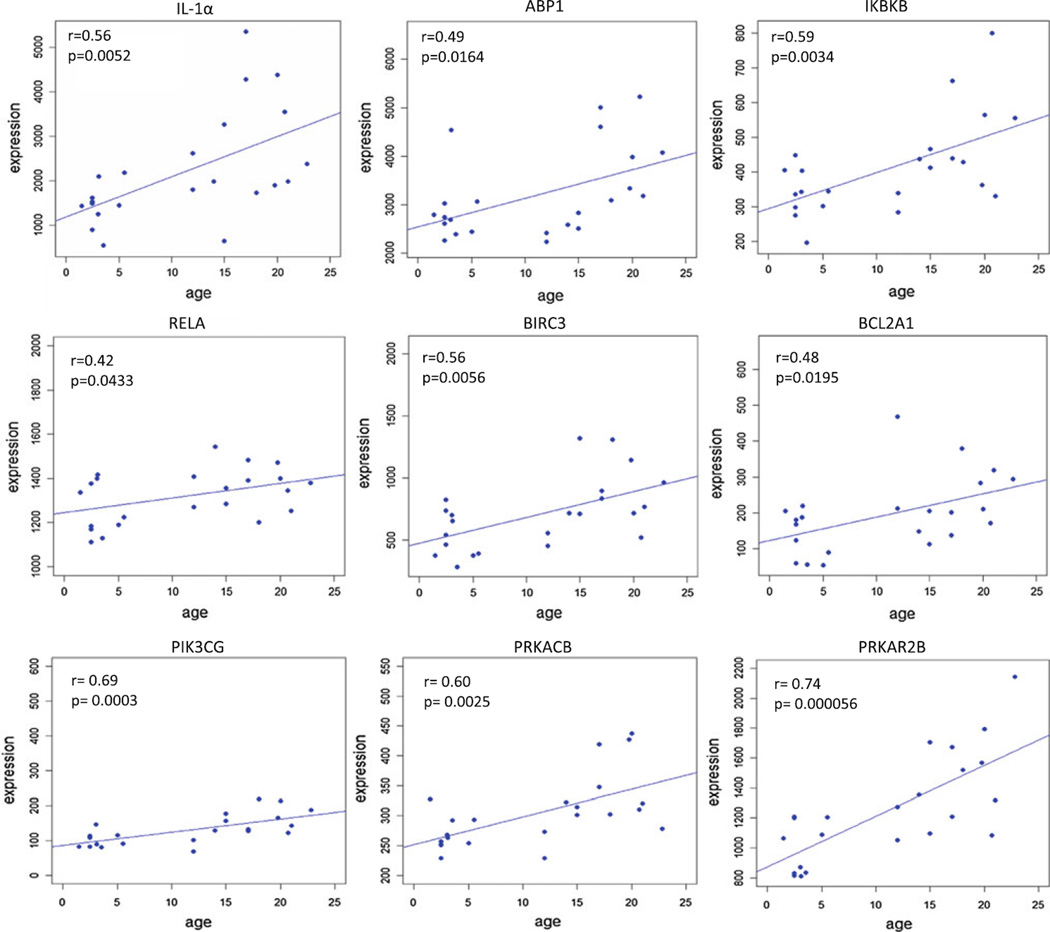
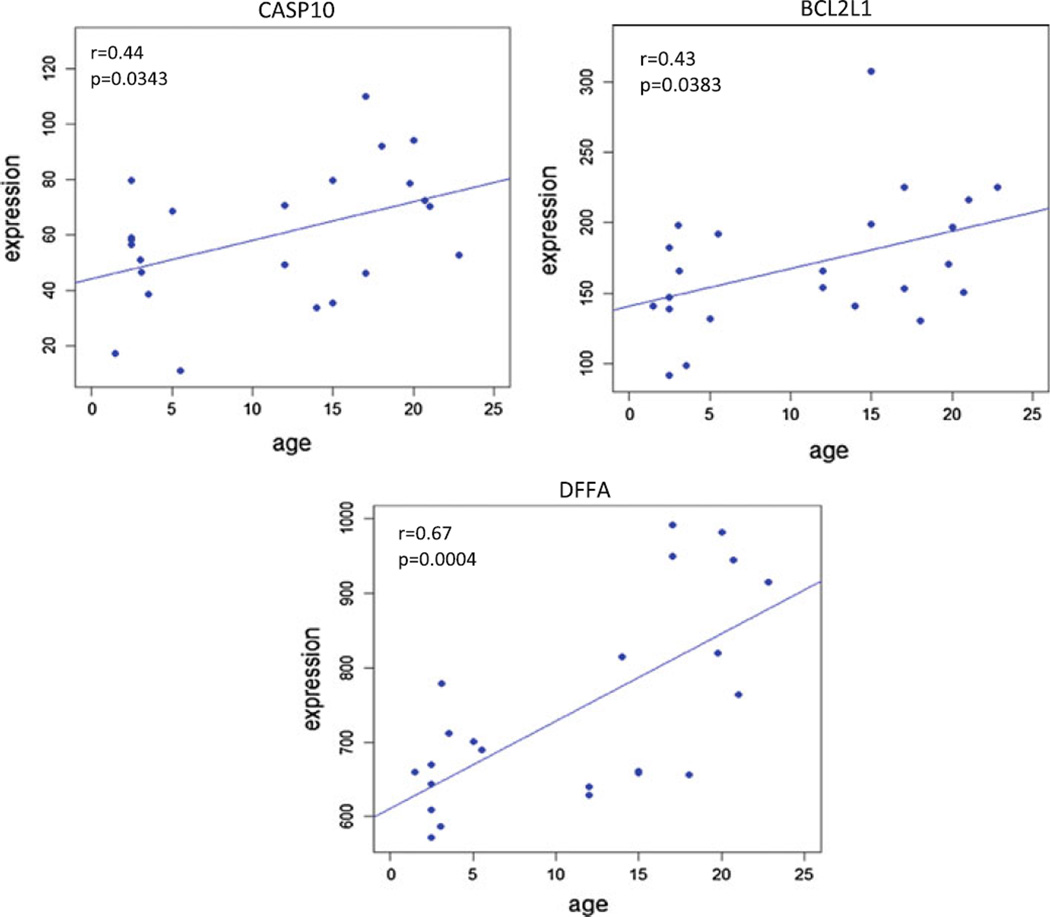
A total of 17 genes involved in various apoptotic pathways, were found to be significantly correlated with aging in healthy gingival tissues (Table 1). Figure 1 provides a KEGG pathway summation of the genes in the various apoptotic pathways that were significantly altered with aging in healthy gingival tissues and highlights the changes that generally reflect an overall decrease in the capacity of cells in the aging tissues to undergo apoptosis. Figure 2 depicts the correlations among interleukin (IL)-1α, amiloride binding protein 1(ABP1)—orthologous to MyD88, inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta (IKBKB), V-rel avian reticuloendotheliosis viral oncogene-like protein (RELA), baculoviral IAP repeat-containing 3 (BIRC3), and Bcl-2-related protein A1 (BCL2A1). These alterations in the transcriptome with aging represent elevated gene expression patterns linked to the presence of an inflammatory stimulus (e.g., IL-1α) that are anti-apoptotic and would be expected to increase cell survival and decrease normal cell turnover. In addition, this figure includes the correlation curves for phosphoinositide-3-kinase catalytic, gamma polypeptide (PIK3CG), protein kinase cAMP-dependent, catalytic, beta (PRKACB), and protein kinase, cAMP-dependent, regulatory, type II, beta (PRKAR2B). As can be noted in the KEGG pathway diagram, elevations in these genes and products would also create a more anti-apoptotic environment and contribute to enhanced cell survival. Figure 3 depicts the data regarding gene expression of caspase 10, apoptosis-related cysteine protease (CASP10), Bcl-2-like 1 isoform 1 (BCL2L1), and DNA fragmentation factor alpha polypeptide (DFFA).While increases in caspase 10 would be expected to enhance extrinsic or mitochondria-independent apoptotic events in the tissue, concomitant elevations in the BCL2L1, IAP (e.g., BIRC3, Fig. 1), and DFFA, which all function as block points for apoptosis, would again be expected to favor an overall decrease in apoptotic potential of cells in the aging gingival tissues.
Table 1.
Apoptosis-related genes whose expression significantly correlated with age
| Overexpressed genes in oral mucosal tissue with age | Gene ID | Probe ID |
| PRKAR2B protein kinase, cAMP-dependent, regulatory, type II, beta | PRKAR2B | MmugDNA.21586.1.S1 |
| Phosphoinositide-3-kinase, catalytic, gamma polypeptide | PIK3CG | MmuSTS.1136.1.S1_at |
| DNA fragmentation factor, 45 kDa, alpha polypeptide | DFFA | MmuSTS.4221.1.S1_at |
| Orthologous to protein kinase, cAMP-dependent, catalytic, beta | PRKACB | MmugDNA.21559.1.S1_at |
| Orthologous to Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta | IKBKB | MmugDNA.8188.1.S1_at |
| Interleukin 1 alpha | IL-1A | Mmu.1167.1.S1_at |
| BIRC3 baculoviral IAP repeat-containing 3 | BIRC3 | Mmu.16393.1.S1_at |
| Amiloride binding protein 1—orthologous to (MyD88) | ABP1 | MmugDNA.10008.1.S1_at |
| Orthologous to Bcl-2-related protein A1 | BCL2A1 | MmugDNA.5658.1.S1_at |
| Caspase 10, apoptosis-related cysteine protease | CASP10 | MmugDNA.13788.1.S1_s_at |
| Similar to Bcl-2-like 1 isoform 1 | BCL2L1 | MmugDNA.12133.1.S1_at |
| V-rel avian reticuloendotheliosis viral oncogene-like protein | RELA | MmuSTS.3377.1.S1_at |
| Underexpressed genes in oral mucosal tissue with age | Gene ID | Probe ID |
| Orthologous to ataxia telangiectasia mutated | ATM | MmugDNA.12666.1.S1_at |
| Orthologous to protein phosphatase 3, catalytic subunit, beta isoform (Calcineurin A beta) | PPP3CB | MmugDNA.25748.1.S1_at |
| Orthologous to Insulin-like growth factor 1 receptor | IGFR1 | MmugDNA.12698.1.S1_at |
| Tubulin tyrosine ligase-like family, member 4 | TTLL4 | Mmu.9989.1.S1_s_at |
| TNF receptor superfamily, member 6 | FAS | MmuSTS.4665.1.S1_at |
Fig. 1.
Aging-related variation in the expression of apoptotic genes in healthy oral mucosal tissues. Apoptotic genes were identified and mapped using KEGG pathway analysis. Genes that were significantly positively correlated with aging are highlighted in red and those that were negatively correlated are highlighted in yellow. Expression of genes shown in purple was unchanged at p ≤ 0.05 significance. +p = phosphorylation event; −p = de-phosphorylation event
Fig. 2.
Apoptotic genes related to cytokine signaling and subsequent intracellular pathways in apoptotic processes that were significantly positively correlated with aging (p ≤ 0.05). The correlation (r) and p-values are shown for each gene
Fig. 3.
Apoptotic genes related to extrinsic pathway signaling in apoptotic processes that were significantly positively correlated with aging (p ≤ 0.05). The correlation (r) and p-values are shown for each gene
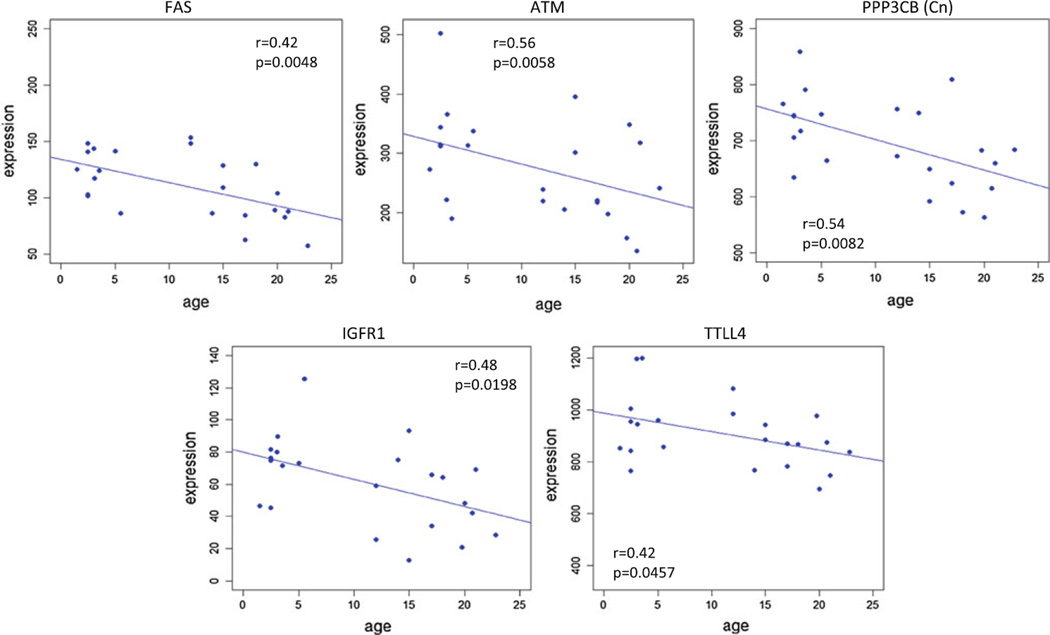
In contrast to the previously described gene expression alterations that occurred in healthy aged gingival tissues, select genes in the apoptotic pathways were significantly negatively correlated with aging in these tissues. The TNF receptor superfamily, member 6 (FAS), which is a major signal transducer for the adapter Fas-associated protein with death domain (FADD) within the extrinsic pathway, decreases with age (Fig. 4). Similarly, calcineurin A beta (CN) that is critical to the Ca+2-induced cell death pathway is decreased with aging, thus potentially being a bottleneck in this pathway of apoptosis. Additionally, the ataxia telangiectasia mutated (ATM) gene expression is significantly decreased with aging and would be expected to minimize physiological apoptotic events that result from DNA damage during cell cycling. Also, as shown in Fig. 4, gene expression of the insulin-like growth factor receptor 1 (IGFR1) and tubulin tyrosine ligase-like family, member 4 (TTLL4) demonstrate significant negative correlations with age in the gingival tissues. The IGFR1 is frequently overexpressed in neoplastic cells, and mediates proliferation as an anti-apoptotic molecule in the cells. TTLL4 is a member of a larger family of enzymes reported to have polyglutamylase activity. The action of these enzymes has been linked to coordination of chromatin remodeling with potential role as anti-proliferative and pro-apoptotic. Thus, its decrease with aging would help to contribute to cellular activities in healthy aging tissues that do not express normal physiologic apoptosis.
Fig. 4.
Apoptotic genes that were significantly negatively correlated with aging (p ≤ 0.05). The correlation (r) and p-values are shown for each gene
Discussion
Apoptosis is a process that results in cell death in the absence of inducing an inflammatory response. Beyond being a central biological mechanism for maintaining tissue homeostasis by balancing mitosis, alterations in the level of apoptotic events play a critical role in multiple diseases including cancer, neurological disorders, cardiovascular disorders, and autoimmune diseases [20]. Most recently, a role for apoptosis as a regulator of the immuno-inflammatory response has also been shown. Thus, the detection and clearance of apoptotic cells by phagocytes, occurs during the continual interaction of host cells with a diverse array of microorganisms at mucosal surfaces, in order to maintain tissue homeostasis [21]. The primary difference of ingestion of apoptotic cells versus engulfment of microbes by phagocytes is the generation of an inflammatory response that is exclusively associated with microbial phagocytosis. This biological feature is in contrast to an anti-inflammatory phagocytic activity that occurs with ingestion of apoptotic cells [7, 22].
In general, studies addressing the potential role of apoptosis in the chronic inflammation of gingival tissues have shown increased expression of apoptosis biomolecules, that appear primarily in phagocytic cells that have emigrated into the gingival tissues in response to bacterial challenge [23]. However, the physiologic or pathophysiologic role of apoptosis in the chronic inflammatory response and tissue destruction of periodontitis remains inconclusive [24]. Historical data have indicated that a higher prevalence and more severe periodontal disease occurs in aged individuals [25, 26]. However, the cellular and molecular changes of the periodontium (including apoptotic events) associated with a higher prevalence of oral diseases in aging remain unclear. Our initial report described that the expression of genes in the apoptotic pathways are altered in aged healthy and periodontitis-affected gingival tissue with regards to age using a nonhuman primate model of periodontitis [14]. The results examining young, adult, and aged animals showed a generally lower expression of anti-apoptotic and higher expression of pro-apoptotic genes associated with healthy gingival tissue from young compared to aged animals.
This report expands on this data by identifying a set of genes in a larger group of nonhuman primates including adolescents through correlation analysis, from within the larger cadre of genes that comprise the range of extrinsic, cytokine, Ca+2 dependent, and survival signaling aspects of apoptosis. Importantly, we found that variation in the expression of several apoptotic genes was reproducible in this larger sample size, re-enforcing the concept that survival pathways are up-regulated in aged-gingival tissues when compared with the young counterpart, whereby gene expression of pro-apoptotic pathways were significantly reduced with aging. Noteworthy, in this report new genes were identified to significantly correlate with age in healthy gingival tissue (e.g., ABP1/MyD88, Fas, and ATM). We identified that a pathway of response related to IL-1, and including MyD88 and the NF-κB transcription activities was up-regulated with aging in the tissues. IL-1α is produced by many cell types including constitutively by epithelial cells and induced in inflammatory cells, particularly activated macrophages. It plays an important central role in the regulation of the immune responses. In tissues, the release of IL-1α from dying cells can initiate sterile inflammation by inducing recruitment of neutrophils, whereas another member of this cytokine family, IL-1β, promotes the recruitment and retention of macrophages [27, 28]. The ABP1 is an orthologous of MyD88 and is a membrane glycoprotein that is expressed in many epithelium-rich and/or hematopoietic tissues. While it can catalyze degradation of a range of polyamines involved in DNA synthesis and that are involved in allergic and immune responses, it is also linked to cell proliferation and apoptosis [29, 30]. The inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta (IKBKB) phosphorylates the inhibitor in the IκB/NF-κB complex, causing dissociation of the inhibitor and activation of NF-κB. This process could have both pro- and anti-inflammatory activities, dependent upon the downstream genes that are transcribed by binding NF-κB [31]. RELA also identified as transcription factor p65 and is part of the NF-κB complex [32–35]. NF-κB is a transcription factor, which is present in most cell types and is involved in diverse biological processes such as inflammation, immunity, differentiation, cell growth, neoplastic changes, and apoptosis. NF-κB complexes are maintained in the cytoplasm in an inactive state complexed with inhibitors (I-κB family). In a conventional activation pathway, I-κB is phosphorylated by I-κB kinases (IKKs) in response to different activators, subsequently degraded thus liberating the active NF-κB complex, which translocates to the nucleus leading to transcription of gene with NF-κB binding domains in their promoters. The BCL2A1 encodes a member of the BCL-2 protein family and acts as an anti-apoptotic regulator. The protein reduces the release of pro-apoptotic cytochrome c from mitochondria and blocks caspase activation, thus, retarding apoptotic events. Additionally, this gene is a direct transcription target of NF-κB in response to inflammatory mediators such as TNFα and IL-1 that are increased with inflammation in gingival tissues [36, 37]. Finally, within the context of the genes related to these pathways of regulation of apoptosis, the BIRC3 is a member of a family of inhibitors of apoptosis (IAP) that interfere with the activation of caspases. This protein inhibits apoptosis induced by serum deprivation, but does not affect apoptosis resulting from increases in free radicals. As importantly, with respect to responses in gingival tissues apoptosis processes can be inhibited by binding to TRAF1/TRAF2 (TNF receptor-associated factors) that are recruited to the TNFR2 (TNF receptor 2) and would interfere with activation of IL-1β converting enzyme (ICE)-like proteases that are a family of mammalian cysteine proteases [38]. Members of this family of proteases are believed to be critical for mammalian cell apoptosis because inhibitors of ICE-like proteases can block apoptosis [39]. This process appears to occur via binding of Fas ligand to Fas or TNFα to the TNFR, which induce apoptosis. This is triggered by recruitment of an ICE-like protease to the activated receptor [40, 41]. Of note, the substrates for ICE-like proteases are catalytic proteins involved in homeostatic pathways in the cell, which suggests that undermining cellular homeostasis is a basic feature that leads to irreversibility of the apoptotic process and cell death [42].
Related to this cytokine pathway of anti-apoptosis, we identified fingerprints of gene regulation that specifically targeted survival factor pathways in the tissues. Phosphoinositide-3-kinase, catalytic, gamma polypeptide (PIK3CG) is a gene that encodes a protein belonging to the pi3/pi4-kinase family of proteins. This enzyme phosphorylates phosphoinositides and is crucial as an important modulator of extracellular signals. Thus, not only does it contribute to the maintenance of the structural and functional integrity of epithelia, but is a signal transducer to cellular engagement of molecules, such as nerve growth factor (NGF) and IL-3 that provide signals for growth, maintenance, and survival of cells, by interfering with intracellular pro-apoptotic molecules [43–45]. The actions of Pi3K molecules synergize with the cAMP-dependent kinases in limiting apoptosis. The Protein kinase, cAMP-dependent, catalytic, beta (PRKACB) is a catalytic subunit of protein kinase A that mediates cAMP-dependent signaling triggered by receptor binding to G-protein-coupled receptors (GPCRs) that comprise a transmembrane receptor family whose functions are to sense molecules outside the cell and activate signal transduction pathways and cellular responses [44, 46–50]. Binding of cAMP to the regulatory subunit of PKA, releases the catalytic subunits, which then phosphorylate a diverse set of proteins, including transcription factors, ion channels and metabolic enzymes. We also noted a relationship of increasing level of protein kinase, cAMP-dependent, regulatory, type II, beta (PRKAR2B) mRNA with aging in the gingival tissues. Four different regulatory subunits and three catalytic subunits of PKA have been identified [50–52], with this being one of the regulatory subunits. This subunit has been shown to interact with and suppress the transcriptional activity of the cAMP-response element binding protein 1 (CREB1) in activated cells [53]. Results from mice suggest that CREB1 may contribute to type 2 diabetes [54] and also appears to regulate some aging-related genes like ATM [55]. Importantly, this combination of genes whose expression is increased with aging in healthy gingival tissues is consistent with a more anti-apoptotic environment in the aged tissues.
Caspase 10, apoptosis-related cysteine protease is one of a family of proteases whose sequential activation plays a central role in the execution-phase of apoptosis. This protein cleaves and activates caspases 3 and 7, and the protein itself is processed by caspase 8. Mutations in this gene are associated with apoptosis defects in some autoimmune syndromes [56]. It has been shown that caspase-10 can function in initiating Fas- and TNF-related apoptosis-inducing ligand-receptor-mediated apoptosis [57]. As such, caspase 10 is recruited to the native TRAIL death-inducing signaling complex (DISC) and to the native Fas (CD95) DISC, and that FADD is necessary for recruitment and activation of these complexes [58]. Related to its potential role at mucosal surfaces, host cells engage in targeted antiviral immune responses by inducing type I IFN and inflammatory cytokines via activation NF-κB through cytoplasmic factors that recognize dsRNA generated during viral replication. Caspase-10 is involved in these pathways, since knockdown of caspase 10 in a human cell line resulted in the reduction of inflammatory cytokine production in antiviral signaling [59]. Thus, the up-regulation of caspase 10 gene expression in aging tissues would be a benefit for enhancing protection of the tissues from viral challenge, but could increase the opportunity for the enzymatic apoptotic signaling cascade. However, also noted in the altered gene expression profiles were increases in both Bcl-2-like 1 isoform 1 (BCL2L1), BIRC3 (IAP containing), and DFFA. The BCL2L1 is another member of the Bcl family that can regulate an array of cell functions. These proteins are located at the outer mitochondrial membrane, and regulate mitochondrial membrane channel opening/potential that controls the production of reactive oxygen species and release of cytochrome C [37]. Each of these is considered potent inducers of cell apoptosis [60, 61]. As noted above, the BIRC3 is a member of a family of IAP that interfere with the activation of caspases [62, 63]. Finally, DFFA is an inhibitor of caspase-activated DNase [64]. The apoptotic process is accompanied by shrinkage and fragmentation of cell nuclei and degradation of chromosomal DNA. DFFA is the substrate for caspase 3 and a portion of the pathway that triggers DNA fragmentation during apoptosis. DFFA when bound to DFFB, which triggers both DNA fragmentation and chromatin condensation during apoptosis, inhibits the apoptotic fragmentation of DNA [65, 66]. However, when DFFA is cleaved by caspase-3, the cleaved fragments of DFFA dissociate from DFFB, enabling action on the cellular DNA. Thus, the combination of increases in the expression of these genes additionally supports an increased anti-apoptotic environment in the aging gingiva, albeit, still enabling functional capacity for antiviral responses to remain relatively intact.
While the above pathways of apoptosis would be expected to be altered via increases of anti-apoptotic factors with aging, we also observed in this model a potential effect of selected pro-apoptotic genes. The TNF receptor superfamily, member 6 (FAS) is a receptor containing a death domain and plays a major role in the physiological regulation of programmed cell death. This receptor-ligand engagement forms a DISC that includes FADD, caspase 8, and caspase 10. This receptor has been also shown to activate NF-kB [67]. Protein phosphatase 3, catalytic subunit, beta isoform (Calcineurin A beta; CN) is a Ca+2- and calmodulin-dependent protein phosphatase [68]. Inactivating calcineurin slows aging in Caenorhabditis elegans and the anti-aging effects of lowered calcineurin activity decrease CREB transcriptional responses [53]. The decreased calcineurin would also lower the Ca+2-induced pathway of apoptosis. This molecule also activates the T cells through the transcription factor, nuclear factor of activated T cell (NFAT). As with NF-κB, the activated NFAT is translocated into the nucleus and up-regulates the expression of IL-2, stimulating growth and differentiation of T cells, thus influencing the characteristics and level of immune responses [69–71]. Lastly, the ATM is a protein kinase that is recruited and activated by DNA double-strand breaks. It phosphorylates proteins that initiate activation of the DNA damage blockade that can trigger cell cycle arrest, DNA repair, or apoptosis [72, 73]. Consequently, the decreased levels of expression of these pro-apoptotic genes that affect multiple pathways resulting in apoptosis would contribute to the overall anti-apoptotic environment in aged gingival tissues. Interestingly, similar variations in the expression of apoptotic genes shown in this study for healthy gingival tissues have also been reported in oral squamous cellular carcinoma (OSCC) [74, 75]. Specifically, significant reduction in ATM expression was found here in aged healthy gingival tissues and also in OSCC, when compared with healthy control biopsies. Since ATM promotes apoptosis and suppresses tumorigenesis through DNA damage detection, down-regulation of ATM could increase the likelihood of aged oral mucosa demonstrating neoplastic changes.
Two additional genes that were significantly negatively correlated with aging in healthy gingival tissues are less clear in how they contribute to differences in the tissue milieu between younger and older individuals. Insulin-like growth factor 1 receptor (IGFR1) is a transmembrane receptor with kinase functions that is activated by insulin-like growth factors 1 and 2 and is frequently overexpressed by neoplasia, and mediates proliferation and apoptosis protection. IGF1R activation protects cells from various apoptosis-inducing extrinsic factors, including oxidative stress [76, 77] and the level appears to be an essential determinant of resistance to apoptosis [78]. The IGF1R suppresses apoptosis primarily through the PI3K pathway. It has also been suggested that the IGFR1 pathway could play an important role in immune function, and lower levels or functions of this receptor may help increase susceptibility to infection [79]. Thus, in the overall picture of the intracellular apoptotic milieu, decreased levels of IGFR1 may be marginalized by the range of other gene expression changes resulting in decreased apoptosis in aged tissues. The last gene whose expression was observed to decrease significantly with aging was the TTLL4, which is a polyglutamylase belonging to the larger TTLL family of proteins [80]. Polyglutamylation is a new class of posttranslational modification in which glutamate side chains are formed in proteins. TTLL4 appears to have some selective effect on histone chaperones, NAP1 and NAP2 suggesting that TTLL4 could play a role in coordination of chromatin remodeling [81]. Knockdown of TTLL4 or exogenous introduction of TTLL4 can enhance tumor cell growth [82]. Thus, decrease in the expression of this gene in aging gingival tissues would be consistent with an anti-apoptotic predilection for the cells.
These results suggested that apoptotic events normally occurring in gingival tissues could be reduced with aging. Based on the anti-inflammatory and immunomodulatory role of apoptosis and evidence showing the majority of apoptotic events occurring in the inflammatory cellular infiltrate during periodontitis, it is tempting to hypothesize that in contrast to young gingival tissues, reduced apoptotic responses in aged gingival tissues could involve failure to control the inflammatory response against the chronic bacterial challenge even in clinical healthy tissues. Unique aspects of apoptotic pathways are potentially involved in the pathophysiology of periodontal disease in aging gingival tissues that are predisposed to a dysregulated clearance of the inflammatory infiltrate. A better understanding of the role of apoptosis in periodontal disease is clearly needed, which will open new possibilities for therapeutic strategies oriented to re-establish the physiological pro-apoptotic mechanisms in diseased gingival tissues, specifically within the inflammatory cell population, as has been proposed for other chronic inflammatory disorders [83].
Age-related variations in apoptotic processes have also been reported in different human tissues (e.g., skeletal muscles, neurons, spermatozoa, hepatocytes, intestinal epithelial cells, and coronary arterial wall), whereby apoptotic events appear to increase or decrease in a tissue-specific manner with aging [84–88]. Most recently, a reduced global apoptosis with aging in humans was determined by serum levels of sFas, FasL and total cytochrome c, which could increase the incidence of diseases whose pathophysiology involves apoptosis dysregulation (e.g., cancer, arthritis and cardiovascular disease) [89]. The high phylogenetic similarity of nonhuman primates to humans, provides an essential translational advantage to understand the cellular and molecular changes of the host associated with aging (e.g., apoptosis), and how those changes could be risk modifiers for age-related diseases [90]. Therefore, future reproduction of these studies using human oral mucosa tissues would be worthwhile.
Acknowledgments
This work was supported by National Institute of General Medical Sciences (NIGMS) grant 8P20GM103538-09. We express our gratitude to the Caribbean Primate Research Center (CPRC) for its invaluable technical support.
Footnotes
Conflicts of interest The authors report no conflicts of interest related to this study.
Contributor Information
Octavio A. Gonzalez, Email: octavio.gonzalez@uky.edu, Center for Oral Health Research, College of Dentistry, University of Kentucky, 1095 VA Drive. HSRB 414, Lexington, KY 40536-0305, USA.
M. John Novak, Center for Oral Health Research, College of Dentistry, University of Kentucky, 1095 VA Drive. HSRB 414, Lexington, KY 40536-0305, USA.
Sreenatha Kirakodu, Center for Oral Health Research, College of Dentistry, University of Kentucky, 1095 VA Drive. HSRB 414, Lexington, KY 40536-0305, USA.
Arnold J. Stromberg, Department of Statistics, College of Arts and Sciences, University of Kentucky, Lexington, KY 40506, USA
Shu Shen, Department of Statistics, College of Arts and Sciences, University of Kentucky, Lexington, KY 40506, USA.
Luis Orraca, School of Dental Medicine, University of Puerto Rico, San Juan, PR 00936, USA.
Janis Gonzalez-Martinez, Caribbean Primate Research Center, University of Puerto Rico, Toa Baja, PR 00949, USA.
Jeffrey L. Ebersole, Center for Oral Health Research, College of Dentistry, University of Kentucky, 1095 VA Drive. HSRB 414, Lexington, KY 40536-0305, USA
References
- 1.Stower H. Microbiology: the human microbiome project. Nat Rev Genet. 2012;13:518. [Google Scholar]
- 2.Torchinsky MB, Garaude J, Blander JM. Infection and apoptosis as a combined inflammatory trigger. Curr Opin Immunol. 2010;22:55–62. doi: 10.1016/j.coi.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuranaga E. Caspase signaling in animal development. Dev Growth Differ. 2011;53:137–148. doi: 10.1111/j.1440-169X.2010.01237.x. [DOI] [PubMed] [Google Scholar]
- 4.Yuan J, Kroemer G. Alternative cell death mechanisms in development and beyond. Genes Dev. 2010;24:2592–2602. doi: 10.1101/gad.1984410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryoo HD, Bergmann A. The role of apoptosis-induced proliferation for regeneration and cancer. Cold Spring Harb Perspect Biol. 2012;4(8) doi: 10.1101/cshperspect.a008797. a008797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cario E. Innate immune signalling at intestinal mucosal surfaces: a fine line between host protection and destruction. Curr Opin Gastroenterol. 2008;24:725–732. doi: 10.1097/MOG.0b013e32830c4341. [DOI] [PubMed] [Google Scholar]
- 7.Fadok VA, Bratton DL, Konowal A, Freed PW, Westcott JY, Henson PM. Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-beta, PGE2, and PAF. J Clin Invest. 1998;101:890–898. doi: 10.1172/JCI1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim S, Elkon KB, Ma X. Transcriptional suppression of interleukin-12 gene expression following phagocytosis of apoptotic cells. Immunity. 2004;21:643–653. doi: 10.1016/j.immuni.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. 2000. [DOI] [PubMed] [Google Scholar]
- 10.Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, Socransky SS, Hasturk H, Van Dyke TE, Dewhirst F, Paster BJ. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80:1421–1432. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolenbrander PE. Multispecies communities: interspecies interactions influence growth on saliva as sole nutritional source. Int J Oral Sci. 2011;3:49–54. doi: 10.4248/IJOS11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Periasamy S, Kolenbrander PE. Mutualistic biofilm communities develop with Porphyromonas gingivalis and initial, early, and late colonizers of enamel. J Bacteriol. 2009;191:6804–6811. doi: 10.1128/JB.01006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez OA, Stromberg AJ, Huggins PM, Gonzalez-Martinez J, Novak MJ, Ebersole JL. Apoptotic genes are differentially expressed in aged gingival tissue. J Dent Res. 2011;90:880–886. doi: 10.1177/0022034511403744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willershausen-Zonnchen B, Gleissner C. Periodontal disease in elderly patients. Eur J Med Res. 1998;3:55–64. [PubMed] [Google Scholar]
- 16.Garcia RI, Krall EA, Vokonas PS. Periodontal disease and mortality from all causes in the VA Dental Longitudinal Study. Ann Periodontol. 1998;3:339–349. doi: 10.1902/annals.1998.3.1.339. [DOI] [PubMed] [Google Scholar]
- 17.Ship JA, Crow HC. Diseases of periodontal tissues in the elderly. Description, epidemiology, aetiology and drug therapy. Drugs Aging. 1994;5:346–357. doi: 10.2165/00002512-199405050-00004. [DOI] [PubMed] [Google Scholar]
- 18.Ebersole JL, Steffen MJ, Gonzalez-Martinez J, Novak MJ. Effects of age and oral disease on systemic inflammatory and immune parameters in nonhuman primates. Clin Vaccine Immunol. 2008;15:1067–1075. doi: 10.1128/CVI.00258-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meka A, Bakthavatchalu V, Sathishkumar S, Lopez MC, Verma RK, Wallet SM, Bhattacharyya I, Boyce BF, Handfield M, Lamont RJ, Baker HV, Ebersole JL, Kesavalu L. Porphyromonas gingivalis infection-induced tissue and bone transcriptional profiles. Mol Oral Microbiol. 2010;25:61–74. doi: 10.1111/j.2041-1014.2009.00555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Favaloro B, Allocati N, Graziano V, Di Ilio C, De Laurenzi V. Role of apoptosis in disease. Aging. 2012;4:330–349. doi: 10.18632/aging.100459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaux DL, Korsmeyer SJ. Cell death in development. Cell. 1999;96:245–254. doi: 10.1016/s0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- 22.Voll RE, Herrmann M, Roth EA, Stach C, Kalden JR, Girkontaite I. Immunosuppressive effects of apoptotic cells. Nature. 1997;390:350–351. doi: 10.1038/37022. [DOI] [PubMed] [Google Scholar]
- 23.Gamonal J, Bascones A, Acevedo A, Blanco E, Silva A. Apoptosis in chronic adult periodontitis analyzed by in situ DNA breaks, electron microscopy, and immunohistochemistry. J Periodontol. 2001;72:517–525. doi: 10.1902/jop.2001.72.4.517. [DOI] [PubMed] [Google Scholar]
- 24.Koulouri O, Lappin DF, Radvar M, Kinane DF. Cell division, synthetic capacity and apoptosis in periodontal lesions analysed by in situ hybridisation and immunohistochemistry. J Clin Periodontol. 1999;26:552–559. doi: 10.1034/j.1600-051x.1999.260810.x. [DOI] [PubMed] [Google Scholar]
- 25.Albandar JM, Tinoco EM. Global epidemiology of periodontal diseases in children and young persons. Periodontol. 2002;29:153–176. doi: 10.1034/j.1600-0757.2002.290108.x. 2000. [DOI] [PubMed] [Google Scholar]
- 26.Streckfus CF, Parsell DE, Streckfus JE, Pennington W, Johnson RB. Relationship between oral alveolar bone loss and aging among African-American and Caucasian individuals. Gerontology. 1999;45:110–114. doi: 10.1159/000022072. [DOI] [PubMed] [Google Scholar]
- 27.Rider P, Carmi Y, Guttman O, Braiman A, Cohen I, Voronov E, White MR, Dinarello CA, Apte RN. IL-1alpha and IL-1beta recruit different myeloid cells and promote different stages of sterile inflammation. J Immunol. 2011;187:4835–4843. doi: 10.4049/jimmunol.1102048. [DOI] [PubMed] [Google Scholar]
- 28.Berda-Haddad Y, Robert S, Salers P, Zekraoui L, Farnarier C, Dinarello CA, Dignat-George F, Kaplanski G. Sterile inflammation of endothelial cell-derived apoptotic bodies is mediated by interleukin-1α. Proc Natl Acad SciUSA. 2011;108:20684–20689. doi: 10.1073/pnas.1116848108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen-Sfady M, Pevsner-Fischer M, Margalit R, Cohen IR. Heat shock protein 60, via MyD88 innate signaling, protects B cells from apoptosis, spontaneous and induced. J Immunol. 2009;183:890–896. doi: 10.4049/jimmunol.0804238. [DOI] [PubMed] [Google Scholar]
- 30.Busca A, Saxena M, Kryworuchko M, Kumar A. Antiapoptotic genes in the survival of monocytic cells during infection. Curr Genomics. 2009;10:306–317. doi: 10.2174/138920209788920967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmid JA, Birbach A. IkappaB kinase beta (IKKbeta/IKK2/IKBKB)—a key molecule in signaling to the transcription factor NF-kappaB. Cytokine Growth Factor Rev. 2008;19:157–165. doi: 10.1016/j.cytogfr.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 32.Sethi G, Ahn KS, Aggarwal BB. Targeting nuclear factor-kappa B activation pathway by thymoquinone: role in suppression of antiapoptotic gene products and enhancement of apoptosis. Mol Cancer Res. 2008;6:1059–1070. doi: 10.1158/1541-7786.MCR-07-2088. [DOI] [PubMed] [Google Scholar]
- 33.Reubold TF, Eschenburg S. A molecular view on signal transduction by the apoptosome. Cell Signal. 2012;24:1420–1425. doi: 10.1016/j.cellsig.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 34.Lawrence T, Fong C. The resolution of inflammation: anti-inflammatory roles for NF-kappaB. Int J Biochem Cell Biol. 2010;42:519–523. doi: 10.1016/j.biocel.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 35.Brown KD, Claudio E, Siebenlist U. The roles of the classical and alternative nuclear factor-kappaB pathways: potential implications for autoimmunity and rheumatoid arthritis. Arthr Res Ther. 2008;10:212. doi: 10.1186/ar2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vogler M. BCL2A1: the underdog in the BCL2 family. Cell Death Differ. 2012;19:67–74. doi: 10.1038/cdd.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou F, Yang Y, Xing D. Bcl-2 and Bcl-xL play important roles in the crosstalk between autophagy and apoptosis. FEBS J. 2011;278:403–413. doi: 10.1111/j.1742-4658.2010.07965.x. [DOI] [PubMed] [Google Scholar]
- 38.Kumar S. ICE-like proteases in apoptosis. Trends Biochem Sci. 1995;20:198–202. doi: 10.1016/s0968-0004(00)89007-6. [DOI] [PubMed] [Google Scholar]
- 39.Nicholson DW, Ali A, Thornberry NA, Vaillancourt JP, Ding CK, Gallant M, Gareau Y, Griffin PR, Labelle M, Lazebnik YA, et al. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 40.Wallach D, Boldin M, Goncharov T, Goltsev Y, Mett I, Malinin N, Adar R, Kovalenko A, Varfolomeev E. Exploring cell death mechanisms by analyzing signaling cascades of the TNF/NGF receptor family. Behring Inst Mitt. 1996;(97):144–155. [PubMed] [Google Scholar]
- 41.Muzio M, Chinnaiyan AM, Kischkel FC, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz JD, Zhang M, Gentz R, Mann M, Krammer PH, Peter ME, Dixit VM. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 42.Rosen A, Casciola-Rosen L. Macromolecular substrates for the ICE-like proteases during apoptosis. J Cell Biochem. 1997;64:50–54. doi: 10.1002/(sici)1097-4644(199701)64:1<50::aid-jcb8>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 43.Sabatel H, Pirlot C, Piette J, Habraken Y. Importance of PIKKs in NF-kappaB activation by genotoxic stress. Biochem Pharmacol. 2011;82:1371–1383. doi: 10.1016/j.bcp.2011.07.105. [DOI] [PubMed] [Google Scholar]
- 44.Bononi A, Agnoletto C, De Marchi E, Marchi S, Patergnani S, Bonora M, Giorgi C, Missiroli S, Poletti F, Rimessi A, Pinton P. Protein kinases and phosphatases in the control of cell fate. Enzyme Res. 2011;2011:329098. doi: 10.4061/2011/329098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duronio V. The life of a cell: apoptosis regulation by the PI3K/PKB pathway. Biochem J. 2008;415:333–344. doi: 10.1042/BJ20081056. [DOI] [PubMed] [Google Scholar]
- 46.Insel PA, Zhang L, Murray F, Yokouchi H, Zambon AC. Cyclic AMP is both a pro-apoptotic and anti-apoptotic second messenger. Acta Physiol (Oxf) 2012;204:277–287. doi: 10.1111/j.1748-1716.2011.02273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yonezawa T, Kurata R, Kimura M, Inoko H. Which CIDE are you on? Apoptosis and energy metabolism. Mol BioSyst. 2011;7:91–100. doi: 10.1039/c0mb00099j. [DOI] [PubMed] [Google Scholar]
- 48.Kfir-Erenfeld S, Sionov RV, Spokoini R, Cohen O, Yefenof E. Protein kinase networks regulating glucocorticoid-induced apoptosis of hematopoietic cancer cells: fundamental aspects and practical considerations. Leuk Lymphoma. 2010;51:1968–2005. doi: 10.3109/10428194.2010.506570. [DOI] [PubMed] [Google Scholar]
- 49.Taylor SS, Kim C, Vigil D, Haste NM, Yang J, Wu J, Anand GS. Dynamics of signaling by PKA. Biochim Biophys Acta. 2005;1754:25–37. doi: 10.1016/j.bbapap.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 50.Taylor SS, Yang J, Wu J, Haste NM, Radzio-Andzelm E, Anand G. PKA: a portrait of protein kinase dynamics. Biochim Biophys Acta. 2004;1697:259–269. doi: 10.1016/j.bbapap.2003.11.029. [DOI] [PubMed] [Google Scholar]
- 51.Bossis I, Voutetakis A, Bei T, Sandrini F, Griffin KJ, Stratakis CA. Protein kinase A and its role in human neoplasia: the Carney complex paradigm. Endocr Relat Cancer. 2004;11:265–280. doi: 10.1677/erc.0.0110265. [DOI] [PubMed] [Google Scholar]
- 52.Cho-Chung YS, Nesterova M, Becker KG, Srivastava R, Park YG, Lee YN, Cho YS, Kim MK, Neary C, Cheadle C. Dissecting the circuitry of protein kinase A and cAMP signaling in cancer genesis: antisense, microarray, gene overexpression, and transcription factor decoy. Ann N Y Acad Sci. 2002;968:22–36. doi: 10.1111/j.1749-6632.2002.tb04324.x. [DOI] [PubMed] [Google Scholar]
- 53.Mair W, Morantte I, Rodrigues AP, Manning G, Montminy M, Shaw RJ, Dillin A. Lifespan extension induced by AMPK and calcineurin is mediated by CRTC-1 and CREB. Nature. 2011;470:404–408. doi: 10.1038/nature09706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cypess AM, Zhang H, Schulz TJ, Huang TL, Espinoza DO, Kristiansen K, Unterman TG, Tseng YH. Insulin/IGF-I regulation of necdin and brown adipocyte differentiation via CREB- and FoxO1-associated pathways. Endocrinology. 2011;152:3680–3689. doi: 10.1210/en.2011-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fernandes ND, Sun Y, Price BD. Activation of the kinase activity of ATM by retinoic acid is required for CREB-dependent differentiation of neuroblastoma cells. J Biol Chem. 2007;282:16577–16584. doi: 10.1074/jbc.M609628200. [DOI] [PubMed] [Google Scholar]
- 56.Chowdhury I, Tharakan B, Bhat GK. Caspases—an update. Comp Biochem Physiol B Biochem Mol Biol. 2008;151:10–27. doi: 10.1016/j.cbpb.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 57.Valmiki MG, Ramos JW. Death effector domain-containing proteins. Cell Mol Life Sci. 2009;66:814–830. doi: 10.1007/s00018-008-8489-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sheikh MS, Huang Y. Death receptor activation complexes: it takes two to activate TNF receptor 1. Cell Cycle. 2003;2:550–552. [PubMed] [Google Scholar]
- 59.Takahashi K, Kawai T, Kumar H, Sato S, Yonehara S, Akira S. Roles of caspase-8 and caspase-10 in innate immune responses to double-stranded RNA. J Immunol. 2006;176:4520–4524. doi: 10.4049/jimmunol.176.8.4520. [DOI] [PubMed] [Google Scholar]
- 60.Kulikov AV, Shilov ES, Mufazalov IA, Gogvadze V, Nedospasov SA, Zhivotovsky B. Cytochrome c: the Achilles’ heel in apoptosis. Cell Mol Life Sci. 2012;69:1787–1797. doi: 10.1007/s00018-011-0895-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mates JM, Segura JA, Alonso FJ, Marquez J. Oxidative stress in apoptosis and cancer: an update. Arch Toxicol. 2012;86(11):1649–1665. doi: 10.1007/s00204-012-0906-3. [DOI] [PubMed] [Google Scholar]
- 62.Smolewski P, Robak T. Inhibitors of apoptosis proteins (IAPs) as potential molecular targets for therapy of hematological malignancies. Curr Mol Med. 2011;11:633–649. doi: 10.2174/156652411797536723. [DOI] [PubMed] [Google Scholar]
- 63.Gyrd-Hansen M, Meier P. IAPs: from caspase inhibitors to modulators of NF-kappaB, inflammation and cancer. Nat Rev Cancer. 2010;10:561–574. doi: 10.1038/nrc2889. [DOI] [PubMed] [Google Scholar]
- 64.Widlak P, Garrard WT. Discovery, regulation, and action of the major apoptotic nucleases DFF40/CAD and endonuclease G. J Cell Biochem. 2005;94:1078–1087. doi: 10.1002/jcb.20409. [DOI] [PubMed] [Google Scholar]
- 65.Widlak P, Garrard WT. Roles of the major apoptotic nuclease-DNA fragmentation factor-in biology and disease. Cell Mol Life Sci. 2009;66:263–274. doi: 10.1007/s00018-008-8472-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang JH, Xu M. DNA fragmentation in apoptosis. Cell Res. 2000;10:205–211. doi: 10.1038/sj.cr.7290049. [DOI] [PubMed] [Google Scholar]
- 67.Lavrik IN, Krammer PH. Regulation of CD95/Fas signaling at the DISC. Cell Death Differ. 2012;19:36–41. doi: 10.1038/cdd.2011.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kurji K, Sharma RK. Potential role of calcineurin in pathogenic conditions. Mol Cell Biochem. 2010;338:133–141. doi: 10.1007/s11010-009-0346-4. [DOI] [PubMed] [Google Scholar]
- 69.Manicassamy S, Gupta S, Huang Z, Molkentin JD, Shang W, Sun Z. Requirement of calcineurin a beta for the survival of naive T cells. J Immunol. 2008;180:106–112. doi: 10.4049/jimmunol.180.1.106. [DOI] [PubMed] [Google Scholar]
- 70.Srinivasan M, Frauwirth KA. Reciprocal NFAT1 and NFAT2 nuclear localization in CD8+ anergic T cells is regulated by suboptimal calcium signaling. J Immunol. 2007;179:3734–3741. doi: 10.4049/jimmunol.179.6.3734. [DOI] [PubMed] [Google Scholar]
- 71.Sieber M, Karanik M, Brandt C, Blex C, Podtschaske M, Erdmann F, Rost R, Serfling E, Liebscher J, Patzel M, Radbruch A, Fischer G, Baumgrass R. Inhibition of calcineurin–NFAT signaling by the pyrazolopyrimidine compound NCI3. Eur J Immunol. 2007;37:2617–2626. doi: 10.1002/eji.200737087. [DOI] [PubMed] [Google Scholar]
- 72.Morio T, Kim H. Ku, Artemis, and ataxia-telangiectasia-mutated: signalling networks in DNA damage. Int J Biochem Cell Biol. 2008;40:598–603. doi: 10.1016/j.biocel.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 73.Lavin MF, Kozlov S. ATM activation and DNA damage response. Cell Cycle. 2007;6:931–942. doi: 10.4161/cc.6.8.4180. [DOI] [PubMed] [Google Scholar]
- 74.Lo Muzio L, Sartini D, Santarelli A, Rocchetti R, Morganti S, Pozzi V, Rubini C, Bambini F, Emanuelli M. Expression and prognostic significance of apoptotic genes in oral squamous cell carcinoma. Mol Carcinog. 2012 doi: 10.1002/mc.21960. [DOI] [PubMed] [Google Scholar]
- 75.Lo Muzio L, Santarelli A, Emanuelli M, Pierella F, Sartini D, Staibano S, Rubini C, De Rosa G. Genetic analysis of oral squamous cell carcinoma by cDNA microarrays focused apoptotic pathway. Int J Immunopathol Pharmacol. 2006;19:675–682. doi: 10.1177/039463200601900323. [DOI] [PubMed] [Google Scholar]
- 76.Dunn SE, Hardman RA, Kari FW, Barrett JC. Insulin-like growth factor 1 (IGF-1) alters drug sensitivity of HBL100 human breast cancer cells by inhibition of apoptosis induced by diverse anticancer drugs. Cancer Res. 1997;57:2687–2693. [PubMed] [Google Scholar]
- 77.Peretz S, Jensen R, Baserga R, Glazer PM. ATM-dependent expression of the insulin-like growth factor-I receptor in a pathway regulating radiation response. Proc Natl Acad Sci USA. 2001;98:1676–1681. doi: 10.1073/pnas.041416598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Resnicoff M, Abraham D, Yutanawiboonchai W, Rotman HL, Kajstura J, Rubin R, Zoltick P, Baserga R. The insulin-like growth factor I receptor protects tumor cells from apoptosis in vivo. Cancer Res. 1995;55:2463–2469. [PubMed] [Google Scholar]
- 79.Puzik A, Rupp J, Troger B, Gopel W, Herting E, Hartel C. Insulin-like growth factor-I regulates the neonatal immune response in infection and maturation by suppression of IFN-gamma. Cytokine. 2012;60(2):369–376. doi: 10.1016/j.cyto.2012.07.025. [DOI] [PubMed] [Google Scholar]
- 80.Janke C, Rogowski K, Wloga D, Regnard C, Kajava AV, Strub JM, Temurak N, van Dijk J, Boucher D, van Dorsselaer A, Suryavanshi S, Gaertig J, Edde B. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science. 2005;308:1758–1762. doi: 10.1126/science.1113010. [DOI] [PubMed] [Google Scholar]
- 81.Ikegami K, Horigome D, Mukai M, Livnat I, MacGregor GR, Setou M. TTLL10 is a protein polyglycylase that can modify nucleosome assembly protein 1. FEBS Lett. 2008;582:1129–1134. doi: 10.1016/j.febslet.2008.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kashiwaya K, Nakagawa H, Hosokawa M, Mochizuki Y, Ueda K, Piao L, Chung S, Hamamoto R, Eguchi H, Ohigashi H, Ishikawa O, Janke C, Shinomura Y, Nakamura Y. Involvement of the tubulin tyrosine ligase-like family member 4 polyglutamylase in PELP1 polyglutamylation and chromatin remodeling in pancreatic cancer cells. Cancer Res. 2010;70:4024–4033. doi: 10.1158/0008-5472.CAN-09-4444. [DOI] [PubMed] [Google Scholar]
- 83.Lugering A, Lebiedz P, Koch S, Kucharzik T. Apoptosis as a therapeutic tool in IBD? Ann N Y Acad Sci. 2006;1072:62–77. doi: 10.1196/annals.1326.013. [DOI] [PubMed] [Google Scholar]
- 84.Marzetti E, Lees HA, Manini TM, Buford TW, Aranda JM, Jr, Calvani R, Capuani G, Marsiske M, Lott DJ, Vandenborne K, Bernabei R, Pahor M, Leeuwenburgh C, Wohlgemuth SE. Skeletal muscle apoptotic signaling predicts thigh muscle volume and gait speed in community-dwelling older persons: an exploratory study. PLoS ONE. 2012;7:e32829. doi: 10.1371/journal.pone.0032829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Higami Y, Shimokawa I. Apoptosis in the aging process. Cell Tissue Res. 2000;301:125–132. doi: 10.1007/s004419900156. [DOI] [PubMed] [Google Scholar]
- 86.Boddaert J, Mallat Z, Fornes P, Esposito B, Lecomte D, Verny M, Tedgui A, Belmin J. Age and gender effects on apoptosis in the human coronary arterial wall. Mech Ageing Dev. 2005;126:678–684. doi: 10.1016/j.mad.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 87.Colin A, Barroso G, Gomez-Lopez N, Duran EH, Oehninger S. The effect of age on the expression of apoptosis biomarkers in human spermatozoa. Fertil Steril. 2010;94:2609–2614. doi: 10.1016/j.fertnstert.2010.04.043. [DOI] [PubMed] [Google Scholar]
- 88.Nooteboom M, Johnson R, Taylor RW, Wright NA, Lightowlers RN, Kirkwood TB, Mathers JC, Turnbull DM, Greaves LC. Age-associated mitochondrial DNA mutations lead to small but significant changes in cell proliferation and apoptosis in human colonic crypts. Aging Cell. 2010;9:96–99. doi: 10.1111/j.1474-9726.2009.00531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kavathia N, Jain A, Walston J, Beamer BA, Fedarko NS. Serum markers of apoptosis decrease with age and cancer stage. Aging. 2009;1:652–663. doi: 10.18632/aging.100069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Haberthur K, Engelman F, Barron A, Messaoudi I. Immune senescence in aged nonhuman primates. Exp Gerontol. 2010;45:655–661. doi: 10.1016/j.exger.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]